Abstract
RidA, the archetype member of the widely conserved RidA/YER057c/UK114 family of proteins, prevents reactive enamine/imine intermediates from accumulating in Salmonella enterica by catalyzing their hydrolysis to stable keto acid products. In the absence of RidA, endogenous 2-aminoacrylate persists in the cellular environment long enough to damage a growing list of essential metabolic enzymes. Prior studies have focused on the dehydration of serine by the pyridoxal 5′-phosphate (PLP)-dependent serine/threonine dehydratases, IlvA and TdcB, as sources of endogenous 2-aminoacrylate. The current study describes an additional source of endogenous 2-aminoacrylate derived from cysteine. The results of in vivo analysis show that the cysteine sensitivity of a ridA strain is contingent upon CdsH, the predominant cysteine desulfhydrase in S. enterica. The impact of cysteine on 2-aminoacrylate accumulation is shown to be unaffected by the presence of serine/threonine dehydratases, revealing another mechanism of endogenous 2-aminoacrylate production. Experiments in vitro suggest that 2-aminoacrylate is released from CdsH following cysteine desulfhydration, resulting in an unbound aminoacrylate substrate for RidA. This work expands our understanding of the role played by RidA in preventing enamine stress resulting from multiple normal metabolic processes.
INTRODUCTION
The RidA/YER057c/UK114 family of proteins is highly conserved across all domains of life. In Salmonella enterica, RidA catalyzes the hydrolysis of reactive enamine/imine intermediates produced as a consequence of serine/threonine dehydratase activity (1). The pyridoxal 5′-phosphate (PLP)-dependent serine/threonine dehydratases (EC 4.3.1.19), IlvA and TdcB, dehydrate serine and threonine to generate the enamine intermediates 2-aminoacrylate (2AA) and 2-aminocrotonate, respectively (2, 3). The unstable enamine intermediates tautomerize to their respective imine forms prior to a hydration event that releases ammonia and generates a stable keto acid product (Fig. 1) (2, 4). Despite the relatively short half-life of 2AA in aqueous solution, RidA increased the rate of IlvA-dependent pyruvate formation from serine in vitro (1). This result suggested that RidA had an affinity for 2AA and may impact accumulation of this toxic metabolite in vivo, where molecular crowding and the lack of abundant free water could increase the half-life of 2AA (5). In fact, removal of RidA from the metabolic network in S. enterica led to pleotropic phenotypes that were attributed to 2AA accumulation (6–10).
FIG 1.
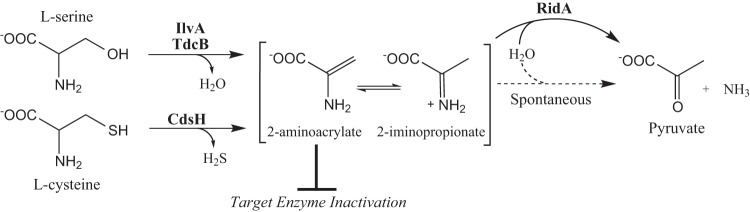
RidA catalyzes the hydrolysis of reactive intermediates. The PLP-dependent α,β-elimination of serine and cysteine proceeds through an unbound aminoacrylate intermediate. RidA protein enhances the rate of intermediate hydrolysis.
A number of PLP enzymes can be inactivated by 2AA in vitro by a mechanism that modifies the PLP cofactor in the active site (11–15). Previous studies suggested that inactivation by 2AA and related species was restricted to the active site of origin (5, 16). In vivo analysis showed that in the absence of RidA, endogenously generated free 2AA persists in the cell long enough to damage distinct PLP enzymes in S. enterica, including those that are involved in isoleucine biosynthesis, one-carbon metabolism, and cell wall synthesis (9, 10, 17). Prior to this study, the characterized sources of endogenous 2AA were the PLP-dependent serine/threonine dehydratases (17). It remains to be determined if the enamine stress caused by serine/threonine dehydratases is unique, or if this stress is a general feature of metabolic enzymes with similar PLP-dependent catalytic mechanisms.
Many prokaryotic and eukaryotic organisms are sensitive to high concentrations of cysteine (18–24). Various mechanisms have been proposed to explain cysteine toxicity, including inhibition of electron transport, inactivation of anabolic enzymes, and stimulation of Fenton chemistry, resulting in hydroxyl radical production (18, 19, 23, 25). In Escherichia coli, cysteine has been shown to cause transient amino acid starvation as a result of threonine deaminase (IlvA) inhibition, which can be overcome by the addition of branched-chain amino acids (BCAAs) (19). The specific mechanism of cysteine toxicity in S. enterica is less clear, but the toxicity seems to be due in part to the generation of reactive oxygen species leading to DNA damage (18, 23). The enzymatic desulfhydration of cysteine has been described previously as a method of cysteine detoxification in many bacterial species and higher organisms, including mammals (26). S. enterica and E. coli have several metabolic enzymes with cysteine desulfhydrase activity, including cystathionine β-lyase (MetC), cysteine synthase A (CysK), cysteine synthase B (CysM), β-cystathionase (MalY), and tryptophanase (TnaA; E. coli only) (25, 27). In contrast to E. coli, S. enterica has a dedicated cysteine desulfhydrase activity that is strongly induced in response to cysteine (26, 28, 29). It was recently demonstrated that this dedicated cysteine desulfhydrase (CDS; EC 2.5.1.47) is encoded by stm0458, which was renamed cdsH (25). CDS is thought to participate in the detoxification of excess cysteine (26), and recent reports suggest an additional role for this enzyme in maintaining sulfide concentrations at a high enough level to support antibiotic resistance and pathogenesis (25, 30, 31). In general, a PLP-dependent CDS performs chemistry similar to that of IlvA, catalyzing β-elimination of the sulfhydryl group from cysteine, yielding sulfide, ammonia, and pyruvate (26, 29). 2-Aminoacrylate has been implicated as an intermediate in CDS-mediated cysteine degradation in vitro (29), making the enzyme mechanism reminiscent of the dehydratase enzymes that contribute to metabolic phenotypes observed for ridA mutants of S. enterica (1–3). This study was initiated to address the impact of cysteine on strains that lack RidA and are unable to respond to 2AA stress.
MATERIALS AND METHODS
Bacterial strains, media, and chemicals.
Bacterial strains used in this study were derived from Salmonella enterica serovar Typhimurium LT2 and are listed with their genotypes in Table 1. Tn10d refers to the transposition-defective mini-Tn10 (Tn10Δ16Δ17) described by Way et al. (32). MudJ refers to the Mud1734 transposon described previously (33).
TABLE 1.
Bacterial strainsa
| Strain | Genotypeb |
|---|---|
| DM9404 | ridA+ in DM3480 background (wild-type) |
| DM3480 | ridA3::MudJ |
| DM4748 | ilvA595::Tn10d(Tc) |
| DM5062 | ridA3::MudJ ilvA595::Tn10d(Tc) |
| DM12740 | BL21AI pET20b-ridA |
| DM13827 | ridA3::MudJ cdsH3 |
| DM14240 | cdsH1::Cm |
| DM14254 | ridA3::MudJ cdsH1::Cm |
| DM14317 | cdsH1::Cm ilvA595::Tn10d(Tc) |
| DM14319 | ridA3::Mud cdsH::Cm ilvA595::Tn10d(Tc) |
| DM14430 | BL21AI pET14b-cdsH |
| DM14497 | ampG::Tn10d(Tc) |
| DM14498 | ampG::Tn10d(Tc) cdsH3 (CdsHW295Stop) |
| DM14499 | ampG::Tn10d(Tc) ridA::MudJ cdsH3 (CdsHW295Stop) |
| DM14500 | ampG::Tn10d(Tc) ridA::MudJ |
Minimal medium was no-carbon E medium (NCE) supplemented with 1 mM MgSO4 (34), trace minerals (35), and 11 mM d-glucose as the sole carbon source. Bacto vitamin-free Casamino Acids (CAAs) were added at 0.1% (wt/vol) when necessary. Difco nutrient broth (NB) (8 g/liter) with NaCl (5 g/liter) was used as rich medium. Superbroth consisting of tryptone (32 g/liter), yeast extract (20 g/liter), sodium chloride (5 g/liter), and sodium hydroxide (0.2 g/liter) was used when high cell densities were required. Difco BiTek agar (15 g/liter) was added for solid medium. Antibiotics were added as necessary to reach the following concentrations in rich and minimal medium, respectively: tetracycline, 20 and 10 μg/ml; ampicillin, 150 and 30 μg/ml; and chloramphenicol, 20 and 5 μg/ml. l-Cysteine was prepared fresh for each experiment and added at the appropriate concentrations. l-Isoleucine was added to a final concentration of 1 mM when needed. Unless otherwise stated, all chemicals were purchased from Sigma-Aldrich Chemical Company, St. Louis, MO.
Genetic techniques and growth analysis.
Transductional crosses were carried out using the high-frequency general transducing mutant of bacteriophage P22 (HT105/1, int-201) (36). Methods for performing transductional crosses, purifying transductants from phage, and identifying phage-free recombinants were described previously (37, 38). All mutant strains listed in Table 1 were constructed using standard genetic techniques. Gene replacements were made using the λ-Red recombinase system described by Datsenko and Wanner (39).
Growth phenotypes were determined in solid medium by using agar overlays and in liquid medium using growth curves as described previously (6). Briefly, strains to be analyzed in liquid culture were grown to full density in NB at 37°C. Cells were pelleted and resuspended in an equal volume of saline solution (85 mM), and 5 μl of the resuspension was used to inoculate 195 μl of the relevant defined growth medium contained in 96-well microtiter plates. Microtiter plates were incubated at 37°C with shaking using the Biotek EL808 ultra microplate reader. Growth was monitored as the change in absorbance at 650 nm over time. Data were plotted using GraphPad Prism 5.0f.
Mutant isolation.
A culture of DM3480 (ridA3::MudJ) was grown overnight in NB medium. Cells were pelleted and suspended in an equal volume of saline solution (85 mM). Approximately 108 cells were spread onto solid glucose medium containing 5 mM l-cysteine. Diethyl sulfate (DES) was spotted in the center of each plate (5 μl), which was incubated at 37°C for 24 to 48 h. Colonies that arose were streaked onto nonselective medium prior to confirming their cysteine-resistant phenotype in liquid minimal medium.
Genome sequencing.
Whole-genome sequencing was used to identify the causative suppressor mutation in DM13827 (ridA3::MudJ cdsH3). High-molecular-weight genomic DNA was isolated using a phenol-chloroform extraction method. A 1-ml culture was grown to full density in superbroth, pelleted in a microcentrifuge tube, and resuspended in buffer (0.1 M Tris [pH 8], 0.15 M NaCl, and 0.1 M EDTA). Lysozyme (0.5 mg) was added, followed by incubation at 37°C for 10 min. Proteinase K (1 mg) and SDS (1%) were added and incubation was allowed to continue at 37°C for 30 min. One milliliter of Tris-saturated phenol-chloroform was added and mixed gently, followed by centrifugation at 17,000 × g for 1 min. The aqueous layer was removed, washed twice with 1 ml of chloroform, and transferred to a clean microcentrifuge tube. The sample was overlaid with 1 ml of ice-cold 100% ethanol and the DNA was spooled out using a hooked Pasteur pipet. The spooled DNA was washed by submerging in ethanol, air dried for 5 min, and suspended overnight in 1 ml of Tris-EDTA buffer (10 mM Tris [pH 8.0] and 1 mM EDTA). The concentration of recovered DNA was determined using a NanoDrop 2000 spectrophotometer (Thermo Scientific). DNA gel electrophoresis was performed to ensure that high-molecular-weight DNA (>10,000 kbp) was abundant prior to sequencing.
Genomic DNA was submitted to the Georgia Genomics Facility (GGF) at the University of Georgia for paired-end (2 × 250 bp) sequencing using the Illumina MiSeq platform. DNA samples were fragmented and tagged with sequencing adapters using the Nextera XT DNA sample preparation kit (Illumina). Processing and assembly of the sequencing data were done by the Georgia Advanced Computing Resource Center (GACRC) at the University of Georgia. Briefly, the raw sequencing data were cleaned up using Trimmomatic (Usadel) with a read length cutoff of 100 bp, resulting in >300-fold coverage of the 4.95-Mb S. enterica LT2 genome (40). Trimmed reads were mapped to the published genome using Bowtie 2 (Source Forge). Variant calling was performed using the Genome Analysis Toolkit (Broad Institute), and single nucleotide polymorphisms (SNPs) were identified using the Integrative Genomics Viewer (Broad Institute).
Molecular methods.
The cdsH gene was amplified by PCR with Herculase II Fusion DNA polymerase (Agilent) using primers STM0458_NdeI_F (5′-GAGACATATGATGAGTAGCAATTGGGTTAA-3′) and STM0458_XhoI _R (5′-GAGACTCGAGCTAGTCGCCGGTAAGTAATT-3′). The resulting PCR product was used for either sequence analysis or cloning. In the latter case, the PCR product was gel purified, digested with NdeI (New England BioLabs) and XhoI (New England BioLabs), and ligated into NdeI/XhoI-cut pET14b (Novagen), forming pDM1375. Constructs were transformed into Escherichia coli strain DH5α and screened for vectors containing inserts. Plasmid inserts were confirmed by sequence analysis, performed by Genewiz.
Isoleucine transaminase (IlvE) assays.
Permeabilized cells were used to assay IlvE activity as previously described (41). Strains were grown to stationary phase in 5 ml minimal glucose medium containing 0.1% CAAs, 1 mM isoleucine, and cysteine added as indicated for each experiment. Cells were pelleted, washed once with NCE medium, and frozen at −20°C. Frozen cell pellets were thawed, resuspended in 50 mM potassium phosphate buffer (pH 8.0), and permeabilized by PopCulture (Novagen). PLP (50 μM) and 10 mM 2-ketoglutarate were mixed with an aliquot (30 μl) of the permeabilized cell suspension, and isoleucine was added (20 mM) to initiate reactions. Reaction mixtures were incubated for 20 min at 37°C, and activity was determined based on the amount of 2-keto-3-methylvalerate (2KMV) formed. 2KMV was derivatized with 2,4-dinitrophenylhydrazine (DNPH) to enable hydrazone formation, followed by organic extraction. The organic layer was washed once with 0.5 N HCl, separated, and then mixed with 1.5 N NaOH to allow chromophore formation. The absorbance of the aqueous layer (containing the chromophore) was measured at 540 nm using a Spectramax M2. The protein content of each cell extract was determined using the bicinchoninic acid (BCA) assay (Pierce). Activity is reported as nmol 2KMV/min/mg protein in permeabilized cells. GraphPad Prism 5.0f was used to perform one-way analysis of variance (ANOVA), and Tukey's test was used to assess significant changes in IlvE activity (P < 0.01).
Purification of CdsH.
Wild-type cdsH cloned into the pET14b vector (Novagen) was transformed into E. coli BL21AI (DM14430) for overexpression and His6 tag purification. Cells were inoculated into 10 ml of superbroth containing ampicillin and grown overnight at 37°C. Overnight cultures were inoculated into 3 liters of superbroth containing ampicillin and grown at 37°C until an A650 of 0.4 to 0.7 was reached. Arabinose (0.02%) was added to induce expression, and cultures were shifted to 22°C for 18 h. Cells were harvested at 4°C by centrifugation (15 min at 8,000 × g) and resuspended in 50 mM Tris-HCl (pH 7.5) containing 200 mM sodium chloride, 5 mM imidazole, 10 μM PLP, and 10% glycerol. Lysozyme (1 mg/ml), phenylmethylsulfonyl fluoride (100 μg/ml), and DNase (25 μg/ml) were added to the cell suspension, which then sat on ice for 1 h. Cells were mechanically lysed using a French pressure cell (5 passes at 10,342 kPa). The resulting lysate was clarified by centrifugation (45 min at 48,000 × g) and filtered through a 0.45-μm membrane. The filtered lysate was loaded onto a nickel-nitrilotriacetic acid (Ni-NTA) Superflow resin (5 ml), and CdsH was purified according to the manufacturer's protocol (Qiagen). Purified protein was concentrated by centrifugation with a 10,000-molecular-weight-cutoff filter unit (Millipore), and the buffer was replaced with 50 mM Tris-HCl (pH 7.5) containing 10 μM PLP and 10% glycerol using a PD-10 desalting column (GE Healthcare). Protein recovery as determined by the BCA assay (Pierce) was approximately 5.5 mg/ml. Protein aliquots were frozen in liquid nitrogen and stored at −80°C.
Purification of RidA.
RidA protein was purified from a BL21AI strain harboring the pET20b-ridA overexpression construct previously described (DM12740) (8). Briefly, an overnight culture of DM12740 grown in superbroth containing ampicillin was inoculated into 3 liters of superbroth with ampicillin. Cultures were grown for ∼3 h at 37°C with aeration until an A650 of 0.6 was reached. Fresh arabinose was added to a final concentration of 0.2%, and cultures were incubated with shaking at 37°C for an additional 10 h. Cells were harvested by centrifugation and resuspended in 50 mM Tris-HCl (pH 8) containing 100 mM sodium chloride, 5 mM imidazole, and 12.5% glycerol. Following lysis, the extract was clarified, filtered, and injected onto a Ni-NTA Superflow resin (5 ml), and RidA was purified according to the manufacturer's protocol (Qiagen). Purified protein was concentrated by centrifugation with a 4,000-molecular-weight-cutoff filter unit (Millipore), and the concentrated protein sample (∼5 ml) was sequentially dialyzed in 1 liter of 10 mM HEPES (pH 8) containing 10 mM EGTA for 1 h, 1 liter of HEPES buffer for 1 h, and 1 liter of HEPES buffer containing 20% glycerol overnight. Protein recovery as determined by the BCA assay (Pierce) was approximately 15.2 mg/ml. Protein aliquots were frozen in liquid nitrogen and stored at −80°C.
Cysteine desulfhydrase assay.
Cysteine desulfhydrase activity was measured by coupling pyruvate formation to NADH oxidation using lactate dehydrogenase (Sigma) and monitoring the decrease in absorbance at 340 nm as described previously (29). Assay mixtures consisted of 100 mM Tris-HCl (pH 8.5) containing 0.25 mM NADH, 30 μM PLP, and 15 U/ml lactate dehydrogenase. CdsH and RidA were added to the reaction mixture at final monomeric concentrations of 0.19 μM and 0.27 μM, respectively. Because RidA is a trimer (42) and CdsH is a hexamer (43), the oligomeric ratio of RidA to CdsH was ∼3:1. Concentrations of fresh l-cysteine ranging from 0 to 20 mM were added to initiate the reaction. Reactions (300 μl) were monitored continuously at 22°C in a 96-well quartz plate using a Spectramax M2 microplate reader. Initial rates were calculated from the change in A340 due to the consumption of NADH over the initial 20 s. The extinction coefficient of NADH at 340 nm (6,220 M−1 cm−1) was used to calculate enzyme activity. Initial activity is reported as the initial rate of pyruvate formation in μmol/min. Experiments were performed in triplicate, and the resulting data were plotted using GraphPad Prism 5.0f.
RESULTS AND DISCUSSION
Growth of a ridA strain is affected by exogenous cysteine.
Growth of wild-type S. enterica in glucose minimal medium is unaffected by cysteine concentrations up to 0.1 mM. Concentrations above that affect growth by increasing the lag phase before the culture enters logarithmic growth (Fig. 2A). The concentration of cysteine that prevented recovery after an extended lag was not determined. In contrast to the wild type, the ridA strain was sensitive to a concentration of cysteine as low as 0.1 mM and displayed a concentration-dependent extension of the lag phase prior to entering exponential growth. The ridA strain had no significant growth within 20 h when cysteine concentrations were 0.25 mM or higher (Fig. 2B). The growth inhibition caused by 0.25 mM cysteine was transient, and after 24 h, the ridA strain grew exponentially, ultimately reaching the same final cell density as the wild-type strain. Together, these data show that a strain lacking RidA was substantially more sensitive to exogenous cysteine than a wild-type strain.
FIG 2.
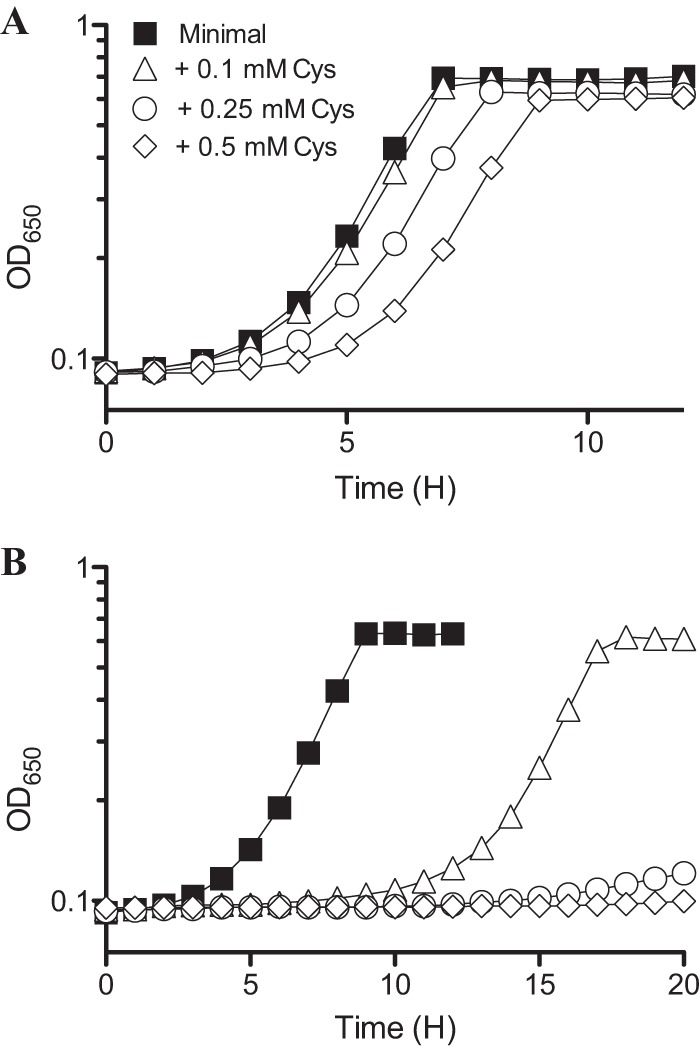
Growth inhibition increases with cysteine concentration. Wild-type (DM9404) (A) and ridA (DM3480) (B) strains were grown at 37°C in minimal glucose medium with cysteine excluded were added to a final concentration of 0.1 mM, 0.25 mM, or 0.5 mM. Error bars representing the standard errors of the means (SEM) of three replicates are excluded, as the replicates deviated less than 5% from the average value.
Serine/threonine dehydratase is not required for the cysteine sensitivity of a ridA strain.
Strains lacking RidA are sensitive to the accumulation of 2-aminoacrylate generated from serine by the serine/threonine dehydratase IlvA (9, 10, 17). Given that cysteine is a β-substituted amino acid akin to serine, it was formally possible that the cysteine sensitivity of a ridA strain would directly or indirectly depend on IlvA activity. Cysteine was shown to have a mixed inhibitory effect on the threonine dehydratase activity of IlvA isolated from E. coli, suggesting that it may serve as a competitive substrate of the enzyme (19). IlvA is the only serine/threonine dehydratase expressed in S. enterica under aerobic conditions (44). Two experiments ruled out a role for IlvA in the cysteine sensitivity of a ridA strain. The growth data in Fig. 3 showed that in the presence of 1 mM cysteine, neither isoleucine nor threonine (data not shown) restored growth to a strain lacking RidA. In contrast, both isoleucine and threonine suppressed the growth defects of a ridA mutant caused by the activity of IlvA (6, 41, 45). Further, the deletion of ilvA did not affect the growth of ridA mutant strains in the presence of cysteine. These data ruled out a role for IlvA in mediating cysteine sensitivity in a ridA strain and indicated that a distinct cellular component was involved in the cysteine sensitivity of ridA strains.
FIG 3.
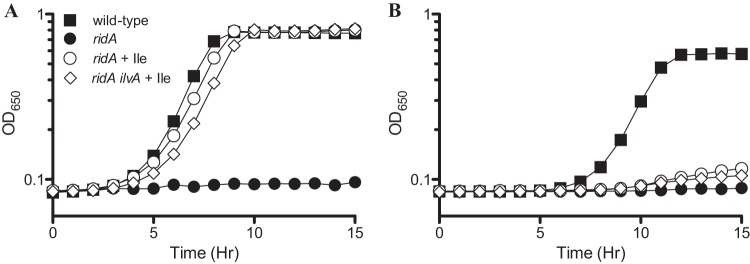
The cysteine sensitivity of a ridA strain does not require IlvA. Strains were grown at 37°C in glucose minimal medium containing (A) 5 mM serine or (B) 1 mM cysteine. Open symbols indicate the addition of 1 mM isoleucine. Growth is displayed for wild-type (DM9404), ridA (DM3480), and ridA ilvA (DM5062) strains. Error bars representing the SEM of three replicates are excluded, as the replicates deviated less than 5% from the average value.
Exogenous cysteine increases the level of free 2AA in the cell.
Based on the in vitro activity associated with RidA (1), a simple scenario was that a non-IlvA enzyme generated 2AA or a related enamine from cysteine that resulted in the cysteine sensitivity observed. In the absence of RidA, endogenous 2AA can react with the PLP cofactor of isoleucine transaminase B (IlvE), forming a covalent active-site modification that renders the enzyme inactive (17, 41, 45). Therefore, the activity of IlvE has been used as a proxy for the levels of 2AA in the cells of strains lacking RidA (17, 41, 45). Wild-type and ridA mutant strains were grown in minimal medium containing Casamino Acids and isoleucine, with and without the addition of cysteine, and IlvE activity was assayed. The addition of 0.1% Casamino Acids to the growth medium resulted in similar growth patterns for each strain. The data in Fig. 4 showed that cysteine did not affect the level of IlvE activity in strains with a functional RidA. In contrast, the transaminase B activity was decreased by ∼35% in a ridA mutant strain when 0.25 mM cysteine was present in the growth medium. Consistent with the growth data, the impact of cysteine on the activity of IlvE was not affected by the presence or absence of IlvA. Additionally, in an in vitro system, neither cysteine alone nor IlvA and cysteine decreased the activity of IlvE (data not shown). In total, these data suggested that an indirect consequence of cysteine or a metabolite of cysteine was responsible for the decreased IlvE activity and growth defect of a ridA mutant.
FIG 4.
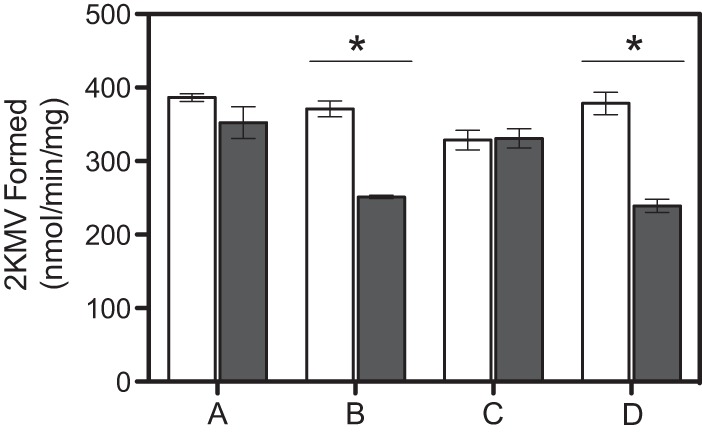
IlvE activity is reduced in ridA strains grown in the presence of exogenous cysteine. Cultures were grown to stationary phase in glucose minimal medium containing 0.1% Casamino Acids and 1 mM isoleucine, without exogenous cysteine (white bars) or with 0.25 mM cysteine (gray bars). The specific activity of IlvE from three independent cultures was determined based on 2-ketomethylvalerate (2KMV) formation in crude extracts. Error bars represent the SEM. (A) Wild type (DM9404); (B) ridA strain (DM3480); (C) ilvA strain (DM4748); (D) ridA ilvA strain (DM5062). An asterisk indicates a significant difference based on one-way ANOVA and Tukey's test (P < 0.01).
Multiple enzymes in S. enterica use cysteine as a substrate and have the potential to generate 2AA as a reaction intermediate. Based on E. coli, three enzymes were identified that may have cysteine desulfhydrase activity in S. enterica: CysK, CysM, and STM1557 (34% identical to E. coli MalY) (27). These PLP-dependent enzymes generate pyruvate, hydrogen sulfide, and ammonia from cysteine, and it was plausible that these reactions would proceed through a 2AA intermediate (29, 46, 47). Deletions of cysK, cysM, and stm1557 were generated by gene replacement (39), and strains that contained each mutation alone or in combination with a ridA mutation were constructed. None of the ridA cysK, ridA cysM, or ridA stm1557 double mutants grew in the presence of cysteine (0.25 mM), while the growth of ridA+ derivatives of each strain was unaffected by the presence of cysteine (data not shown). CysK and CysM are isozymes involved in cysteine biosynthesis, and a cysK cysM double mutant strain is a cysteine auxotroph. A concentration of cysteine exceeding 0.5 mM was needed to satisfy the cysteine requirement of this strain. If either CysK or CysM contributed to 2AA stress arising from cysteine, a cysK cysM ridA triple mutant would be expected to grow in the presence of cysteine despite the ridA mutation. The triple mutant strain failed to grow in the presence of 0.5 mM or 1 mM cysteine. These data suggested that CysK, CysM, and STM1557 did not contribute significantly to the production of 2AA in S. enterica.
Mutations in cdsH partially relieve cysteine sensitivity.
Mutations that restored growth of a ridA mutant in the presence of cysteine were isolated. Approximately 108 cells of the ridA mutant strain DM3480 were spread on minimal cysteine (5 mM) plates. Despite numerous attempts, no spontaneous mutants that allowed growth were recovered after 3 to 5 days of incubation. When the same process was repeated and diethyl sulfate (DES) was spotted in the middle of the cysteine plate, multiple colonies arose. The cysteine-resistant phenotypes were verified, and two of these mutants were selected for further analysis. Whole-genome sequencing of a representative mutant, DM13827, revealed a G-to-A substitution in the gene encoding STM0458, changing residue 295 from a tryptophan to a stop codon (W295Stop). The same mutation was observed in both of the suppressor strains. In the course of our studies, it was reported that stm0458 encoded the major CDS in Salmonella, and the gene was renamed cdsH to reflect this finding (25). The cdsH mutation (cdsH3) was reconstructed to confirm that it was the causative lesion. Subsequently, an insertion linked to cdsH3 was used to generate an isogenic pair of strains in both the ridA (DM3480) and wild-type strain (DM9404) background.
Anticipating that a nonsense mutation would be recessive and display the phenotype of a null allele, a deletion of cdsH was generated (ΔcdsH1::Cm). Growth analysis showed that while both alleles of cdsH increased growth of a ridA mutant in the presence of cysteine, they displayed different levels of suppression (Fig. 5). Both cdsH alleles significantly reduced the growth lag of the ridA mutant in the presence of 0.1 mM cysteine (data not shown). In the presence of 0.25 mM cysteine, when a ridA mutant had no growth after 20 h, the cdsH3 allele restored growth, while the cdsH1::Cm deletion had a lesser effect (Fig. 5). The difference between the two alleles was presumed to be due to the periodic read-through of the UGA (opal) nonsense codon (48) or reduced activity of the partial protein. The positive growth response allowed by decreasing or eliminating CdsH activity was consistent with the general hypothesis that CDS generated 2AA, which was toxic in the absence of RidA. However, a few observations about growth of the mutant strains indicated that additional complexities were present in the cell. First, if the generation of 2AA were the only deleterious consequence of cysteine, deletion of cdsH would have the best suppressing effect. In addition, if the generation of 2AA from cysteine was by CdsH alone, a ridA cdsH1::Cm double mutant (DM14254) should grow similarly to the cdsH1::Cm single mutant (DM14240). The growth discrepancy between the ridA+ and ridA strains suggested that an additional enzyme generates a RidA substrate from cysteine.
FIG 5.
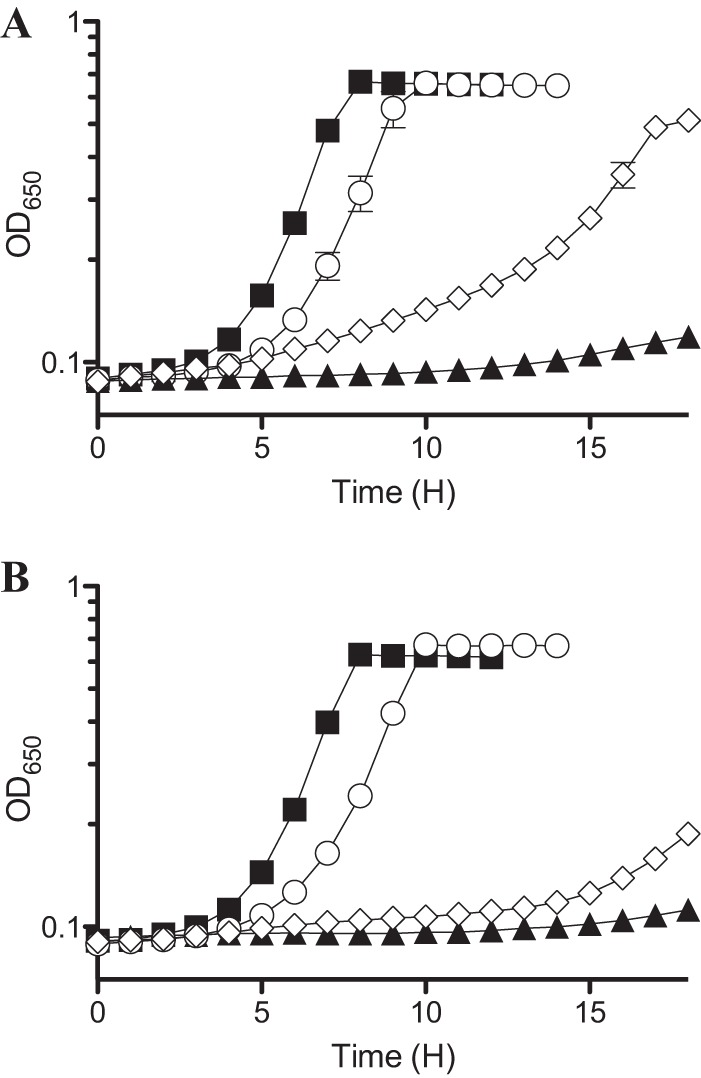
Lesions in cdsH improve growth of a ridA strain in the presence of cysteine. Strains were grown in glucose minimal medium containing 0.25 mM cysteine. (A) Growth of wild-type (DM14497; closed squares), ridA (DM14500; closed triangles), cdsH3 (DM14498; open circles), and ridA cdsH3 (open diamonds) strains. (B) Growth of wild-type (DM9404; closed squares), ridA (DM3480; closed triangles), cdsH1::Cm (DM14240; open circles) and ridA cdsH1::Cm (DM14254; open diamonds) strains. Experiments were done in triplicate. Error bars represent the SEM.
Loss of CdsH exacerbates cysteine toxicity in wild-type Salmonella.
The inability of the cdsH deletion to restore wild-type growth to a ridA mutant in the presence of cysteine indicated that 2AA was not the only metabolic problem generated by the presence of cysteine. During this study, another group reported that the deletion of cdsH resulted in a 10-fold decrease in CDS activity and caused increased sensitivity to cysteine (25). Consistently, both the cdsH deletion mutant and the nonsense mutant had an extended lag phase when grown in medium containing 0.25 mM cysteine (Fig. 5). When 0.5 mM cysteine was present, the defect was more severe (data not shown).
Collectively, these results revealed the opposing consequences of eliminating cdsH and the complexity of dissecting the physiological status of strains lacking both RidA and CdsH. On the one hand, deletion of cdsH affected the ability of the cell to remove, or detoxify, cysteine. This led to the accumulation of cysteine, which inhibited growth by poorly characterized mechanisms. On the other hand, cells without CdsH produce less 2AA, because the desulfhydrase reaction does not proceed. Therefore, the toxic effects of 2AA derived from cysteine in a ridA mutant are negated. Taken together, these data suggest that the detoxification of cysteine by CDS enzymes proceeds through a reactive intermediate, which itself is toxic and must be quenched by RidA. The results herein indicated that the decreased growth of S. enterica in the presence of cysteine was a consequence of both features.
RidA increases the rate of cysteine desulfhydrase-dependent pyruvate formation in vitro.
The scenario above predicted RidA would have a detectable influence on the products of the CdsH protein in vitro. CdsH activity was assessed by a coupled assay with lactate dehydrogenase to measure the formation of pyruvate. The rate of the reaction was calculated over time in the presence or absence of RidA protein (Fig. 6). From these data, a number of points were noted. In the absence of RidA, the rate of pyruvate formation decreased at concentrations of cysteine above 1 mM. This behavior of CdsH was observed previously and was attributed to the spontaneous reaction of l-cysteine with a 2AA intermediate, leading to the formation of the cyclized compound 2-methyl-2,4-thiazolidine-dicarboxylate (MTD) (29). The relevant reaction is schematically shown in Fig. 7. An alternative assay monitoring sulfide release from cysteine determined the Km of CdsH for l-cysteine was 0.17 to 0.21 mM and found sulfide inhibited the reaction with a Ki of 0.010 mM (26, 29). The data presented in Fig. 6 displayed sigmoidal kinetics of CdsH, consistent with the positive cooperatively (n = 1.9) reported for this enzyme (29).
FIG 6.
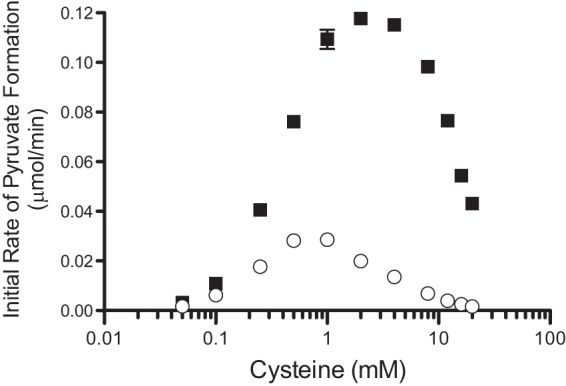
The initial rate of CdsH-catalyzed pyruvate formation is improved by RidA. The initial rate of pyruvate formation versus cysteine concentration is plotted for CdsH alone (circles) and CdsH plus RidA (squares). Experiments were performed in triplicate, and the resulting data are displayed with error bars representing the SEM.
FIG 7.
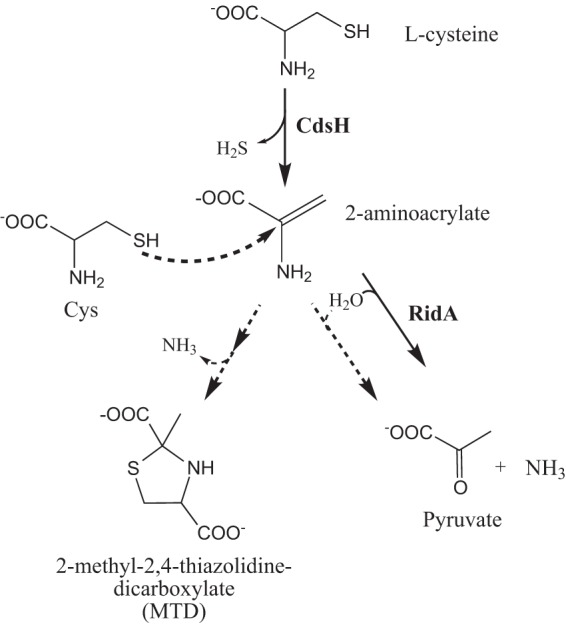
Nucleophilic attack by cysteine reduces the amount of 2-aminoacrylate converted to pyruvate. The PLP-dependent α,β-elimination reaction catalyzed by CdsH converts cysteine to 2-aminoacrylate and hydrogen sulfide. In solvent water, 2-aminoacrylate can be hydrolyzed to pyruvate spontaneously or enzymatically through the activity of RidA. The nucleophilic sulfhydryl group of cysteine can also attack the C=C double bond of 2-aminoacrylate, ultimately leading to the formation of the cyclized thiazolidine derivative 2-methyl-2,4-thiazolidine-dicarboxylate (MTD) (29). Solid arrows indicate enzyme-catalyzed steps, and dashed arrows indicate spontaneous reactions.
The addition of RidA to the CdsH reaction increased the rate of pyruvate formation at all cysteine concentrations (Fig. 6). The CdsH-dependent rate of pyruvate formation was greatest at 1 mM cysteine, where including RidA in the reaction increased the rate 3.8-fold. Increasing the concentration of cysteine to 2 mM decreased the rate of CdsH-catalyzed pyruvate formation, which is attributed to the diversion of 2AA by cysteine, as depicted in Fig. 7. However, the addition of RidA had the greatest relative effect at 2 mM cysteine, increasing the rate 5.8-fold. At concentrations of cysteine above 4 mM, the rate of pyruvate formation decreased in a cysteine-dependent manner, despite the presence of RidA (Fig. 6). Additional controls substituting bovine serum albumin (10 μg/μl) for RidA had no effect on CDS activity. These data, when viewed in light of the reaction mechanism previously described, suggest that at high concentrations of cysteine, there is competition between cysteine and RidA for the 2AA intermediate (Fig. 7).
Conclusions.
The purpose of this study was to describe the mechanism of cysteine sensitivity provoked by removing RidA from the metabolic network in S. enterica. Previous reports focused on the dehydration of serine by PLP-dependent serine/threonine dehydratases (IlvA and TdcB) as sources of endogenous 2AA (7, 9, 10, 17, 41). The work here describes an additional mechanism of endogenous 2AA formation that is dependent on the cysteine desulfhydrase activity of CdsH. In vitro analysis showed that RidA enhanced the rate of CdsH-dependent pyruvate formation from cysteine, consistent with the hypothesis that an unbound 2AA intermediate is formed by CdsH and subsequently acted upon by RidA to increase the rate of enamine/imine hydrolysis. Eliminating CdsH partially decreased the cysteine sensitivity of ridA mutants while increasing the sensitivity of the wild-type strain.
The findings presented here introduce an interesting dichotomy in which the detoxification of cysteine, which is itself toxic to wild-type Salmonella when present at high concentrations, relies on an enzyme-catalyzed reaction that proceeds through a dangerous reactive intermediate, 2-aminoacrylate. S. enterica has a built-in defense against 2AA stress in the form of RidA. Therefore, it seems that the benefit of detoxifying cysteine via a toxic intermediate outweighs the potential consequence given the robust protection afforded to the cell by RidA. Salmonella relies on the dedicated cysteine desulfhydrase encoded by cdsH to prevent cysteine from accumulating to toxic levels. The PLP-dependent mechanism used by CdsH dictates that 2AA be generated as a deliberate intermediate in the cysteine detoxification pathway and demands the presence of RidA to prevent 2AA stress.
This is the first example of a role for RidA in a dedicated detoxification pathway and potentially represents a conserved role for RidA proteins in cysteine detoxification in other organisms. In contrast, the dehydration of serine by IlvA is a side reaction of an enzyme that acts primarily to convert threonine to 2-ketobutyrate in isoleucine biosynthesis. The generation of 2AA in this context is therefore a consequence of substrate promiscuity, as serine degradation by IlvA does not play a crucial role in metabolism. In fact, the S. enterica genome encodes three Fe-S-dependent serine deaminases (EC 4.3.1.17), SdaA, SdaB, and TdcG, which specifically prevent the accumulation of toxic levels of serine (49). The serine deaminase reactions catalyzed by these enzymes are thought to proceed through a 2AA intermediate (50), yet current data suggest that these enzymes do not contribute to free 2AA accumulation in Salmonella (our unpublished data).
The unusual kinetics we observed for CdsH in vitro are in agreement with the data obtained by Kredich et al. (29). They reported that under similar assay conditions, the amount of free pyruvate generated by CdsH in the presence of 2 mM cysteine was less than 10% of that expected based on the relative amount of sulfide produced (29). The remaining ∼90% of unrecovered pyruvate had been diverted to MTD (Fig. 7). The relevance of this side reaction forming MTD in vivo is questionable given the differences between the assay conditions and cell environment. The neutral pH and relatively low concentrations of cysteine found in the cell may prevent MTD from being formed (29), especially when RidA is present. Furthermore, the physiological relevance of MTD in any organism remains unclear (21, 51). However, there is interest in the use of thiazolidine derivatives in therapeutic applications (21). MTD and other thiazolidine derivatives can be converted to cysteine nonenzymatically or enzymatically and are viewed as a means of delivering adequate doses of cysteine to mammalian systems without causing cysteine toxicity (21). Given that cysteine behaves as a nucleophile when attacking 2AA, future studies will address the possibility that additional nucleophilic species react with 2AA in vitro. The results herein describe a new role for RidA in metabolism and raise the possibility that other detoxification pathways could proceed through reactive intermediates that are quenched by RidA.
ACKNOWLEDGMENTS
We thank Lauren Palmer for constructing the pET14b-cdsH plasmid for protein overproduction.
This work was supported by competitive grant GM095837 from the National Institutes of Health.
Footnotes
Published ahead of print 7 July 2014
REFERENCES
- 1.Lambrecht JA, Flynn JM, Downs DM. 2012. Conserved YjgF protein family deaminates reactive enamine/imine intermediates of pyridoxal 5′-phosphate (PLP)-dependent enzyme reactions. J. Biol. Chem. 287:3454–3461. 10.1074/jbc.M111.304477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chargaff E, Sprinson DB. 1943. Studies on the mechanism of deamination of serine and threonine in biological systems. J. Biol. Chem. 151:273–280 [Google Scholar]
- 3.Phillips AT, Wood WA. 1965. The mechanism of action of 5′-adenylic acid-activated threonine dehydrase. J. Biol. Chem. 240:4703–4709 [PubMed] [Google Scholar]
- 4.Layer RW. 1963. The chemistry of imines. Chem. Rev. 63:489–510. 10.1021/cr60225a003 [DOI] [Google Scholar]
- 5.Hillebrand GG, Dye JL, Suelter CH. 1979. Formation of an intermediate and its rate of conversion to pyruvate during the tryptophanase-catalyzed degradation of So-nitrophenyl-l-cysteine. Biochemistry 18:1751–1755. 10.1021/bi00576a018 [DOI] [PubMed] [Google Scholar]
- 6.Enos-Berlage JL, Langendorf MJ, Downs DM. 1998. Complex metabolic phenotypes caused by a mutation in yjgF, encoding a member of the highly conserved YER057c/YjgF family of proteins. J. Bacteriol. 180:6519–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christopherson MR, Schmitz GE, Downs DM. 2008. YjgF is required for isoleucine biosynthesis when Salmonella enterica is grown on pyruvate medium. J. Bacteriol. 190:3057–3062. 10.1128/JB.01700-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambrecht JA, Browne BA, Downs DM. 2010. Members of the YjgF/YER057c/UK114 family of proteins inhibit phosphoribosylamine synthesis in vitro. J. Biol. Chem. 285:34401–34407. 10.1074/jbc.M110.160515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flynn JM, Christopherson MR, Downs DM. 2013. Decreased coenzyme A levels in ridA mutant strains of Salmonella enterica result from inactivated serine hydroxymethyltransferase: ridA mutants are deficient in one carbon metabolism. Mol. Microbiol. 89:751–759. 10.1111/mmi.12313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flynn JM, Downs DM. 2013. In the absence of RidA, endogenous 2-aminoacrylate inactivates alanine racemases by modifying the pyridoxal 5′-phosphate cofactor. J. Bacteriol. 195:3603–3609. 10.1128/JB.00463-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arfin SM, Koziell DA. 1971. Inhibition of growth of Salmonella typhimurium and of threonine deaminase and transaminase B by β-chloroalanine. J. Bacteriol. 105:519–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silverman RB, Abeles RH. 1976. Inactivation of pyridoxal phosphate dependent enzymes by mono-and polyhaloalanines. Biochemistry 15:4718–4723. 10.1021/bi00666a028 [DOI] [PubMed] [Google Scholar]
- 13.Likos JJ, Ueno H, Feldhaus RW, Metzler DE. 1982. A novel reaction of the coenzyme of glutamate decarboxylase with L-serine O-sulfate. Biochemistry 21:4377–4386. 10.1021/bi00261a029 [DOI] [PubMed] [Google Scholar]
- 14.Ueno H, Likos JJ, Metzler DE. 1982. Chemistry of the inactivation of cytosolic aspartate aminotransferase by serine O-sulfate. Biochemistry 21:4387–4393. 10.1021/bi00261a030 [DOI] [PubMed] [Google Scholar]
- 15.Walsh C. 1982. Suicide substrates: mechanism-based enzyme inactivators. Tetrahedron 38:871–909. 10.1016/0040-4020(82)85068-0 [DOI] [PubMed] [Google Scholar]
- 16.Walsh CT. 1984. Suicide substrates, mechanism-based enzyme inactivators: recent developments. Annu. Rev. Biochem. 53:493–535. 10.1146/annurev.bi.53.070184.002425 [DOI] [PubMed] [Google Scholar]
- 17.Lambrecht JA, Schmitz GE, Downs DM. 2013. RidA proteins prevent metabolic damage inflicted by PLP-dependent dehydratases in all domains of life. mBio 4:e00033–13. 10.1128/mBio.00033-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomez RF, Montville T, Blais K. 1980. Toxic effect of cysteine against Salmonella typhimurium. Appl. Environ. Microbiol. 39:1081–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris CL. 1981. Cysteine and growth inhibition of Escherichia coli: threonine deaminase as the target enzyme. J. Bacteriol. 145:1031–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cowman RA, Baron SS, Fitzgerald RJ. 1983. Cysteine toxicity for oral streptococci and effect of branched-chain amino acids. Infect. Immun. 39:1107–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wlodek L, Rommelspacher H, Susilo R, Radomski J, Höfle G. 1993. Thiazolidine derivatives as source of free L-cysteine in rat tissue. Biochem. Pharmacol. 46:1917–1928. 10.1016/0006-2952(93)90632-7 [DOI] [PubMed] [Google Scholar]
- 22.Sørensen MA, Pedersen S. 1991. Cysteine, even in low concentrations, induces transient amino acid starvation in Escherichia coli. J. Bacteriol. 173:5244–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park S, Imlay JA. 2003. High levels of intracellular cysteine promote oxidative DNA damage by driving the Fenton reaction. J. Bacteriol. 185:1942–1950. 10.1128/JB.185.6.1942-1950.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar A, John L, Alam MM, Gupta A, Sharma G, Pillai B, Sengupta S. 2006. Homocysteine- and cysteine-mediated growth defect is not associated with induction of oxidative stress response genes in yeast. Biochem. J. 396:61. 10.1042/BJ20051411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oguri T, Schneider B, Reitzer L. 2012. Cysteine catabolism and cysteine desulfhydrase (CdsH/STM0458) in Salmonella enterica serovar Typhimurium. J. Bacteriol. 194:4366–4376. 10.1128/JB.00729-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collins JM, Monty KJ. 1973. The cysteine desulfhydrase of Salmonella typhimurium kinetic and catalytic properties. J. Biol. Chem. 248:5943–5949 [PubMed] [Google Scholar]
- 27.Awano N, Wada M, Mori H, Nakamori S, Takagi H. 2005. Identification and functional analysis of Escherichia coli cysteine desulfhydrases. Appl. Environ. Microbiol. 71:4149–4152. 10.1128/AEM.71.7.4149-4152.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guarneros G, Ortega MV. 1970. Cysteine desulfhydrase activities of Salmonella typhimurium and Escherichia coli. Biochim. Biophys. Acta 198:132–142. 10.1016/0005-2744(70)90041-0 [DOI] [PubMed] [Google Scholar]
- 29.Kredich NM, Foote LJ, Keenan BS. 1973. The stoichiometry and kinetics of the inducible cysteine desulfhydrase from Salmonella typhimurium. J. Biol. Chem. 248:6187–6196 [PubMed] [Google Scholar]
- 30.Turnbull AL, Surette MG. 2008. L-Cysteine is required for induced antibiotic resistance in actively swarming Salmonella enterica serovar Typhimurium. Microbiology 154:3410–3419. 10.1099/mic.0.2008/020347-0 [DOI] [PubMed] [Google Scholar]
- 31.Shatalin K, Shatalina E, Mironov A, Nudler E. 2011. H2S: a universal defense against antibiotics in bacteria. Science 334:986–990. 10.1126/science.1209855 [DOI] [PubMed] [Google Scholar]
- 32.Way JC, Davis MA, Morisato D, Roberts DE, Kleckner N. 1984. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene 32:369–379. 10.1016/0378-1119(84)90012-X [DOI] [PubMed] [Google Scholar]
- 33.Castilho B, Olfson P, Casadaban MJ. 1984. Plasmid insertion mutagenesis and lac gene fusion with mini-mu bacteriophage transposons. J. Bacteriol. 158:488–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogel HJ, Bonner DM. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97–106 [PubMed] [Google Scholar]
- 35.Balch WE, Wolfe RS. 1976. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl. Environ. Microbiol. 32:781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmieger H. 1972. Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 119:75. 10.1007/BF00270447 [DOI] [PubMed] [Google Scholar]
- 37.Chan RK, Botstein D, Watanabe T, Ogata Y. 1972. Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II. Properties of a high-frequency-transducing lysate. Virology 50:883–898 [DOI] [PubMed] [Google Scholar]
- 38.Downs DM, Petersen L. 1994. apbA, a new genetic locus involved in thiamine biosynthesis in Salmonella typhimurium. J. Bacteriol. 176:4858–4864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Gene 1:H2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856. 10.1038/35101614 [DOI] [PubMed] [Google Scholar]
- 41.Schmitz G, Downs DM. 2004. Reduced transaminase B (IlvE) activity caused by the lack of yjgF is dependent on the status of threonine deaminase (IlvA) in Salmonella enterica serovar Typhimurium. J. Bacteriol. 186:803. 10.1128/JB.186.3.803-810.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burman JD, Stevenson CE, Sawers RG, Lawson DM. 2007. The crystal structure of Escherichia coli TdcF, a member of the highly conserved YjgF/YER057c/UK114 family. BMC Struct. Biol. 7:30. 10.1186/1472-6807-7-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kredich NM, Keenan BS, Foote LJ. 1972. The purification and subunit structure of cysteine desulfhydrase from Salmonella typhimurium. J. Biol. Chem. 247:7157–7162 [PubMed] [Google Scholar]
- 44.Umbarger HE, Brown B. 1957. Threonine deamination in E. coli II: evidence for two l-threonine deaminases. J. Bacteriol. 73:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Christopherson MR, Lambrecht JA, Downs D, Downs DM. 2012. Suppressor analyses identify threonine as a modulator of ridA mutant phenotypes in Salmonella enterica. PLoS One 7:e43082. 10.1371/journal.pone.0043082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miles EW, Hatanaka M, Crawford IP. 1968. A new thiol-dependent transamination reaction catalyzed by the B protein of Escherichia coli tryptophan synthetase. Biochemistry 7:2742–2753. 10.1021/bi00848a008 [DOI] [PubMed] [Google Scholar]
- 47.Ågren D, Schnell R, Schneider G. 2009. The C-terminal of CysM from Mycobacterium tuberculosis protects the aminoacrylate intermediate and is involved in sulfur donor selectivity. FEBS Lett. 583:330–336. 10.1016/j.febslet.2008.12.019 [DOI] [PubMed] [Google Scholar]
- 48.Bertram G, Innes S, Minella O, Richardson JP, Stansfield I. 2001. Endless possibilities: translation termination and stop codon recognition. Microbiology 147:255–269 [DOI] [PubMed] [Google Scholar]
- 49.Zhang X, El-Hajj ZW, Newman E. 2010. Deficiency in L-serine deaminase interferes with one-carbon metabolism and cell wall synthesis in Escherichia coli K-12. J. Bacteriol. 192:5515–5525. 10.1128/JB.00748-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grabowski R, Hofmeister A, Buckel W. 1993. Bacterial L-serine dehydratases: a new family of enzymes containing iron-sulfur clusters. Trends Biochem. Sci. 18:297–300. 10.1016/0968-0004(93)90040-T [DOI] [PubMed] [Google Scholar]
- 51.Wlodek L. 1984. Formation of MTD from L-cysteine in rat tissues. Acta Biochim. Pol. 31:279–288 [PubMed] [Google Scholar]


