Abstract
Methanopterin (MPT) and its analogs are coenzymes required for methanogenesis and methylotrophy in specialized microorganisms. The methyl groups at C-7 and C-9 of the pterin ring distinguish MPT from all other pterin-containing natural products. However, the enzyme(s) responsible for the addition of these methyl groups has yet to be identified. Here we demonstrate that a putative radical S-adenosyl-l-methionine (SAM) enzyme superfamily member encoded by the MJ0619 gene in the methanogen Methanocaldococcus jannaschii is likely this missing methylase. When MJ0619 was heterologously expressed in Escherichia coli, various methylated pterins were detected, consistent with MJ0619 catalyzing methylation at C-7 and C-9 of 7,8-dihydro-6-hydroxymethylpterin, a common intermediate in both folate and MPT biosynthesis. Site-directed mutagenesis of Cys77 present in the first of two canonical radical SAM CX3CX2C motifs present in MJ0619 did not inhibit C-7 methylation, while mutation of Cys102, found in the other radical SAM amino acid motif, resulted in the loss of C-7 methylation, suggesting that the first motif could be involved in C-9 methylation, while the second motif is required for C-7 methylation. Further experiments demonstrated that the C-7 methyl group is not derived from methionine and that methylation does not require cobalamin. When E. coli cells expressing MJ0619 were grown with deuterium-labeled acetate as the sole carbon source, the resulting methyl group on the pterin was predominantly labeled with three deuteriums. Based on these results, we propose that this archaeal radical SAM methylase employs a previously uncharacterized mechanism for methylation, using methylenetetrahydrofolate as a methyl group donor.
INTRODUCTION
Methanopterin (MPT) (Fig. 1A) and its derivatives are one-carbon (C1) carrier coenzymes involved in the essential biochemical processes of methanogenesis and methylotrophy performed by specialized archaea and bacteria (1–4). MPT is structurally and functionally similar to folate (Fig. 1B), the canonical C1 carrier involved in several important biosynthetic processes. Both coenzymes are biologically active in their 5,6,7,8-tetrahydro (H4) (Fig. 1D) forms and function as C1 carriers between formyl and methyl oxidation states (1, 5). Generally, methanogenic archaea use tetrahydromethanopterin (H4MPT) for all C1 metabolism, while methylotrophic bacteria contain an H4MPT-related carrier for their C1 energy metabolism and H4folate presumably for biosynthetic purposes (3). The one known exception to this is Methanosarcina species, which use both folate and MPT (6). Despite the similarities between MPT and folate, most of the enzymes that employ each respective coenzyme (7) and the enzymes involved in their biosynthesis (27) are not homologous, indicating that these C1 carriers evolved independently.
FIG 1.
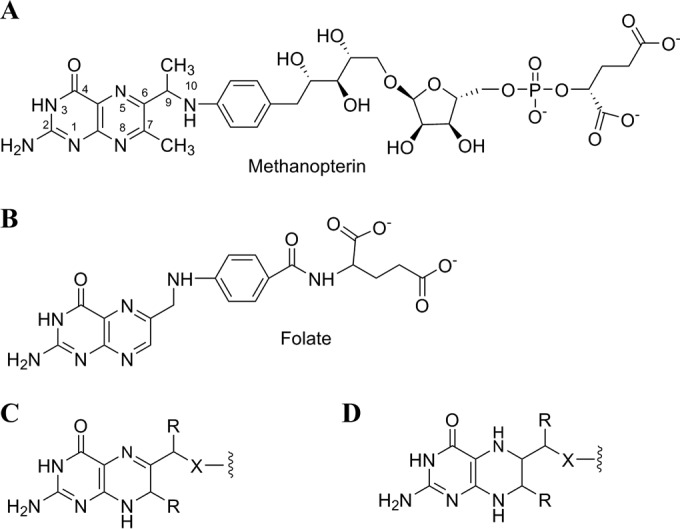
Structures of MPT (A), folate (B), 7,8-H2pterins (C), and 5,6,7,8-H4pterins (D). R is H or CH3, and X is O or NH.
During the biosynthesis of MPT, two methyl groups are introduced at the C-7 and C-9 positions (Fig. 1A) of the pterin ring. These methyl groups distinguish MPT from folate and all other known pterin-containing natural products. Early work suggested that both methyl groups were derived from the methyl group of methionine (8, 9), and it was proposed that the methylations occurred via traditional S-adenosyl-l-methionine (SAM)-dependent nucleophilic substitution chemistry (10). However, attempts to identify these putative SAM-dependent methyltransferases associated with MPT biosynthesis in the genomes of methanogens or methylotrophs have not been successful.
An alternate mechanism for the methylations in MPT biosynthesis could involve radical-dependent chemistry. Examples of radical-dependent enzymatic methylations catalyzed by radical SAM superfamily members have recently appeared, and other putative radical SAM methyltransferases have a widespread distribution (11). Radical SAM enzymes generally have a CX3CX2C amino acid motif that ligates three of the irons in a four-iron, four-sulfur ([4Fe-4S]) cluster (12, 13). SAM acts as the fourth ligand to the cluster during catalysis (14, 15). The first step in a radical SAM enzyme reaction is homolytic cleavage of SAM initiated by electron transfer from the reduced [4Fe-4S]+1 cluster to the sulfonium group of SAM to produce a 5′-deoxyadenosyl radical (Ado-CH2·) (see Fig. S1 in the supplemental material). Ado-CH2· then abstracts a hydrogen atom from the respective substrate, generating 5′-deoxyadenosine (Ado-CH3) and a substrate-based radical that undergoes further chemistry (44).
Through genomic comparisons, we identified a gene in some methanogens that encodes a radical SAM enzyme and is in the neighborhood of the gene encoding beta-ribofuranosylaminobenzene 5′-phosphate synthase, an enzyme known to be involved in MPT biosynthesis (16). Here we demonstrate that the homologous radical SAM enzyme from Methanocaldococcus jannaschii, MJ0619, methylates both the C-7 and C-9 positions of a folate biosynthetic intermediate when heterologously expressed in Escherichia coli, suggesting that MJ0619 is likely the methylase involved in MPT biosynthesis in M. jannaschii.
MATERIALS AND METHODS
Chemicals.
7-Methylpterin, 6,7-dimethylpterin, 6-ethylpterin, 6-ethyl-7-methylpterin, and 6-hydroxyethyl-7-methylpterin were prepared as described previously (9, 17). 7-Methylfolate and 7,9-dimethylfolate were prepared as described previously (18) except that the product was purified on an anion exchange column. All other reagents were obtained from Sigma-Aldrich.
Cloning of the MJ0619 gene, generation of MJ0619 variants, and recombinant MJ0619 overexpression.
The MJ0619 gene (Swiss-Prot accession number Q58036) was amplified by PCR from M. jannaschii genomic DNA using oligonucleotide primers MJ0619-Fwd (5′-GGTGGTCATATGGAGAAAAAAACG-3′) and MJ0619-Rev (5′-GATCGGATCCTTAATCTTCTC-3′). For the generation of cysteine-to-alanine variants, the primers pairs used were MJ0619-M1(C77A)-Fwd (5′-CTGCCCTTATGATGCTGGTCTTTGCCCCAATC-3′) and MJ0619-M1(C77A)-Rev (5′-GATTGGGGCAAAGACCAGCATCATAAGGGCAG-3′) and MJ0619-M2(C102A)-Fwd (5′-GATGTAATTTAAACGCCCCTATATGTTTTG-3′) and MJ0619-M2(C102A)-Rev (5′-CAAAACATATAGGGGCGTTTAAATTACATC-3′). PCR amplification was performed by using an annealing temperature of 55°C. Purified PCR products were digested with NdeI and BamHI restriction enzymes and ligated into compatible sites in plasmid pET19b. Sequencing confirmed the presence of the desired gene in the resulting plasmids, designated pMJ0619, pMJ0619-M1, and pMJ0619-M2. These plasmids were transformed into E. coli strain BL21-CodonPlus(DE3)-RIL (Stratagene), which contains extra copies of genes encoding specific tRNAs that allow increased expression of recombinant proteins from organisms with AT-rich genomes. The transformed cells were grown at 37°C with shaking in Luria-Bertani (LB) broth or M9 broth (200 ml) supplemented with 100 μg/ml ampicillin and 400 μM ferrous ammonium sulfate. When the cells were grown with l-[methyl-2H3]methionine (C2H3-Met), it was added to LB or M9 medium at a concentration of 5 mM. When M9 medium was used, glucose (2%) or [methyl-2H3]acetate (C2H3-acetate) (0.3%) was added as a carbon source. When the culture reached an optical density at 600 nm (OD600) of 1.0, recombinant protein expression was induced by the addition of 28 mM lactose. After culturing for an additional 4 to 5 h for cells grown in LB broth or 18 h for cells grown in M9 broth, the cells were harvested by centrifugation (4,000 rpm for 10 min) and stored at −20°C. Expression of MJ0619 was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoreses (SDS-PAGE) of total cellular proteins and subsequent matrix-assisted laser desorption ionization (MALDI) mass spectral analysis of the band that migrated consistent with the expected molecular weight of 57,351.
Isolation and purification of pterins from E. coli cells.
E. coli cell pellets (200 to 400 mg [wet weight]) were resuspended in 1 ml 50% methanol. The suspension was incubated at 100°C for 10 min, followed by centrifugation (14,000 × g for 5 min) to pellet insoluble cell debris. The resulting soluble extract was concentrated by evaporation under a stream of nitrogen gas and applied to a Dowex 50W-8X-H+ column (1 by 5 mm). The column was washed with water (300 μl), and the pterins were eluted with 6 M aqueous ammonia (300 μl). After concentration and evaporation, the desired pterins were further purified by preparative thin-layer chromatography (TLC) on Silica Gel 60 TLC plates (E. Merck) and detected by their fluorescence. In a solvent system consisting of acetonitrile, water, and 88% formic acid (8:2:1), the Rf values of the pterins were as follows: 0.56 for 7-methylpterin, 0.36 for 6-hydroxymethylpterin, and 0.69 for 6-hydroxyethyl-7-methylpterin. The area of the plate containing the pterin of interest was removed from the plate and eluted with the TLC solvent. After removal of the solvents by evaporation with a stream of nitrogen gas, the sample was dissolved in 80 μl of water for high-pressure liquid chromatography (HPLC) and liquid chromatography-mass spectrometry (LC-MS) analyses.
Isolation and purification of folates from E. coli cells.
After extraction with 50% methanol as described above, the methanol was evaporated, and 200 μl rat plasma was added in order to deglutamylate the folylpolyglutamates to folate (19). The mixture was incubated overnight at 37°C and then centrifuged to remove precipitants. The supernatant was applied to a DEAE-Sephadex A-25 column (2 by 10 mm) equilibrated with water. The column was washed with water and 0.5 M ammonium bicarbonate, and folates were eluted with 2 M ammonium bicarbonate. The ammonium bicarbonate was removed, and the sample was dissolved in water for LC-MS analysis.
Reductive cleavage of pterins and folates.
E. coli cells were extracted with 50% methanol as described above. After centrifugation, the methanol was evaporated from the soluble extract, and the resulting aqueous solution was adjusted to 1 M HCl in a total volume of 1 ml. Zinc dust (∼10 mg) was added, and the solution was incubated with shaking at room temperature for 10 min (9, 20). The excess zinc was pelleted by centrifugation (14,000 × g for 5 min), and the resulting pterins were purified as described above. After reduction, further manipulation under aerobic conditions produces the oxidized pterin, which is the form that is subsequently detected via fluorescence and LC-MS. The Rf values of the pterins on the TLC plate were 0.66 for 6,7-dimethylpterin, 0.77 for 6-ethyl-7-methylpterin, and 0.58 for 6-methylpterin.
HPLC analysis.
A Shimadzu HPLC system with a Pursuit XRs 5 C18 column (250 by 4.6 mm, 2.6-μm particle size; Agilent) equipped with a photodiode array (PDA) detector and a fluorescence detector was utilized for the initial identification of various pterins. The elution profile consisted of 5 min at 95% buffer A (25 mM sodium acetate [pH 6.0], 0.02% sodium azide) and 5% buffer B (methanol), followed by a linear gradient from 5% to 50% buffer B over 25 min at 1 ml/min. The pterins were detected by excitation at 356 nm and emission at 450 nm.
LC-MS analysis.
An AB Sciex 3200 Q Trap mass spectrometry system attached to an Agilent 1200 series liquid chromatograph with a Zorbax Eclipse XDB-C18 column (4.6 by 50 mm, 1.8-μm particle size; Agilent) was used for the identification of pterins and folates. For pterins, the elution profile consisted of a 10-min linear gradient from 95% solvent A (25 mM ammonium acetate) and 5% solvent B (methanol) to 35% solvent A and 65% solvent B at 0.5 ml/min. For folates, the elution profile consisted of a 10-min linear gradient from 95% solvent A (0.1% formic acid in water) and 5% solvent B (0.1% formic acid in methanol) to 90% solvent A and 10% solvent B at 0.5 ml/min. MS data were acquired in the positive mode for pterins and in the negative mode for folates. For pterins, electrospray ionization (ESI) was employed at 4,500 V at a temperature of 400°C. For folates, ESI was employed at −4,500 V at a temperature of 500°C. The curtain gas was set at 35, and ion source gas 1 and ion source gas 2 were 60 and 50, respectively. Standards were used to develop sensitive multiple-reaction monitoring (MRM) methods for the detection of specific pterins and folates (see Table S1 in the supplemental material). Analyst software (Applied Biosystems/MDS SCIEX) was used for system operation and data processing.
Amino acid analysis.
The E. coli cell pellet (200 mg) was resuspended in 50% methanol and incubated at 100°C for 10 min, followed by centrifugation (14,000 × g for 5 min). The supernatant was removed, the pellet was resuspended in 1 ml 6 M HCl, and the mixture was incubated at 100°C overnight. HCl was removed by evaporation with a stream of nitrogen gas. Amino acids were purified on a Dowex 50W-8X-H+ column (1 by 5 mm) and then converted to their N-trifluoroacetyl methyl ester derivatives for gas chromatography-mass spectrometry (GC-MS) analysis.
Genomic analysis.
Analysis of the M. jannaschii genome and comparison to other designated methanogens were carried out by using STRING (21) and EDGAR (Efficient Database Framework for Comparative Genome Analyses Using BLAST) (22).
RESULTS AND DISCUSSION
Methylation activity of MJ0619.
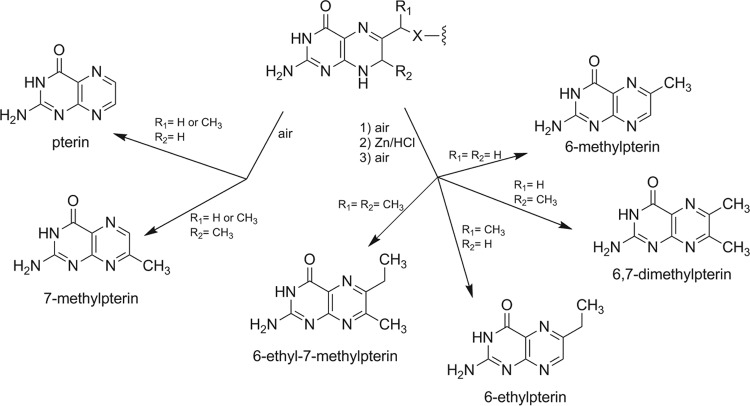
Since M. jannaschii is not yet readily amenable to genetic manipulation, we decided to test our proposal that MJ0619 is the enzyme responsible for the methylation reactions in MPT biosynthesis by cloning and heterologous expression. The MJ0619 gene was cloned into pET19b and was overexpressed in E. coli. Expression of MJ0619, which has an expected molecular weight of 57,351, was confirmed by SDS-PAGE (see Fig. S2 in the supplemental material) and MALDI mass spectral analysis of the tryptic peptides derived from the SDS protein band. Due to the structural similarity between MPT and folate, we reasoned that MJ0619 may methylate folate and/or a folate precursor when expressed in E. coli. To test this, the pterins from E. coli cells containing heterologously expressed MJ0619 (E. coli_MJ0619 cells) were extracted, partially purified, and analyzed by HPLC with fluorescence detection and LC-MS. When isolated under oxidative conditions, a large percentage of H4folate undergoes oxidative cleavage to produce pterin as one of the major reaction products (23). Similarly, 7-methylpterin is observed as a result of oxidative degradation of H4MPT (24). During the biosynthesis of MPT and folate, the pterin is present in the 7,8-dihydro (H2) pterin form (Fig. 1C). The oxidative degradation of H2pterin-containing molecules is more complex and depends on many variables, but the oxidized pterins are also a common product, as seen for H4pterin oxidation (Fig. 2) (25, 26). Only these oxidized forms of pterins were assayed in this work. Based on the above-described logic, if MJ0619 methylated the pterin of H4folate or a H4folate precursor at the C-7 position, 7-methylpterin would be present in the E. coli cell extract.
FIG 2.
H2pterin degradation products from exposure to air and Zn/HCl reduction. X is O or NH.
Initial observations of the pterins from E. coli_MJ0619 cells by HPLC with fluorescence detection showed a peak with the same retention time as that of authentic 7-methylpterin (see Fig. S3 in the supplemental material). To confirm the identity of 7-methylpterin in the cell extracts, we used LC-MS. An MRM method was developed, which specifically detects 7-methylpterin from the fragment ions generated from the collision-induced dissociation (CID) of the MH+ ion. Figure 3 shows the MRM ion chromatogram for authentic 7-methylpterin compared to partially purified pterins from E. coli_MJ0619 cells and E. coli cells containing another recombinant protein from M. jannaschii (MJ0815) unrelated to MPT biosynthesis (E. coli_control cells). E. coli containing an empty pET19b vector was also used as a negative control for the experiments described here (data not shown). The peak at 6.1 min corresponds to 7-methylpterin (Fig. 3), demonstrating that MJ0619 methylates a pterin substrate at the C-7 position when heterologously expressed in E. coli.
FIG 3.
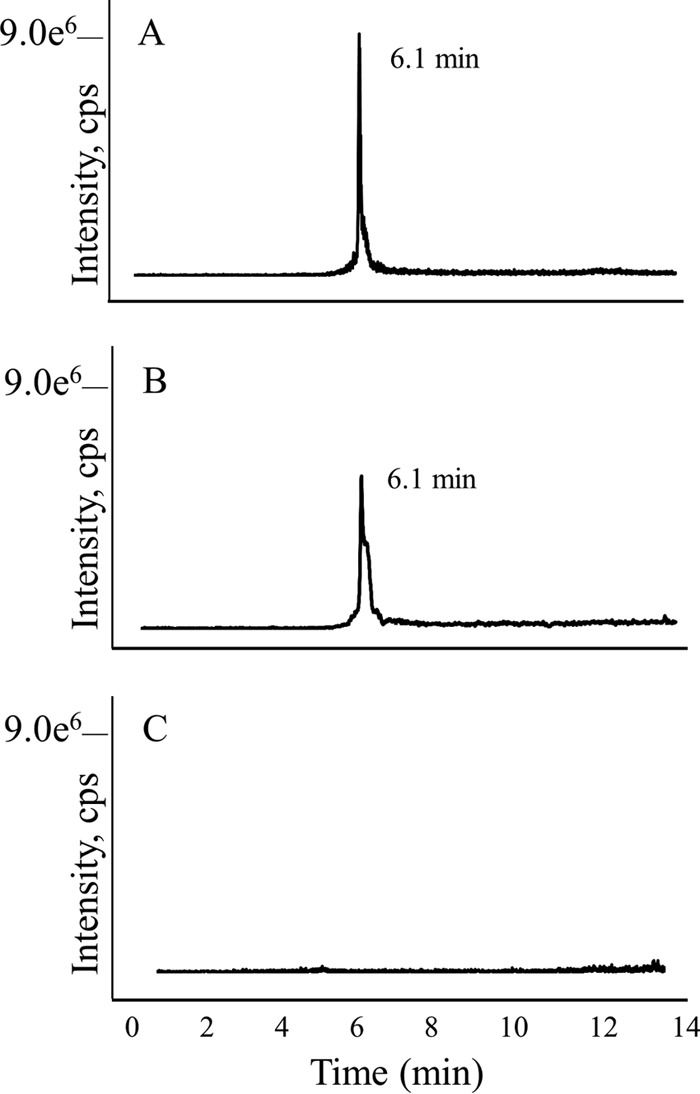
LC-MS MRM trace for 7-methylpterin. (A) Authentic 7-methylpterin; (B) partially purified pterin extract from E. coli_MJ0619 cells; (C) partially purified pterin extract from E. coli_control cells.
In order to determine whether MJ0619 also methylates the C-9 position of a folate precursor in addition to the C-7 position, we performed a zinc reduction on the cell extract and then isolated the resulting pterins. This procedure reductively cleaves the bond between the 9 and 10 positions associated with the pterin ring (Fig. 2). LC-MS analysis showed the presence of 6-ethyl-7-methylpterin (Fig. 2) in the zinc-treated E. coli_MJ0619 extracts (Fig. 4), indicating that MJ0619 methylates both the C-7 and C-9 positions of a pterin substrate. We also assayed for monomethylated pterins in the samples that had been treated with zinc. If MJ0619 methylated the C-7 position first and released the product, 6,7-dimethylpterin would be observed. Similarly, if the C-9 position were methylated first, we would expect to detect 6-ethylpterin after zinc reduction (Fig. 3). In our analysis, we did not observe 6,7-dimethylpterin or 6-ethylpterin, suggesting that MJ0619 may methylate both positions consecutively.
FIG 4.
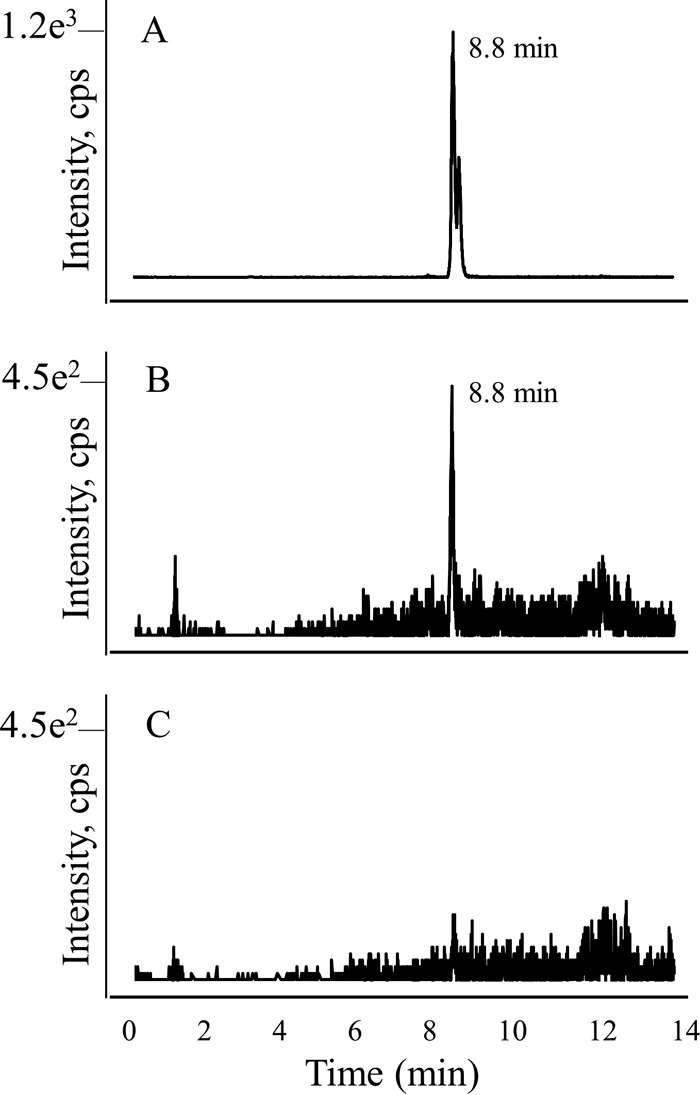
LC-MS MRM trace for 6-ethyl-7-methylpterin. (A) Authentic 6-ethyl-7-methylpterin; (B) partially purified zinc-reduced pterin extract from E. coli_MJ0619 cells; (C) partially purified zinc-reduced pterin extract from E. coli_control cells.
MPT biosynthetic pathway and determination of the MJ0619 substrate for methylation.
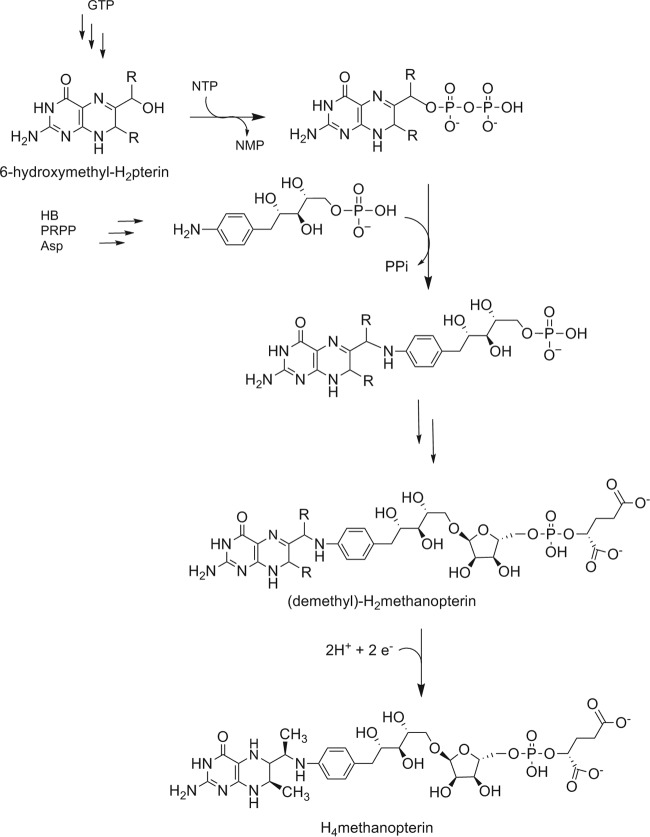
The biosynthesis of MPT involves two converging pathways, which are partially depicted in Fig. 5. Similarly to folate biosynthesis, GTP acts as a precursor to the pterin portion of the coenzyme (27, 28). The arylamine [5-(4-aminophenyl)-1,2,3,4-tetrahydroxypentane] of MPT is derived from 4-hydroxybenzoate (HB), the nitrogen of aspartate, and 5-phospho-α-d-ribose-1-diphosphate (PRPP) (27, 29). When growing cultures are fed 4-aminobenzoic acid (AB), methanogens can incorporate AB directly into the arylamine. The two pathways converge to produce the functional portion of the coenzyme, and a final series of reactions leads to the remainder of the biologically active molecule (30). The side chain containing the phosphate and α-hydroxyglutarate is absent from the MPT found in the methylotrophic bacterium Methylobacterium extorquens, and this truncated form of the coenzyme has been termed dephospho-MPT (3).
FIG 5.
Proposed H4MPT biosynthetic pathway. R is H or CH3.
The point in the pathway at which the methylations occur remains unclear and appears to vary between different methanogens (8–10). Before this study, existing evidence from experiments with extracts from methanogens not closely related to M. jannaschii, Methanobacterium thermoautotrophicum strain ΔH, M. thermoautotrophicum strain Marburg, and Methanosarcina thermophila, was consistent with methylation occurring in one of the final steps of H4MPT biosynthesis (10). Specifically, the substrate was suggested to be demethylated H2MPT (Fig. 5). This prompted us to speculate that the MJ0619 substrate for methylation in E. coli would be H2folate, with 7,9-dimethyl-H2folate as a product. To test this, we isolated intact folates from E. coli_MJ0619 cells and compared the sample to synthetic 7-methylfolate and 7,9-dimethylfolate by LC-MS (see Fig. S4 and S5 in the supplemental material). In our analysis, folate and N10-formylfolate were observed in the cells, but no methylated folates were detected. The folate profile of the E. coli_MJ0619 cells was analogous to that of the E. coli_control cells (see Fig. S4 in the supplemental material). We also did not detect methylated pteroic acid (folate precursor lacking the glutamate side chain) in the E. coli_MJ0619 cell extracts, indicating that the methylations must occur before the pterin is condensed with AB.
One common intermediate in folate and MPT biosynthesis is 6-hydroxymethyl-H2pterin (Fig. 5). Therefore, we decided to assay for 6-hydroxyethyl-7-methylpterin in E. coli_MJ0619 cells in order to determine if the methylations were taking place at this stage in the pathway. The LC-MS MRM chromatogram for chemically synthesized 6-hydroxyethyl-7-methylpterin compared to the pterin extract from E. coli_MJ0619 cells and E. coli_control cells (Fig. 6) demonstrates the presence of 6-hydroxyethyl-7-methylpterin only in the E. coli_MJ0619 cells. Therefore, the substrate for methylation by MJ0619 in E. coli is likely 6-hydroxymethyl-H2pterin. These results indicate that the methylation reactions occur earlier in the MPT biosynthetic pathway in M. jannaschii than reported for other methanogens. Since we have not identified any methylated folates in the E. coli cell extracts, the methylated intermediate is likely not a substrate for the subsequent enzyme(s) in folate biosynthesis.
FIG 6.
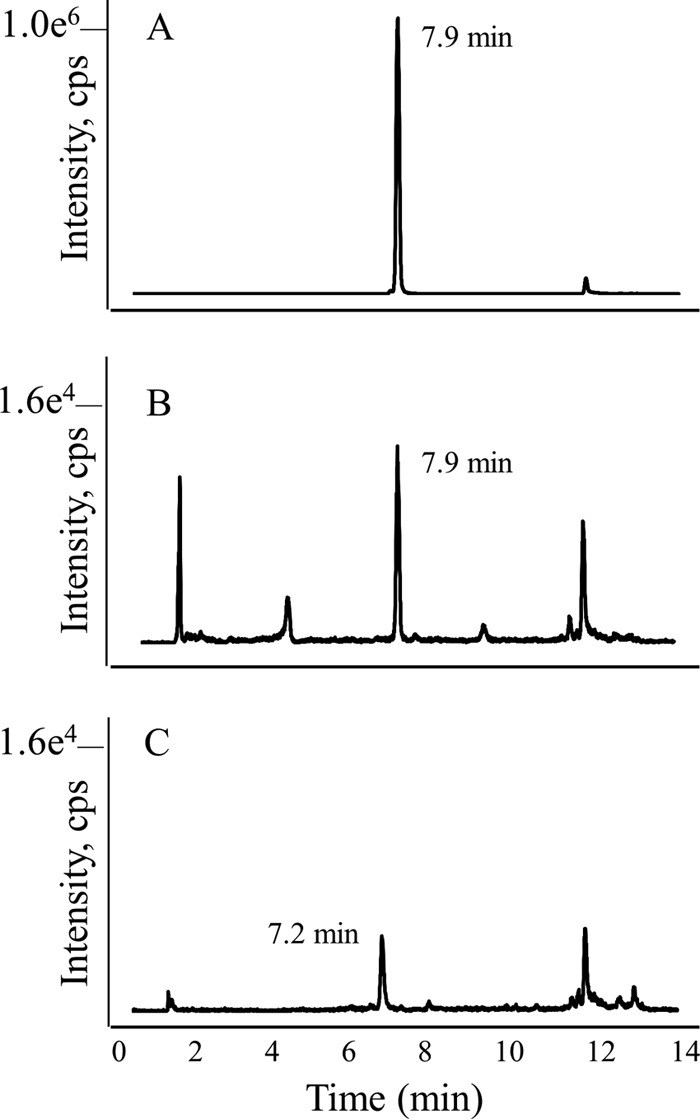
LC-MS MRM trace for 6-hydroxyethyl-7-methylpterin. (A) Authentic 6-hydroxyethyl-7-methylpterin; (B) partially purified pterin extract from E. coli_MJ0619 cells; (C) partially purified pterin extract from E. coli_control cells.
Cysteine-to-alanine variants in the radical SAM motif.
The MJ0619 gene codes for two canonical CX3CX2C radical SAM amino acid motifs (C73, C77, and C80, and C98, C102, and C105). To determine the role of each motif, we generated two variants in which the middle cysteine in each motif was separately converted to an alanine. In the C77A variant, 7-methylpterin was still observed in the E. coli cell extract (see Fig. S6A in the supplemental material). However, in the C102A variant, no methylation activity was observed (see Fig. S6B in the supplemental material), indicating that the latter position is involved in binding the requisite [4Fe-4S] cluster for radical SAM-dependent methylation at the C-7 position. Importantly, we did not observe C-9 methylation with either of these variants. This suggests that the most N-terminal cluster (C73, C77, and C80) could be involved in C-9 methylation and that C-7 methylation must occur before C-9 methylation.
Methylation mechanism.
A subgroup of recently characterized radical SAM methyltransferases involved in secondary metabolite biosynthesis in bacteria contain a vitamin B12 (cobalamin)-binding motif and require cobalamin for activity (13, 31–34). These enzymes are thought to use one molecule of SAM in typical radical SAM fashion to produce Ado-CH2· for substrate radical formation (see Fig. S1 in the supplemental material) and another molecule of SAM as a methyl group donor with a methylcobalamin intermediate that mediates the transfer of a methyl radical to the respective substrate. MJ0619 does not contain a cobalamin-binding motif, but to confirm that it does not require cobalamin for activity, we grew E. coli_MJ0619 cells in M9 minimal medium. Under these conditions, E. coli does not have access to cobalamin, since it cannot synthesize the coenzyme de novo (35). 7-Methylpterin was detected in E. coli extracts grown under these conditions (see Fig. S7 in the supplemental material). Therefore, MJ0619 does not require cobalamin for activity.
The best-characterized radical SAM methyltransferases are RlmN and Cfr, homologous bacterial enzymes that methylate an adenosine residue in 23S rRNA. Initial work on these enzymes showed that SAM acts both as a source of Ado-CH2· and as a methyl group donor in the methylation reactions (36). Later, extensive mechanistic and crystallographic studies revealed that RlmN catalyzes an initial SN2 nucleophilic substitution reaction to methylate a cysteine residue on the enzyme and uses subsequent radical SAM-dependent chemistry to generate a methyl group on the rRNA substrate (37–39). In the latter reaction, the methyl group that is appended to the adenosine residue retains only two hydrogens from the original SAM-derived methyl group. Early work on MPT methylation in two methanogens, Methanobrevibacter ruminantium (40) and Methanococcus voltae, grown with C2H3-Met, indicated that both methyl groups at C-7 and C-9 were derived from methionine with the retention of all three deuteriums (8, 9). These observations led us to the conclusion that methylation in MPT biosynthesis does not proceed by a mechanism analogous to that of RlmN. In order to test this idea, we grew E. coli_MJ0619 cells in M9 medium supplemented with C2H3-Met and analyzed deuterium incorporation into 7-methylpterin by LC-MS. Under these conditions, completely unlabeled 7-methylpterin was observed, but surprisingly, no deuterium incorporation was detected. The amount of labeled methionine incorporated into the E. coli proteins was about 40% based on GC-MS analysis, proving that the C2H3-Met was indeed incorporated into the cells. We also performed several experiments to ensure that the deuteriums in chemically synthesized 7-C2H3-pterin do not exchange with the solvent.
E. coli_MJ0619 cells grown with C2H3-acetate as the sole carbon source, however, readily led to 7-methylpterins containing the following distribution of deuterium: 21% 2H0, 12% 2H1, 28% 2H2, and 39% 2H3. These labeling experiments indicate that the source of the methyl group is not methionine. Therefore, the methylation reaction mechanism catalyzed by MJ0619 from M. jannaschii is completely different from the methylations in MPT biosynthesis in M. ruminantium and M. voltae. Genomic comparison of M. jannaschii to these two methanogens revealed that only about 40% of M. jannaschii genes are homologous to genes in M. ruminantium or M. voltae. Some of the nonhomologous genes encode known methanogenic coenzyme biosynthesis proteins in M. jannaschii, including genes for MPT biosynthesis, indicating that these methanogens use different enzymes and different chemistries for some essential biochemical processes. Therefore, the mechanism and enzyme(s) required for methylation in MPT biosynthesis likely vary among different methanogens.
At this point, our data demonstrate that the methyl group is not derived from methionine and that cobalamin is not involved in the pterin methylation reaction catalyzed by MJ0619. We next wanted to determine whether CH3H4folate was acting as the methyl group donor when MJ0619 was expressed in E. coli. To test this, we analyzed deuterium labeling in the methyl group of methionine isolated from E. coli_MJ0619 cells grown with C2H3-acetate as the sole carbon source. Methionine is synthesized from homocysteine with a methyl group from N5-methyl-tetrahydrofolate (CH3H4folate). GC-MS analysis showed that the methyl group of methionine in these cells was mostly unlabeled, with the following distribution of deuterium: 46% 2H0, 35% 2H1, 14% 2H2, and 5% 2H3. This result is strikingly different from the labeling distribution that we observed for the methyl group of 7-methylpterin isolated from the same cells, in which the majority of the molecules contained three deuterium atoms. This result indicates that the methyl group added by MJ0619 is not derived from CH3H4folate.
The distribution of deuterium labeling on the methyl group of the 7-methylpterin isolated from E. coli_MJ0619 cells grown in medium containing deuterated acetate as a carbon source is comparable to the distribution of labeling that we previously observed for the methyl group of thymine when E. coli was grown with deuterated serine as a sole carbon source (41). Thymine is synthesized from dUMP to generate dTMP by thymidylate synthase. In this reaction, N5,N10-methylene-H4folate (CH2H4folate) serves as a methylene group donor as well as a hydride ion donor to generate the final methyl group on dTMP. dUMP is activated for nucleophilic attack through the formation of a covalent cysteine adduct. We propose that MJ0619 catalyzes a similar type of methylation reaction using CH2H4folate as a methyl group source. Since the H2pterin substrate in the MJ0619-catalyzed reaction cannot be activated by a covalent cysteine intermediate, we propose that activation is achieved instead via substrate radical formation.
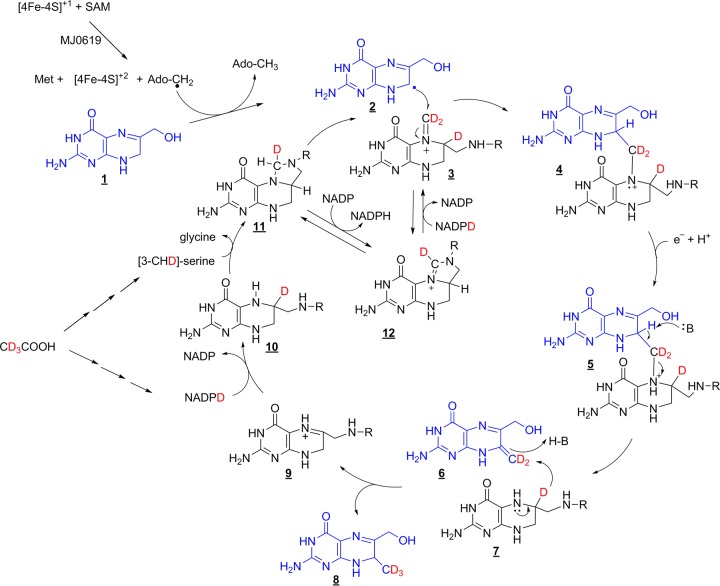
In our proposed mechanism depicted in Fig. 7, Ado-CH2·, generated via typical radical SAM chemistry, abstracts a hydrogen atom from the C-7 position of 6-hydroxymethyl-H2pterin (compound 1) to produce a substrate-based radical (compound 2). The substrate radical then attacks the methylene of the iminium form of CH2H4folate (compound 3) to generate a radical cation and a covalent bond between the two substrates (compound 4). Addition of a proton and an electron generates compound 5. Abstraction of a proton initiates the collapse of this intermediate to generate a methylene at the C-7 position of the pterin (compound 6). Hydride from the C-6 position of H4folate (compound 7) then attacks the methylene to give the final methylated product (compound 8). The folate product (compound 9) can reenter normal folate metabolism, in which reduction by H2folate reductase yields H4folate (compound 10) and catalysis by serine hydroxymethyltransferase yields CH2H4folate (compound 11).
FIG 7.
Proposed mechanism for H2pterin methylation at the C-7 position catalyzed by MJ0619. C-9 methylation would occur by analogous chemistry. The substrate pterin is shown in blue, and the folate methyl group donor is shown in black. D is 2H (deuterium), and R is remainder of the folate or MPT coenzyme (Fig. 1).
This mechanism is well supported by our isotopic labeling data obtained with E. coli_MJ0619 cells grown in C2H3-acetate medium. GC-MS analysis showed that serine generated from C2H3-acetate contains primarily one deuterium at the C-3 position, which is used to generate CH2H4folate (compound 11). This methylene group, which mostly contains one deuterium, can be further enriched with deuterium by its oxidation to methenyl-H4folate (compound 12) and reduction back to CH2H4folate with NADP2H. This reversible reaction represents the source of the second deuterium incorporated into the methyl group of the observed 7-methylpterin. The NADP2H generated by isocitrate dehydrogenase when E. coli is grown with C2H3-acetate contains a deuterium that is used to reduce H2folate to generate H4folate (compound 9 to compound 10). The C-2-labeled isocitrate for this reaction arises via deuterated acetyl coenzyme A and deuterated succinate after two turns of the citric acid cycle. The hydride at the C-6 position of compound 7 produces the final methyl group in our proposed mechanism and is the source of the third deuterium. These pathways explain our observation that most 7-methylpterin isolated under conditions with C2H3-acetate as the sole carbon source contains 3 deuteriums. The pathway from C2H3-acetate to serine and finally to CH2H4folate contains intermediate metabolites where deuterium is in a solvent-exchangeable position; therefore, a range of deuterium labeling is observed in the isolated 7-methylpterin.
Here we have shown that CH2H4folate is likely the methyl group donor for the MJ0619-catalyzed methylation reaction when the enzyme is heterologously expressed in E. coli. Since M. jannaschii and other methanogens do not have folate, this implies that MPT, in the CH2H4MPT form, may be a cofactor in its own biosynthesis. Precedent for this phenomenon is found in pyridoxal 5′-phosphate biosynthesis (42) and thiamine biosynthesis (43). This use of CH2H4folate as a methyl group donor is unusual in biochemistry and has been described previously only for the thymidylate synthase-catalyzed reaction.
Future studies will shed light on this previously undescribed radical mechanism for methylation and determine the true methyl group donor. We have purified recombinant MJ0619, and experiments are being carried out to test the ability of the enzyme to catalyze the methylation of MPT precursors.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Science Foundation grant MCB0722787 awarded to R.H.W.
We thank Walter Niehaus for assistance with editing the manuscript, Kim Harich (Department of Biochemistry, Virginia Tech) for mass spectral assistance, W. Keith Ray (Virginia Tech Mass Spectrometry Incubator, Virginia Tech) for MALDI analysis, Rebecca Wattam (Virginia Bioinformatics Institute, Virginia Tech) for bioinformatics assistance, and Jochen Blom (Bioinformatics and Systems Biology, Justus Liebig University Giessen) for EDGAR genomic analysis development.
Footnotes
Published ahead of print 7 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01903-14.
REFERENCES
- 1.Escalante-Semerena JC, Rinehart KL, Jr, Wolfe RS. 1984. Tetrahydromethanopterin, a carbon carrier in methanogenesis. J. Biol. Chem. 259:9447–9455 [PubMed] [Google Scholar]
- 2.Fischer R, Thauer RK. 1989. Methyltetrahydromethanopterin as an intermediate in methanogenesis from acetate in Methanosarcina barkeri. Arch. Microbiol. 151:459–465. 10.1007/BF00416607 [DOI] [Google Scholar]
- 3.Chistoserdova L, Vorholt JA, Thauer RK, Lidstrom ME. 1998. C1 transfer enzymes and coenzymes linking methylotrophic bacteria and methanogenic Archaea. Science 281:99–102. 10.1126/science.281.5373.99 [DOI] [PubMed] [Google Scholar]
- 4.Vorholt JA, Chistoserdova L, Stolyar SM, Thauer RK, Lidstrom ME. 1999. Distribution of tetrahydromethanopterin-dependent enzymes in methylotrophic bacteria and phylogeny of methenyl tetrahydromethanopterin cyclohydrolases. J. Bacteriol. 181:5750–5757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Escalante-Semerena JC, Leigh JA, Rinehart KL, Wolfe RS. 1984. Formaldehyde activation factor, tetrahydromethanopterin, a coenzyme of methanogenesis. Proc. Natl. Acad. Sci. U. S. A. 81:1976–1980. 10.1073/pnas.81.7.1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchenau B, Thauer RK. 2004. Tetrahydrofolate-specific enzymes in Methanosarcina barkeri and growth dependence of this methanogenic archaeon on folic acid or p-aminobenzoic acid. Arch. Microbiol. 182:313–325. 10.1007/s00203-004-0714-0 [DOI] [PubMed] [Google Scholar]
- 7.Maden BE. 2000. Tetrahydrofolate and tetrahydromethanopterin compared: functionally distinct carriers in C1 metabolism. Biochem. J. 350(Part 3):609–629. 10.1042/0264-6021:3500609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White RH. 1986. Biosynthesis of the 7-methylated pterin of methanopterin. J. Bacteriol. 165:215–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White RH. 1990. Biosynthesis of methanopterin. Biochemistry 29:5397–5404. 10.1021/bi00474a027 [DOI] [PubMed] [Google Scholar]
- 10.White RH. 1998. Methanopterin biosynthesis: methylation of the biosynthetic intermediates. Biochim. Biophys. Acta 1380:257–267. 10.1016/S0304-4165(97)00148-7 [DOI] [PubMed] [Google Scholar]
- 11.Zhang Q, van der Donk WA, Liu W. 2012. Radical-mediated enzymatic methylation: a tale of two SAMS. Acc. Chem. Res. 45:555–564. 10.1021/ar200202c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marsh EN, Patwardhan A, Huhta MS. 2004. S-Adenosylmethionine radical enzymes. Bioorg. Chem. 32:326–340. 10.1016/j.bioorg.2004.06.001 [DOI] [PubMed] [Google Scholar]
- 13.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. 2001. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 29:1097–1106. 10.1093/nar/29.5.1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krebs C, Broderick WE, Henshaw TF, Broderick JB, Huynh BH. 2002. Coordination of adenosylmethionine to a unique iron site of the [4Fe-4S] of pyruvate formate-lyase activating enzyme: a Mossbauer spectroscopic study. J. Am. Chem. Soc. 124:912–913. 10.1021/ja017562i [DOI] [PubMed] [Google Scholar]
- 15.Walsby CJ, Ortillo D, Yang J, Nnyepi MR, Broderick WE, Hoffman BM, Broderick JB. 2005. Spectroscopic approaches to elucidating novel iron-sulfur chemistry in the “radical-Sam” protein superfamily. Inorg. Chem. 44:727–741. 10.1021/ic0484811 [DOI] [PubMed] [Google Scholar]
- 16.Scott JW, Rasche ME. 2002. Purification, overproduction, and partial characterization of beta-RFAP synthase, a key enzyme in the methanopterin biosynthesis pathway. J. Bacteriol. 184:4442–4448. 10.1128/JB.184.16.4442-4448.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White RH. 1985. 7-Methylpterin and 7-methyllumizine: oxidative degradation products of 7-methyl-substituted pteridines in methanogenic bacteria. J. Bacteriol. 162:516–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boothe JH, Mowat JH, Waller CW, Angier RB, Semb J, Gazzola AL. 1952. 7-Methylpteroylglutamic acid and some related compounds. J. Am. Chem. Soc. 74:5407–5409. 10.1021/ja01141a055 [DOI] [Google Scholar]
- 19.Pribat A, Blaby IK, Lara-Nunez A, Gregory JF, III, de Crecy-Lagard V, Hanson AD. 2010. FolX and FolM are essential for tetrahydromonapterin synthesis in Escherichia coli and Pseudomonas aeruginosa. J. Bacteriol. 192:475–482. 10.1128/JB.01198-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foo SK, Cichowicz DJ, Shane B. 1980. Cleavage of naturally occurring folates to unsubstituted p-aminobenzoylpoly-gamma-glutamates. Anal. Biochem. 107:109–115. 10.1016/0003-2697(80)90499-6 [DOI] [PubMed] [Google Scholar]
- 21.Snel B, Lehmann G, Bork P, Huynen MA. 2000. STRING: a Web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 28:3442–3444. 10.1093/nar/28.18.3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blom J, Albaum SP, Doppmeier D, Puhler A, Vorholter FJ, Zakrzewski M, Goesmann A. 2009. EDGAR: a software framework for the comparative analysis of prokaryotic genomes. BMC Bioinformatics 10:154. 10.1186/1471-2105-10-154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reed LS, Archer MC. 1980. Oxidation of tetrahydrofolic acid by air. J. Agric. Food Chem. 28:801–805. 10.1021/jf60230a044 [DOI] [Google Scholar]
- 24.White RH. 1997. Structural characterization of modified folates in Archaea. Methods Enzymol. 281:391–401. 10.1016/S0076-6879(97)81046-4 [DOI] [PubMed] [Google Scholar]
- 25.Dantola ML, Vignoni M, Capparelli AL, Lorente C, Thomas AH. 2008. Stability of 7,8-dihydropterins in air-equilibrated aqueous solutions. Helv. Chim. Acta 91:411–425. 10.1002/hlca.200890046 [DOI] [Google Scholar]
- 26.Vignoni M, Cabrerizo FM, Lorente C, Claparols C, Oliveros E, Thomas AH. 2010. Photochemistry of dihydrobiopterin in aqueous solution. Org. Biomol. Chem. 8:800–810. 10.1039/b913095k [DOI] [PubMed] [Google Scholar]
- 27.Grochowski LL, White RH. 2010. Biosynthesis of the methanogenic coenzymes, p 711–748 In Mander L, Liu H-W. (ed), Comprehensive natural products II: chemistry and biology, vol 7 Elsevier, Oxford, United Kingdom [Google Scholar]
- 28.Bermingham A, Derrick JP. 2002. The folic acid biosynthesis pathway in bacteria: evaluation of potential for antibacterial drug discovery. Bioessays 24:637–648. 10.1002/bies.10114 [DOI] [PubMed] [Google Scholar]
- 29.White RH. 2011. The conversion of a phenol to an aniline occurs in the biochemical formation of the 1-(4-aminophenyl)-1-deoxy-D-ribitol moiety in methanopterin. Biochemistry 50:6041–6052. 10.1021/bi200362w [DOI] [PubMed] [Google Scholar]
- 30.Xu H, Aurora R, Rose GD, White RH. 1999. Identifying two ancient enzymes in Archaea using predicted secondary structure alignment. Nat. Struct. Biol. 6:750–754. 10.1038/11525 [DOI] [PubMed] [Google Scholar]
- 31.Werner WJ, Allen KD, Hu K, Helms GL, Chen BS, Wang SC. 2011. In vitro phosphinate methylation by PhpK from Kitasatospora phosalacinea. Biochemistry 50:8986–8988. 10.1021/bi201220r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pierre S, Guillot A, Benjdia A, Sandstrom C, Langella P, Berteau O. 2012. Thiostrepton tryptophan methyltransferase expands the chemistry of radical SAM enzymes. Nat. Chem. Biol. 8:957–959. 10.1038/nchembio.1091 [DOI] [PubMed] [Google Scholar]
- 33.Kim HJ, McCarty RM, Ogasawara Y, Liu YN, Mansoorabadi SO, Levieux J, Liu HW. 2013. GenK-catalyzed C-6′ methylation in the biosynthesis of gentamicin: isolation and characterization of a cobalamin-dependent radical SAM enzyme. J. Am. Chem. Soc. 135:8093–8096. 10.1021/ja312641f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allen KD, Wang SC. 2014. Initial characterization of Fom3 from Streptomyces wedmorensis: the methyltransferase in fosfomycin biosynthesis. Arch. Biochem. Biophys. 543:67–73. 10.1016/j.abb.2013.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Volcani BE, Toohey JI, Barker HA. 1961. Detection of cobamide coenzymes in microorganisms by the ionophoretic bioautographic method. Arch. Biochem. Biophys. 92:381–391. 10.1016/0003-9861(61)90376-9 [DOI] [PubMed] [Google Scholar]
- 36.Yan F, LaMarre JM, Rohrich R, Wiesner J, Jomaa H, Mankin AS, Fujimori DG. 2010. RlmN and Cfr are radical SAM enzymes involved in methylation of ribosomal RNA. J. Am. Chem. Soc. 132:3953–3964. 10.1021/ja910850y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boal AK, Grove TL, McLaughlin MI, Yennawar NH, Booker SJ, Rosenzweig AC. 2011. Structural basis for methyl transfer by a radical SAM enzyme. Science 332:1089–1092. 10.1126/science.1205358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grove TL, Benner JS, Radle MI, Ahlum JH, Landgraf BJ, Krebs C, Booker SJ. 2011. A radically different mechanism for S-adenosylmethionine-dependent methyltransferases. Science 332:604–607. 10.1126/science.1200877 [DOI] [PubMed] [Google Scholar]
- 39.Grove TL, Radle MI, Krebs C, Booker SJ. 2011. Cfr and RlmN contain a single [4Fe-4S] cluster, which directs two distinct reactivities for S-adenosylmethionine: methyl transfer by SN2 displacement and radical generation. J. Am. Chem. Soc. 133:19586–19589. 10.1021/ja207327v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lovley DR, Greening RC, Ferry JG. 1984. Rapidly growing rumen methanogenic organism that synthesizes coenzyme M and has a high affinity for formate. Appl. Environ. Microbiol. 48:81–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.White RH. 1983. Proton exchange on carbons 2 and 3 of serine during their conversion into methyl groups of methionine and thymine in Escherichia coli. Biochemistry 22:1883–1888. 10.1021/bi00277a022 [DOI] [PubMed] [Google Scholar]
- 42.Drewke C, Klein M, Clade D, Arenz A, Muller R, Leistner E. 1996. 4-O-Phosphoryl-L-threonine, a substrate of the pdxC (serC) gene product involved in vitamin B6 biosynthesis. FEBS Lett. 390:179–182. 10.1016/0014-5793(96)00652-7 [DOI] [PubMed] [Google Scholar]
- 43.Sprenger GA, Schorken U, Wiegert T, Grolle S, de Graaf AA, Taylor SV, Begley TP, Bringer-Meyer S, Sahm H. 1997. Identification of a thiamin-dependent synthase in Escherichia coli required for the formation of the 1-deoxy-D-xylulose 5-phosphate precursor to isoprenoids, thiamin, and pyridoxol. Proc. Natl. Acad. Sci. U. S. A. 94:12857–12862. 10.1073/pnas.94.24.12857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frey PA, Hegeman AD, Ruzicka FJ. 2008. The radical SAM superfamily. Crit. Rev. Biochem. Mol. Biol. 43:63–88. 10.1080/10409230701829169 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.