Abstract
The expression pattern of the Escherichia coli genome is controlled in part by regulating the utilization of a limited number of RNA polymerases among a total of its approximately 4,600 genes. The distribution pattern of RNA polymerase changes from modulation of two types of protein-protein interactions: the interaction of core RNA polymerase with seven species of the sigma subunit for differential promoter recognition and the interaction of RNA polymerase holoenzyme with about 300 different species of transcription factors (TFs) with regulatory functions. We have been involved in the systematic search for the target promoters recognized by each sigma factor and each TF using the newly developed Genomic SELEX system. In parallel, we developed the promoter-specific (PS)-TF screening system for identification of the whole set of TFs involved in regulation of each promoter. Understanding the regulation of genome transcription also requires knowing the intracellular concentrations of the sigma subunits and TFs under various growth conditions. This report describes the intracellular levels of 65 species of TF with known function in E. coli K-12 W3110 at various phases of cell growth and at various temperatures. The list of intracellular concentrations of the sigma factors and TFs provides a community resource for understanding the transcription regulation of E. coli under various stressful conditions in nature.
INTRODUCTION
Single-cell bacteria are directly exposed to frequently changing environments in nature and thus carry sophisticated genetic systems for adaptation to environmental changes (1, 2). On the basis of the complete genome sequence of several model Escherichia coli strains, the whole set of about 4,600 genes have been predicted to exist in E. coli K-12 (3, 4). Even for this best-characterized model organism E. coli, the gene functions remain unidentified or unpredicted for approximately one-fourth, because expression conditions of those uncharacterized genes have not been established under laboratory culture conditions. In rapidly growing E. coli cells under laboratory culture conditions, only one-fourth to one-third of the protein-coding genes on its genome are expressed, but by using more sensitive RNA-Seq methods, low-level expression has been identified for the increased number of genes, in particular those encoding varieties of regulatory RNA, such as approximately 1,000 species of anti-sense RNA (5). The majority of the uncharacterized silent genes may be expressed and utilized for adaptation and survival of E. coli under stressful conditions present in nature. In fact, the high-throughput modern technology for detection of gene expression such as transcriptomics and RNA-Seq analyses indicated marked changes in the genome expression pattern upon exposure to stressful conditions, such as changes in nutrients (6), exposure to heat shock (7) or cold shock (8), in the presence of hydrogen peroxide (9) or external metals (10), under anaerobic conditions (11), and within biofilms (12). Along this line, RNA-Seq analysis is becoming the method of choice (13–15). Exploration of regulation mechanisms for genome expression remains an important research subject in modern genetics.
We proposed a model in which changes in genome-wide transcription patterns take place mainly through controlling the utilization of RNA polymerase (RNAP) among the 4,600 genes on the genome (1, 2, 16). Two groups of regulatory proteins are involved in modulation of gene selectivity of RNAP through protein-protein interactions: sigma factors and transcription factors. In the first step, the sigma subunit binds to the RNAP core enzyme, leading to the formation of the RNAP holoenzyme. In E. coli K-12, seven species of the sigma subunit exist, each recognizing a specific set of promoters.
Gene selectivity of RNAP is further modulated, in the second step, through interaction with a second group of regulatory proteins, herein referred to as transcription factors (TFs). A decision about gene utilization is therefore executed by both sigma factors and TFs. In E. coli K-12, a total of about 300 species of TFs have been identified (1, 2, 17). Extensive efforts have been devoted to identifying the target genes under the control of each TF by using high-throughput experimental systems, such as transcriptomics and RNA-Seq methods of TF-defective E. coli mutants (5–15), as well as chromatin immunoprecipitation with microarray technology (ChIP-chip) analysis of TF-associated sites along the E. coli genome (18, 19). These in vivo analyses together provide information about regulated targets of TFs on the E. coli genome under the culture conditions employed. However, they are not sufficient to identify the whole set of TF recognition sites, because the functional forms of TFs are not always present in E. coli under laboratory culture conditions and because the TF-binding sites are often interfered with by other DNA-binding proteins.
For the identification in vitro of direct targets by each TF, we developed an improved method of the Genomic SELEX screening system (20) and successfully applied this to the identification of regulation targets by more than 200 TFs (for the current state of screening, see Table S1 in the supplemental material and also reference 2). Using this Genomic SELEX screening system, we also succeeded in identifying the set of constitutive promoters recognized in vitro by the RpoD holoenzyme alone in the absence of supporting TFs (21). In parallel, we have developed the promoter-specific transcription factor (PS-TF) screening system for identification of the set of promoter-specific TFs working on one specific promoter (22). Concomitant with the advance in Genomic SELEX and PS-TF screening systems, the concept of transcription regulation has changed remarkably in two aspects: (i) a single TF is generally involved in regulation of a number of genes and (ii) one promoter is often regulated by a number of TFs (2). The number of regulation targets for each TF exceeds those listed in databases such as RegulonDB (23) and EcoCyc (24).
However, prediction of the pattern of genome transcription also requires knowledge of the intracellular concentrations of the functional forms of the TFs. At present, only fragmentary information is available on the intracellular concentrations of TFs (for instance, see references 25 and 26). Systematic determination of large numbers of TF levels has not been performed. Previously, we determined the intracellular concentrations of the seven sigma subunits in one E. coli K-12 W3110 strain under one set of culture conditions (27–29). In this study, we apply the same quantitative immunoblot assay system to determine the intracellular concentrations of TFs in E. coli growing in a rich medium at different times and at different temperatures. This is the first report that describes the levels of 65 TF species with known functions at high accuracy. The results herein described provide a community resource for understanding the regulation of transcription in E. coli genome-wide under various stressful conditions present in nature.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli K-12 W3350 type A containing the intact rpoS gene (30) was used for measurement of TFs throughout this study. E. coli BW25113 and a set of otherwise isogenic single-gene deletion mutants, each lacking one specific TF gene, are the products of the Keio collection (31) and obtained from the E. coli Stock Center (National Bio-Resource Center, Mishima, Japan). E. coli DH5a was used for plasmid amplification, while E. coli BL21 was used for expression and purification of TFs.
Purification of TFs.
TF expression plasmids were constructed as described previously (32). E. coli BL21 transformed with each of the TF expression plasmids was grown in LB broth in the presence of 50 μg/ml ampicillin, and TF expression was induced in mid-log phase by adding 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After 3 h of induction, cells were harvested and TF purification was carried out according to the standard procedure in this laboratory (32). TFs used throughout this study were more than 95% pure as analyzed by SDS-PAGE.
Production of anti-TF antibodies.
Antibodies against TFs were produced in rabbits by injecting purified TF proteins. The protocol for antibody production was raised, following the ethical guidelines proposed by the Science Council of Japan and the Japanese Government, in the Animal Laboratory of Mitsubishi Chemical Mediemce Co. (Kumamoto, Japan) and the Nippon Institute for Biological Science (Tokyo, Japan). Anti-TF antibodies were raised in rabbits after repeated injection of purified TFs, and the production of antibodies was monitored by enzyme-linked immunosorbent assays (ELISAs). For most of the anti-TF antibodies used in this study, the immunization was performed using two rabbits. The specificity and activity of anti-TF antibodies were checked by Western blotting.
Preparation of cell lysates for TF measurement.
Wild-type E. coli K-12 W3110 type A was grown in LB medium under aeration with constant shaking at 140 rpm. The cell culture was carried out at five different temperatures (22, 27, 32, 37, and 42°C) and for various times from exponential growth to stationary phase. Cell growth was monitored by measuring the culture turbidity at 600 nm.
Cell lysate was prepared according to the standard procedure used for the determination of sigma subunits in E. coli (27–29). In brief, cells were collected by centrifugation and resuspended in 40 mM Tris-HCl (pH 8.1 at 4°C) containing 25% sucrose. After treatment with 1 mM EDTA and 0.5 mg of lysozyme per ml at 0°C for 10 min, cells were lysed by adding 0.5% nonionic detergent Brij-58. The lysate was supplemented with 0.01 M MgCl2 and 0.2 M KCl, digested at 37°C for 10 min with 0.02 mg of RNase A per milliliter and 0.1 mg of DNase I per milliliter in the presence of 1 mM phenylmethylsulfonyl fluoride (PMSF), and sonicated for 1 min with a Cosmo Bio Bioruptor. After centrifugation for 30 min at 15,000 rpm, aliquots of the cell lysate were stored frozen until use. To avoid protein degradation arisen from freeze-thawing, the measurement of each TF was performed using each aliquot.
Quantitative Western blot analysis of TFs.
For the measurement of TFs, a quantitative immunoblotting was used under the standard conditions (27–29). In brief, cell lysates were treated with an SDS sample buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 1% 2-mercaptoethanol, 10% glycerol, and 0.025% bromophenol blue) and separated on SDS-polyacrylamide gels (5.0, 7.5, or 10%). Proteins in the gels were directly electroblotted onto polyvinylidene difluoride membranes. Blots were blocked overnight at 4°C in 3% bovine serum albumin in phosphate-buffered saline (PBS), probed with the anti-TF antibody, washed with 0.5% Tween 20 in PBS, and incubated with goat-rabbit immunoglobulin G conjugated with hydroxyperoxidase. The blots were developed with 3,3′-diaminobenzine tetrahydrochloride. Staining intensity was measured with a PDI image analyzer system. Some TF antibodies cross-reacted against other E. coli proteins in cell lysates, and some TF antibody preparations used contained antibodies against contaminating proteins in purified TF preparations used as antigens. For measurement of TF levels, antigen-antibody complexes were subjected to SDS-PAGE, and after immunostaining, only the TF bands were measured, which migrated to the same positions with the authentic TFs. Test samples and standard TFs were subjected to the same gels to achieve immunostaining under the same conditions. The level of test TF was measured using the standard curve prepared using various amounts of purified TFs (see Fig. S1 in the supplemental material). Since the test TFs are within the background of whole-cell lysates, we prepared the standard curves of purified TFs in the presence of whole-cell lysates from E. coli lacking each test TF. The fluctuation level between different measurements was less than 10% for all the test TFs described in this report (see Table S1 in the supplemental material).
RESULTS AND DISCUSSION
Determination of the intracellular concentrations of TFs: overall research strategy and tactics.
In the simplest model, transcription of one gene or operon would be under the control of one gene-specific TF, while each TF would regulate one specific target gene or operon (32, 33). However, we now know that the promoters for stress response genes can be controlled by multiple sigma factors and multiple TFs, each sensing a specific external signal and condition (2). Previously, we determined the intracellular concentrations of the 7 species of the RNA polymerase sigma subunit (27–29). To understand the transcription regulation pattern of the E. coli genome, it is necessary to know the intracellular concentrations of the 300 TFs.
Using the quantitative immunoblot system, we have now determined the intracellular concentrations of most of the 300 transcription factors (1, 2). In this report, we describe the intracellular concentrations of the 65 species of TF with known regulatory functions (see Table S1 in the supplemental material). For this group of TFs, measurements were repeated at least three times using independent cultures, yielding values with less than 10% fluctuation. This set includes 9 major nucleoid proteins, i.e., 5 universal nucleoid proteins (UNP) (CbpA, H-NS, HU, IHF, Rob, StpA) and 3 growth-phase-specific nucleoid proteins (GNP) (Dan, Dps, and Fis) (34). All these nucleoid proteins are bifunctional, playing both architectural roles in folding and packaging genomic DNA and regulatory roles of genome functions. In addition, 17 species of the response regulator of the two-component system (TCS), one TF-enzyme fusion PutA, 27 one-component TFs with known effectors, and 9 one-component TFs with unidentified effector were analyzed in this study. Some target promoters, genes, and operons regulated by these TFs have been deposited in E. coli databases such as RegulonDB (23) and EcoCyc (24), but the number of regulated targets increased remarkably from a systematic search using the newly developed Genomic SELEX (20) and PS-TF (promoter-specific transcription factor) (22) screening systems, ranging from a single target to more than 1,000 (see Table S1; also described in reference 2).
The DNA-binding TFs express their regulatory functions through direct interaction with RNA polymerase. The affinity of these DNA-binding TFs to RNA polymerase is generally weak, and thus DNA binding is needed to increase the local concentration of the TFs near target promoters for effective interaction with RNAP. The level of DNA-bound TFs per a single copy of the genome should vary, because the total DNA concentration varies in response to changes in growth rate. For determination of both free and DNA-associated TFs, cell lysates were prepared after treatment with both DNase and RNase. Since the amount of genome DNA per cell varies depending on the growth rate and growth phase, the intracellular concentrations of TFs measured as described above were converted into the number of TF molecules per genome equivalent DNA. The intracellular level of the RNAP alpha subunit is relatively constant throughout growth (16), so TF levels were determined relative to the internal reference RpoA.
The intracellular concentrations of transcription factors in growing cells.
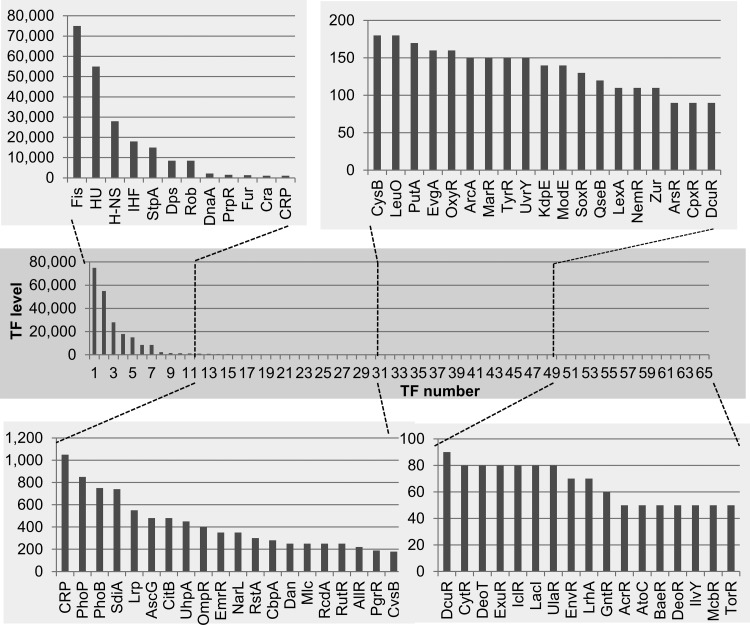
The intracellular concentrations of the 65 TFs in exponentially growing cells are shown in Fig. 1 (for details, see Table S1 in the supplemental material). The level was the highest (75,000 molecules per genome) for Fis, a growth-phase-specific nucleoid protein (GNP). Fis regulates a number of growth-related genes, such as the rRNA and tRNA genes needed for translation, a set of the genes needed for carbon and nitrogen metabolism, and groups of genes needed for motility, chemotaxis, biofilm formation, stress responses, and virulence (35, 36). Correlating with its high level, Fis binds to as many as 1,300 sites in vitro, as measured by Genomic SELEX (1) and to about 900 sites in vivo as measured by ChIP-chip analysis (37). Some of the potential Fis-binding sites may be masked in vivo by other DNA-binding proteins. High concentrations were also detected for the universal nucleoid proteins (UNP) HU, H-NS, IHF, StpA, and Rob (or CbpB). The intracellular levels were low for the stationary-phase-specific nucleoid-associated DNA-binding protein under anaerobic condition (Dan) (38) and DNA-binding protein in starved cells (Dps) (39). DnaA, a key factor for the initiation of DNA replication, is at a high concentration, correlating with enhanced replication in growing cells, likely forming oligomers at DnaA boxes in the ori region (40). The levels of TFs other than the nucleoid proteins are less than 1,000 molecules per genome, of which about two-thirds are present at less than 200 molecules per genome and about one-third are present at less than 100 copies per gnome.
FIG 1.
Intracellular concentration of TFs in exponential-phase cells of E. coli K-12 W3110. Cells were grown in LB-glucose medium at 37°C with shaking. In the middle of exponential phase (optical density at 600 nm [OD600], 0.4), cells were harvested and the cell lysate was prepared by the standard method (27–29). The intracellular concentration was determined for a total of 65 species of TF by the quantitative immunoblot method as described in Materials and Methods. The TF concentration was calculated as the relative value to that of RNA polymerase RpoA subunit and is represented as the number of molecules per genome equivalent of DNA. TF level (y axis) represents the number of TF molecules per genome.
A set of TFs facilitating a large number of genes has been classified as the global regulators, including cAMP receptor protein (CRP), fumarate and nitrate reduction (FNR), anoxic redox control (ArcA), ferric uptake regulation (Fur), leucine-responsive regulatory protein (Lrp), and nitrate/nitrite response regulator (NarL) (41). The levels of the global regulators are generally high (Table 1 and Fig. 1). The high concentrations of these global TFs allow them to be spread around the nucleoid surface so as to access target genes. In addition to these previously recognized global regulators, a larger number of regulation targets was also identified by the Genomic SELEX and PS-TF screens for TFs, such as catabolite repressor activator (Cra), leucine operon regulator (LeuO), leucine-responsive regulatory protein (Lrp), and superoxide response protein (SoxR) (see Table S1 in the supplemental material).
The environmental signal response global regulators Fur and PhoP are at high levels even during exponential growth phase (Fig. 1; see also Table S1 in the supplemental material). Fur is a key regulator for maintaining the homeostasis of not only Fe(II) but also different divalent metals. In addition, Fur is involved in control of wide varieties of cellular functions, including respiration and energy metabolism (42). Likewise, high levels of FNR (43) and NarL (44), which mediate the transition from aerobic to anaerobic growth through activation of the genes for anaerobic energy metabolism and repression of the genes for aerobic metabolism, were detected. PhoP is a dual transcriptional regulator that is activated in response to low extracellular levels of the divalent cations, Mg(II) and Ca(II), and activates transcription of a large collection of genes involved in Mg2+ homeostasis, acid resistance, and lipopolysaccharide (LPS) modification (45). PhoB, a dual-transcription regulator that activates expression of the Pho regulon for phosphate uptake and metabolism (46), is also one of the abundant regulators for response to environmental conditions. These findings indicate that E. coli cells monitor the lack of Fe(II), Mg(II), phosphate, and oxygen even during exponential growth.
The intracellular levels are also high for the key regulators of metabolism for the essential components for cell growth. Two coordinated regulators, CRP and Cra, control carbon source utilization pathways. CRP, perhaps the best-characterized E. coli regulator, regulates as many as 378 operons for carbon utilization (47), while Cra regulates as many as 164 target operons (48). CRP and Cra play different roles: Cra regulates genes for the TCA cycle, whereas CRP regulates genes for import of carbon sources, genes for metabolism downstream of the TCA cycle, and gluconeogenesis (47, 48). Lrp and LeuO, two coordinated regulators that are employed for nitrogen source utilization, are also abundant in growing cells. Genes involved in amino acid biosynthesis are positively regulated by the global regulator Lrp, whereas genes involved in catabolic reactions are negatively regulated by Lrp. Accordingly, the expression of Lrp is high in minimal medium and lower in rich medium (49). LeuO was originally identified as a regulator of leucine biosynthesis, but it was later found to control a variety of stress response processes, including biofilm formation and virulence expression. Under normal growth conditions, these stress response genes are silenced by H-NS, and LeuO antisilences H-NS (50). Two key regulators, pyrimidine utilization regulator (RutR) (51) and allantoin repressor (AllR) (52), which are involved in control of pyrimidine and purine degradation, respectively, are also abundant (see Table S1 in the supplemental material), presumably because nucleotides are degraded for reutilization as nitrogen sources under the culture conditions employed. An abundant protein, propionate regulator (PrpR), might also be involved in degradation and utilization of fatty acids (53).
E. coli contains about 30 two-component systems (TCSs), each of which has a sensor of an external signal or condition as well as a response regulator (32). Among the TCS response regulators, high levels were detected for citrate utilization (CitB), uptake of hexose phosphate (UhpA), response to osmolarity change (OmpR), and acid tolerance (RstA), in addition to PhoP and PhoB. The TCS TFs are converted to their active forms only when phosphorylated by their respective sensor kinases, and the levels of the active forms remain to be determined. Two key regulators in the control of cell division and cell differentiation, suppressor of the cell division inhibitor (SdiA) and regulator of csgD (RcdA), also exist at high levels (54, 55).
Growth-phase-dependent changes in the intracellular concentrations of transcription factors.
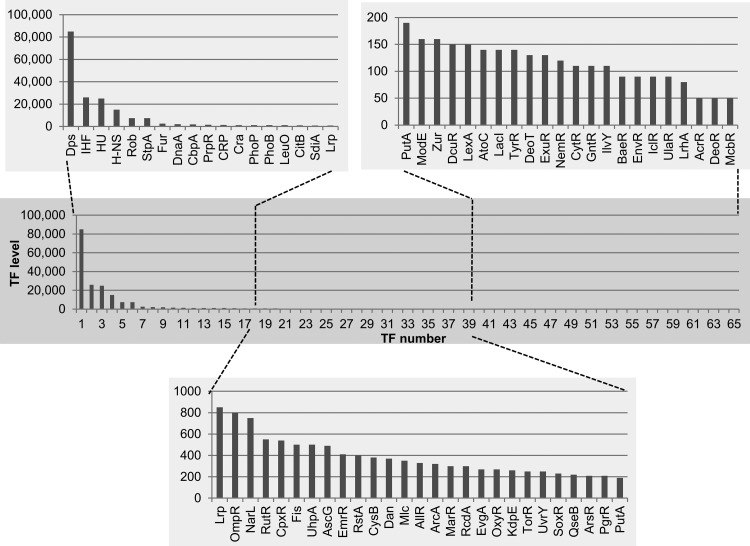
TFs play key roles in growth condition-coupled control of genome expression. We analyzed possible fluctuations of TF levels at various phases of cell growth and upon exposure to various stresses. The intracellular concentrations of the 65 TFs were measured at various phases of cell growth at 37°C. For most TFs, the rank order in intracellular level stayed the same under these conditions (Fig. 2), but there were marked changes in the levels of a small number of TFs, most notably the nucleoid-associated proteins (Fig. 2; see also the details in Table S1 in the supplemental material). The level of Fis decreased from 75,000 molecules per genome in log phase to 150 in stationary phase (Fig. 2; see also Table S1). In contrast, the stationary-phase-specific nucleoid protein Dps increased to 85,000 molecules per genome, enough to cover the whole genome surface (39, 56, 57). The relative composition of the nucleoid proteins as visualized in Fig. 3 is essentially the same as that determined previously using the same E. coli strain but under different culture conditions and using a different method of protein determination (56).
FIG 2.
Intracellular concentration of TFs in stationary-phase cells of E. coli K-12 W3110. Cells were grown in LB-glucose medium at 37°C with shaking. In the stationary phase (OD600, 1.5), cells were harvested and the cell lysate was prepared by the standard method (27–29). The intracellular concentration was determined for a total of 65 species of TF as described fpr Fig. 1. The TF concentration was calculated as the relative value to that of RNA polymerase RpoA subunit and is represented as the number of molecules per genome equivalent of DNA. TF level (y axis) represents the number of TF molecules per genome.
FIG 3.
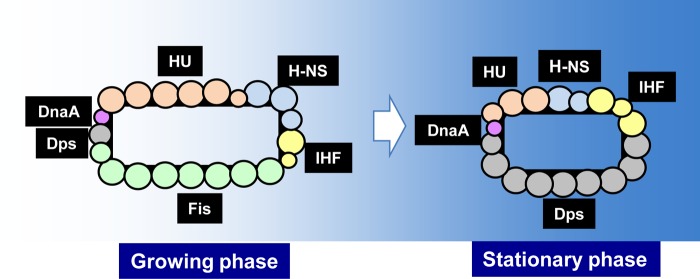
The protein composition of nucleoid. The concentration of nucleoid proteins was determined at various growth phases according to the standard procedure as described in Fig. 1 and 2. The protein composition of nucleoid is illustrated for both exponential growing phase and stationary phase. Two growth-phase-specific nucleoid proteins (GNP), Fis and Dps, are most abundant for log-phase and stationary-phase nucleoids, respectively. One large circle represents a total of 10,000 protein molecules per genome.
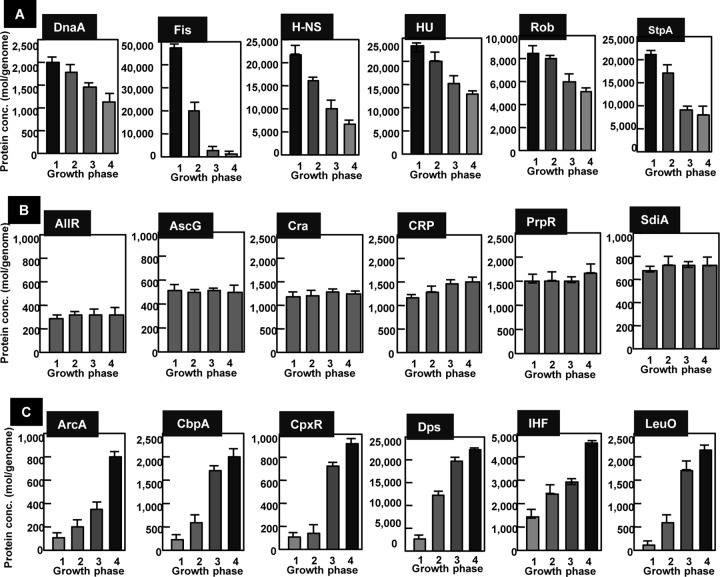
With respect to the growth-phase-dependent variation pattern, TFs can be classified into three types (Fig. 4): type GA, in which the TF level is high in exponential growth and thereafter decreases upon entry into stationary phase; type GB, in which the TF level stays constant throughout growth; and type GC, in which the TF level is low in log phase but increases in stationary phase. The nucleoid proteins DnaA, Fis, H-NS, HU, Rob (CbpB), and StpA belonged to type GA. DNA-binding protein under anaerobic conditions (Dan) can be detected in cells grown under anaerobic conditions (40). The level of Dan is as low as 150 to 250 molecules per genome in aerobic growth but in anaerobic growth increases to 7,000 to 9,000, as high as the nucleoid proteins IHF, H-NS, and HU. There are as many as 700 Dan-binding sites on the E. coli genome, including the only known regulation target, which is within the ttdAB operon encoding tartrate dehydrogenase (58).
FIG 4.
Growth-phase-dependent variation of TF concentration. The intracellular concentrations of 65 species of TF were determined at various growth phases from early exponential phase to stationary phase. OD600: 0.1, lane 1; 0.4, lane 2; 1.0, lane 3; and 1.5, lane 4. (A) GA group TFs. The intracellular level decreases upon entry to the stationary phase. (B) GB group TFs. The intracellular concentration stays constant throughout the progress of cell growth. (C) GC group TFs. The intracellular level increases upon entry into the stationary phase. Error bars show standard errors of the means from triplicate or quadruplicate determinations.
The intracellular levels of TFs involved in regulation of the genes for metabolism stayed constant throughout growth phase (Fig. 4, type GB pattern). AllR for nitrogen metabolism and AscG, Cra, CRP, and PrpR for carbon metabolism were all classified in this type GB group. The key regulator for cell division, SdiA (22, 59), is also present at a constant level throughout cell growth. Finally, a set of TFs classified in type GC increased concomitant with the growth transition from log to stationary phases. Another set of nucleoid proteins, such as CbpA, Dps, and IHF, are classified as type GC. Stress response regulators aerobic respiration control (ArcA) and conjugate plasmid gene expression (CpxR) increase in the stationary phase and thus are classified in the type GC group (Fig. 4C). ArcA is involved in repression of the genes for aerobic respiration, while CpxR regulates a set of stress response genes.
Influence of culture temperature on the intracellular concentrations of TFs for thermal adaptation.
The upper thermal limit of bacterial survival is defined as the maximal temperature at which the cells survived but did not grow, whereas the upper thermal limit of cell growth is defined as the maximal temperature at which the cells increased in number over time. In the case of E. coli, the upper limit of survival is between 45 and 50°C and the optimal growth temperature is 37°C (60). We determined the intracellular concentrations of the same set of 65 TFs in E. coli cells grown at various temperatures, at 5°C intervals from 22°C to 42°C.
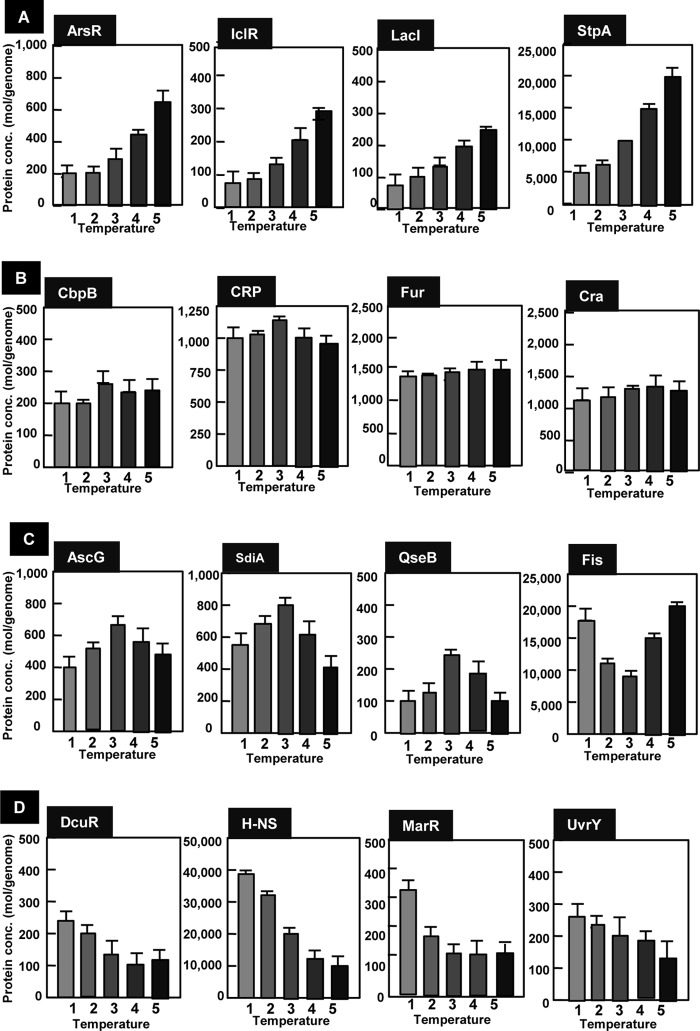
The levels were constant for most of these 65 TFs, but some showed marked changes (Fig. 5). Based on the temperature-dependent change, the pattern was classified into four types. Type TA TFs, including arsenate inducibility regulator (ArsR), isocitrate lyase regulator (IclR), LacI, and StpA, increased with the increase in culture temperature (Fig. 5A). The response of bacteria to high temperature has often been studied by applying a short temperature upshift and thus is called the heat shock response. The heat shock response consists of the induction of a number of heat shock proteins (HSPs) (61). The majority of heat shock proteins are either molecular chaperones that assist the refolding of denatured proteins or proteases that degrade misfolded and abnormal proteins. Following the acclimation phase, E. coli becomes adapted to high temperature and resumes growth at the rate characteristic of the elevated temperature. In contrast to the accumulation of research focusing the regulation of heat shock response, little is known on the regulation of genome expression after the final adaptation to high temperature. A set of genes under the control of type TA TFs might be needed for survival and growth at high temperature.
FIG 5.
Culture temperature-dependent variation of TF concentration. E. coli K-12 W3110 was grown in LB-glucose medium at various temperatures (22, 27, 32, 37, and 42°C). Cells were harvested in the middle of exponential phase (OD600, 0.4). Cell lysate was prepared by method A (see Materials and Methods), and TF concentration was determined by the quantitative immunoblot method as described in Materials and Methods. The temperature-dependent change was classified into four types: (A) TA group, TF level increased with the increase in culture temperate within the range of 22 to 42°C; (B) TB group, TF level stays constant between the culture temperature 22 and 42°C; (C) TC group, the intracellular level is either maximum or minimum at intermediate temperature (32 to 37°C); (D) TD group, TF level increases with the decrease in culture temperature from 42 to 22°C. Error bars show standard errors of the means from triplicate or quadruplicate determinations.
One striking finding is that the level of StpA (62), a homolog of H-NS but with fewer characterized functions, increased at high temperature, whereas the general silencer H-NS decreased with increased culture temperature (Fig. 5D). As a result, the molar ratio between two homologous nucleoid proteins showed a marked change, from an H-NS-abundant state at low temperature to an StpA-abundant state at high temperature. This finding implies, for the first time, that there might be a difference in the physiological roles of H-NS and StpA. In addition to H-NS, the type TD TFs, C4-dicarboxylate regulator (DcuR), multiple antibiotic resistance (MarR), and regulator of UV repair (UvrY), decreased with increased growth temperature (Fig. 5D). From the genome-wide analysis and the SELEX-chip screening, these TFs appear to be global regulators controlling a number of stress response genes.
Thermal niches for E. coli are different at the high and low ends of the temperature scale (63, 64). Cold shock leads to an inhibition of growth and overall repression of gene expression in E. coli; however, a set of cold shock genes are induced transiently to modify cell metabolism and readjust expression to the unfavorable change in temperature. Following an acclimation phase, however, E. coli adapts to the low temperature and resumes growth at a lower rate. The expression of the cold shock proteins declines in this phase, and bulk protein synthesis restarts at levels characteristic of the lower growth rate (65). The group of genes that are relatively highly expressed at low temperatures includes the genes encoding proteins needed for modulation of cell membrane, the genes for RNA metabolism and degradation, and the genes coding for some of the Csp family of transcription factors (CspA and CspE), the transcription termination factor NusA, and the stationary-phase-specific sigma factor RpoS, which regulates more than half of the genes expressed at low temperatures (66). Some of these genes are under the control of StpA, DcuR, MarR, or UvrY, as identified by Genomic SELEX screening (A. Ishihama, unpublished data).
Several TFs showed a unique pattern with respect to the change in culture temperature. The level of AscG was the highest at the physiological temperature 37°C, while the levels of Fis, SdiA, and QseB were the lowest at 37°C (Fig. 5C). Both SdiA and QseB play key roles in monitoring environmental signals, like quorum signals, and control a set of genes involved in cell division and differentiation (55).
Functional interconversion of TFs: discrimination between active and inactive TFs.
In this study, we determined the intracellular concentration of 65 TFs within a single bacterial species (E. coli K-12 W3110) grown under the same culture conditions. We emphasize that the specificity and activity of many TFs are modulated by effector ligands such as inducers or corepressors or by covalent modification such as phosphorylation, acetylation, or methylation. Many E. coli TFs can function as both repressors and activators, depending on the effector bound (67) or the position of the DNA-binding site relative to the RNAP-binding site (1, 2). The effectors affecting the mode of action of the 65 TF species analyzed in this study are listed in Table S1 in the supplemental material.
The majority of E. coli TFs are in the one-component signal transduction class, in which a single polypeptide contains both an effector-binding sensory domain and a DNA-binding domain. The activity of single-component TFs is controlled by a single species of the effector ligand, i.e., an inducer or corepressor, as listed in Table S1 in the supplemental material. Recently, however, cases have been identified in which there are two effectors (allantoin and glyoxylate for AllR, arginine and lysine for ArgP or IciA, glyoxylate and pyruvate for IclR, hypoxanthine and guanine for PurR, ATP and maltotriose for MalT, and uracil and thymine for RutR). There are also TFs under the control of three or more effectors. That is, TyrR, a regulator of a set of genes for synthesis and transport of aromatic amino acids, is regulated by binding tyrosine, phenylalanine, and tryptophan, and SdiA, the master regulator of cell division and differentiation, is also controlled by multiple effectors.
A total of about 30 TCS TFs are functional only when phosphorylated by the respective protein kinases that sense environmental signals and activate their cognate response regulators. Both the TF phosphorylation rates and the metabolic stabilities of the phosphorylated TFs differ among these TCS TFs (32), indicating that the intracellular concentrations of the functional forms can vary depending on the environmental conditions. Regulation of protein function by acetylation is one of the major control systems in eukaryotes, but recently this type of control has been identified for some E. coli TFs as well, including McbR, RcsB, RutR, CRP, Rho, and Rsd (68, 69). Protein methylation takes place for a number of ribosomal proteins (70). The TF Ada is methylated as an intermediate during its function of removing the methyl group from alkylated DNA (71). And posttranslational modifications of ribosomal proteins, including addition of amino acids to the polypeptide chain (72), have also been found. Enzymes with iron-sulfur centers are generally inactivated by nitrosylation in the presence of nitric oxide (NO), while redox-sensitive TFs such as SoxR and OxyR are activated by nitrosylation at their iron-sulfur centers (73, 74). The functional and nonfunctional forms of these TFs remain to be determined.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge Rick Gourse for valuable comments and proofreading of the manuscript.
This work was supported by Grant-in-Aid for Scientific Research A (21241047), B (18310133), and C (25430173) to A.I. from MEXT (Ministry of Education, Culture, Sports, Science and Technology of Japan) and MEXT-Supported Program for the Strategic Research Foundation at Private Universities 208-2012 (S0801037).
Footnotes
Published ahead of print 16 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01579-14.
REFERENCES
- 1.Ishihama A. 2010. Prokaryotic genome regulation: multi-factor promoters, multi-target regulators and hierarchic networks. FEMS Microbial. Rev. 34:628–645. 10.1111/j.1574-6976.2010.00227.x [DOI] [PubMed] [Google Scholar]
- 2.Ishihama A. 2012. Prokaryotic genome regulation: a revolutionary paradigm. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 88:485–508. 10.2183/pjab.88.485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Chi S, Ohtsubo E, Baba T, Wanner BL, Mori H, Horiuchi T. 2006. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol. Syst. Biol. 2:2006.0007. 10.1038/msb4100049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riley M, Abe T, Arnaud MB, Berlyn MKB, Blattner FR, Chaudhuri RR, Glasner JD, Horiuchi T, Keseler IM, Kosuge T, Mori H, Perna NT, Plunkett G, III, Rudd KE, Serres MH, Thomas GH, Thomson NR, Wishart D, Wanner BL. 2006. Escherichia coli K-12: a cooperatively developed annotation snapshot—2005. Nucleic Acids Res. 34:1–9. 10.1093/nar/gkj405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dornenburg JE, DeVita AM, Palumbo MJ, Wade JT. 2010. Widespread antisense transcription in Escherichia coli. mBio 1(1):e00024-10. 10.1128/mBio.00024-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh M-K, Rohlin L, Kao KC, Liao JC. 2002. Global expression profiling of acetate-grown Escherichia coli. J. Biol. Chem. 277:13175–13183. 10.1074/jbc.M110809200 [DOI] [PubMed] [Google Scholar]
- 7.Richmond CS, Glasner JD, Mau R, Jin H, Blattner F. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27:3821–3835. 10.1093/nar/27.19.3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phadtare S, Inouye M. 2004. Genome-wide transcriptional analysis of the cold-shock response in wild-type and cold-sensitive, quadruple-csp-deletion strains of Escherichia coli. J. Bacteriol. 186:7007–7914. 10.1128/JB.186.20.7007-7014.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng M, Wang X, Templeton LJD, Smulski R, LaRossa RA, Storz G. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562–4570. 10.1128/JB.183.15.4562-4570.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee LJ, Barrett JA, Poole RK. 2005. Genome-wide transcriptional response to chemostat-cultured Escherichia coli to zinc. J. Bacteriol. 187:1124–1134. 10.1128/JB.187.3.1124-1134.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Overton TW, Griffiths L, Patel MD, Hobman JL, Penn CW, Cole JA, Constantinidou C. 2006. Microarray analysis of gene regulation by oxygen, nitrate, nitrite, FNR, NarL and NarP during anaerobic growth of Escherichia coli: new insights into microbial physiology. Biochem. Soc. Transact. 34:104–107. 10.1042/BST0340104 [DOI] [PubMed] [Google Scholar]
- 12.Ren D, Bedzyk LA, Thomas SM, Ye RW, Wood TK. 2004. Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 64:515–524. 10.1007/s00253-003-1517-y [DOI] [PubMed] [Google Scholar]
- 13.Cho BK, Zengler K, Qiu Y, Park YS, Knight EM, Barrett CL, Gao Y, Palsson BO. 2009. The transcriptome unit architecture of the Escherichia coli genome. Nat. Biotechnol. 27:1043–1049. 10.1038/nbt.1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raghavan R, Sage A, Ochman H. 2011. Genome-wide identification of transcription start sites yields a novel thermosensing RNA and new cyclic AMP receptor protein-regulated genes in Escherichia coli. J. Bacteriol. 193:2871–2874. 10.1128/JB.00398-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ginnoukos G, Ciulla DM, Huang K, Haas BJ, Izard J, Levin JZ, Livny J, Earl AM, Gevers D, Yard DV, Husbaum C, Birren BW, Gnirke A. 2012. Efficient and robust RNA-seq process for cultured bacteria and complex community transcriptomes. Genome Biol. 13:r23.70. 10.1186/gb-2012-13-3-r23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishihama A. 2000. Functional modulation of Escherichia coli RNA polymerase. Annu. Rev. Microbiol. 54:499–518. 10.1146/annurev.micro.54.1.499 [DOI] [PubMed] [Google Scholar]
- 17.Peruz-Rueda E, Collado-Vides J. 2000. The repertoire of DNA-binding transcription regulators in Escherichia coli K-12. Nucleic Acids Res. 28:1838–1847. 10.1093/nar/28.8.1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herring CD, Raffaelle M, Allen TE, Kanin EI, Landick R, Ansari AZ, Palsson BO. 2005. Immobilization of Escherichia coli RNA polymerase and location of binding sites by use of chromatin immunoprecipitation and microarrays. J. Bacteriol. 187:6166–6174. 10.1128/JB.187.17.6166-6174.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mooney RA, Davis SE, Peters JM, Rowland JL, Ansari AZ, Landick R. 2009. Regulator trafficking on bacterial transcription units in vivo. Mol. Cell 33:97–108. 10.1016/j.molcel.2008.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimada T, Fujita N, Maeda M, Ishihama A. 2005. Systematic search for the Cra-binding promoters using genomic SELEX. Genes Cells 10:907–918. 10.1111/j.1365-2443.2005.00888.x [DOI] [PubMed] [Google Scholar]
- 21.Shimada T, Yamazaki Y, Tanaka K, Ishihama A. 2014. The whole set of constitutive promoters recognized by RNA polymerase RpoD holoenzyme of Escherichia coli. PLoS One 9(3):e90447. 10.1371/journal.pone.0090447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimada K, Ogasawara H, Yamada K, Shimura M, Kori A, Shimada T, Yamanaka Y, Yamamoto K, Ishihama A. 2013. Screening of promoter-specific transcription factors: multiple regulators for the sdiA gene involved in cell division control and quorum sensing. Microbiology 159:2501–2512. 10.1099/mic.0.067538-0 [DOI] [PubMed] [Google Scholar]
- 23.Salgado H, Peralta-Gil M, Gama-Castro S, Santos-Zavaleta A, Munz-Rascado L, Garcia-Sotelo JS, Wess V, Solano-Lira H, Martinez-Flores I, Medina-Rivera A, Salgado-Osorio S, Alqicira-Hermandez S, Alquicira-Hernandez K, Lopez-Fuentes A, Porron-Sotelo L, Huerta AM, Bonavides-Martinez C, Balderas-Martinez YI, Pannier L, Olvera M, Labastida A, Jimenez-Jacinto V, Vega-Alvarado L, del Moral-Chavex V, Hernandez-Alvarez A, Morett E, Collado-Vides J. 2012. RegullonDB v8.0: omics data sets, evolutionary conservation, regulatory phrases, cross-validated gold standards and more. Nucleic Acids Res. 41:D203–D213. 10.1093/nar/gks1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keseler IM, Kackie A, Peralta-Gil M, Santos-Zavalete A, Gama-Castro S, Bonavides-Martinez C, Fulcher C, Huerta AM, Kothari A, Krummenacker M, Latendresse M, Muniz-Rascado L, Ong Q, Paley S, Schroder I, Shearer AG, Subhraveti P, Travers M, Weerasinghe D, Weiss V, Collado-Vides J, Gunslus RP, Paulsen I, Karp PD. 2012. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 41:D605–D612. 10.1093/nar/gks1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gunsalus RP, Miguel AG, Gunsalus GL. 1986. Intracellular Trp repressor levels in Escherichia coli. J. Bacteriol. 167:272–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borst DW, Blumenthal RM, Mattews RG. 1996. Use of an in vitro titration method to study a global regulator: effects of varying Lrp levels on expression of gltBDF in Escherichia coli. J. Bacteriol. 178:6904–6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jishage M, Ishihama AA. 1995. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of σ70 and σ38. J. Bacteriol. 177:6832–6835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jishage M, Iwata A, Ueda S, Ishihama A. 1996. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J. Bacteriol. 178:5447–5451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maeda H, Jishage M, Nomura T, Fujita N, Ishihama A. 2000. Promoter selectivity of the RNA polymerase holoenzyme containing the extracytoplasmic function (ECF) sigma subunit, sigma-E and FecI. J. Bacteriol. 182:1181–1184. 10.1128/JB.182.4.1181-1184.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jishage M, Ishihama A. 1997. Variation in RNA polymerase sigma subunit composition within different stocks of Escherichia coli W3110. J. Bacteriol. 179:959–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baba T, Ara T, Hasgawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:20006.0006. 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamamoto K, Hirao K, Ohshima T, Aiba H, Utsumi R, Ishihama A. 2005. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J. Biol. Chem. 280:1448–1456. 10.1074/jbc.M410104200 [DOI] [PubMed] [Google Scholar]
- 33.Martinez-Antonio A, Collado-Vides J. 2003. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 6:482–489. 10.1016/j.mib.2003.09.002 [DOI] [PubMed] [Google Scholar]
- 34.Ishihama A. 2009. The nucleoid: an overview. In Boek A, Curtiss R, III, Kaper JB, Karp PD, Neidhardt FC, Nystrom T, Slauch JM, Squires CL, Ussery D. (ed). EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC [Google Scholar]
- 35.Finkel ES, Johnson RC. 1992. The Fis protein: it's not just for DNA inversion anymore. Mol. Microbiol. 6:3257–3265. 10.1111/j.1365-2958.1992.tb02193.x [DOI] [PubMed] [Google Scholar]
- 36.Cho BK, Knight EM, Barrett CL, Palsson BO. 2008. Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts. Genome Res. 18:900–910. 10.1101/gr.070276.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Browning DF, Grainger DC, Busby SJ. 2010. Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr. Opin. Microbiol. 13:773–780. 10.1016/j.mib.2010.09.013 [DOI] [PubMed] [Google Scholar]
- 38.Teramoto J, Yoshimura SH, Takeysu K, Ishihama A. 2010. A novel nucleoid protein of Escherichia coli induced under anaerobic growth conditions. Nucleic Acids Res. 38:3605–3618. 10.1093/nar/gkq077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Almiron M, Link AJ, Furlong D, Kolter R. 1992. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 6:2646–2654. 10.1101/gad.6.12b.2646 [DOI] [PubMed] [Google Scholar]
- 40.Leonard AC, Grimwade JE. 2011. Regulation of DnaA assembly and activity: taking directions from the genome. Annu. Rev. Microbiol. 65:19–35. 10.1146/annurev-micro-090110-102934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balderas-Martinez YI, Savageau M, Saldago H, Perez-Rueda E, Morett E, Collado-Vides J. 2013. Transcription factors in Escherichia coli prefer the holo conformation. PLoS One 8:e65723. 10.1371/journal.pone.0065723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hantke K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172–177. 10.1016/S1369-5274(00)00184-3 [DOI] [PubMed] [Google Scholar]
- 43.Kang Y, Weber KD, Qiu Y, Kiley PJ, Blattner FR. 2005. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J. Bacteriol. 187:1135–1160. 10.1128/JB.187.3.1135-1160.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iuchi S, Weiner L. 1996. Cellular and molecular physiology of Escherichia coli in the adaptation to aerobic environments. J. Biochem. 120:1055–1063. 10.1093/oxfordjournals.jbchem.a021519 [DOI] [PubMed] [Google Scholar]
- 45.Minagawa S, Ogasawara H, Kato A, Yamamoto K, Eguchi Y, Oshima T, Mori H, Ishihama A, Utsumi R. 2003. Identification and characterization of the Mg2+ stimulon of Escherichia coli. J. Bacteriol. 185:3696–3702. 10.1128/JB.185.13.3696-3702.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wanner BL. 1993. Gene regulation by phosphate in enteric bacteria. J. Cell. Biochem. 51:47–54. 10.1002/jcb.240510110 [DOI] [PubMed] [Google Scholar]
- 47.Shimada T, Fujita N, Yamamoto K, Ishihama AA. 2011. Novel roles of cAMP receptor protein (CRP) in regulation of transport and metabolism of carbon sources. PLoS One 6:e20081. 10.1371/journal.pone.0020081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimada T, Yamamoto K, Ishihama A. 2011. Novel members of the Cra regulon involved in carbon metabolism in Escherichia coli. J. Bacteriol. 193:649–659. 10.1128/JB.01214-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Calvo JM, Matthews RG. 1994. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 58:466–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimada T, Bridier A, Briandet R, Ishihama A. 2011. Novel roles of LeuO in transcription regulation in E. coli: antagonistic interplay with the universal silencer H-NS. Mol. Microbiol. 82:376–397. 10.1111/j.1365-2958.2011.07818.x [DOI] [PubMed] [Google Scholar]
- 51.Shimada T, Hirao K, Kori A, Yamamoto K, Ishihama A. 2007. RutR is the uracil/thymine-sensing master regulator of a set of genes for synthesis and degradation of pyrimidines. Mol. Microbiol. 66:744–757. 10.1111/j.1365-2958.2007.05954.x [DOI] [PubMed] [Google Scholar]
- 52.Rintoul MR, Cusa E, Baldoma L, Badia J, Reitzer L, Aguilar J. 2002. Regulation of the Escherichia coli allantoin regulon: coordinated function of the repressor AllR and the activator AllS. J. Mol. Biol. 324:599–610. 10.1016/S0022-2836(02)01134-8 [DOI] [PubMed] [Google Scholar]
- 53.Masiewicz P, Brzostek A, Wolanski M, Dziadek J, Zakrzewska-Czerwinska J. 2012. A novel role of the RppR as a transcription factor involved in the regulation of methylcitrate pathway in Mycobacterium tuberculosis. PLoS One 8:e43651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shimada T, Shimada K, Matsui M, Kitai Y, Igarashi J, Suga H, Ishihama A. 2014. Roles of cell division control factor SdiA: recognition of quorum sensing signals and modulation of transcription regulation targets. Genes Cells 19:405–418. 10.1111/gtc.12139 [DOI] [PubMed] [Google Scholar]
- 55.Shimada T, Katayama Y, Kawakita S, Ogasawara H, Nakano M, Yamanaka K, Ishihama A. 2012. A novel regulator RcdA of the csgD gene encoding the master regulator of biofilm formation in Escherichia coli. Microbiologyopen 1:381–394. 10.1002/mbo3.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Azam TA, Iwata A, Nishimura A, Ueda S, Ishihama A. 1999. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 181:6361–6370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nair S, Finkel SE. 2004. Dps protects cells against multiple stresses during stationary phase. J. Bacteriol. 186:4192–4198. 10.1128/JB.186.13.4192-4198.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oshima T, Biville F. 2006. Functional identification of ygiP as a positive regulator of the ttdA-ttdB-ygjE operon. Microbiology 152:2129–2135. 10.1099/mic.0.28753-0 [DOI] [PubMed] [Google Scholar]
- 59.Garcia-Lara J, Shang LH, Rothfield LI. 1996. An extracellular factor regulates expression of sdiA, a transcriptional activator of cell division genes in Escherichia coli. J. Bacteriol. 178:2742–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rudolph B, Gebendorfer KM, Buchner J, Winter J. 2010. Evolution of Escherichia coli for growth at high temperatures. J. Biol. Chem. 285:19029–19034. 10.1074/jbc.M110.103374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guisbert E, Yura T, Rhodius VA, Gross CA. 2008. Convergence of molecular, modeling, and systems approaches for an understanding of the Escherichia coli heat shock response. Microb. Mol. Biol. Rev. 72:545–554. 10.1128/MMBR.00007-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang A, Rimsky S, Reaban ME, Buc H, Belfort M. 1996. Escherichia coli protein analogs StpA and H-NS: regulatory loops, similar and disparate effects on nucleic acid dynamics. EMBO J. 15:1340–1349 [PMC free article] [PubMed] [Google Scholar]
- 63.Cullum AJ, Bennett AF, Lenski RE. 2001. Evolutionary adaptation to temperature. IX. Preadaptation to novel stressful environments of Escherichia coli adapted to high temperature. Evolution 55:2194–2202 [DOI] [PubMed] [Google Scholar]
- 64.Travisano M, Lenski RE. 1996. Long-term experimental evolution in Escherichia coli. IV. Targets of selection and the specificity of adaptation. Genetics 143:15–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barria C, Malecki M, Arraiano CM. 2013. Bacterial adaptation to cold. Microbiology 159:2437–2443. 10.1099/mic.0.052209-0 [DOI] [PubMed] [Google Scholar]
- 66.White-Ziegler CA, Um S, Perez NM, Berns AL, Malhowski AJ, Young S. 2008. Low temperature (23°C) increases expression of biofilm-, cold-shock-, and RpoS-dependent genes in Escherichia coli K-12. Microbiology 154:148–166. 10.1099/mic.0.2007/012021-0 [DOI] [PubMed] [Google Scholar]
- 67.Browning DF, Busby SJ. 2004. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2:57–65. 10.1038/nrmicro787 [DOI] [PubMed] [Google Scholar]
- 68.Thao S, Chen C-S, Zhu H, Escalante-Semerena JC. 2010. N-Lysine acetylation of a bacterial transcription factor inhibits its DNA-binding activity. PLoS One 5:e15123. 10.1371/journal.pone.0015123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu BJ, Kim JA, Moon JH, Ryu SE, Pan J-G. 2008. Diversity of lysine-acetylated proteins in Escherichia coli. J. Microbiol. Biotechnol. 18:1529–1536 [PubMed] [Google Scholar]
- 70.Alix JH. 1988. Post-translational methylations of ribosomal proteins. Adv. Exp. Med. Biol. 231:371–385 [DOI] [PubMed] [Google Scholar]
- 71.Sakumi K, Igarashi K, Sekiguchi M, Ishihama A. 1993. The Ada protein is a class I transcription factor of Escherichia coli. J. Bacteriol. 175:2455–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kang WK, Icho T, Isono S, Kitakawa M, Isono K. 1989. Characterization of the gene rimK responsible for the addition of glutamic acid residues to the C-terminus of ribosomal protein S6 in Escherichia coli K-12. Mol. Gen. Genet. 217:281–288. 10.1007/BF02464894 [DOI] [PubMed] [Google Scholar]
- 73.Ding H, Demple B. 2011. Direct nitric oxide signal transduction via nitrosylation of iron-sulfur centers in the SoxR transcription activator. Proc. Natl. Acad. Sci. U. S. A. 97:5146–5150. 10.1073/pnas.97.10.5146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seth D, Hausladen A, Wang Y-J, Stampler JS. 2012. Endogenous protein S-nitrosylation in E. coli: regulation by OxyR. Science 336:470–473. 10.1126/science.1215643 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.