Abstract
Chemosensory systems are complex, highly modified two-component systems (TCS) used by bacteria to control various biological functions ranging from motility to sporulation. Chemosensory systems and TCS both modulate phosphorelays comprised of histidine kinases and response regulators, some of which are single-domain response regulators (SD-RRs) such as CheY. In this study, we have identified and characterized the Che7 chemosensory system of Myxococcus xanthus, a common soil bacterium which displays multicellular development in response to stress. Both genetic and biochemical analyses indicate that the Che7 system regulates development via a direct interaction between the SD-RR CheY7 and a HEAT repeat domain-containing protein, Cpc7. Phosphorylation of the SD-RR affects the interaction with its target, and residues within the α4-β5-α5 fold of the REC domain govern this interaction. The identification of the Cpc7 interaction with CheY7 extends the diversity of known targets for SD-RRs in biological systems.
INTRODUCTION
Chemosensory systems are complex, highly modified variations of two-component systems (TCS) used to control bacterial behavior. While the canonical chemosensory system regulates flagellum-based motility (1, 2), it has become clear that these systems can also regulate alternative cellular functions (ACFs) (3). Several examples are known in which ACF systems affect multicellular development, including biofilm (4–7), cyst (8), and fruiting-body (9, 10) formation. Like systems that regulate chemotaxis (1, 2), chemosensory systems are composed of a core phosphorelay (defined by the presence of a CheA homolog and its cognate response regulator partner) and an associated adaptation module conferring sensitivity to gradients in the environment.
For chemosensory systems, stimuli are perceived by methyl-accepting chemotaxis proteins (MCPs), and the signal is transmitted via CheW coupling proteins to a CheA kinase. CheA autophosphorylates on a conserved His residue and subsequently passes the phosphoryl group to a conserved Asp residue within a receiver (REC) domain of a response regulator (RR) to control the output of the system. The domain organization of the RR can vary. For prototypical TCS, ∼85% of REC domains are covalently linked to DNA binding domains (DBD), whereas many REC domains encoded within chemosensory systems exist as single domain response regulators (SD-RR), including CheY. During chemotaxis, Escherichia coli CheY interacts directly with the flagellar components FliM and FliN to bias the rotation of the flagella, thereby regulating behavior (11–13). The CheY-FliM/N affinity is dependent on the phosphorylation state of CheY (14).
Despite the extensive knowledge gained through many studies on E. coli CheY, there are few examples of the roles and targets of SD-RRs for ACF systems. The search for those roles and targets is complicated by the fact that the E. coli chemotaxis system contains a second response regulator, CheB, responsible for demethylation of the MCPs to bring about adaptation in the presence of a stimulus. However, in the case of CheB, the REC domain is covalently linked to a methylesterase domain that becomes activated upon phosphorylation of the REC domain. Additionally, CheY∼P interacts with CheZ, which promotes dephosphorylation of the response regulator to reset the system. Thus, two REC domains with very similar topologies interact with multiple, distinct targets, including the CheA histidine kinase, the flagellar switch complex, a methylesterase, and a phosphatase. Similarly, SD-RRs are likely to have a large array of potential interaction partners for the regulation of alternative cellular functions.
The soil bacterium Myxococcus xanthus has over 40 SD-RRs, 6 of which are genetically associated with chemosensory systems (15, 16). M. xanthus is a model organism for the study of signal transduction and development. Cells coordinate their movement to mount predatory attacks (17). When cells sense low nutrient availability, they begin a developmental program that results in multicellular fruiting-body formation and differentiation into resistant myxospores (18). Regulation of these complex processes is governed in part by a large repertoire of signaling proteins, including 127 TCS and 8 chemosensory systems (15, 16). In this report, we characterize the Che7 ACF chemosensory system for its role in M. xanthus development. We identify the immediate output for the SD-RR CheY7 to be a HEAT repeat domain-containing protein, Cpc7, required for the appropriate coupling of aggregation and sporulation by M. xanthus.
MATERIALS AND METHODS
Strains and growth conditions.
Wild-type M. xanthus DZ2 was used in this study. Bacterial strains and plasmids are listed in Table S1 in the supplemental material. M. xanthus was grown routinely in Casitone-yeast extract (CYE) media at 32°C (19). Kanamycin and spectinomycin were used at 100 μg/ml and 800 μg/ml, respectively. E. coli strains were grown in Luria broth (LB) at 37°C. Kanamycin (40 μg/ml), spectinomycin (100 μg/ml), and ampicillin (100 μg/ml) were used for maintenance of plasmids.
DNA manipulations and strain construction.
Standard cloning procedures were used. Primers are listed in Table S2 in the supplemental material. Site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Agilent), and constructs were verified by sequencing (Nevada Genomics).
In-frame deletions were made in M. xanthus using counterselection plasmid pBJ114 as previously reported (20). Briefly, 1-kb flanking regions were PCR amplified and cloned into pBJ114 using restriction enzymes. Plasmids were electroporated into M. xanthus and transformants plated in soft agar (0.7%) containing 40 μg/ml of kanamycin. Resulting merodiploid colonies were grown for several generations and plated on CTT (1% Casitone, 10 mM morpholinepropanesulfonic acid [MOPS; pH 7.6], 1 mM KH2PO4) media containing 2% galactose for counterselection. Galactose-resistant, kanamycin-sensitive colonies were screened by colony PCR for deletions using gene-specific primers.
Complementation constructs were integrated at the Mx8 site using standard protocols (21). Plasmids were electroporated into M. xanthus and transformants plated in soft agar containing 40 μg/ml of kanamycin. Resulting colonies were grown for several generations, and integration was verified by site-specific PCR.
Developmental assays.
Strains were grown to mid-log phase and harvested by centrifugation (8,000 × g). Cells were washed in deionized water (dH2O) and concentrated to 8 × 108 cells/ml (see Fig. 2A, 4, and 5; see also Fig. S2 and S3 in the supplemental material) or 2 × 108 cells/ml (see Fig. 2B). Ten-microliter spots were plated on CFL agar [10 mM MOPS (pH 7.6), 1 mM KH2PO4, 8 mM MgSO4, 0.02% (NH4)2SO4, 0.02% citrate, 0.02% pyruvate, and 15 mg/liter of Casitone] (22) and incubated at 32°C. Pictures were taken using a Nikon Eclipse 50i phase-contrast microscope and a Nikon SMZ1500 dissecting microscope with a Q Imaging MicroPublisher 5.0 RTV camera and software. For spore counts, 10 spots were inoculated per plate. At various time points, cells were scraped using a razor blade and resuspended in 1 ml of dH2O. Samples were incubated at 55°C and sonicated using Branson sonifier 150 set at intensity 4 for 10 s to kill vegetative cells. Dilutions were plated on CYE media in soft agar, and CFU were counted.
FIG 2.
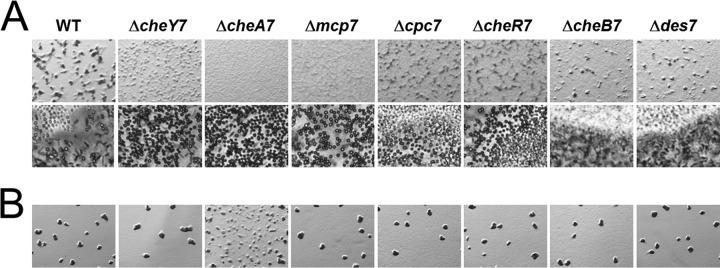
che7 mutant cells display decoupled aggregation and sporulation during development. Strains were plated on CFL starvation agar at concentrations of 8 × 108 cells/ml (A) and 2 × 108 cells/ml (B). Images were taken at 72 h at magnifications of ×30 (A, top row) and ×200 (A, bottom row) and at 120 h at a magnification of ×30 (B). Development is characterized by cells aggregating to form fruiting bodies. Light-refractile spores can be seen within the aggregates.
FIG 4.
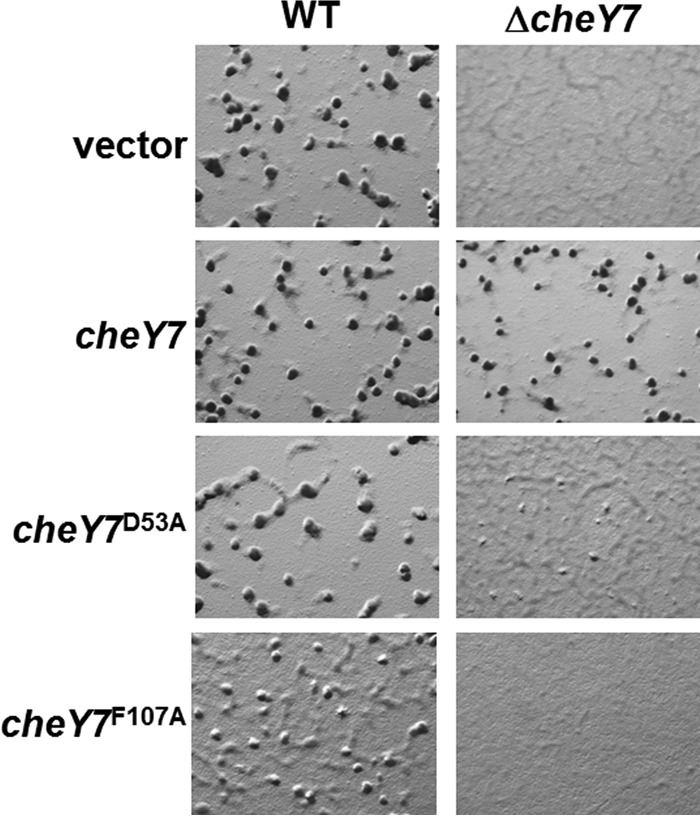
In vivo analysis of developmental defects for cheY7 mutants. cheY7 mutant alleles predicted to affect phosphorylation (D53A) or interaction with downstream targets (F107A) were assayed for defects in development. Cells were plated on CFL starvation agar and assessed for aggregation. Each allele is under the control of a constitutive pilA promoter and was integrated at the Mx8 phage attachment site. The vector control represents an insertion at the ectopic locus without a corresponding cheY allele.
FIG 5.
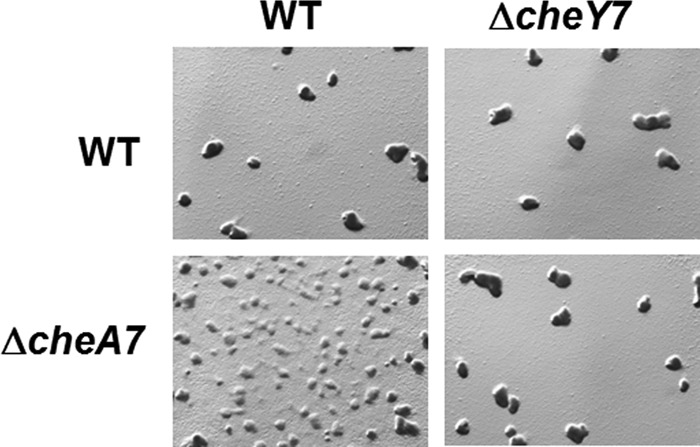
Epistasis analysis of ΔcheA7 and ΔcheY7 strains. Strains were plated on CFL agar at a concentration of 2 × 108 cells/ml. Images were taken at ×30 at 120 h. The data indicate that CheY7 is the downstream target for CheA7.
Protein purification.
CheY7, CheY7D53A, CheY7F107A, and Cpc7 were cloned into pET28a expression vector to generate a T7-6× His N-terminal tag. For acetyl phosphate (AcP) experiments, proteins were purified by standard batch procedure as previously described (23). For fluorescence binding assays, proteins were affinity purified over a Ni2+ column and a size exclusion column. For all proteins, purity was determined by Coomassie staining.
Acetyl phosphate labeling.
Proteins were radiolabeled with acetyl phosphate ([32P]AcP) as described previously (10, 24). Briefly, [32P]AcP was generated using E. coli acetate kinase (Sigma). Protein was removed by centrifugation in an Amicon Ultra filter unit (Millipore). A 5 μM concentration of CheY7 or CheY7D53A was incubated with AcP for 1 h and cleaned over a Zeba spin desalting column (Thermo Scientific). Samples were separated by SDS-PAGE and visualized using a phosphor screen and Typhoon imager (GE).
Bacterial two-hybrid analysis.
Fusion proteins were generated in pUT18c and pKT25 plasmids (see Table S2 in the supplemental material for primers). Reporter strain BTH101 was transformed and colonies grown in LB overnight at 30°C. Samples were resuspended in Z buffer, sonicated (intensity 4 for 10 s), and centrifuged to remove debris (16,000 × g). One hundred microliters of lysate was assayed in a standard Miller assay. Protein concentrations were determined by Bradford assay (Bio-Rad).
Fluorescence binding assay.
Intrinsic tryptophan fluorescence was measured at 25°C using a Fluorolog 3 spectrofluorimeter (Jobin Yvon, Horiba) in a 1.5-ml sample of 0.5 μM Cpc7 in Tris buffer (25 mM Tris-HCl [pH 7.6], 125 mM NaCl, 5% glycerol). For each measurement, excitation was at 291 nm (14.7-nm slit width). The fluorescence emission spectrum was collected in the range of 300 to 400 nm (14.7-nm slit width). CheY7 or CheY7F107A was titrated to a final concentration of 15 μM. Each titration point was stirred for 30 s. Peak fluorescence was normalized to a buffer control titration. The Kd (dissociation constant) for each titration was determined by fitting buffer-corrected data to the following equation by nonlinear regression (Sigma Plot; SPPS, Inc.), where A is fluorescence at each step during the titration, Amin is the initial fluorescence, Bmax is maximum fluorescence at saturation, and [RR] is the total concentration of response regulator in solution.
The resulting dissociation constant was the average of three independent experiments.
Circular dichroism.
CheY7 and CheY7F107A proteins were purified as described for the fluorescence binding assay. Samples were diluted to a final concentration of 20 μM in Tris buffer (25 mM Tris-HCl [pH 7.6], 125 mM NaCl, 5% glycerol) and analyzed using a 1-mm cuvette in a Jasco J-815 circular-dichroism (CD) spectrometer.
RESULTS
Identification of the che7 cluster.
M. xanthus encodes eight chemosensory systems, four of which have been described in some detail: Frz (25–27), Dif (28, 29), Che3 (9, 10), and Che4 (30). The central processing component for all chemosensory systems is CheA. In previous work, we analyzed the M. xanthus genome for cheA homologs and identified four additional clusters, including Che5 to -8 (31, 32). Because each cluster contains unique genes predicted to encode novel protein functionality regarding inputs and/or outputs, we further characterized the Che7 system (genes MXAN_6965 to MXAN_ 6957) for its role in development in M. xanthus. One unique component encoded within the che7 cluster is a HEAT repeat domain-containing protein, Cpc7. This protein has homology to phycocyanobilin lyases found in cyanobacteria. As determined by reverse transcription-PCR (RT-PCR), the Che7 cluster encodes CheY7, CheA7, CheW7, Mcp7, Cpc7, CheB7, CheR7, and Des7 (Fig. 1; see also Fig. S1 in the supplemental material).
FIG 1.
The M. xanthus che7 locus and phyletic tree of che7 clusters. (A) The che7 cluster contains six chemosensory genes (cheY7, cheA7, cheW7, mcp7, cheR7, and cheB7) as well as accessory genes cpc7 (HEAT repeat protein) and des7 (fatty acid [FA] desaturase). (B) Shown is a phylogenetic tree based on CheA-CheR-CheB concatenated proteins from cyanobacterial and myxobacterial species that possess Cpc homologs (see the text). Gene order for the corresponding clusters is shown at right. Blue shaded boxes indicate cyanobacterial species containing che7. Orange shaded boxes indicate myxobacteria containing che7. cpc7 is conserved in most che7 clusters, whereas des7 is specific to Myxococcus species.
An in silico analysis of Che7-like systems based on cheA7, mcp7, and cpc7 sequences revealed loci with the same gene order. A phylogenetic tree was produced based on a CheA-CheR-CheB concatenated protein, as described previously (33) (Fig. 1B). Notably, che7 was found largely within two clades, the myxobacteria and cyanobacteria. Within the myxobacteria, che7 was found only in species with a developmental cycle: Myxococcus fulvus, Corallococcus coralloides, Stigmatella aurantiaca, and Sorangium cellulosum. In contrast, che7 was not found in the closely related species Anaeromyxobacter dehalogens, a myxobacterium that does not undergo development, or within other members of the Deltaproteobacteria clade. Importantly, each che7 cluster contains a cpc7 homolog. Together, these results suggest that Che7 may regulate a process specific to myxobacteria capable of development and that Cpc7 may be involved in regulation of that process.
Cyanobacteria that possess a che7-like locus, including a Cpc7-like HEAT repeat gene annotated as a phycocyanobilin lyase, include Arthrospira species, Crinalium episammum, and Oscillartoria acuminata (15). Phycocyanobilin lyase homologs have been described and have roles in maintenance of light-harvesting phycobilisomes (34, 35). Thus, it is not surprising to find these genes in cyanobacteria. However, HEAT repeat proteins can mediate protein-protein interactions (36), bind iron (37), stabilize [Fe-S] clusters (38, 39), and interact with chemotaxis proteins (40). In addition to cyanobacteria, che7 loci were found in Gemmata obscuriglobus, Opitutus terrae, and Anaerolinea thermophila. Based on the occurrence of the cpc7 gene in chemosensory clusters (Fig. 1B), Che7 may have originated in cyanobacteria with a role in harvesting light, whereas in myxobacteria the system appears to have evolved to regulate some aspect of development (see below).
Mutations in the Che7 system alter aggregation and development into spores.
To determine the role of Che7 during the M. xanthus life cycle, we constructed in-frame deletions in genes within the che7 cluster. We did not observe any defects in growth, motility, or exopolysaccharide (EPS) production during vegetative growth (see Fig. S1 in the supplemental material; other data not shown). Because the che7 system is found in developing myxobacteria, we tested each strain for development on CFL starvation media. We found that che7 mutants had severe aggregation defects relative to the parent strain, DZ2 (Fig. 2A). For example, the ΔcheY7 mutant cells formed poorly defined aggregates that never matured into discrete fruiting bodies. Cells sporulated nonetheless, both within mounds and between the aggregates as individual cells; this is in contrast to the wild type, which sporulates almost exclusively within fruiting bodies (Fig. 2). Despite aberrant sporulation by the ΔcheY7 mutant, motile rods could be found at the periphery of the colony (see Fig. S2B in the supplemental material), indicating that sporulation occurs only in a subset of cells, similar to findings for the wild type. Strains with mutations in cpc7 and cheR7 also aggregated poorly compared to the wild type. The ΔcheA7 and the Δmcp7 mutants further decoupled aggregation from sporulation, forming lawns of spores at the initial spot of inoculation. The ΔcheB7 and Δdes7 mutant cells formed normal fruiting bodies, although the timing of aggregation was delayed compared to that for the parent.
To determine if the Δche7 mutant spores were viable, we harvested cells over time, sonicated and heat killed vegetative cells, and plated resistant spores for CFU determination. The results for the parent, ΔcheY7, ΔcheA7, and Δcpc7 strains are shown in Table 1. At 72 h, we saw an increase of 2 orders of magnitude in the che7 strains relative to the parent. However, by 120 h, the parent and the che7 mutants had reached similar CFU. Therefore, while che7 strains sporulate earlier than the parent and outside aggregates, ultimately the same number of cells sporulate as observed for the parent. We also assessed sporulation efficiency for the remaining che7 mutants at 72 h and found that sporulation levels were inversely correlated with aggregation (Fig. 2; see also Table S3 in the supplemental material) such that early sporulation relative to the parent occurred without aggregation. Together, the data suggest that Che7 is involved in coupling aggregation to sporulation.
TABLE 1.
Viable spore count
| Genotype | Viable sport count, CFU (mean ± SD) |
||
|---|---|---|---|
| Day 3 | Day 4 | Day 5 | |
| WTa | (1.50 ± 1.93) × 103 | (2.23 ± 0.46) × 106 | (1.44 ± 0.59) × 107 |
| ΔcheA7 | (2.13 ± 0.31) × 105 | (3.63 ± 0.15) × 106 | (1.20 ± 0.61) × 107 |
| ΔcheY7 | (2.20 ± 1.14) × 105 | (5.00 ± 1.67) × 106 | (6.84 ± 8.06) × 106 |
| Δcpc7 | (8.03 ± 2.89) × 104 | (2.57 ± 1.70) × 106 | (8.63 ± 10.00) × 106 |
WT, wild type.
M. xanthus development depends on cell density (41). We tested if density played a role for the phenotypes described above by lowering the concentration of cells plated (Fig. 2B). At the lower cell density, we saw that the che7 mutants formed fruiting bodies indistinguishable from those of the parent, with the exception of the ΔcheA7 strain. The ΔcheA7 strain formed immature aggregates, indicating a partial rescue. Therefore, che7 mutant strains are capable of developing normally under certain conditions, and high cell density is tied to Che7 coordination of aggregation and sporulation.
CheY7 functions as a single domain response regulator.
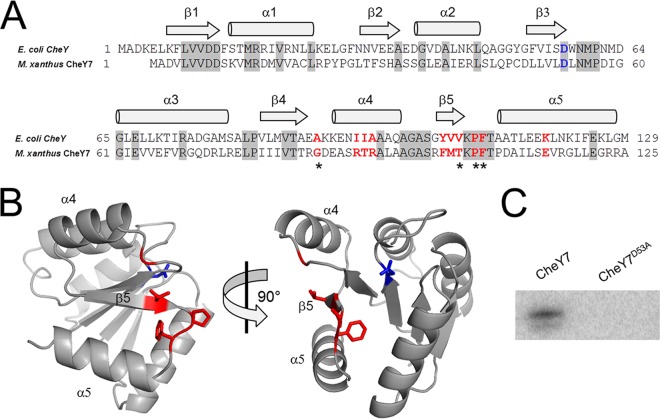
Based on homology to E. coli CheY, we predicted that M. xanthus CheY7 is the RR for the Che7 system. As an SD-RR, CheY7 would likely interact with other protein(s) to generate a response to the signal sensed by the Che7 system. Therefore, we studied CheY7 in more detail. An alignment between M. xanthus CheY7 and E. coli CheY revealed that the residues required for phosphorylation and substrate binding are present in the M. xanthus homolog (Fig. 3A) (42–44). Furthermore, the predicted (β-α)5 structure appeared to be conserved when modeled using Phyre2 to thread CheY7 onto Vibrio cholerae CheY3 (Fig. 3A and B) (45).
FIG 3.
CheY7 is a phosphorylatable SD-RR. (A) Amino acid alignment of E. coli CheY and M. xanthus CheY7. Residues important for interaction between E. coli CheY7 and FliM/CheZ substrates are indicated in red. The conserved aspartate residue that is the site of phosphorylation in REC domains is highlighted in blue. Gray shading indicates identity between E. coli and M. xanthus homologs. Regions of the proteins predicted to encode alpha helices or beta sheets are indicated above the alignment. (B) Depiction of two views of M. xanthus CheY7 threaded onto V. cholerae CheY3. Residues affecting CheY7-Cpc7 interactions as determined in the bacterial two-hybrid assay (Table 2) are shown in red. The putative site of phosphorylation is shown in blue. (C) In vitro phosphorylation of CheY7 and CheY7D53A using acetyl phosphate (see Materials and Methods) is shown. The D53A mutation eliminates phosphorylation of CheY7.
We first complemented the ΔcheY7 mutant using an epitope-tagged version of cheY7 under the control of the M. xanthus pilA promoter, which is known to constitutively drive gene expression when cells are plated on agar (46). The construct was integrated at the Mx8 phage attachment site in both the parent and ΔcheY7 strains (21). Expression of a second copy of cheY7 in the parent strain did not have any apparent impact on fruiting-body formation (Fig. 4). Expression of tagged cheY7 was sufficient to restore wild-type aggregation and sporulation within fruiting bodies in the ΔcheY7 mutant (Fig. 4).
Response regulators are typically phosphorylated on a conserved Asp residue. CheY7 contains an Asp at position 53, as well as other residues known to be important for phosphorylation (Fig. 3A) (42–44). To test if the conserved Asp53 was important for cellular function, we changed the residue to Ala. We integrated the tagged alleles at the Mx8 site and assayed for the ability to complement the ΔcheY7 strain (Fig. 4). Importantly, CheY7D53A was unable to complement the aggregation phenotype, suggesting that phosphorylation of CheY7 is important under these conditions. We also generated a CheY7D53E construct to mimic the activated state of CheY7. However, we found that the D53E variant was unable to complement the ΔcheY7 strain under conditions tested (data not shown), indicating that CheY7D53E is a null allele similar to CheY7D53A.
To further support a role for phosphorylation of Asp53, we determined if CheY7 could be phosphorylated in vitro by the high-energy phosphodonor acetyl phosphate (AcP), a technique used routinely for phosphorylating response regulators (47). We purified the wild-type and D53A versions of CheY7 using affinity purification. We then generated radiolabeled AcP using purified E. coli acetate kinase (Sigma). Incubation of wild-type CheY7 with AcP yielded a radiolabeled band when separated by SDS-PAGE (Fig. 3C). Thus, CheY7 can be phosphorylated in vitro. In contrast, the D53A construct did not yield a phosphorylated product, indicating the requirement of Asp53 for phosphorylation. Together, the in vivo and in vitro data support a role for CheY7 as a bona fide phosphorylatable SD-RR that is modified at the conserved position typically found in REC domains.
CheY7 interacts with and functions downstream of CheA7.
We sought to identify a kinase that modulates the CheY7 phosphorylation state in vivo. Based on the paradigm for chemotaxis, we predicted that CheY7 would function downstream of CheA7. To determine this, we performed epistasis analysis between mutations resulting in deletions for cheY7 and cheA7. At low cell density, a phenotypic difference between the ΔcheY7 and ΔcheA7 strains is clear: ΔcheA7 cells are defective at aggregation, and ΔcheY7 cells appear more similar to the parent strain (Fig. 2B). To determine if CheY7 is downstream of CheA7, we constructed a ΔcheY7 ΔcheA7 double mutant strain and assayed for development at low cell density (Fig. 5). Consistent with the predicted model, the ΔcheY7 ΔcheA7 strain phenocopied the ΔcheY7 parent, indicating that the ΔcheY7 mutation is epistatic to the ΔcheA7 mutation. These data support a model in which CheY7 is the cognate RR for CheA7.
To test if the interaction between CheA7 and CheY7 was direct, we conducted further studies. Cognate kinases and response regulators interact to transfer phosphoryl groups. We attempted to purify CheA7 from E. coli but were unable to produce stable, soluble protein for in vitro phosphorylation assays. In lieu of direct phosphotransfer assays, we assessed if CheA7 and CheY7 could interact using the Bordetella bacterial two-hybrid system, in which interacting proteins couple two halves of adenylate cyclase (T18 and T25) to drive a LacZ reporter (Table 2) (48). Expression of T18-CheY7 and T25-CheA7 in the E. coli BTH101 reporter strain resulted in an ∼60-fold increase in β-galactosidase activity compared to the vector controls. Based on these data, CheA7 and CheY7 interact in vivo. Together with the epistasis analysis, these data lead us to conclude that CheA7 and CheY7 comprise the core signaling pathway for the Che7 system in M. xanthus.
TABLE 2.
Results of bacterial two-hybrid analysis
| Protein coupled to plasmid |
β-Galactosidase activity/μg of protein (mean ± SD) | % of WT | |
|---|---|---|---|
| pUT18c | pKT25 | ||
| 0.53 ± 0.05 | |||
| CheY7 | 0.43 ± 0.09 | ||
| CheA7 | 0.54 ± 0.05 | ||
| CheY7 | CheA7 | 29.93 ± 1.82 | 100.0 |
| Cpc7 | 0.65 ± 0.08 | ||
| CheY7 | Cpc7 | 15.21 ± 1.01 | 100.0 |
| D53A | Cpc7 | 5.26 ± 3.17 | 34.6 |
| D53E | Cpc7 | 7.40 ± 2.49 | 48.7 |
| G86A | Cpc7 | 4.25 ± 0.11 | 28.0 |
| R91A | Cpc7 | 13.35 ± 1.86 | 87.8 |
| T92A | Cpc7 | 17.23 ± 4.81 | 113.3 |
| R93A | Cpc7 | 10.65 ± 1.94 | 70.0 |
| F102A | Cpc7 | 13.19 ± 1.82 | 86.7 |
| M103A | Cpc7 | 12.74 ± 1.77 | 83.8 |
| T104A | Cpc7 | 5.73 ± 3.75 | 37.7 |
| P106A | Cpc7 | 0.65 ± 0.25 | 4.3 |
| F107A | Cpc7 | 0.87 ± 0.64 | 5.7 |
| E115A | Cpc7 | 8.12 ± 0.66 | 53.4 |
Complementation of CheY7 requires Cpc7.
Based on studies from other ACF chemosensory systems, we predicted that either the Cpc7 or Des7 proteins would act as an output for the Che7 system and possibly interact directly with CheY7. To test if either Cpc7 or Des7 functions downstream of CheY7, we employed epistasis analyses based on the differential phenotypes displayed during development (as shown in Fig. 2A). To do so, we constructed additional mutant strains (ΔcheY7 Δdes7 and ΔcheY7 Δcpc7) and assayed them for their capacity to sporulate outside aggregation centers or fruiting bodies. Comparison with the ΔcheY7 and Δdes7 mutant strains and the ΔcheY7 Δdes7 strain revealed that the double mutant phenocopied the ΔcheY7 parent strain, indicating that CheY7 functions downstream of Des7 or that Des7 is not directly part of the Che7 pathway (Fig. 6, top row). In contrast, the ΔcheY7 Δcpc7 strain was able to aggregate poorly, resembling the Δcpc7 strain rather than the ΔcheY7 strain (Fig. 2A and 6). This result suggests that Cpc7 likely acts downstream of CheY7.
FIG 6.
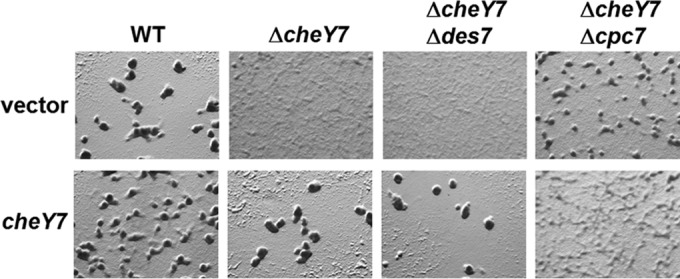
Epistasis analysis of ΔcheY7, Δdes7, and Δcpc7 strains. cheY7 was expressed from the Mx8 phage attachment site (see text) in various mutant backgrounds and assayed for defects in development. Cells were plated on CFL starvation agar and assessed for aggregation. The cpc7 mutation is epistatic to the mutation in cheY7 and could not be complemented by expression of cheY7 in the absence of cpc7.
The results shown in Fig. 4 demonstrate that the ΔcheY7 mutant could be complemented with the wild-type allele of cheY7. Based on the phenotypes of the double mutant strains listed above, we predicted that expressing cheY7 from an ectopic locus would result in complementation of the ΔcheY7 Δdes7 mutant but not the ΔcheY7 Δcpc7 mutant. Developmental assays revealed that the ΔcheY7 Δdes7 strain was complemented for its ability to aggregate and sporulate, indicating that CheY7 does not require Des7 for function during starvation-induced development (Fig. 6, bottom row). In contrast, expression of CheY7 was unable to complement the ΔcheY7 Δcpc7 strain. In fact, addition of CheY7 resulted in an exaggerated phenotype with very poor aggregation and a corresponding lawn of spores. A possible explanation for the difference in aggregation between the ΔcheY7 Δcpc7 strain and the ΔcheY7 Δcpc7 strain expressing cheY7 could be cross talk due to removal of a CheY7 target. Alternatively, CheY7 could have an additional unidentified target. Nonetheless, the data indicate that CheY7 requires Cpc7 to coordinate aggregation and sporulation in M. xanthus.
CheY7 interacts with Cpc7.
Response regulators have been shown to interact with a variety of other domains to control the output of both two-component and chemosensory systems. Since CheY7 requires Cpc7 to complement the aggregation and sporulation phenotypes associated with ΔcheY7 mutant cells, we hypothesized that CheY7 and Cpc7 interact directly. To test this possibility, we used the bacterial two-hybrid system to determine if an interaction occurs between CheY7 and Cpc7. When expressed in the reporter BTH101, the T18-CheY7 and T25-Cpc7 fusion products produced an ∼30-fold increase in β-galactosidase reporter activity over that of the vector-based controls, indicating a relatively strong protein-protein interaction, similar to that shown for CheA7 and CheY7 (Table 2). We also generated a T18-CheY7D53A fusion construct to determine if phosphorylation was important for this interaction. Interestingly, the D53A version of CheY7 was able to promote binding; however, the allele did not result in wild-type levels of activity (Table 2). These results are consistent with E. coli CheY and CheY∼P both displaying the capacity to interact with FliM, though the latter has a higher affinity (14, 49, 50). Therefore, it is likely that phosphorylation of CheY7 enhances binding with Cpc7 in vivo.
As a further verification of the CheY7-Cpc7 interaction, we designed an in vitro assay. We purified tagged versions of wild-type CheY7 and Cpc7 using affinity and size exclusion chromatography. Using the intrinsic fluorescence of the three tryptophan residues in Cpc7 (W48, W296, and W531), we measured the fluorescence spectrum as a function of CheY7 concentration. Titration of Cpc7 with CheY7 produced a change in fluorescence (Fig. 7) corresponding to an apparent Kd of 0.30 ± 0.05 μM, indicating that a relatively tight interaction occurs in vitro. On the basis of the in vivo bacterial two-hybrid results, the epistasis analyses, and the in vitro fluorescence binding data, we conclude that CheY7 and Cpc7 make a direct interaction to promote development in M. xanthus. Moreover, we have identified an interacting partner for SD-RR or CheY-like proteins not previously described.
FIG 7.

Intrinsic tryptophan fluorescence assay for binding affinity between Cpc7 and CheY7. The fluorescence spectrum for purified epitope-tagged Cpc7 (0.5 μM) was obtained (see Materials and Methods). CheY7WT (▼) or CheY7F107A (●) was titrated in and spectra were normalized to a buffer control. The change in fluorescence is reported as relative fluorescence over the concentration of ligand. The fitted curve for CheY7WT was calculated using nonlinear regression to reveal a Kd of 0.30 ± 0.05 μM. Each titration was done in triplicate and a representative experiment is presented.
The α4-β5-α5 region of CheY7 is required for interaction with Cpc7.
E. coli CheY interacts with both FliM and CheZ via its C-terminal α4-β5-α5 fold (43, 49–51). To assess whether the same region of CheY7 is required for interaction with Cpc7, we performed site-directed mutagenesis on residues known to affect the E. coli CheY-FliM and CheY-CheZ interactions (see residues in red in Fig. 3A). Based on that alignment and using site-directed mutagenesis, we generated mutations corresponding to alanine substitutions in the following residues in M. xanthus CheY7: G86, R91, T92, R93, F102, M103, T104, P106, F107, and E115. Each mutant allele was cloned into the bacterial two-hybrid system and assayed for interaction with Cpc7. While some alleles retained wild-type levels of reporter activity when expressed with the Cpc7 fusion, several displayed reduced activity, including G86A, T104A, P106A, and F107A (Table 2). The mutation corresponding to P106A likely affects secondary structure, whereas G86, T104, and F107 may be required for maintaining contacts with Cpc7. Of note is residue F107, as it is conserved in both E. coli and M. xanthus CheY homologs. The F107A mutant was severely reduced for reporter activity, suggesting a prominent role for interaction with the target Cpc7.
To further evaluate the interaction between Cpc7 and CheY7, we purified the tagged-CheY7F107A variant and tested it using the tryptophan fluorescence binding assay. Indeed, the mutant CheY7 protein was unable to alter the fluorescence profile of Cpc7 (Fig. 7). To determine if the F107A mutant protein was properly folded, we generated circular dichroism spectra for CheY7F107A and wild-type CheY7. Both proteins maintained secondary structures consistent with proper folding (see Fig. S3 in the supplemental material). Lastly, we assayed the CheY7F107A protein for its effect on development. We expressed the tagged CheY7F107A in the parent and the ΔcheY7 mutant. CheY7F107A was unable to complement the ΔcheY7 mutant (Fig. 4). In addition, expression of CheY7F107A in the wild-type background reduced the ability of cells to aggregate properly, indicating partial dominance in trans over the wild-type CheY7 protein. One possible explanation is that the F107A variant is phosphorylated in vivo, titrating phosphoryl groups from wild-type CheY7, but cannot interact with the Cpc7 target to regulate development. From these results, we conclude that the C-terminal α4-β5-α5 region of CheY7 is required for its interaction with Cpc7 to regulate fruiting-body formation in M. xanthus. Additionally, as described for other REC-protein interactions, we conclude that the conserved C-terminal α4-β5-α5 fold governs interactions between SD-RRs and their targets.
DISCUSSION
In this report, we describe a protein-protein interaction between an SD-RR (CheY7) and a HEAT repeat protein (Cpc7) required for the proper regulation of development in M. xanthus. Due to the structural and functional diversity of domains known to interact with REC domains, it has been difficult to identify targets for SD-RR regulation. The identification of the Cpc7 interaction with CheY7 extends the diversity of known targets for SD-RRs in nature. Nevertheless, the requirements for SD-RR (or REC) interaction with a given target appear to be conserved: phosphorylation of the SD-RR affects interaction with its target and the α4-β5-α5 fold within the REC domain governs the interaction. Phosphorylation of CheY7 at position D53 is required to complement a ΔcheY7 strain, while residues in the α4-β5-α5 fold, including G86, T104, P106, and F107, are essential for interaction with Cpc7. Residues corresponding to those positions in E. coli CheY are required for proper interaction with CheZ. Furthermore, the predicted structure of CheY7 places F107 within the linker region between β5 and α5 and facing the cleft created by the α4-β5-α5 fold (Fig. 3C). Replacement of residue F107 with alanine abolished CheY7's ability to complement the ΔcheY7 cells for aggregation, indicating that this region is required for function. Therefore, the results indicate that CheY7 acts like a typical REC domain yet interacts with a previously unknown target.
HEAT repeat domains are alpha helix folds found in proteins in bacteria, archaea, and eukaryotes. Two anti-parallel alpha helices comprise each HEAT domain, and they are often found in tandem repeats, creating large, solenoid structures (36). A model of Cpc7 threaded onto known HEAT repeat proteins using Phyre2 indicates that Cpc7 is made of 27 tandem alpha helices (45). Proteins containing HEAT repeats can vary in function and have been described as having roles in scaffolding (36), lyase function (34, 35), iron binding (37), and [Fe-S] cluster stabilization (38, 39). While the mechanism of Cpc7-mediated regulation of development in M. xanthus is still unknown, we predict that it is involved in production of a developmental signal. The che7 mutant phenotypes were observed under a relatively high cell density (Fig. 2), suggesting that a density-dependent factor accumulates in the parent strain. Che7 may be involved in maintaining levels of this factor, and we are currently investigating this possibility.
Lastly, this work describes an alternative cellular function for a chemosensory system. The Che7 signal transduction system impacts the coordination between aggregation and sporulation during starvation-induced development (Fig. 8). Sporulation occurs in che7 mutants prior to aggregation, and viable spores mature earlier in che7 mutants than in the parent. The data suggest a model in which Che7 provides a repressive checkpoint for sporulation, possibly to ensure aggregation occurs prior to sporulation, and may be related to phenotypes previously described for the EspABC and RedABCD systems in M. xanthus (52–55). Together, our data support a model in which a chemosensory system with a prototypical SD-RR output regulates starvation induced development through interaction with a HEAT repeat protein.
FIG 8.
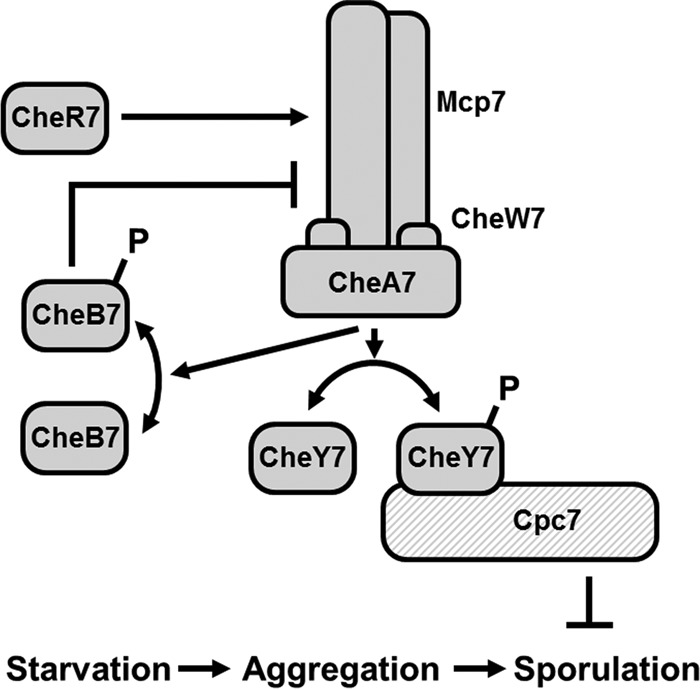
Model for Che7 regulation of development. M. xanthus responds to starvation stress by aggregating into fruiting bodies. Subsequently, a subpopulation of cells convert to resistant spores. Che7 regulates the phosphorylation state of CheY7 via the CheA7 histidine kinase. CheY7∼P interacts with HEAT repeat protein Cpc7 to inhibit sporulation prior to aggregation.
Supplementary Material
ACKNOWLEDGMENTS
Support for this work was provided by NSF MCB-1244021 to J.R.K. Additional support for C.L.D. was provided by the Center for Biocatalysis and Bioprocessing. Start-up funds to E.J.F. were provided by the Carver College of Medicine and the Department of Biochemistry.
We thank Dan Kearns for the gift of BTH101 and David Weiss for the gift of pUT18c and pKT25 and the reviewers for critical reading of the manuscript.
The content is the responsibility of the authors and does not represent the official views of the NSF. The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 23 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01866-14.
REFERENCES
- 1.Sourjik V, Wingreen NS. 2012. Responding to chemical gradients: bacterial chemotaxis. Curr. Opin. Cell Biol. 24:262–268. 10.1016/j.ceb.2011.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourret RB, Borkovich KA, Simon MI. 1991. Signal transduction pathways involving protein phosphorylation in prokaryotes. Annu. Rev. Biochem. 60:401–441. 10.1146/annurev.bi.60.070191.002153 [DOI] [PubMed] [Google Scholar]
- 3.Kirby JR. 2009. Chemotaxis-like regulatory systems: unique roles in diverse bacteria. Annu. Rev. Microbiol. 63:45–59. 10.1146/annurev.micro.091208.073221 [DOI] [PubMed] [Google Scholar]
- 4.Güvener ZT, Harwood CS. 2007. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol. Microbiol. 66:1459–1473. 10.1111/j.1365-2958.2007.06008.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. U. S. A. 102:14422–14427. 10.1073/pnas.0507170102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huangyutitham V, Güvener ZT, Harwood CS. 2013. Subcellular clustering of the phosphorylated wspr response regulator protein stimulates its diguanylate cyclase activity. mBio 4(3):e00242–13. 10.1128/mBio.00242-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Connor JR, Kuwada NJ, Huangyutitham V, Wiggins PA, Harwood CS. 2012. Surface sensing and lateral subcellular localization of WspA, the receptor in a chemosensory-like system leading to c-di-GMP production. Mol. Microbiol. 86:720–729. 10.1111/mmi.12013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berleman JE, Bauer CE. 2005. Involvement of a Che-like signal transduction cascade in regulating cyst cell development in Rhodospirillum centenum. Mol. Microbiol. 56:1457–1466. 10.1111/j.1365-2958.2005.04646.x [DOI] [PubMed] [Google Scholar]
- 9.Kirby JR, Zusman DR. 2003. Chemosensory regulation of developmental gene expression in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 100:2008–2013. 10.1073/pnas.0330944100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willett JW, Kirby JR. 2011. CrdS and CrdA comprise a two-component system that is cooperatively regulated by the Che3 chemosensory system in Myxococcus xanthus. mBio 2(4):e00110–11. 10.1128/mBio.00110-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bren A, Eisenbach M. 1998. The N terminus of the flagellar switch protein, FliM, is the binding domain for the chemotactic response regulator, CheY. J. Mol. Biol. 278:507–514. 10.1006/jmbi.1998.1730 [DOI] [PubMed] [Google Scholar]
- 12.Sarkar MK, Paul K, Blair D. 2010. Chemotaxis signaling protein CheY binds to the rotor protein FliN to control the direction of flagellar rotation in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 107:9370–9375. 10.1073/pnas.1000935107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker MD, Wolanin PM, Stock JB. 2006. Signal transduction in bacterial chemotaxis. Bioessays 28:9–22. 10.1002/bies.20343 [DOI] [PubMed] [Google Scholar]
- 14.Welch M, Oosawa K, Aizawa S, Eisenbach M. 1993. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc. Natl. Acad. Sci. U. S. A. 90:8787–8791. 10.1073/pnas.90.19.8787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ulrich LE, Zhulin IB. 2010. The MiST2 database: a comprehensive genomics resource on microbial signal transduction. Nucleic Acids Res. 38:D401–D407. 10.1093/nar/gkp940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Müller S, Willett JW, Bahr SM, Darnell CL, Hummels KR, Dong CK, Vlamakis HC, Kirby JR. 2013. Draft genome sequence of Myxococcus xanthus wild-type strain DZ2, a model organism for predation and development. Genome Announc. 1(3):e00217–13. 10.1128/genomeA.00217-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berleman JE, Scott J, Chumley T, Kirby JR. 2008. Predataxis behavior in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 105:17127–17132. 10.1073/pnas.0804387105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kroos L. 2007. The Bacillus and Myxococcus developmental networks and their transcriptional regulators. Annu. Rev. Genet. 41:13–39. 10.1146/annurev.genet.41.110306.130400 [DOI] [PubMed] [Google Scholar]
- 19.Bretscher AP, Kaiser D. 1978. Nutrition of Myxococcus xanthus, a fruiting myxobacterium. J. Bacteriol. 133:763–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Julien B, Kaiser AD, Garza A. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 97:9098–9103. 10.1073/pnas.97.16.9098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magrini V, Creighton C, Youderian P. 1999. Site-specific recombination of temperate Myxococcus xanthus phage Mx8: genetic elements required for integration. J. Bacteriol. 181:4050–4061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berleman JE, Kirby JR. 2007. Multicellular development in Myxococcus xanthus is stimulated by predator-prey interactions. J. Bacteriol. 189:5675–5682. 10.1128/JB.00544-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willett JW, Tiwari N, Muller S, Hummels KR, Houtman JC, Fuentes EJ, Kirby JR. 2013. Specificity residues determine binding affinity for two-component signal transduction systems. mBio 4(6):e00420–13. 10.1128/mBio.00420-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willett JW, Kirby JR. 2012. Genetic and biochemical dissection of a HisKA domain identifies residues required exclusively for kinase and phosphatase activities. PLoS Genet. 8:e1003084. 10.1371/journal.pgen.1003084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mauriello EM, Astling DP, Sliusarenko O, Zusman DR. 2009. Localization of a bacterial cytoplasmic receptor is dynamic and changes with cell-cell contacts. Proc. Natl. Acad. Sci. U. S. A. 106:4852–4857. 10.1073/pnas.0810583106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McBride MJ, Weinberg RA, Zusman DR. 1989. “Frizzy” aggregation genes of the gliding bacterium Myxococcus xanthus show sequence similarities to the chemotaxis genes of enteric bacteria. Proc. Natl. Acad. Sci. U. S. A. 86:424–428. 10.1073/pnas.86.2.424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun H, Zusman DR, Shi W. 2000. Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr. Biol. 10:1143–1146. 10.1016/S0960-9822(00)00705-3 [DOI] [PubMed] [Google Scholar]
- 28.Black WP, Xu Q, Yang Z. 2006. Type IV pili function upstream of the Dif chemotaxis pathway in Myxococcus xanthus EPS regulation. Mol. Microbiol. 61:447–456. 10.1111/j.1365-2958.2006.05230.x [DOI] [PubMed] [Google Scholar]
- 29.Yang Z, Geng Y, Xu D, Kaplan HB, Shi W. 1998. A new set of chemotaxis homologues is essential for Myxococcus xanthus social motility. Mol. Microbiol. 30:1123–1130. 10.1046/j.1365-2958.1998.01160.x [DOI] [PubMed] [Google Scholar]
- 30.Vlamakis HC, Kirby JR, Zusman DR. 2004. The Che4 pathway of Myxococcus xanthus regulates type IV pilus-mediated motility. Mol. Microbiol. 52:1799–1811. 10.1111/j.1365-2958.2004.04098.x [DOI] [PubMed] [Google Scholar]
- 31.Kirby JR, Berleman JE, Muller S, Li D, Scott J, Wilson J. 2008. Chemosensory signal transduction systems in Myxococcus xanthus, p 135–147 In Whitworth DE. (ed), Myxobacteria: multicellularity and differentiation. ASM Press, Washington, DC [Google Scholar]
- 32.Moine A, Agrebi R, Espinosa L, Kirby JR, Zusman DR, Mignot T, Mauriello EM. 2014. Functional organization of a multimodular bacterial chemosensory apparatus. PLoS Genet. 10:e1004164. 10.1371/journal.pgen.1004164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wuichet K, Zhulin IB. 2010. Origins and diversification of a complex signal transduction system in prokaryotes. Sci. Signal. 3:ra50. 10.1126/scisignal.2000724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou J, Gasparich GE, Stirewalt VL, de Lorimier R, Bryant DA. 1992. The cpcE and cpcF genes of Synechococcus sp. PCC 7002. Construction and phenotypic characterization of interposon mutants. J. Biol. Chem. 267:16138–16145 [PubMed] [Google Scholar]
- 35.Fairchild CD, Zhao J, Zhou J, Colson SE, Bryant DA, Glazer AN. 1992. Phycocyanin alpha-subunit phycocyanobilin lyase. Proc. Natl. Acad. Sci. U. S. A. 89:7017–7021. 10.1073/pnas.89.15.7017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobe B, Gleichmann T, Horne J, Jennings IG, Scotney PD, Teh T. 1999. Turn up the HEAT. Structure 7:R91–R97. 10.1016/S0969-2126(99)80060-4 [DOI] [PubMed] [Google Scholar]
- 37.Kim YS, Kang KR, Wolff EC, Bell JK, McPhie P, Park MH. 2006. Deoxyhypusine hydroxylase is an Fe(II)-dependent, HEAT-repeat enzyme. Identification of amino acid residues critical for Fe(II) binding and catalysis. J. Biol. Chem. 281:13217–13225. 10.1074/jbc.M601081200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morimoto K, Sato S, Tabata S, Nakai M. 2003. A HEAT-repeats containing protein, IaiH, stabilizes the iron-sulfur cluster bound to the cyanobacterial IscA homologue, IscA2. J. Biochem. 134:211–217. 10.1093/jb/mvg131 [DOI] [PubMed] [Google Scholar]
- 39.Morimoto K, Nishio K, Nakai M. 2002. Identification of a novel prokaryotic HEAT-repeats-containing protein which interacts with a cyanobacterial IscA homolog. FEBS Lett. 519:123–127. 10.1016/S0014-5793(02)02736-9 [DOI] [PubMed] [Google Scholar]
- 40.Schlesner M, Miller A, Streif S, Staudinger WF, Muller J, Scheffer B, Siedler F, Oesterhelt D. 2009. Identification of Archaea-specific chemotaxis proteins which interact with the flagellar apparatus. BMC Microbiol. 9:56. 10.1186/1471-2180-9-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim SK, Kaiser D, Kuspa A. 1992. Control of cell density and pattern by intercellular signaling in Myxococcus development. Annu. Rev. Microbiol. 46:117–139. 10.1146/annurev.mi.46.100192.001001 [DOI] [PubMed] [Google Scholar]
- 42.Birck C, Mourey L, Gouet P, Fabry B, Schumacher J, Rousseau P, Kahn D, Samama JP. 1999. Conformational changes induced by phosphorylation of the FixJ receiver domain. Structure 7:1505–1515. 10.1016/S0969-2126(00)88341-0 [DOI] [PubMed] [Google Scholar]
- 43.Cho HS, Lee S-Y, Yan D, Pan X, Parkinson JS, Kustu S, Wemmer DE, Pelton JG. 2000. NMR structure of activated CheY. J. Mol. Biol. 297:543–551. 10.1006/jmbi.2000.3595 [DOI] [PubMed] [Google Scholar]
- 44.Lewis RJ, Brannigan JA, Muchová K, Barák I, Wilkinson AJ. 1999. Phosphorylated aspartate in the structure of a response regulator protein. J. Mol. Biol. 294:9–15. 10.1006/jmbi.1999.3261 [DOI] [PubMed] [Google Scholar]
- 45.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371. 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- 46.Wu SS, Kaiser D. 1997. Regulation of expression of the pilA gene in Myxococcus xanthus. J. Bacteriol. 179:7748–7758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lukat GS, McCleary WR, Stock AM, Stock JB. 1992. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc. Natl. Acad. Sci. U. S. A. 89:718–722. 10.1073/pnas.89.2.718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U. S. A. 95:5752–5756. 10.1073/pnas.95.10.5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dyer CM, Dahlquist FW. 2006. Switched or not?: the structure of unphosphorylated CheY bound to the N terminus of FliM. J. Bacteriol. 188:7354–7363. 10.1128/JB.00637-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McAdams K, Casper ES, Matthew Haas R, Santarsiero BD, Eggler AL, Mesecar A, Halkides CJ. 2008. The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY help to explain their binding affinities to the FliM and CheZ peptides. Arch. Biochem. Biophys. 479:105–113. 10.1016/j.abb.2008.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guhaniyogi J, Robinson VL, Stock AM. 2006. Crystal structures of beryllium fluoride-free and beryllium fluoride-bound CheY in complex with the conserved C-terminal peptide of CheZ reveal dual binding modes specific to CheY conformation. J. Mol. Biol. 359:624–645. 10.1016/j.jmb.2006.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Higgs PI, Jagadeesan S, Mann P, Zusman DR. 2008. EspA, an orphan hybrid histidine protein kinase, regulates the timing of expression of key developmental proteins of Myxococcus xanthus. J. Bacteriol. 190:4416–4426. 10.1128/JB.00265-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee B, Higgs PI, Zusman DR, Cho K. 2005. EspC is involved in controlling the timing of development in Myxococcus xanthus. J. Bacteriol. 187:5029–5031. 10.1128/JB.187.14.5029-5031.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Higgs PI, Cho K, Whitworth DE, Evans LS, Zusman DR. 2005. Four unusual two-component signal transduction homologs, RedC to RedF, are necessary for timely development in Myxococcus xanthus. J. Bacteriol. 187:8191–8195. 10.1128/JB.187.23.8191-8195.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jagadeesan S, Mann P, Schink CW, Higgs PI. 2009. A novel “four-component” two-component signal transduction mechanism regulates developmental progression in Myxococcus xanthus. J. Biol. Chem. 284:21435–21445. 10.1074/jbc.M109.033415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.