Abstract
Transcriptional regulators of the AraC/XylS family have been associated with multidrug resistance, organic solvent tolerance, oxidative stress, and virulence in clinically relevant enterobacteria. In the present study, we identified four homologous AraC/XylS regulators, Rob, SoxS, PliA, and OpiA, from the fire blight pathogen Erwinia amylovora Ea1189. Previous studies have shown that the regulators MarA, Rob, and SoxS from Escherichia coli mediate multiple-antibiotic resistance, primarily by upregulating the AcrAB-TolC efflux system. However, none of the four AraC/XylS regulators from E. amylovora was able to induce a multidrug resistance phenotype in the plant pathogen. Overexpression of rob led to a 2-fold increased expression of the acrA gene. However, the rob-overexpressing strain showed increased resistance to only a limited number of antibiotics. Furthermore, Rob was able to induce tolerance to organic solvents in E. amylovora by mechanisms other than efflux. We demonstrated that SoxS from E. amylovora is involved in superoxide resistance. A soxS-deficient mutant of Ea1189 was not able to grow on agar plates supplemented with the superoxide-generating agent paraquat. Furthermore, expression of soxS was induced by redox cycling agents. We identified two novel members of the AraC/XylS family in E. amylovora. PliA was highly upregulated during the early infection phase in apple rootstock and immature pear fruits. Multiple compounds were able to induce the expression of pliA, including apple leaf extracts, phenolic compounds, redox cycling agents, heavy metals, and decanoate. OpiA was shown to play a role in the regulation of osmotic and alkaline pH stress responses.
INTRODUCTION
Erwinia amylovora, a plant-pathogenic member of the family Enterobacteriaceae, causes the devastating disease fire blight on rosaceous plants, with economic importance for the disease on apple and pear plants (1). During pathogenesis in the plant, microbes are exposed to a variety of antimicrobial compounds produced by the host. Successful pathogens utilize a variety of different systems to circumvent the toxic effects of these substrates. Important among these mechanisms are multidrug efflux pumps that are able to recognize and efficiently expel a wide range of structurally diverse compounds from the cell (2). In Gram-negative bacteria, efflux pumps that form a large tripartite complex spanning both the inner and outer membranes play a key role in multidrug resistance (3–5). These pumps consist of an inner membrane transporter (ABC, MFS, or RND family transporter), a membrane fusion protein, and an outer membrane channel (6, 7). In E. amylovora, the RND-type efflux pump AcrAB-TolC has been demonstrated to play an important role in virulence (8, 9). Mutants defective in this efflux system were impaired in colonization of apple rootstock. Furthermore, the mutants were susceptible to apple leaf extracts as well as to the apple phytoalexins phloretin, naringenin, quercetin, and (+)-catechin (8).
Bacteria are able to utilize multidrug efflux pumps in a very efficient, precise, and complex way by specifically regulating the expression of the drug transporter genes. Many genes are under either local or global transcriptional control. For example, the multidrug transporter AcrAB is locally controlled by the transcriptional repressor AcrR in Escherichia coli (10, 11) and Salmonella enterica (12) and by global transcriptional activators of the AraC/XylS family, such as MarA, SoxS, and Rob (13–15).
Transcriptional regulators belonging to the AraC/XylS family are the most common positive regulators (16–18). The main characteristic of this family is a conserved 100-amino-acid sequence, constituting a helix-turn-helix (HTH) DNA-binding domain, which is important for the activation of transcription upon binding to the target sequence of the respective promoter region. However, the DNA-binding domain is not involved in binding of effector molecules. Chemical signals may be recognized by additional, nonconserved domains located in the same polypeptide as the DNA-binding domain (e.g., AraC and XylS), or transcription of the AraC/XylS family member may be controlled by another regulator. Binding of effector molecules to this additional regulator induces transcription of the AraC/XylS protein gene and often leads to overexpression of the AraC/XylS regulator (e.g., MarA and SoxS) (reviewed in reference 17).
Most members of this family are involved in the control of carbon metabolism, pathogenicity, or stress responses (15, 17). In E. coli, the homologous regulators MarA, SoxS, and Rob control about 40 genes involved in resistance to antibiotics, heavy metals, organic solvents, and oxidative stress (19–22), and together they form the so-called mar/sox/rob regulon, whose function is associated with binding to the so-called marbox, a 20-bp asymmetric sequence (23). Due to the high levels of sequence similarity of the three regulators, an overlapping regulon is observed, albeit with different efficiencies for activation of particular promoter regions, depending on the transcriptional activator concentration (24), DNA-binding affinity for the specific region (23–25), and RNA polymerase attraction (26, 27).
Expression of marA, rob, and soxS is increased upon exposure to a wide variety of signals. Transcription of MarA (multiple-antibiotic resistance) is controlled by the local repressor MarR and can be derepressed through various compounds, such as salicylate, benzoate, and plumbagin (28, 29). Expression of soxS (superoxide stress) is activated by the oxidative stress sensor SoxR. Upon exposure to superoxides, nitric oxide, or redox cycling compounds, such as paraquat and plumbagin, the [2Fe-2S] cluster of SoxR becomes oxidized, allowing it to activate the transcription of soxS (22, 30).
Rob (right oriC binding) is different from MarA and SoxS in that it is a larger protein consisting of two domains. The N-terminal, DNA-binding domain shows high degrees of sequence similarity to the small activators MarA and SoxS, whereas the C-terminal domain can bind low-molecular-weight effectors (31). Rob is constitutively present at high concentrations in cells and is posttranslationally activated by a sequestering-dispersal mechanism (32–34). Under noninducing conditions, Rob is inactive because it is sequestered into aggregates. Several compounds, including decanoate, unconjugated bile salts, and dipyridyl, bind to the C-terminal domain of Rob and induce its dispersal from the sequestered state, thus enabling Rob to activate transcription of its target genes (14, 35).
MarA, SoxS, and Rob are known for their role in promoting antibiotic resistance due to the upregulation of the multidrug efflux pump AcrAB-TolC in E. coli (13, 36–38). Despite the fact that these activators are able to induce the expression of acrAB under stress conditions in E. coli, they appear to be less important in regulation of multidrug resistance in S. enterica (39). Interestingly, in S. enterica, RamA (resistance antibiotic multiple), another member of the AraC/XylS family, was found to be involved in conferring multidrug resistance by directly increasing the expression of acrAB (39). Furthermore, it has also been demonstrated that mutational inactivation of the AcrAB-TolC pump leads to increased expression of ramA in S. enterica (40).
The aim of this study was to identify members of the AraC/XylS family involved in regulation of multidrug efflux in E. amylovora. We identified genes for four transcriptional regulators of the AraC/XylS family with homology to MarA, SoxS, and Rob of E. coli in the genome sequence of E. amylovora. Two regulators were homologous to either Rob or SoxS, whereas the other two proteins, PliA and OpiA, did not show significant similarity to characterized members of the AraC/XylS family. We characterized the roles of these regulators in antibiotic resistance, organic solvent tolerance, and oxidative stress. Moreover, their contributions to virulence in apple rootstock and immature pear fruits were investigated.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1 and in Table S1 in the supplemental material, respectively. E. amylovora strains were cultured at 28°C in lysogeny broth (LB), double yeast tryptone (dYT), or asparagine minimal medium 2 (AMM2; 10 g fructose, 4 g l-asparagine, 12.8 g Na2HPO4 · 7H2O, 3 g K2HPO4, 3 g NaCl, 0.2 g MgSO4 · 7H2O, 0.25 g nicotinic acid, 0.2 g thiamine per liter) (8). E. coli XL-1 Blue was used as the cloning host. E. coli DH5α λpir was used as the host for the replacement vectors. E. coli cells were routinely maintained at 37°C in dYT medium. Cultures harboring individual vectors were supplemented with 50 μg/ml ampicillin (Ap) for E. coli or 250 μg/ml Ap for E. amylovora, 25 μg/ml chloramphenicol (Cm), 2 μg/ml gentamicin (Gm), or 25 μg/ml kanamycin (Km), where necessary. Bacterial growth was monitored using a spectrophotometer to measure the optical density at 600 nm (OD600).
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant characteristics or genotypea | Reference or source |
|---|---|---|
| Escherichia coli strains | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tcr)] | Stratagene |
| S17-1 | Tcr Smr recA pro hsdR (RP4-2-Tc::Mu-Km::Tn7) | 90 |
| S17-1 λpir | λpir phage lysogen of S17-1 | 90 |
| DH5α λpir | F− supE44 ΔlacU169 (ϕlacZΔM15) recA1 endA1 hsdR17(rK− mK+) thi-1 gyrA96 relA1 λpir phage lysogen | D. Lies, Caltech |
| Erwinia amylovora strains | ||
| Ea1189 | Wild type | GSPBb |
| Ea1189-3 | Ea1189 with Kmr cassette in acrB | 8 |
| Ea1189.opiA− | Ea1189 with Cmr cassette in opiA | This study |
| Ea1189.pliA− | Ea1189 with Cmr cassette in pliA | This study |
| Ea1189.rob− | Ea1189 with Cmr cassette in rob | This study |
| Ea1189.soxS− | Ea1189 with Cmr cassette in soxS | This study |
Cmr, chloramphenicol resistant; Kmr, kanamycin resistant; Smr, streptomycin resistant; Tcr, tetracycline resistant.
GSPB, Göttinger Sammlung Phytopathogener Bakterien, Göttingen, Germany.
PCR amplifications, modifications, and protein purification.
PCR primers are listed in Table S2 in the supplemental material. Primers were designed based on the genome sequence of E. amylovora CFBP1430 available from NCBI (accession no. NC_013961.1). Screening PCRs were carried out using DreamTaq DNA polymerase (Thermo Scientific) in accordance with the manufacturer's instructions. For high-fidelity PCRs, Phusion DNA polymerase (Thermo Scientific) was used.
Restriction enzyme (Thermo Scientific) and T4 DNA ligase (Thermo Scientific) reactions were performed following the manufacturer's instructions, with restriction enzyme reactions performed at the appropriate temperature and all ligation reaction mixtures incubated at room temperature. DNA purifications were performed using either a GeneJET PCR purification kit (Thermo Scientific) or a GeneJET gel extraction kit (Thermo Scientific) according to the manufacturer's instructions.
Protein purification was carried out using a Ni-nitrilotriacetic acid (Ni-NTA) spin kit (Qiagen) following the manufacturer's instructions.
Construction of pliA- and rob-deficient mutants of E. amylovora.
For mutagenesis of the pliA gene, a 1,013-bp fragment was amplified using the primer pair pliA_ko_fwd and pliA_ko_rev. For deletion of the rob gene, a 1,087-bp fragment was amplified using the primer pair rob_ko_fwd and rob_ko_rev. Obtained PCR products were verified by sequencing. Next, a chloramphenicol cassette flanked by Flp-FRT sites was cut from plasmid pFCm1 (96) and inserted into the PstI-digested pliA fragment as well as into the AatII-digested, blunt-ended rob fragment. The deletion alleles were further cloned into the final replacement vector pCAM-Km by use of SpeI and EcoRI, yielding plasmids pCAM-Km.pliA-ko.Cm and pCAM-Km.rob-ko.Cm. The plasmids were transformed into electrocompetent cells of E. amylovora Ea1189, which subsequently were grown for 3 h at 28°C in dYT broth. Putative mutants were screened for homologous recombination events by testing their antibiotic resistance. Mutants that resulted from single-crossover events were identified by the ability to grow on plates containing Km. In order to confirm gene deletion through a double-crossover event in Cm-resistant and Km-sensitive colonies, primers binding up- and downstream of the knockout fragments were designed (pliA_out and rob_out primers). PCRs were done using these locus-specific primers in combination with outward-facing primers binding within the Cm cassette (cat_out primers). PCR products were verified by sequencing.
Construction of soxS- and opiA-deficient mutants of E. amylovora.
For generation of the soxS knockout vector, a 1,268-bp fragment was amplified using the primer pair soxS_ko_fwd and soxS_ko_rev. A chloramphenicol cassette flanked by Flp-FRT sites was cut from plasmid pFCm1 (96) and inserted into the EcoRV-digested soxS fragment. The deletion allele was further cloned into the final replacement vector pCAM-Km via BglII, yielding plasmid pCAM-Km.soxS-ko.Cm.
The construction of the opiA knockout vector was based on the protocol described by Zumaquero et al. (41). Briefly, a 1,220-bp fragment flanking the 5′ region and a 777-bp fragment flanking the 3′ region of the opiA gene were PCR amplified using primer pairs opiA-A5-1–opiA-A6 and opiA-B3–opiA-B4, respectively. Primers opiA-A6 and opiA-B3 share a 20-nucleotide homologous sequence at their 5′ ends, consisting of the T7 primer sequence and a KpnI restriction site (41). The obtained fragments were gel purified, and approximately 40 ng (each) of A and B fragments was used for a fusion PCR with primers opiA-A5 and opiA-B4. The resulting fusion product was gel purified and confirmed by sequencing. Next, a chloramphenicol cassette flanked by Flp-FRT sites was cut from plasmid pFCm1 and subsequently inserted into the KpnI-digested opiA fragment. The deletion allele was cut with BglII and further ligated into BglII-digested pCAM-Km, yielding the final replacement plasmid pCAM-Km.opiA-ko.Cm.
Next, knockout constructs were transformed into electrocompetent cells of E. amylovora Ea1189, and double-crossover mutants were selected as described above.
Cloning of the pliA, rob, soxS, and opiA regions of E. amylovora.
The pliA, rob, soxS, and opiA genes, including the respective promoter regions, were PCR amplified using the primer pairs pliA-P-fwd-KpnI–pliA_rev_BamHI (594 bp), rob-P-fwd-ApaI–rob_rev_SacII (1,160 bp), soxS-P-fwd-KpnI–soxS_rev_BamHI (606 bp), and opiA-P-fwd-ApaI–opiA_rev_BamHI (720 bp), respectively. The obtained PCR products were sequenced and subsequently cloned into pBlueScript II SK(+) in the opposite orientation with respect to the lac promoter to obtain expression from the native promoters, yielding plasmids pBlueSK.pliA-Pro, pBlueSK.rob-Pro, pBlueSK.soxS-Pro, and pBlueSK.opiA-Pro. In order to obtain expression from the lac promoter, the respective genes were amplified using the forward primers pliA_fwd_KpnI, rob_fwd_ApaI, soxS_fwd_KpnI, and opiA_fwd_KpnI in combination with the above-mentioned reverse primers. The PCR products were cloned into pBlueScript II KS(+), yielding plasmids pBlueKS.pliA, pBlueKS.rob, pBlueKS.soxS, and pBlueKS.opiA. E. amylovora does not carry a homologue of the lac repressor. Therefore, the lac promoter shows full activity in E. amylovora without the addition of inducers.
Drug susceptibility tests.
The MICs of antimicrobial compounds on E. amylovora strains were determined by a 2-fold dilution assay using 96-well plates and Mueller-Hinton broth (MHB). All tests were performed at least in triplicate following the recommendations of the Clinical and Laboratory Standards Institute (42). Growth of bacteria at 28°C was examined by visual inspection after 48 h of incubation. The MIC was defined as the lowest concentration of an antibiotic that completely prevented visible cell growth.
Solvent tolerance assay.
Organic solvent tolerance levels of bacteria were visually determined by the appearance of colony formation on agar plates overlaid with n-hexane and cyclohexane. Therefore, an overnight culture was diluted to an OD600 of 0.1, and 20 μl of the diluted cell suspension was spotted onto an agar plate and kept until the spot was completely dried. Next, plates were overlaid with 2 ml n-hexane or 1 ml cyclohexane, sealed, and incubated at 28°C for 48 to 60 h. Growth was visually determined by means of colony formation.
Paraquat-induced superoxide resistance.
Superoxide resistance was tested on MHB II agar plates supplemented with different concentrations of paraquat (10, 20, 30, 40, and 50 μg/ml) as previously described (43). Briefly, overnight cultures grown in MHB broth were diluted to an OD600 of 0.1 and further cultivated until an OD600 of approximately 1.0. Next, 10-μl aliquots of the bacterial cultures were spread on agar plates supplemented with paraquat. After 48 to 60 h of incubation at 28°C, the plates were visually monitored for bacterial growth by means of colony formation.
Determination of intracellular ROS levels.
In order to measure intracellular peroxide levels, we utilized the reactive oxygen species (ROS)-sensitive probe 2′,7′-dichlorofluorescein diacetate (DCF; Sigma-Aldrich) as previously described (44). Briefly, overnight cultures grown in LB broth were diluted to an OD600 of 0.1 and further incubated at 28°C until an OD600 of 0.5 was obtained. Cells were harvested by centrifugation and washed with phosphate-buffered saline (PBS). Next, the cell suspension was incubated with 10 μM DCF at 28°C with shaking at 220 rpm for 30 min. Fluorescence intensity was measured using an Infinite M1000 Pro microplate reader (Tecan, Crailsheim, Germany), with an excitation wavelength of 485 nm and emission detection at 528 nm.
Effects of pH and osmotic stress on growth of Erwinia amylovora.
Overnight cultures grown in AMM2 medium were diluted to an OD600 of 0.1 and further cultured in AMM2 medium at pH 5.5, 7.0, or 8.5 or in AMM2 medium supplemented with 300 mM NaCl. Cultures were incubated at 28°C for 24 h in a shaking incubator at 220 rpm. Growth of the bacteria was determined using a spectrophotometer to measure the OD600.
Promoter activity of the RND efflux pump AcrAB in vitro.
A transcriptional fusion between the promoter region of acrAB and the reporter gene egfp was employed to study the impact of overexpression of pliA, rob, soxS, and opiA on expression of the multidrug efflux pump AcrAB in E. amylovora. The overexpression plasmids carrying the AraC/XylS regulator genes under the control of the lac promoter were transformed into Ea1189 harboring plasmid pBBR.acrA-Pro.egfp (45, 46). Transformed cells of Ea1189 were grown at 28°C in LB broth until stationary phase, harvested by centrifugation, resuspended in phosphate-buffered saline, and adjusted to an OD600 of 0.5. Next, enhanced green fluorescent protein (EGFP) fluorescence was measured using an Infinite M1000 Pro microplate reader (Tecan, Crailsheim, Germany) set to an excitation wavelength of 470 nm, with emission detection at 516 nm.
Virulence assay on apple rootstock.
Apple plants (rootstock Malus MM106) were grown in a greenhouse at 20 to 25°C with 60% humidity and a 12-h photoperiod (15,000 lx). E. amylovora strains were grown on LB agar for 24 h and then resuspended and diluted to an OD600 of 1.0 in sterile demineralized water. Apple plants were inoculated by the prick technique (47). Each bacterial strain was inoculated into one shoot each of a minimum of five single plants. A bacterial suspension (5 μl) was placed onto each wound on the shoot tip. Plants were monitored for symptom development daily. Survival of bacteria in plant tissue was examined by reisolation of bacterial cells 1, 3, and 7 days after inoculation, from 1 cm of the shoot tip around the inoculation area.
In order to analyze mRNA transcript abundances of pliA, rob, soxS, and opiA during growth of E. amylovora in apple rootstock MM106, total RNAs were isolated from infected shoots at 1 and 7 days postinoculation. Five individual wound regions were pooled, homogenized in sterile water (1 ml/wound), and centrifuged for 2 min at 4,000 rpm. The supernatant was transferred to 15 ml killing buffer (20 mM Tris-HCl, pH 7.5; 20 mM NaN3) (48) and centrifuged for 20 min at 4,000 rpm. The supernatant was decanted and the pellet frozen at −80°C for further RNA extraction. Total RNA was isolated by acid phenol-chloroform extraction (48).
Virulence assay on immature pears.
Virulence of E. amylovora Ea1189 and pliA, rob, soxS, and opiA mutants was determined on immature pears (Pyrus communis L. cv. Bartlett). Bacteria grown on LB agar plates at 28°C for 24 h were resuspended and adjusted to an OD600 of 1.0 in sterile demineralized water for inoculation. Immature pear fruits were surface sterilized and pricked with a sterile needle (49). Wounds were inoculated with 5 × 106 CFU/ml, and fruits were incubated in a humidified chamber at 18°C for 14 days. Disease symptoms were visually recorded as detection of necrosis surrounding the infection site.
To analyze gene expression of E. amylovora during growth on pear fruits, immature fruits were cut in slices (approximately 0.5 cm), and five slices were inoculated with 100 μl of a bacterial suspension adjusted to an OD600 of 1.0 in sterile demineralized water. The suspension was evenly distributed on the slice. After 12 h of incubation in a humidified chamber at room temperature, the upper layer of the surface was scratched from the five slices, resuspended in 25 ml of PBS, and centrifuged for 2 min at 4,000 rpm (45). The supernatant was transferred to 15 ml killing buffer and further processed as described above.
RNA isolation and quantitative reverse transcription-PCR (qRT-PCR).
Cell cultures were grown in LB broth until the desired optical densities were reached. An aliquot containing 15 × 109 CFU (equivalent of 15 ml at an OD600 of 1.0) was transferred to 15 ml killing buffer (48) and centrifuged for 20 min at 4,000 rpm. The supernatant was decanted and the pellet frozen at −80°C for further RNA extraction.
Total RNA was isolated using a GeneJET RNA purification kit (Thermo Scientific) following the manufacturer's instructions. The obtained RNA was treated with DNase (Ambion/Life Technologies) and subsequently checked for purity by gel electrophoresis and determination of the A260/A280 and A260/A230 ratios, using a Nanodrop ND-2000 spectrophotometer (Thermo Fischer Scientific). High-quality RNA was reverse transcribed and amplified with a OneStep RT-PCR kit according to the manufacturer's protocol (Qiagen). Template RNA (5 ng) was used in a standard 25-μl qRT-PCR mixture with specific primers (see Table S2 in the supplemental material). As a control, RNA samples without reverse transcriptase were included to detect possible DNA contamination.
For analysis, a Mastercycler ep realplex2 gradient S machine (Eppendorf, Hamburg, Germany) was used. Cycling parameters included a 15-min initial denaturation at 95°C to activate the DNA polymerase followed by 40 cycles consisting of 15 s at 95°C, 30 s at 55°C, and 30 s at 72°C. The final step consisted of 1 min at 95°C and 30 s at 55°C. A melting curve analysis with a temperature ramp from 25°C to 95°C in 20 min was performed at the end of each run to determine the specificity of amplified qPCR products.
Each sample was analyzed for gene expression in triplicate. Quantification of mRNA transcripts was performed by the comparative threshold cycle (CT) method. Briefly, the CT values of the samples of interest were compared with that of a nontreated sample. All CT values were normalized to the housekeeping gene recA, which shows constant expression at different ODs and with different medium compositions, as well as an amplification efficiency similar to that of the target gene (45, 50). The comparative CT method was calculated as 2−(ΔCT,sample − ΔCT,reference), where the ΔCT value was normalized to that for the endogenous housekeeping gene recA. Subsequently, fold changes between the samples were determined based on the calculated CT method.
RESULTS
Bioinformatic analysis of MarA/Rob/Sox homologues in E. amylovora.
Searches with the BLASTP program (NCBI), using the amino acid sequences of MarA, Rob, and SoxS from E. coli K-12 as queries, identified four homologous sequences in the genome of E. amylovora CFBP1430 (see Table S3 in the supplemental material). One homologue (EAMY_2956) shared 72% amino acid sequence identity with Rob from E. coli. A sequence alignment between Rob from E. amylovora and Rob from E. coli showed that the N-terminal, DNA-binding HTH domains share 77% identity with each other, whereas the C-terminal, effector-binding GyrI-like domains share only 59% identity. Analysis of the up- and downstream regions flanking the rob homologues from E. amylovora and E. coli using the Artemis comparison tool (91) revealed several differences. The upstream region of rob from E. coli contains the creABCD locus, including the two-component system creBC, involved in catabolic regulation (51). In E. amylovora, only creA is present upstream of rob (see Fig. S1). The downstream regions are highly similar. However, an additional small hypothetical protein gene is carried downstream of rob in E. amylovora.
The genome sequence of E. amylovora also contains genes for a homologue of the oxidative stress regulator system SoxRS from E. coli, which consists of the oxidative stress sensor SoxR, a member of the MerR family, and the AraC/XylS family member SoxS. SoxS from E. amylovora (EAMY_3287) shares 64% identity with SoxS from E. coli, and SoxR from E. amylovora (EAMY_3286) shares 72% identity with SoxR from E. coli (see Table S3 in the supplemental material). The up- and downstream regions flanking the soxRS locus in E. amylovora CFBP1430 and E. coli K-12 are similar (see Fig. S1). However, both regions of E. amylovora contain insertions. The gstA gene, encoding a glutathione S-transferase, is located upstream of soxR in E. amylovora. The downstream region of soxS from E. amylovora contains an insertion of about 5.8 kb, encoding several small hypothetical proteins, and hecB, encoding a member of the two-partner secretion family involved in protein export.
BLASTP searches identified genes for two additional members of the AraC/XylS family in the genome of E. amylovora, with the proteins sharing high levels of amino acid sequence identity with MarA, Rob, and SoxS from E. coli K-12. The proteins EAMY_1795 (annotated Rob1) and EAMY_3560 (annotated SoxS3) were found to share about 50% identity to MarA, Rob, and SoxS from E. coli (see Table S3 in the supplemental material). Based on our findings, we propose renaming EAMY_1795 “PliA” (plant-inducible activator) and renaming EAMY_3560 “OpiA” (osmotically and pH-inducible activator).
Phylogenetic analysis of all 13 AraC/XylS family members from E. amylovora showed that Rob, SoxS, PliA, and OpiA are closely related (92, 93) (see Fig. S2 in the supplemental material). The close relationship of the four proteins is based on the conserved HTH domain responsible for specific DNA binding (23, 92, 94, 95) (see Fig. S3).
Inactivation of the transcriptional regulators PliA, Rob, SoxS, and OpiA has no effect on antibiotic resistance of E. amylovora.
Transcriptional regulators of the AraC/XylS family are well known for their role in coordinating the responses of enterobacteria to various chemical stresses and antimicrobial compounds. In order to investigate the impact of the transcriptional regulators PliA, Rob, SoxS, and OpiA of E. amylovora Ea1189 on antibiotic resistance, the respective genes were disrupted by insertion of a chloramphenicol cassette. Antibiotic susceptibility tests against a variety of antimicrobial agents were performed. However, disruption of the transcriptional regulators resulted in no change in sensitivity to any tested compound (see Table S4 in the supplemental material).
Overexpression of transcriptional regulators increases resistance to fusidic acid, nalidixic acid, tetracycline, and novobiocin.
Previous studies have shown that high-level expression of AraC/XylS regulators is required to induce antibiotic resistance in enteric bacteria (52–54). Therefore, we generated high-copy-number plasmids overexpressing the pliA, rob, soxS, and opiA genes from their native promoters. Levels of overexpression determined by qRT-PCR were 69-fold for pliA, 34-fold for rob, 48-fold for soxS, and 56-fold for opiA in cells grown in LB broth to an OD600 of 0.5.
The overexpression plasmids were mobilized into the E. amylovora Ea1189 wild type as well as into the acrB-deficient mutant Ea1189-3, which is hypersensitive to many drugs (8), and the sensitivities of the transformants to various substrates were determined (Table 2). Overexpression of pliA caused elevated resistance of the wild type to fusidic acid (4-fold), nalidixic acid (4-fold), and tetracycline (4-fold), whereas overexpression of rob increased the resistance of the wild type to fusidic acid (4-fold) and tetracycline (4-fold). The resistance profiles of transformants overexpressing soxS and opiA in the wild-type background were not altered. In the acrB-deficient mutant, overexpression of rob and opiA caused a 4-fold increase in resistance to novobiocin.
TABLE 2.
Antimicrobial susceptibility profiles of E. amylovora Ea1189 and the acrB-deficient mutant Ea1189-3, harboring the plasmids pBlueScript II SK, pBlueSK.pliA, pBlueSK.rob, pBlueSK.soxS, and pBlueSK.opiAa
| Drug or compound | MIC (μg/ml)b |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ea1189 |
Ea1189-3 |
|||||||||
| pBlueScript II SK | pBlueSK.pliA | pBlueSK.rob | pBlueSK.soxS | pBlueSK.opiA | pBlueScript II SK | pBlueSK.pliA | pBlueSK.rob | pBlueSK.soxS | pBlueSK.opiA | |
| Flavones | ||||||||||
| Daidzein | >5,000 | >5,000 | >5,000 | >5,000 | >5,000 | 125 | 125 | 125 | 125 | 125 |
| Genistein | >1,000 | >1,000 | >1,000 | >1,000 | >1,000 | 62.5 | 62.5 | 62.5 | 62.5 | 62.5 |
| Naringenin | 1,000 | 1,000 | 1,000 | 1,000 | 1,000 | 250 | 250 | 250 | 250 | 250 |
| Phloretin | 5,000 | 5,000 | 5,000 | 5,000 | 5,000 | 312 | 312 | 312 | 312 | 312 |
| Antibiotics | ||||||||||
| Amikacin | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 12.5 | 12.5 | 6.25 |
| Azithromycin | 0.63 | 0.63 | 1.25 | 0.63 | 0.63 | 0.31 | 0.31 | 0.31 | 0.31 | 0.31 |
| Cefepime | 12.5 | 12.5 | 12.5 | 12.5 | 12.5 | 5 | 5 | 10 | 10 | 5 |
| Chloramphenicol | 3.13 | 6.25 | 6.25 | 6.25 | 3.13 | 1.56 | 1.56 | 1.56 | 1.56 | 1.56 |
| Ciprofloxacin | 0.03 | 0.06 | 0.03 | 0.03 | 0.03 | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 |
| Erythromycin | 1.25 | 1.25 | 1.25 | 1.25 | 1.25 | 1.25 | 1.25 | 1.25 | 2.50 | 1.25 |
| Fusidic acid | 250 | 1,000 | 1,000 | 250 | 250 | 3.13 | 3.13 | 6.25 | 3.13 | 3.13 |
| Nalidixic acid | 2.5 | 10 | 2.5 | 2.5 | 2.5 | 0.63 | 0.63 | 0.63 | 0.63 | 0.63 |
| Norfloxacin | 0.16 | 0.16 | 0.16 | 0.16 | 0.16 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 |
| Novobiocin | 250 | 500 | 250 | 250 | 250 | 6.25 | 6.25 | 25 | 6.25 | 25 |
| Tetracycline | 1.25 | 5 | 5 | 2.5 | 1.25 | 0.63 | 0.63 | 0.63 | 0.63 | 0.63 |
| Heavy metals | ||||||||||
| Cadmium acetate | 50 | 50 | 50 | 100 | 50 | 100 | 100 | 100 | 100 | 100 |
| Copper sulfate | 1,250 | 1,250 | 1,250 | 1,250 | 1,250 | 1,250 | 1,250 | 1,250 | 1,250 | 1,250 |
| Silver nitrate | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 |
| Zinc sulfate | 500 | 500 | 500 | 500 | 500 | 500 | 500 | 500 | 500 | 500 |
| Antimicrobials | ||||||||||
| Bile | 5,000 | 5,000 | 5,000 | 5,000 | 5,000 | 2,500 | 2,500 | 2,500 | 2,500 | 2,500 |
| Phenazine | 50 | 100 | 100 | 100 | 50 | 12.5 | 12.5 | 12.5 | 12.5 | 12.5 |
| Plumbagin | 50 | 100 | 100 | 100 | 100 | 6.25 | 12.5 | 12.5 | 12.5 | 12.5 |
| Tannin | 2,500 | 2,500 | 2,500 | 2,500 | 2,500 | 1,250 | 1,250 | 1,250 | 1,250 | 1,250 |
| Dyes | ||||||||||
| Acriflavine | 125 | 125 | 125 | 125 | 125 | 12.5 | 12.5 | 12.5 | 12.5 | 12.5 |
| Crystal violet | 12.5 | 12.5 | 12.5 | 12.5 | 12.5 | 1.25 | 1.25 | 1.25 | 1.25 | 1.25 |
| Ethidium bromide | 500 | 500 | 1,000 | 1,000 | 500 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 |
The pliA, rob, soxS, and opiA genes, including the respective promoter regions, were cloned into pBlueScript II SK(+) in the opposite orientation with respect to the lac promoter to obtain expression from the native promoters.
MICs were determined by 2-fold dilution assay in three or more independent experiments, with similar results. Numbers in bold indicate increases of >2-fold.
Organic solvent tolerance.
Because overexpression of soxS or robA causes increased organic solvent tolerance in E. coli (53, 55), we were prompted to investigate solvent tolerance levels of E. amylovora mutants deficient in PliA, Rob, SoxS, and OpiA, as well as those of transformants overexpressing these regulators.
Our data show that E. amylovora Ea1189 is able to grow in the presence of n-hexane but not in the presence of cyclohexane. Interestingly, the pliA- and rob-deficient mutants showed decreased growth in the presence of n-hexane compared to the wild-type strain (Table 3), indicating a possible involvement of PliA and Rob in regulation of solvent detoxification mechanisms. In agreement with this observation, we found that overexpression of pliA and rob increased the resistance of E. amylovora Ea1189 to n-hexane. Furthermore, we demonstrated that overexpression of pliA and soxS increased tolerance toward cyclohexane.
TABLE 3.
Organic solvent tolerance of E. amylovora Ea1189 and pliA-, rob-, soxS-, and opiA-deficient mutants, as well as of the wild-type strain and the acrB-deficient mutant Ea1189-3 harboring the plasmids pBlueScript II SK, pBlueSK.pliA, pBlueSK.rob, pBlueSK.soxS, and pBlueSK.opiAa
| Strain | Mutation or plasmid | Organic solvent toleranceb |
|
|---|---|---|---|
| n-Hexane | Cyclohexane | ||
| Ea1189 | Wild type | + | − |
| pliA | − | − | |
| rob | +/− | − | |
| soxS | + | − | |
| opiA | + | − | |
| pBlueScript II SK | + | − | |
| pBlueSK.pliA | ++ | + | |
| pBlueSK.rob | ++ | − | |
| pBlueSK.soxS | + | + | |
| pBlueSK.opiA | + | − | |
| Ea1189-3 | pBlueScript II SK | + | − |
| pBlueSK.pliA | + | − | |
| pBlueSK.rob | ++ | − | |
| pBlueSK.soxS | + | − | |
| pBlueSK.opiA | + | − | |
The pliA, rob, soxS, and opiA genes, including the respective promoter regions, were cloned into pBlueScript II SK(+) in the opposite orientation with respect to the lac promoter to obtain expression from the native promoters.
++, growth; +, single colonies; −, no growth. Data presented are observations from at least three replicates.
It was previously reported that deletion of the multidrug efflux pump AcrAB from E. coli resulted in hypersusceptibility to both n-hexane and cyclohexane (21). Our data revealed that the acrB-deficient mutant of E. amylovora was able to survive on n-hexane but not cyclohexane (Table 3), indicating that resistance mechanisms other than efflux are involved in organic solvent tolerance in the plant pathogen E. amylovora.
Rob affects expression of the RND-type efflux pump AcrAB in E. amylovora.
Since the expression of the multidrug efflux pump AcrAB is modulated by the AraC/XylS family members MarA, SoxS, and Rob in E. coli (13, 14), as well as by RamA in S. enterica (56), we investigated whether the expression of acrAB is regulated by the homologous regulators PliA, Rob, SoxS, and OpiA in E. amylovora Ea1189. Therefore, we used a transcriptional fusion between the promoter region of acrAB and the reporter gene egfp, as previously described (45). Our results showed a 2-fold induction of acrAB in cells harboring the rob overexpression plasmid. However, overexpression of pliA, soxS, or opiA did not affect the promoter activity of acrAB (Table 4).
TABLE 4.
Effects of overexpression of the transcriptional regulators PliA, Rob, SoxS, and OpiA on expression of the multidrug efflux pump AcrABa
| Plasmid | Fold change in acrAB promoter activityb | Fold change in mRNA transcript levelc |
|---|---|---|
| pBlueKS.pliA | 1 | 1.1 |
| pBlueKS.rob | 2 | 2.3 |
| pBlueKS.sox | 0.9 | 1 |
| pBlueKS.opiA | 0.9 | 1 |
The pliA, rob, soxS, and opiA genes were cloned into pBlueScript II KS(+) to obtain expression from the lac promoter.
Fold changes in the acrAB promoter activity were determined by measuring the fluorescence of Ea1189 cells cotransformed with regulator overexpression plasmids and pBBR.acrA-Pro.egfp, harboring a transcriptional fusion of the acrAB promoter to a promoterless egfp gene. Cells were grown in LB until stationary phase.
Total RNA was isolated from cells grown in LB broth to an OD600 of 0.5. Transcript abundances in cells overexpressing the regulators were determined by quantitative RT-PCR and compared to the RT-PCR signal from cells transformed with an empty expression plasmid.
In addition, we used qRT-PCR analysis to elucidate whether overexpression of the transcriptional activators PliA, Rob, SoxS, and OpiA causes induction of the AcrAB efflux pump in E. amylovora. Therefore, overexpression of the transcriptional regulators was achieved from high-copy-number plasmids carrying the genes under the control of the lac promoter (163-fold for pliA, 332-fold for rob, 287-fold for soxS, and 407-fold for opiA). Fold changes in mRNA transcript levels were determined for cells grown in LB broth to an OD600 of 0.5.
In agreement with the fluorescence data, we found a 2.3-fold induction of the acrAB efflux pump in cells overexpressing rob, while the overexpression of the other regulators did not affect the expression of acrAB in E. amylovora (Table 4).
In light of the above findings, we investigated whether Rob is able to bind to the acrAB promoter region under in vitro conditions. Therefore, an electrophoretic mobility shift assay using Cy5-labeled DNA fragments of the acrAB promoter region and His-tag-purified Rob protein was performed. However, no interaction between Rob and the acrAB promoter region was detected (data not shown), indicating that the observed induction of acrAB is not directly mediated by Rob in E. amylovora Ea1189.
Transcriptional analysis of pliA, rob, soxS, and opiA in acrB- and tolC-deficient mutants.
In E. coli, the mar/sox/rob regulon includes the major multidrug efflux pump AcrAB and the outer membrane channel TolC. It has been found that transcription of marA, soxS, and rob is increased in tolC mutants (57), and this prompted us to investigate whether the expression of the regulators pliA, rob, soxS, and opiA is also influenced in an acrB or tolC mutant of E. amylovora Ea1189. However, our analysis revealed that the expression of pliA, rob, soxS, and opiA was not significantly changed in mutants defective in AcrB or TolC (data not shown).
Induction of opiA by osmotic and pH stress.
We analyzed the involvement of the AraC/XylS regulators in response to pH and osmotic stress. First, we investigated the effects of pH and salt concentration on the growth of E. amylovora Ea1189. We found that the growth of Ea1189 was significantly reduced at pH 5.5 and 8.5. The cultures grown in LB broth adjusted to these pH values reached an OD600 of about 1.0 after 24 h, whereas cultures grown at pH 7.0 reached an OD600 of 2.6 (see Fig. S4A in the supplemental material). A similar growth reduction was found in cultures of Ea1189 supplemented with 300 mM NaCl (see Fig. S4B).
The growth of the pliA, rob, soxS, and opiA mutants was not altered compared to the growth of the wild type in response to pH changes or increasing salt concentrations (Fig. 1A). However, overexpression of opiA increased growth of E. amylovora >2-fold at alkaline pH and a high salt concentration (300 mM) (Fig. 1B).
FIG 1.
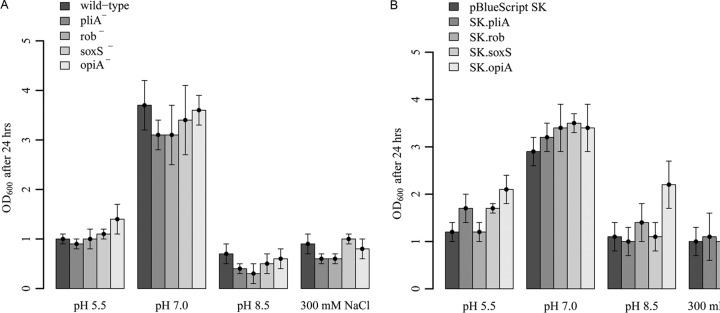
pH and osmolarity sensitivity assays. (A) E. amylovora Ea1189 wild type and pliA-, rob-, soxS-, and opiA-deficient mutants. (B) E. amylovora Ea1189 harboring overexpression plasmids. Tests were performed at pH 5.5, 7.0, and 8.5 and with 300 mM NaCl (stress concentrations were determined prior to assays [see Fig. S4 in the supplemental material]). Optical densities were determined after 24 h of growth in AMM2 medium.
pliA and soxS are induced by phenolic acids and ROS.
We conducted qRT-PCR analysis to explore whether the expression of pliA, rob, soxS, or opiA is induced by phytochemicals, phenolic acids, ROS producers, or heavy metals (Table 5). Expression of pliA was increased upon exposure to a wide variety of signals. Transcript levels of pliA were increased in response to apple leaf extracts (4.4-fold), the phenolic acids gallic acid (6.2-fold) and salicylic acid (3.7-fold), the ROS inducers indole (10-fold), paraquat (3.9-fold), plumbagin (67.8-fold), and phenazine methosulfate (29.9-fold), the fatty acid sodium decanoate (109.6-fold), and the heavy metals copper (2.7-fold) and zinc (2.6-fold). Expression of soxS was induced by gallic acid (3.3-fold) and after treatment with the ROS producers indole (8.8-fold), paraquat (192.2-fold), plumbagin (16.2-fold), and phenazine methosulfate (18-fold). Expression of rob and opiA was not induced by any of the tested compounds (Table 5). It is worth mentioning that expression of rob was downregulated in response to most of the tested compounds. Additionally, a >3-fold downregulation of soxS and opiA was detected upon exposure of E. amylovora Ea1189 to zinc sulfate for 10 min.
TABLE 5.
Fold changes of pliA, rob, soxS, and opiA mRNA transcript levels in E. amylovora Ea1189 after 2 h of incubation (or 10 min, where indicated) with various substrates, as determined by qRT-PCRa
| Substrate (concn) | Fold change in mRNA transcript levelb |
|||
|---|---|---|---|---|
| pliA | rob | soxS | opiA | |
| Phytochemicals | ||||
| Apple leaf extracts (10 μl/ml) | 4.4 | 0.7 | 1.7 | 1.3 |
| Indole-3-acetic acid (2 mM) | 1.4 | 0.7 | 1.6 | 1.2 |
| Phloretin (4 μg/ml) | 1.3 | 0.6 | 1.4 | 0.5 |
| Sodium decanoate (5 mM) | 109.6 | 0.7 | 1.5 | 0.5 |
| Tannin (0.5 mg/ml) | 1.0 | 0.4 | 1.2 | 0.5 |
| Phenolic acids | ||||
| Gallic acid (1 mg/ml) | 6.2 | 0.7 | 3.3 | 1.4 |
| Salicylic acid (5 mM) | 3.7 | 0.5 | 1.0 | 0.9 |
| ROS producers | ||||
| Indole (2 mM) | 10.0 | 0.8 | 8.8 | 1.1 |
| Paraquat (0.2 mM) | 3.9 | 0.7 | 192.2 | 0.7 |
| Plumbagin (0.1 mM) | 67.8 | 0.4 | 16.2 | 1.0 |
| Phenazine methosulfate (0.1 mM) | 29.9 | 0.2 | 18.0 | 0.6 |
| Heavy metals | ||||
| Copper sulfate (1 mM) | 1.3 | 0.6 | 1.3 | 0.6 |
| Copper sulfate (1 mM, 10 min) | 2.7 | 1.2 | 1.5 | 1.8 |
| Zinc sulfate (1 mM) | 1.4 | 0.7 | 1.5 | 0.5 |
| Zinc sulfate (1 mM, 10 min) | 2.6 | 0.9 | 0.2 | 0.3 |
Total RNA was isolated from bacterial cells incubated with the indicated substrates in LB broth. Transcript abundances were determined by quantitative RT-PCR and compared to the RT-PCR signal from cells grown in LB broth containing only the solvent of the respective substrate.
Values in bold indicate an increase or decrease of >2-fold. The presented data values are the means of results from at least three replicates.
Paraquat inhibits growth of a soxS-deficient mutant.
Members of the AraC/XylS family have been found to coordinate the induction of genes for resistance to oxidative stress (e.g., SoxS of E. coli). To investigate a possible involvement of PliA, Rob, SoxS, and OpiA in oxidative stress resistance, we tested the respective mutants for growth on agar plates supplemented with increasing concentrations of the superoxide-generating agent paraquat. Our results showed that the soxS-deficient mutant was not able to grow on MHB II agar plates containing 10 μg/ml paraquat (see Fig. S5 in the supplemental material), indicating that SoxS is involved in superoxide resistance.
Deletion of Rob increases intracellular ROS levels.
To elucidate the effect of disruption or overexpression of the transcriptional regulator genes pliA, rob, soxS, and opiA on the intracellular redox status of E. amylovora, we determined intracellular ROS levels by measuring the oxidation of the nonfluorescent dye 2′,7′-dichlorodihydrofluorescein diacetate to its highly fluorescent derivative 2′,7′-dichlorofluorescein. Our data revealed that cells of the rob mutant possessed an approximately 2-fold higher intracellular ROS level than that in cells of the wild type (Fig. 2).
FIG 2.
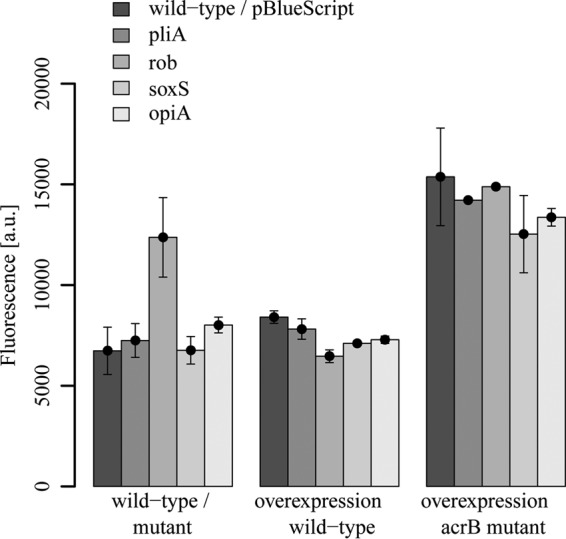
Intracellular ROS levels in the E. amylovora Ea1189 wild type and its pliA-, rob-, soxS-, and opiA-deficient mutants and in strains harboring overexpression plasmids in wild-type and acrB-deficient backgrounds. Cells were grown to an OD600 of 0.5 and incubated with DCF diacetate. Whole-cell fluorescence was determined after 30 min. Fluorescence intensity (absorbance units [a.u.]) was normalized to an OD600 value of 0.1. Experiments were performed in triplicate, with similar results.
Transcriptional analysis of pliA, rob, soxS, and opiA in planta.
In order to analyze the expression of pliA, rob, soxS, and opiA transcripts in planta, E. amylovora was inoculated onto immature pear fruit slices or injected into shoot tips of apple rootstock MM106. Bacteria were reisolated from immature pear fruit slices after 12 h or from apple plants after 1 and 7 days (Table 6). Transcript abundances were determined by qRT-PCR and compared to qRT-PCR signals from cells grown in LB broth to an OD600 of 0.5. Analysis of fold changes in mRNA transcript levels showed 15.3-, 5.5-, and 5.0-fold increases in expression of pliA, soxS, and opiA, respectively, on immature pear fruits. Expression of rob was similar in cells grown in LB broth and cells recovered from infected pear fruits (Table 6).
TABLE 6.
Fold changes of pliA, rob, soxS, and opiA mRNA transcript levels after inoculation of E. amylovora Ea1189 on apple rootstock MM106 and immature pear fruit slicesa
| Gene | Fold change in mRNA transcript level |
||
|---|---|---|---|
| Apple rootstockb |
Immature pearc | ||
| 1 dpi | 7 dpi | 12 hpi | |
| pliA | 97.3 | 1.9 | 15.3 |
| rob | 1.4 | 0.9 | 1.3 |
| soxS | 11.5 | 1.1 | 5.5 |
| opiA | 2.5 | 0.5 | 5.0 |
Total RNAs were isolated from bacterial cells recovered from infected plant tissues. Transcript abundances were determined by quantitative RT-PCR and compared to RT-PCR signals from cells grown in LB broth to an OD600 of 0.5.
Bacteria were reisolated from infected shoots of apple rootstock at 1 and 7 days postinoculation (dpi).
Bacteria were reisolated from infected immature pears at 12 h postinoculation (hpi).
Furthermore, our results demonstrated that expression of pliA was 97.3-fold, expression of soxS 11.5-fold, and expression of opiA 2.5-fold higher during infection of apple rootstock 1 day after inoculation than during growth in LB medium. The expression of the three genes declined to basal levels after 7 days of infection. Like the case for pear fruit infection, expression of rob was not induced during infection of apple rootstock (Table 6).
Contributions of PliA, Rob, SoxS, and OpiA to virulence of E. amylovora on apple rootstock and immature pear fruits.
The development of disease symptoms on apple rootstock MM106 was analyzed to study the impact of the transcriptional regulators PliA, Rob, SoxS, and OpiA on virulence of E. amylovora Ea1189. After 7 days, all shoots showed typical disease symptoms, including shepherd's crook-like bending, tissue necrosis, and ooze formation, regardless of whether the shoots were infected with the wild type or with one of the regulatory mutants. In order to study the establishment of bacterial populations within the tissue, samples were taken at 1, 3, and 7 days postinoculation, and numbers of CFU per stem were determined. The wild type and all regulatory mutants showed no significant differences in growth 3 days after infection. However, the population size of the soxS mutant was approximately 10 times lower than the population size of the wild type after 7 days (Fig. 3; see Table S5 in the supplemental material). This result indicates that soxS contributes to the ability of E. amylovora to colonize apple rootstock.
FIG 3.
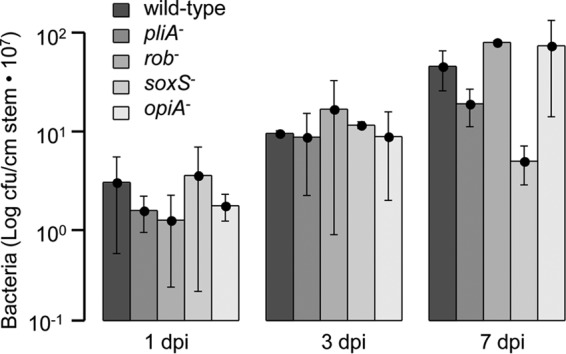
Virulence assay on apple rootstock MM106. Bacteria were inoculated into the shoot tips by the prick technique, with an inoculum of 5 × 106 CFU/shoot. Establishment of populations of the E. amylovora Ea1189 wild type and its pliA-, rob-, soxS-, and opiA-deficient mutants was determined at 1, 3, and 7 days postinoculation (dpi). A minimum of five shoot tips were inoculated for each strain. Data represent the numbers of CFU/cm of stem/shoot tip times 107. Average values and standard deviations are listed in Table S5 in the supplemental material.
Furthermore, since E. amylovora is also able to infect pears, immature pear fruits were used to study the impact of the transcriptional regulator mutants on virulence in this host plant. Fruits were infected with the wild-type strain and the corresponding regulatory mutants and incubated at 18°C for 14 days. However, no significant difference between the wild type and the mutants was observed (see Fig. S6 in the supplemental material).
DISCUSSION
Multidrug efflux systems, such as the AcrAB-TolC complex, play an important role in the intrinsic resistance of bacteria against antimicrobial compounds. The regulation of the acrAB genes has been studied extensively in human-pathogenic enterobacteria. Several members of the AraC/XylS family, including MarA, RamA, Rob, and SoxS, have been identified as positive transcriptional regulators of this efflux system (10, 13, 14, 21, 54, 58).
In this study, we identified four regulators from the plant pathogen E. amylovora that are closely related to the MarA/Rob/SoxS group of AraC/XylS transcriptional activators from E. coli (see Fig. S3 in the supplemental material). Due to the high level of sequence similarity, it was tempting to speculate that the four AraC/XylS regulators from E. amylovora might have a similar function in the regulation of the multidrug efflux pump AcrAB. However, a key finding of this work was that none of the identified regulators is able to induce a multidrug resistance phenotype in E. amylovora. E. amylovora strains overexpressing the regulatory proteins from a high-copy-number vector showed only minor changes in the substrate resistance profiles compared to the wild type (Table 2). Overexpression of rob led to an about 2-fold increase in expression of the acrA gene. However, the strain overexpressing rob showed increased resistance to only a limited number of antibiotics, not to a wide range of antimicrobials as usually associated with induction of the AcrAB-TolC multidrug efflux pump.
Overexpression of rob and of pliA increased the resistance of E. amylovora Ea1189 to fusidic acid and tetracycline and, in the case of pliA, also toward nalidixic acid. The overlap in the resistance patterns of the strains overexpressing Rob and PliA suggests that these regulators induce similar drug resistance mechanisms. However, overexpression of rob and of pliA in the acrB-deficient mutant Ea1189-3 did not increase the resistance to fusidic acid, tetracycline, and nalidixic acid, suggesting that the AcrAB pump is involved in the resistance mechanism promoted by Rob and PliA.
Rob (right origin-binding protein).
There is plenty of evidence that Rob is involved in the regulation of genes conferring a multidrug resistance phenotype in various species of the Enterobacteriaceae family, and the reduced susceptibility to antibiotics has often been linked to the AcrAB-TolC efflux system (21, 59, 60). We also observed a 2-fold increase in expression of acrA upon overexpression of Rob from a high-copy-number vector in E. amylovora (Table 4). However, the increased expression of the acrA gene was not associated with a multidrug resistance phenotype. A similar effect was found in clinical isolates of Klebsiella pneumoniae when the mean expression levels of acrA were correlated with the MIC values for the antibiotic tigecycline. The acrA expression levels differed over the range of MICs for tigecycline (61).
In addition to the role of AcrA as a periplasmic fusion protein in the AcrAB-TolC system, it was shown that AcrA is required for the function of the AcrD efflux pump (62, 63). Thus, the Rob-mediated 2-fold increase in expression of acrA may lead to a higher level of activity of the AcrD pump, which could partially explain the observed resistance phenotype. We recently reported that AcrD of E. amylovora contributes to resistance against fusidic acid, for which the Rob-overexpressing strain had a 4-fold higher MIC value (45). Another possible explanation for why the increased expression of acrA did not lead to an elevated resistance of E. amylovora toward multiple antibiotics may be that the constitutive level of AcrAB is so high that additional expression shows no effect.
Organic solvent tolerance has not been investigated in E. amylovora. Therefore, we assessed the sensitivity of the plant pathogen to n-hexane and cyclohexane and investigated the role of the AraC/XylS regulators in organic solvent tolerance. Disruption of rob caused increased susceptibility to n-hexane, whereas its overexpression resulted in an increased tolerance toward n-hexane (Table 3). Similar phenotypes have been described for E. coli (21). However, overexpression of rob also increased tolerance of E. coli to cyclohexane, a phenotype which was not observed in E. amylovora (21, 53). Furthermore, organic solvent tolerance of E. coli requires a functional AcrAB pump. Deletion of acrAB from the E. coli wild type resulted in a loss of tolerance to n-hexane, and it abolished the induced resistance to cyclohexane in E. coli strains overexpressing rob, marA, and soxS (21). In contrast, overexpression of rob in an acrB-deficient mutant of E. amylovora resulted in an increased n-hexane tolerance. Although organic solvent tolerance mediated by AraC/XylS transcriptional regulators has been associated with the expression of the RND-type efflux pump AcrAB in E. coli and S. enterica (21, 64), our data suggest that Rob of E. amylovora induces mechanisms other than efflux to overcome organic solvent toxicity. Bacteria use a wide variety of mechanisms to increase their organic solvent tolerance (65). Bacteria prevent entry of organic solvents into the cell by modification of lipopolysaccharides (66). To compensate for the damaging membrane fluidity changes imposed by solvents, bacteria allow a denser packing of their membranes by cis/trans-isomerization of unsaturated fatty acids (67). Moreover, activation of general stress responses, including the induction of chaperones that refold proteins denatured by the solvent, and activation of the oxidative stress response have been reported to contribute to bacterial solvent resistance (55, 68). However, the nature of the mechanism(s) of organic solvent tolerance induced by Rob in E. amylovora remains to be identified in future studies.
Our data revealed a general downregulation of rob transcription after addition of various compounds to the growth medium (Table 5). It is known that the transcriptional regulators MarA, SoxS, and Rob regulate each other's expression in E. coli. Both MarA and SoxS were shown to repress the rob promoter (58, 69, 70). Thus, the downregulation of rob might indicate a possible transcriptional cross talk between the homologous regulators of the AraC/XylS family to coordinate the cellular responses to environmental stresses.
SoxS (superoxide stress protein).
Expression of the AraC/XylS family member SoxS has been described to be activated in response to oxidative stress in E. coli and other enterobacteria (15). In the present study, we demonstrated that SoxS from E. amylovora Ea1189 is also involved in superoxide resistance. A soxS-deficient mutant of Ea1189 was not able to grow on agar plates supplemented with the superoxide-generating agent paraquat (see Fig. S5 in the supplemental material). Furthermore, expression of soxS was induced by treatment of the cultures with redox cycling agents, such as phenazine methosulfate, paraquat, and plumbagin. Additionally, soxS expression was increased in the presence of indole, a biological oxidant. Indole is proposed to dissolve in membrane lipids, enabling direct interaction between the redox cycling agents isoprenoid quinones and dioxygen and resulting in the generation of superoxide (71). In the SoxRS regulatory cascade, SoxR, a regulator of the MerR family, senses oxidative stress via the oxidation state of its [2Fe-2S] cluster (72). Upon oxidation, SoxR becomes an activator of soxS transcription, and the resulting increased level of SoxS activates a variety of cellular defense mechanisms against oxidative stress (73).
Our data demonstrated that the expression of soxS was significantly downregulated after exposure of E. amylovora to zinc (Table 5), a finding that has been reported before for E. coli, by Graham et al. (74). Zn(II) is an essential micronutrient for bacteria, although it has significant toxicity at high concentrations (75). Previous results showed that SoxS is a major regulator responsible for growth under Zn-depleted conditions. SoxS increases the expression of the zinc uptake system ZnuACB, and deletion of soxS led to reduced growth of E. coli in Zn-depleted medium (76). We observed a downregulation of soxS in E. amylovora after exposure to 1 mM zinc sulfate, suggesting that the cells of the plant pathogen avoid zinc toxicity by reduction of its import via the ABC transporter ZnuACB. Recently, McDevitt et al. (77) identified a molecular mechanism linking the bacterial susceptibility to zinc with oxidative stress. Zinc interferes with acquisition of manganese by bacterial cells. Mn(II) plays an important role in oxidative stress management, e.g., as a cofactor of the manganese superoxide dismutase or as a substitute for ferrous iron in the enzyme ribulose-5-phosphate 3-epimerase under oxidative stress (78, 79).
The oxidative burst, a rapid production of ROS, is one of the earliest responses of plants to microbial infection (80). The predominant ROS detected in plant-pathogen interactions are superoxide anion (O2−), hydrogen peroxide (H2O2), and hydroxyl radical (OH·) (81). The oxidative burst is usually correlated with incompatible plant-pathogen interactions leading to a rapid and localized programmed cell death known as the hypersensitive response (82). However, it was previously reported that E. amylovora is able to generate an oxidative stress response even in a compatible situation with pear (83). It is suggested that E. amylovora uses the production of ROS as a tool to provoke host cell death during pathogenesis to invade plant tissues.
Our results showed that the promoter activity of soxS was induced during growth of E. amylovora in apple rootstock and immature pear fruits (Table 6). We identified gallic acid, a phenolic compound found in a wide variety of plants, including apple and pear (84, 85), as a plant-derived signal which increased soxS expression 3.3-fold. Furthermore, we found that the soxS-deficient mutant of E. amylovora reached an ∼10-fold-lower population density than that of the wild type on apple rootstock 7 days after infection (Fig. 3). Our results show that SoxS can be induced in response to phenolic acids and the oxidative stress generated by the host during pathogenic interaction and that it is required for successful colonization of a host plant.
PliA (plant-inducible activator protein).
In this work, we characterized a novel regulator of the AraC/XylS family, PliA, in E. amylovora Ea1189. The amino acid sequence of PliA shares about 50% identity to MarA, Rob, and SoxS from E. coli as well as to Rob, SoxS, and OpiA from E. amylovora (see Table S3 in the supplemental material). We demonstrated that plasmid-mediated overexpression of pliA produced resistance to fusidic acid, nalidixic acid, and tetracycline, as well as increased tolerance to n-hexane and cyclohexane, in E. amylovora. The resistance phenotypes required the presence of the functional RND-type efflux pump AcrAB. However, we did not observe an increased expression of acrA upon overexpression of PliA, suggesting that resistance mechanisms other than efflux are induced by the transcriptional activator PliA.
The most striking finding was the high level of induction of pliA expression during the early infection phase in apple rootstock and immature pear fruits (Table 6). Transcription of pliA was induced almost 100-fold 1 day after inoculation of E. amylovora Ea1189 into apple shoot tips. On day 7, the expression level came back to the low expression level of pliA in E. amylovora cells grown in LB broth.
We identified gallic acid and salicylic acid, as well as ROS, as inducers of pliA expression in vitro that may also be responsible for the increased pliA expression in planta. Both gallic and salicylic acids are hydroxybenzoic acids and, as major plant phenolics, play an important role in plant-pathogen interactions (86). The rapid production of ROS is described as one of the earliest responses to pathogen infection (80). Our results strongly suggest that PliA is involved in the immediate response of E. amylovora to contact with its host and that plant phenolics or ROS induce stress responses in E. amylovora via PliA during the early plant infection phase.
Despite the fact that we observed a significant induction of pliA expression in planta, deletion of the pliA gene had no effect on the ability of E. amylovora to colonize its host plants, apple and pear. This inconsistency can be explained by the fact that MarA-like regulators often show redundancy among their regulons. We observed, for instance, that oxidative stress induced SoxS as well as PliA in E. amylovora. Casaz et al. (87) investigated the functions of MarA, SoxS, and Rob from E. coli as virulence factors in a murine model of urinary tract infection. Only an E. coli mutant lacking all three transcriptional regulators was significantly less virulent than the parental strain. Furthermore, complementation of the triple-knockout mutant with marA, soxS, and rob individually restored wild-type virulence.
Multiple compounds were able to induce the expression of pliA, including plant extracts, phenolic compounds, redox cycling agents, heavy metals, and the saturated fatty acid decanoate. However, PliA is a small protein, consisting of 113 amino acids, that contains only a DNA-binding domain. For this protein to exert its regulatory action, the intracellular concentration of PliA must increase. The transcription of other AraC/XylS regulators (e.g., MarA and SoxS) that contain only a DNA-binding motif is controlled by another regulator (17). We identified a gene for a transcriptional regulator of the Rrf2 repressor family upstream of the pliA gene in E. amylovora. One of the best-studied Rrf2 proteins, IscR, represses the iscRSUA operon, encoding Fe-S cluster biosynthesis proteins (88). Several regulators of the widespread but poorly characterized Rrf2 family coordinate an Fe-S cluster that participates in signal transduction (89). The oxidation state of the Fe-S cluster may modulate the function of these regulators, as found with the [2Fe-2S]-containing transcription factor SoxR (72). Future work will examine the role of the Rrf2-type regulator located upstream of pliA in the induction of the AraC/XylS regulator by different environmental signals.
Furthermore, our induction experiments point to a possible transcriptional cross talk between Rob and PliA. The expression of pliA was induced >100-fold by decanoate. Sodium decanoate was originally found to posttranslationally activate Rob in E. coli (14). Whether the observed induction of pliA expression by decanoate is due to binding and activation of Rob in E. amylovora will be a subject of our future studies.
OpiA (osmotically and pH-inducible activator).
OpiA, the fourth MarA-like transcriptional regulator of the AraC/XylS family from E. amylovora, did not show significant sequence similarity to characterized members of this family. The first experiments to characterize this protein did not yield any obvious phenotype, except for a downregulation of opiA transcripts upon exposure to zinc. However, we made an interesting observation during the generation of the opiA mutant. We isolated opiA-deficient mutants only on LB agar without salt. No mutants were found on high-salt LB agar containing 10 g/liter sodium chloride. This finding suggested that OpiA might play a role in the regulation of osmotic stress responses. In order to prove this hypothesis, we tested the opiA mutant for growth at various pHs and salt concentrations. However, we were not able to detect a significant difference in the growth of the mutant and the wild type. Next, we investigated whether the overexpression of opiA might increase the tolerance to pH or osmotic stress, and indeed, we found that opiA overexpression increased tolerance to alkaline pH and high salt concentrations (Fig. 1). Therefore, we concluded that OpiA is involved in the adaptation of E. amylovora to alkaline pH and high salinity. Further experiments need to be conducted to identify which genes are regulated by OpiA in E. amylovora.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by Jacobs University Bremen and by the MOLIFE Research Center, Jacobs University Bremen.
We acknowledge Yvonne Braun for a critical reading of the manuscript, and we thank Anna Elisabeth Oja and Hanna Englert for their contributions in generating several genetic constructs.
Footnotes
Published ahead of print 16 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01838-14.
REFERENCES
- 1.Vanneste JL. 2000. Fire blight: the disease and its causative agent, Erwinia amylovora. CABI Publishing, Wallingford, Oxon, United Kingdom. [Google Scholar]
- 2.Martinez JL, Sanchez MB, Martinez-Solano L, Hernandez A, Garmendia L, Fajardo A, Alvarez-Ortega C. 2009. Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol. Rev. 33:430–449. 10.1111/j.1574-6976.2008.00157.x [DOI] [PubMed] [Google Scholar]
- 3.Nikaido H. 1996. Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178:5853–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piddock LJ. 2006. Multidrug-resistance efflux pumps—not just for resistance. Nat. Rev. Microbiol. 4:629–636. 10.1038/nrmicro1464 [DOI] [PubMed] [Google Scholar]
- 5.Walsh C. 2000. Molecular mechanisms that confer antibacterial drug resistance. Nature 406:775–781. 10.1038/35021219 [DOI] [PubMed] [Google Scholar]
- 6.Zgurskaya HI, Nikaido H. 2000. Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 37:219–225. 10.1046/j.1365-2958.2000.01926.x [DOI] [PubMed] [Google Scholar]
- 7.Pos KM. 2009. Trinity revealed: stoichiometric complex assembly of a bacterial multidrug efflux pump. Proc. Natl. Acad. Sci. U. S. A. 106:6893–6894. 10.1073/pnas.0902837106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burse A, Weingart H, Ullrich MS. 2004. The phytoalexin-inducible multidrug efflux pump AcrAB contributes to virulence in the fire blight pathogen, Erwinia amylovora. Mol. Plant Microbe Interact. 17:43–54. 10.1094/MPMI.2004.17.1.43 [DOI] [PubMed] [Google Scholar]
- 9.Al-Karablieh N, Weingart H, Ullrich MS. 2009. The outer membrane protein TolC is required for phytoalexin resistance and virulence of the fire blight pathogen Erwinia amylovora. Microb. Biotechnol. 2:465–475. 10.1111/j.1751-7915.2009.00095.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma D, Alberti M, Lynch C, Nikaido H, Hearst JE. 1996. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol. Microbiol. 19:101–112. 10.1046/j.1365-2958.1996.357881.x [DOI] [PubMed] [Google Scholar]
- 11.Su CC, Rutherford DJ, Yu EW. 2007. Characterization of the multidrug efflux regulator AcrR from Escherichia coli. Biochem. Biophys. Res. Commun. 361:85–90. 10.1016/j.bbrc.2007.06.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olliver A, Valle M, Chaslus-Dancla E, Cloeckaert A. 2004. Role of an acrR mutation in multidrug resistance of in vitro-selected fluoroquinolone-resistant mutants of Salmonella enterica serovar Typhimurium. FEMS Microbiol. Lett. 238:267–272. 10.1111/j.1574-6968.2004.tb09766.x [DOI] [PubMed] [Google Scholar]
- 13.Randall LP, Woodward MJ. 2002. The multiple antibiotic resistance (mar) locus and its significance. Res. Vet. Sci. 72:87–93. 10.1053/rvsc.2001.0537 [DOI] [PubMed] [Google Scholar]
- 14.Rosenberg EY, Bertenthal D, Nilles ML, Bertrand KP, Nikaido H. 2003. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 48:1609–1619. 10.1046/j.1365-2958.2003.03531.x [DOI] [PubMed] [Google Scholar]
- 15.Duval V, Lister IM. 2013. MarA, SoxS and Rob of Escherichia coli—global regulators of multidrug resistance, virulence and stress response. Int. J. Biotechnol. Wellness Indus. 2:101–124. 10.6000/1927-3037.2013.02.03.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramos JL, Rojo F, Zhou L, Timmis KN. 1990. A family of positive regulators related to the Pseudomonas putida TOL plasmid XylS and the Escherichia coli AraC activators. Nucleic Acids Res. 18:2149–2152. 10.1093/nar/18.8.2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL. 1997. AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallegos MT, Michan C, Ramos JL. 1993. The XylS/AraC family of regulators. Nucleic Acids Res. 21:807–810. 10.1093/nar/21.4.807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin RG, Rosner JL. 2002. Genomics of the marA/soxS/rob regulon of Escherichia coli: identification of directly activated promoters by application of molecular genetics and informatics to microarray data. Mol. Microbiol. 44:1611–1624. 10.1046/j.1365-2958.2002.02985.x [DOI] [PubMed] [Google Scholar]
- 20.Alekshun MN, Levy SB. 1999. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol. 7:410–413. 10.1016/S0966-842X(99)01589-9 [DOI] [PubMed] [Google Scholar]
- 21.White DG, Goldman JD, Demple B, Levy SB. 1997. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J. Bacteriol. 179:6122–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Demple B. 1996. Redox signaling and gene control in the Escherichia coli soxRS oxidative stress regulon—a review. Gene 179:53–57. 10.1016/S0378-1119(96)00329-0 [DOI] [PubMed] [Google Scholar]
- 23.Martin RG, Rosner JL. 2001. The AraC transcriptional activators. Curr. Opin. Microbiol. 4:132–137. 10.1016/S1369-5274(00)00178-8 [DOI] [PubMed] [Google Scholar]
- 24.Li Z, Demple B. 1996. Sequence specificity for DNA binding by Escherichia coli SoxS and Rob proteins. Mol. Microbiol. 20:937–945. 10.1111/j.1365-2958.1996.tb02535.x [DOI] [PubMed] [Google Scholar]
- 25.Martin RG, Gillette WK, Rosner JL. 2000. Promoter discrimination by the related transcriptional activators MarA and SoxS: differential regulation by differential binding. Mol. Microbiol. 35:623–634. 10.1046/j.1365-2958.2000.01732.x [DOI] [PubMed] [Google Scholar]
- 26.Martin RG, Gillette WK, Martin NI, Rosner JL. 2002. Complex formation between activator and RNA polymerase as the basis for transcriptional activation by MarA and SoxS in Escherichia coli. Mol. Microbiol. 43:355–370. 10.1046/j.1365-2958.2002.02748.x [DOI] [PubMed] [Google Scholar]
- 27.Wall ME, Markowitz DA, Rosner JL, Martin RG. 2009. Model of transcriptional activation by MarA in Escherichia coli. PLoS Comput. Biol. 5:e1000614. 10.1371/journal.pcbi.1000614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alekshun MN, Levy SB. 1999. Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J. Bacteriol. 181:4669–4672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chubiz LM, Rao CV. 2010. Aromatic acid metabolites of Escherichia coli K-12 can induce the marRAB operon. J. Bacteriol. 192:4786–4789. 10.1128/JB.00371-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gu M, Imlay JA. 2011. The SoxRS response of Escherichia coli is directly activated by redox-cycling drugs rather than by superoxide. Mol. Microbiol. 79:1136–1150. 10.1111/j.1365-2958.2010.07520.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwon HJ, Bennik MH, Demple B, Ellenberger T. 2000. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nat. Struct. Biol. 7:424–430. 10.1038/75213 [DOI] [PubMed] [Google Scholar]
- 32.Skarstad K, Thony B, Hwang DS, Kornberg A. 1993. A novel binding protein of the origin of the Escherichia coli chromosome. J. Biol. Chem. 268:5365–5370 [PubMed] [Google Scholar]
- 33.Kakeda M, Ueguchi C, Yamada H, Mizuno T. 1995. An Escherichia coli curved DNA-binding protein whose expression is affected by the stationary phase-specific sigma factor sigma S. Mol. Gen. Genet. 248:629–634. 10.1007/BF02423459 [DOI] [PubMed] [Google Scholar]
- 34.Griffith KL, Fitzpatrick MM, Keen EF, 3rd, Wolf RE., Jr 2009. Two functions of the C-terminal domain of Escherichia coli Rob: mediating “sequestration-dispersal” as a novel off-on switch for regulating Rob's activity as a transcription activator and preventing degradation of Rob by Lon protease. J. Mol. Biol. 388:415–430. 10.1016/j.jmb.2009.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosner JL, Dangi B, Gronenborn AM, Martin RG. 2002. Posttranscriptional activation of the transcriptional activator Rob by dipyridyl in Escherichia coli. J. Bacteriol. 184:1407–1416. 10.1128/JB.184.5.1407-1416.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okusu H, Ma D, Nikaido H. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178:306–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jair KW, Yu X, Skarstad K, Thony B, Fujita N, Ishihama A, Wolf RE., Jr 1996. Transcriptional activation of promoters of the superoxide and multiple antibiotic resistance regulons by Rob, a binding protein of the Escherichia coli origin of chromosomal replication. J. Bacteriol. 178:2507–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller PF, Gambino LF, Sulavik MC, Gracheck SJ. 1994. Genetic relationship between soxRS and mar loci in promoting multiple antibiotic resistance in Escherichia coli. Antimicrob. Agents Chemother. 38:1773–1779. 10.1128/AAC.38.8.1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey AM, Ivens A, Kingsley R, Cottell JL, Wain J, Piddock LJ. 2010. RamA, a member of the AraC/XylS family, influences both virulence and efflux in Salmonella enterica serovar Typhimurium. J. Bacteriol. 192:1607–1616. 10.1128/JB.01517-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Webber MA, Bailey AM, Blair JM, Morgan E, Stevens MP, Hinton JC, Ivens A, Wain J, Piddock LJ. 2009. The global consequence of disruption of the AcrAB-TolC efflux pump in Salmonella enterica includes reduced expression of SPI-1 and other attributes required to infect the host. J. Bacteriol. 191:4276–4285. 10.1128/JB.00363-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zumaquero A, Macho AP, Rufian JS, Beuzon CR. 2010. Analysis of the role of the type III effector inventory of Pseudomonas syringae pv. phaseolicola 1448a in interaction with the plant. J. Bacteriol. 192:4474–4488. 10.1128/JB.00260-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. CLSI document M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 43.Fabrega A, Rosner JL, Martin RG, Sole M, Vila J. 2012. SoxS-dependent coregulation of ompN and ydbK in a multidrug-resistant Escherichia coli strain. FEMS Microbiol. Lett. 332:61–67. 10.1111/j.1574-6968.2012.02577.x [DOI] [PubMed] [Google Scholar]
- 44.Perez JM, Arenas FA, Pradenas GA, Sandoval JM, Vasquez CC. 2008. Escherichia coli YqhD exhibits aldehyde reductase activity and protects from the harmful effect of lipid peroxidation-derived aldehydes. J. Biol. Chem. 283:7346–7353. 10.1074/jbc.M708846200 [DOI] [PubMed] [Google Scholar]
- 45.Pletzer D, Weingart H. 2014. Characterization of AcrD, a resistance-nodulation-cell division-type multidrug efflux pump from the fire blight pathogen Erwinia amylovora. BMC Microbiol. 14:13. 10.1186/1471-2180-14-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kovach ME, Phillips RW, Elzer PH, Roop RM, 2nd, Peterson KM. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16:800–802 [PubMed] [Google Scholar]
- 47.May R, Völksch B, Kampmann G. 1997. Antagonistic activities of epiphytic bacteria from soybean leaves against Pseudomonas syringae pv. glycinea in vitro and in planta. Microb. Ecol. 34:118–124. 10.1007/s002489900041 [DOI] [PubMed] [Google Scholar]
- 48.Schenk A, Weingart H, Ullrich MS. 2008. Extraction of high-quality bacterial RNA from infected leaf tissue for bacterial in planta gene expression analysis by multiplexed fluorescent Northern hybridization. Mol. Plant Pathol. 9:227–235. 10.1111/j.1364-3703.2007.00452.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGhee GC, Jones AL. 2000. Complete nucleotide sequence of ubiquitous plasmid pEA29 from Erwinia amylovora strain Ea88: gene organization and intraspecies variation. Appl. Environ. Microbiol. 66:4897–4907. 10.1128/AEM.66.11.4897-4907.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takle GW, Toth IK, Brurberg MB. 2007. Evaluation of reference genes for real-time RT-PCR expression studies in the plant pathogen Pectobacterium atrosepticum. BMC Plant Biol. 7:50. 10.1186/1471-2229-7-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cariss SJ, Tayler AE, Avison MB. 2008. Defining the growth conditions and promoter-proximal DNA sequences required for activation of gene expression by CreBC in Escherichia coli. J. Bacteriol. 190:3930–3939. 10.1128/JB.00108-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gambino L, Gracheck SJ, Miller PF. 1993. Overexpression of the MarA positive regulator is sufficient to confer multiple antibiotic resistance in Escherichia coli. J. Bacteriol. 175:2888–2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakajima H, Kobayashi K, Kobayashi M, Asako H, Aono R. 1995. Overexpression of the robA gene increases organic solvent tolerance and multiple antibiotic and heavy metal ion resistance in Escherichia coli. Appl. Environ. Microbiol. 61:2302–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bailey AM, Paulsen IT, Piddock LJ. 2008. RamA confers multidrug resistance in Salmonella enterica via increased expression of acrB, which is inhibited by chlorpromazine. Antimicrob. Agents Chemother. 52:3604–3611. 10.1128/AAC.00661-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakajima H, Kobayashi M, Negishi T, Aono R. 1995. soxRS gene increased the level of organic solvent tolerance in Escherichia coli. Biosci. Biotechnol. Biochem. 59:1323–1325. 10.1271/bbb.59.1323 [DOI] [PubMed] [Google Scholar]
- 56.Nikaido E, Yamaguchi A, Nishino K. 2008. AcrAB multidrug efflux pump regulation in Salmonella enterica serovar Typhimurium by RamA in response to environmental signals. J. Biol. Chem. 283:24245–24253. 10.1074/jbc.M804544200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosner JL, Martin RG. 2009. An excretory function for the Escherichia coli outer membrane pore TolC: upregulation of marA and soxS transcription and Rob activity due to metabolites accumulated in tolC mutants. J. Bacteriol. 191:5283–5292. 10.1128/JB.00507-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneiders T, Levy SB. 2006. MarA-mediated transcriptional repression of the rob promoter. J. Biol. Chem. 281:10049–10055. 10.1074/jbc.M512097200 [DOI] [PubMed] [Google Scholar]
- 59.Ariza RR, Li Z, Ringstad N, Demple B. 1995. Activation of multiple antibiotic resistance and binding of stress-inducible promoters by Escherichia coli Rob protein. J. Bacteriol. 177:1655–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanaka T, Horii T, Shibayama K, Sato K, Ohsuka S, Arakawa Y, Yamaki K, Takagi K, Ohta M. 1997. RobA-induced multiple antibiotic resistance largely depends on the activation of the AcrAB efflux. Microbiol. Immunol. 41:697–702. 10.1111/j.1348-0421.1997.tb01913.x [DOI] [PubMed] [Google Scholar]
- 61.Ruzin A, Immermann FW, Bradford PA. 2008. Real-time PCR and statistical analyses of acrAB and ramA expression in clinical isolates of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 52:3430–3432. 10.1128/AAC.00591-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elkins CA, Nikaido H. 2002. Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominantly by two large periplasmic loops. J. Bacteriol. 184:6490–6498. 10.1128/JB.184.23.6490-6499.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamasaki S, Nagasawa S, Hayashi-Nishino M, Yamaguchi A, Nishino K. 2011. AcrA dependency of the AcrD efflux pump in Salmonella enterica serovar Typhimurium. J. Antibiot. 64:433–437. 10.1038/ja.2011.28 [DOI] [PubMed] [Google Scholar]
- 64.Barchiesi J, Castelli ME, Soncini FC, Vescovi EG. 2008. mgtA expression is induced by rob overexpression and mediates a Salmonella enterica resistance phenotype. J. Bacteriol. 190:4951–4958. 10.1128/JB.00195-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sardessai Y, Bhosle S. 2002. Tolerance of bacteria to organic solvents. Res. Microbiol. 153:263–268. 10.1016/S0923-2508(02)01319-0 [DOI] [PubMed] [Google Scholar]
- 66.Pinkart HC, Wolfram JW, Rogers R, White DC. 1996. Cell envelope changes in solvent-tolerant and solvent-sensitive Pseudomonas putida strains following exposure to o-xylene. Appl. Environ. Microbiol. 62:1129–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heipieper HJ, Meinhardt F, Segura A. 2003. The cis-trans isomerase of unsaturated fatty acids in Pseudomonas and Vibrio: biochemistry, molecular biology and physiological function of a unique stress adaptive mechanism. FEMS Microbiol. Lett. 229:1–7. 10.1016/S0378-1097(03)00792-4 [DOI] [PubMed] [Google Scholar]
- 68.Segura A, Godoy P, van Dillewijn P, Hurtado A, Arroyo N, Santacruz S, Ramos JL. 2005. Proteomic analysis reveals the participation of energy- and stress-related proteins in the response of Pseudomonas putida DOT-T1E to toluene. J. Bacteriol. 187:5937–5945. 10.1128/JB.187.17.5937-5945.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McMurry LM, Levy SB. 2010. Evidence that regulatory protein MarA of Escherichia coli represses rob by steric hindrance. J. Bacteriol. 192:3977–3982. 10.1128/JB.00103-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Michan C, Manchado M, Pueyo C. 2002. SoxRS down-regulation of rob transcription. J. Bacteriol. 184:4733–4738. 10.1128/JB.184.17.4733-4738.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garbe TR, Kobayashi M, Yukawa H. 2000. Indole-inducible proteins in bacteria suggest membrane and oxidant toxicity. Arch. Microbiol. 173:78–82. 10.1007/s002030050012 [DOI] [PubMed] [Google Scholar]
- 72.Pomposiello PJ, Bennik MH, Demple B. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183:3890–3902. 10.1128/JB.183.13.3890-3902.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pomposiello PJ, Demple B. 2001. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 19:109–114. 10.1016/S0167-7799(00)01542-0 [DOI] [PubMed] [Google Scholar]
- 74.Graham AI, Sanguinetti G, Bramall N, McLeod CW, Poole RK. 2012. Dynamics of a starvation-to-surfeit shift: a transcriptomic and modelling analysis of the bacterial response to zinc reveals transient behaviour of the Fur and SoxS regulators. Microbiology 158:284–292. 10.1099/mic.0.053843-0 [DOI] [PubMed] [Google Scholar]
- 75.Blencowe DK, Morby AP. 2003. Zn(II) metabolism in prokaryotes. FEMS Microbiol. Rev. 27:291–311. 10.1016/S0168-6445(03)00041-X [DOI] [PubMed] [Google Scholar]
- 76.Warner DM, Levy SB. 2012. SoxS increases the expression of the zinc uptake system ZnuACB in an Escherichia coli murine pyelonephritis model. J. Bacteriol. 194:1177–1185. 10.1128/JB.05451-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McDevitt CA, Ogunniyi AD, Valkov E, Lawrence MC, Kobe B, McEwan AG, Paton JC. 2011. A molecular mechanism for bacterial susceptibility to zinc. PLoS Pathog. 7:e1002357. 10.1371/journal.ppat.1002357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sheng Y, Abreu IA, Cabelli DE, Maroney MJ, Miller AF, Teixeira M, Valentine JS. 2014. Superoxide dismutases and superoxide reductases. Chem. Rev. 114:3854–3918. 10.1021/cr4005296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sobota JM, Imlay JA. 2011. Iron enzyme ribulose-5-phosphate 3-epimerase in Escherichia coli is rapidly damaged by hydrogen peroxide but can be protected by manganese. Proc. Natl. Acad. Sci. U. S. A. 108:5402–5407. 10.1073/pnas.1100410108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lamb C, Dixon RA. 1997. The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48:251–275. 10.1146/annurev.arplant.48.1.251 [DOI] [PubMed] [Google Scholar]
- 81.Mehdy MC. 1994. Active oxygen species in plant defense against pathogens. Plant Physiol. 105:467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Greenberg JT. 1996. Programmed cell death: a way of life for plants. Proc. Natl. Acad. Sci. U. S. A. 93:12094–12097. 10.1073/pnas.93.22.12094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Venisse JS, Gullner G, Brisset MN. 2001. Evidence for the involvement of an oxidative stress in the initiation of infection of pear by Erwinia amylovora. Plant Physiol. 125:2164–2172. 10.1104/pp.125.4.2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li X, Gao WY, Huang LJ, Zhang JY, Guo XH. 2011. Antioxidant and antiinflammation capacities of some pear cultivars. J. Food. Sci. 76:C985–C990. 10.1111/j.1750-3841.2011.02302.x [DOI] [PubMed] [Google Scholar]
- 85.Boyer J, Liu RH. 2004. Apple phytochemicals and their health benefits. Nutr. J. 3:5. 10.1186/1475-2891-3-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ananthakrishnan TN. 1997. Gallic and salicylic acids: sentinels of plant defence against insects. Curr. Sci. 73:576–579 [Google Scholar]
- 87.Casaz P, Garrity-Ryan LK, McKenney D, Jackson C, Levy SB, Tanaka SK, Alekshun MN. 2006. MarA, SoxS and Rob function as virulence factors in an Escherichia coli murine model of ascending pyelonephritis. Microbiology 152:3643–3650. 10.1099/mic.0.2006/000604-0 [DOI] [PubMed] [Google Scholar]
- 88.Schwartz CJ, Giel JL, Patschkowski T, Luther C, Ruzicka FJ, Beinert H, Kiley PJ. 2001. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc. Natl. Acad. Sci. U. S. A. 98:14895–14900. 10.1073/pnas.251550898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shepard W, Soutourina O, Courtois E, England P, Haouz A, Martin-Verstraete I. 2011. Insights into the Rrf2 repressor family—the structure of CymR, the global cysteine regulator of Bacillus subtilis. FEBS J. 278:2689–2701. 10.1111/j.1742-4658.2011.08195.x [DOI] [PubMed] [Google Scholar]
- 90.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat. Biotechnol. 1:784–791. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- 91.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. 10.1093/bioinformatics/16.10.944 [DOI] [PubMed] [Google Scholar]
- 92.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gonnet GH, Hallett MT, Korostensky C, Bernardin L. 2000. Darwin v. 2.0: an interpreted computer language for the biosciences. Bioinformatics 16:101–103. 10.1093/bioinformatics/16.2.101 [DOI] [PubMed] [Google Scholar]
- 94.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res. 42:D222–D230. 10.1093/nar/gkt1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. 10.1016/S0378-1119(98)00130-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


