Abstract
Bacteria utilize multiple sigma factors that associate with core RNA polymerase (RNAP) to control transcription in response to changes in environmental conditions. In Escherichia coli and Salmonella enterica, Crl positively regulates the σS regulon by binding to σS to promote its association with core RNAP. We recently characterized the determinants in σS responsible for specific binding to Crl. However, little is known about the determinants in Crl required for this interaction. Here, we present the X-ray crystal structure of a Crl homolog from Proteus mirabilis in conjunction with in vivo and in vitro approaches that probe the Crl-σS interaction in E. coli. We show that the P. mirabilis, Vibrio harveyi, and E. coli Crl homologs function similarly in E. coli, indicating that Crl structure and function are likely conserved throughout gammaproteobacteria. We utilize phylogenetic conservation and bacterial two-hybrid analyses to predict residues in Crl important for the interaction with σS. The results of p-benzoylphenylalanine (BPA)-mediated UV cross-linking studies further support the model in which an evolutionarily conserved central cleft is the surface on Crl that binds to σS. Within this conserved binding surface, we identify a key residue in Crl that is critical for activation of EσS-dependent transcription in vivo and in vitro. Our study provides a physical basis for understanding the σS-Crl interaction.
INTRODUCTION
Bacteria regulate gene expression in order to adjust to changes in environmental conditions. Switching among the σ factors that bind to core RNA polymerase (RNAP) reprograms the cell's transcriptional networks, redistributing RNAP to promoters that are utilized under alternative conditions (reviewed in reference 1). Escherichia coli has 7 σ factors, including σS (σ38; RpoS), the general stress sigma factor (reviewed in references 2 and 3). The concentration of σS is regulated at the levels of transcription, translation, and protein stability (reviewed in references 3 and 4). In response to a variety of stresses, the level of σS begins to rise, but binding of σS to core RNAP to form the holoenzyme (EσS) is weak compared to that of other σ factors (5–7). Therefore, transcription from σS-dependent promoters with poor binding constants for the holoenzyme may be limited by the rate of EσS formation.
The 133-amino-acid protein Crl positively regulates σS-mediated transcription (8). Crl is found in many gammaproteobacteria, although thus far it has been studied only in E. coli and Salmonella enterica (7, 9, 10). Most transcription factors function by binding to specific DNA sites near the promoter and interacting with the RNAP α or σ subunit to regulate gene expression (reviewed in reference 1). However, some transcription factors regulate promoter activity by interacting with the catalytic subunits of RNAP, β and β′, without binding to DNA, and function by altering the kinetics of the transcription initiation mechanism (reviewed in reference 11). Crl is unusual in that it activates transcription by yet another mechanism in which it binds to free σS and stimulates RNAP holoenzyme assembly, raising the concentration of EσS, leading to increased activity of σS-dependent promoters (7, 9, 12–14). Crl might stay associated with σS and also affect later steps in transcription initiation, such as promoter DNA binding and/or open-complex formation (12, 14).
Crl interacts with σS conserved domain 2 (σS2), the domain primarily responsible for melting the −10 element in the promoter (7, 10, 15). Recently, we identified specific features in σS2 that are required for the Crl-σS interaction (7). Crl binds in close proximity to the residues in σS predicted to interact with promoter DNA and directly adjacent to the region where there is a large nonconserved sequence insertion in σ70, the primary sigma factor and closest relative of σS (7). This Crl binding determinant and a DPE sequence found in σS but not in σ70 are necessary and sufficient to account for the ability of Crl to distinguish among the σ factors. However, identification of the determinants on Crl necessary for its interaction with σS has been hampered by a lack of structural information about Crl.
Here, we present the X-ray crystal structure of Proteus mirabilis Crl at 1.90-Å resolution and identify residues in Crl required for its interaction with σS. We utilize a combination of structural information, phylogenetic conservation analysis, bacterial two-hybrid interaction assays, in vivo and in vitro transcription, and site-specific protein-protein cross-linking to determine a patch on Crl that is necessary for the σS-Crl interaction. This study thus provides critical information for understanding how interaction of Crl with σS leads to transcriptional activation.
MATERIALS AND METHODS
Additional details and modifications from previously published methods are provided in the supplemental material.
Strains, plasmids, and oligonucleotides.
Complete lists of strains, plasmids, and oligonucleotides are provided in Tables S3 to S7 in the supplemental material. Site-directed mutagenesis of plasmids was performed using the QuikChange Lightning kit or by DNA synthesis with Pfu Ultra II Fusion DNA Polymerase (Stratagene) (7). Random mutagenesis was performed by PCR as described previously (16). Strains for bacterial two-hybrid (BTH) analysis were created by cotransformation of a strain harboring a promoter-lacZ reporter on an F episome with plasmids encoding bait and prey fusion proteins. The osmY promoter-lacZ transcriptional fusion reporter is encoded by a single-copy λ prophage (7). Gene deletions and allelic replacements were made by λ Red-mediated recombination (17) or were obtained from the Keio collection (18). Mutant alleles were transferred to strains by transduction with P1vir (19).
Protein purification.
E. coli core RNA polymerase was prepared as described previously (20). Untagged E. coli σS was prepared from inclusion bodies and refolded as described previously (21). His6-tagged P. mirabilis Crl with Se-Met substitutions was overexpressed and purified by Ni-nitrilotriacetic acid (NTA) affinity chromatography and the AKTAxpress system (GE Health Systems), as described previously (22), and the affinity tag was cleaved with His-tagged tobacco etch virus protease, followed by an additional Ni-NTA purification to separate the protease, uncut protein, and affinity tag. E. coli His6-Crl was overexpressed and purified using Ni affinity and heparin chromatography, and the tag was removed by thrombin cleavage. His6-Crl (or Crl variants) and His6-σS (or σS variants) with p-benzoylphenylalanine (BPA) substitutions, some containing an N-terminally encoded heart muscle kinase (HMK) recognition site, were expressed and purified by Ni affinity chromatography as described previously (7, 23, 24).
Protein crystallization.
Crl crystals were screened using commercially available screens: MCSG-1-4 (Microlytic, Burlington, MA) and Index Screen (Hampton Research Corp., Aliso Viejo, CA) at 24°C and 16°C. Vapor diffusion sitting drops contained 0.4 μl of protein and 0.4 μl of screening solution and were set up in 96-well CrystalQuick plates (Greiner Bio-One, Monroe, NC) over wells containing 140 μl screening solution using a Mosquito liquid dispenser (TTP Labtech, Cambridge, MA). Crystals were obtained under several conditions, cryoprotected by a brief transfer to the crystallization solution plus 25% ethylene glycol, flash cooled in liquid nitrogen, and analyzed with synchrotron X-ray radiation near the selenium edge at the Structural Biology Center, sector 19-ID beamline, at the Advanced Photon Source, Argonne National Laboratory.
Data collection, structure determination, and refinement.
One crystal was subjected to an X-ray fluorescence scan to determine the selenium K absorption edge. Data were collected at 100 K to 1.90 Å at the peak wavelength (0.9790 Å) for the single best seleno-Met-substituted Crl crystal and processed using the HKL-3000 suite (25). Single-wavelength anomalous diffraction data) were used to solve the structure, as implemented in the HKL-3000 structure solution package. A single selenium site was located with SHELXCD, and phases were calculated and improved with iterative rounds of MLPHARE and DM (29). An initial model was built with ARP/wARP (26). The model was completed manually with Coot (27), and the structure was refined with Refmac5 (28) from the CCP4 suite (29). The structure's all-atom contacts and geometry were analyzed with MolProbity (30) and further refined. The final atomic coordinates and amplitudes have been deposited in the Protein Data Bank (PDB).
BPA-mediated cross-linking assays.
32P radiolabeling of proteins was performed using protein kinase A from bovine heart (Sigma) and an N-terminally encoded HMK recognition site, as previously described (31, 32). Photoactivated cross-linking of proteins with BPA substitutions was performed in duplicate, as described previously (7, 24).
BTH analysis.
Protein-protein interaction was determined using a previously described method utilizing an RNAP α subunit fusion protein, a λcI fusion protein, and a lacZ reporter construct (33, 34). β-Galactosidase assays were performed in 96-well microtiter plates as previously described (7, 35).
In vivo promoter activity assays.
β-Galactosidase activity assays from an osmY promoter-lacZ transcriptional fusion (see above) (7) were performed on cells harvested during the transition from exponential to early stationary phase (optical density at 600 nm [OD600] ≈ 1.4). For the complementation experiments, Crl was encoded on a plasmid under the control of an IPTG-inducible pTrc promoter. Crl variants were constructed in single copies on the chromosome at the native crl location for analysis in vivo.
In vitro promoter activity assays.
Multiple-round transcription was performed on plasmid templates as described previously (7). The osmY promoter sequence endpoints on the plasmid used as a template in the in vitro transcription assays were −107 to +20 with respect to the transcription start site.
Western analysis.
Western analysis was performed in triplicate from cells harvested during the transition from exponential to early stationary phase (OD600 ≈ 1.4) using a Crl polyclonal antibody (36).
Computational analyses.
Sequences were obtained from the Joint Genome Institute-Integrated Microbial Genomes (JGI-IMG) (http://img.jgi.doe.gov) (37) website and aligned using ClustalX (38) or Mult Align (39). Sequence similarities, surface accessibility, and secondary-structure information from aligned sequences were rendered (see Fig. 1B) using ESPript 3.0 (40), which calculates a solvent accessibility value for each residue using DSSP (41). Redundancy in the data set was reduced to 95% similarity using Decrease Redundancy (http://web.expasy.org/decrease_redundancy/). Conservation scores were calculated using ConSurf (http://consurf.tau.ac.il/) (42). A fractional accessible surface area (ASA) of amino acids (see Fig. 3) was calculated using Vadar (43). The E. coli structural homology model used (see Fig. 3 and 6) was created based on the P. mirabilis X-ray crystal structure using Phyre (44). The quality of this model was evaluated using the structure assessment tool of the SWISS-MODEL protein structure homology-modeling server (http://swissmodel.expasy.org) (45). The model had an overall QMEAN6 score of 0.782. The QMEAN6 score is a composite score consisting of a linear combination of 6 terms to estimate model reliability. This quality estimate ranges between 0 and 1, with 1 being ideal (46).
FIG 1.
X-ray crystal structure of P. mirabilis Crl. (A) Ribbon model of P. mirabilis Crl with a color gradient of blue to red from the N to the C terminus. Secondary-structure elements are labeled. (B) Amino acid sequence alignment of Crl homologs from P. mirabilis (YP_002150140.1), E. coli (NP_414775.1), S. enterica (NP_454931.1), Aeromonas hydrophila (YP_857911.1), V. harveyi (YP_001444378.1), Photobacterium profundum (YP_129053.1), and Psychromonas ingrahamii (YP_944255.1). Structural elements are indicated above the sequences, and 100% conserved sequences are indicated by red shading. Residues in red letters are similar within a group. Residues framed in blue are similar across groups. The relative surface accessibility (acc) of each residue is plotted below the sequences as a colored box: dark blue for accessible, cyan for intermediate, and white for buried. Red indicates no prediction. (C) Evolutionarily conserved amino acids on a surface view of P. mirabilis Crl. Conservation was determined using ConSurf and is displayed in greyscale with light to dark colors indicating more to less conserved; 100% conserved residues are shown in red. (D) Predicted electrostatic surface of P. mirabilis Crl generated using Pymol.
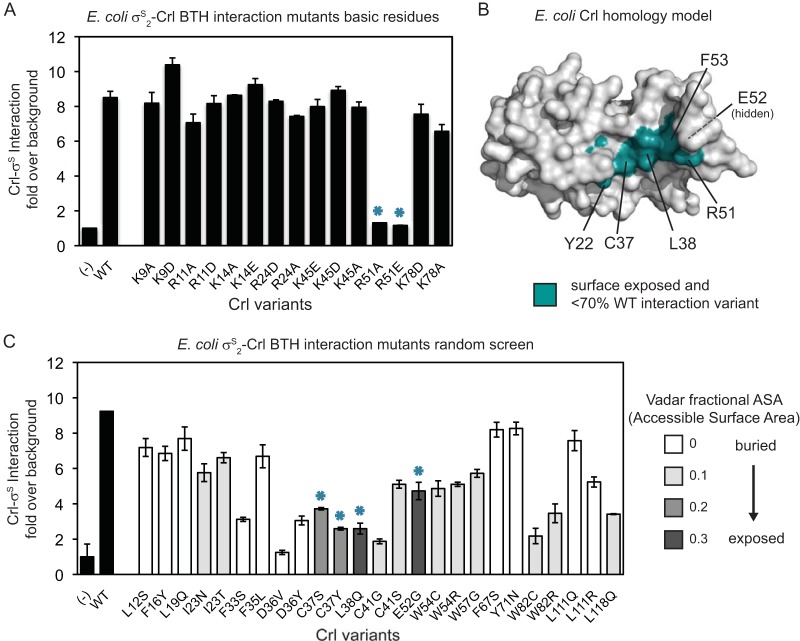
FIG 3.
Crl-σS2 BTH interaction analysis. (A) BTH assay indicating levels of interaction between σS2 and Crl variants generated by site-directed mutagenesis of conserved basic residues. The bars represent β-galactosidase activity in Miller units over background (an unfused control). The error bars show the SD from 2 separate experiments with 2 independent cultures each. The asterisks indicate variants that have reduced interaction. (B) Surface representation of a homology model of E. coli Crl. Surface-exposed conserved residues identified from our BTH analyses are shown in teal, as are F53 and Y22, which were identified in a previous study (48). Residue E52 (labeled “hidden”) is surface exposed but is not visible in this view. (C) BTH assay as in panel A, except with Crl variants generated by random PCR mutagenesis. Black bars, WT Crl and an unfused control (−). The bar shading of the variants indicates the fractional ASA determined by Vadar (43). The asterisks indicate variants that have reduced interaction and more surface exposure.
FIG 6.
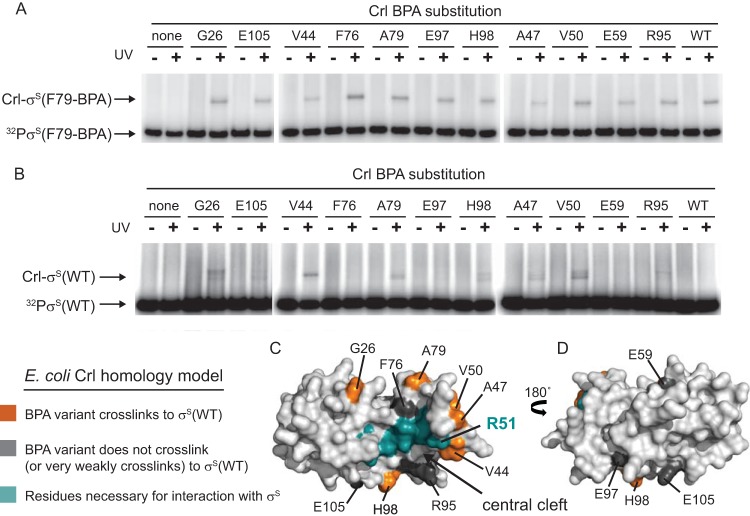
BPA-mediated cross-linking of Crl to σS. (A and B) SDS-PAGE gels showing the results of UV exposure on 32P-σS(F79BPA) (A) or 32P-σS(WT) (B) incubated with either WT Crl or Crl variants with the photoreactive amino acid analog BPA incorporated at the indicated positions. Free 32P-σS and cross-linked 32P-σS-Crl complexes are indicated with arrows. Crl-BPA variants did not produce higher-molecular-weight bands in the absence of UV treatment or without σS. (C and D) Surface representations of the E. coli Crl homology model. Panel D is a 180° rotation of panel C. The positions of BPA substitutions in panels A and B are labeled and colored: orange, Crl-BPA variants that cross-link strongly to σS(WT); dark gray, Crl-BPA variants that cross-link very weakly or not at all to σS(WT); teal, residues in the central cleft patch proposed to interact with σS (Fig. 3B shows the identities of residues in addition to R51).
Protein structure accession number.
The final atomic coordinates and structure factors have been deposited in the PDB with accession number 3RPJ.
RESULTS
Structure of P. mirabilis Crl.
Crl is a small (133 amino acids in P. mirabilis) globular protein. The X-ray crystal structure of P. mirabilis Crl was refined to a resolution of 1.90 Å (PDB 3RPJ; see Materials and Methods) (Fig. 1A). Data collection and structure refinement statistics are provided in Table S1 in the supplemental material. The structure provides a framework for understanding the interaction of Crl with σS and therefore the molecular mechanism by which Crl modulates RNAP assembly.
Crl forms a globular α + β fold consisting of 5 β-strands, 2 α-helices, and two short 310-helices (η). A total of five β-turns occur between helices α1 and η1, helix η1 and strand β1, β1 and η2, strands β2 and β3, and strands β3 and β4. A groove (central cleft) is located on one surface of the protein, the bottom of which is formed by a 4-stranded anti-parallel β-sheet (strands β1 to β4) and rimmed by the intervening loops between these strands. Amino acids within the cleft are evolutionarily conserved, especially those from the first β-turn through β2. The opposite surface of the protein is dominated by two long helices, α-1 and α-2, that consist of largely nonconserved amino acids (Fig. 1B and C). The fold comes to an end with strand β-5, running parallel to β-3, and helix η3. The protein has little or no apparent structural similarity to previously described folds as determined by a three-dimensional (3D) structure search performed by the protein structure comparison service Fold at the European Bioinformatics Institute (http://www.ebi.ac.uk/msd-srv/ssm). No hits were found with a P score above 1.1. The P score is calibrated so that values below 3 are statistically insignificant (47). There are two copies of Crl in the asymmetric unit, with monomers from two adjacent asymmetric units forming a potential dimer interface with a buried surface area of 1,400 Å2 (see Fig. S1 in the supplemental material).
Conservation of Crl.
We identified Crl homologs in 190 of 2,126 nonredundant bacterial genomes analyzed, comprising 4 orders of gammaproteobacteria: Enterobacteriales (20 genera, including Escherichia, Proteus, and Salmonella), Vibrionales (2 genera), Aeromonadales (2 genera), and Alteromonadales (1 genus). We generated an alignment of 51 Crl sequences from the database that were less than 95% identical but were evolutionarily diverse to avoid overrepresentation of closely related groups (see Table S2 in the supplemental material). We then used this alignment to determine the conservation and variation among residues in Crl homologs (see Fig. S2A in the supplemental material). The conserved residues are shown in an alignment of amino acid sequences from representative genera (Fig. 1B) and displayed on a surface representation of the structure in Fig. 1C. Several highly conserved clusters were apparent, including (E. coli numbering) GPYXR (residues 20 to 24), FDCLAXC (35 to 41), and PEXREFWGWW (48 to 57) (Fig. 1B). Because the surface on σS that interacts with Crl is highly conserved (7) (see Fig. S2B in the supplemental material), we speculated that the complementary surface on Crl that interacts with σS might also be highly conserved and include one or more of these conserved clusters. Crl has many conserved charged residues; a predicted electrostatic surface distribution is represented in Fig. 1D.
Evolutionarily diverse Crl homologs interact with E. coli σS and complement Crl function in vivo.
The Crl homolog from P. mirabilis is 47% identical/65% similar to E. coli Crl. In contrast, σS2, the domain of σS that interacts with Crl, is much more highly conserved between P. mirabilis and E. coli (93% identical/98% similar). Crl function has been investigated in detail only in E. coli and S. enterica. Therefore, to address whether the structure of the P. mirabilis Crl homolog was appropriate for modeling the structure of E. coli Crl and its interaction with σS2, we used a complementation assay to determine whether P. mirabilis Crl can function similarly to E. coli Crl in vivo. We also investigated the Crl homolog from Vibrio harveyi, also a gammaproteobacterium but more divergent from E. coli (39% identity/56% similarity) than the enterobacterium P. mirabilis.
E. coli Crl, or the P. mirabilis or V. harveyi Crl homolog, was expressed from a plasmid in an E. coli strain lacking crl. Both the P. mirabilis and V. harveyi Crl homologs increased transcription from the rpoS-dependent osmY promoter-lacZ fusion to about the same level as E. coli Crl or when Crl was expressed from its native locus on the E. coli chromosome (Fig. 2A). Thus, even though the P. mirabilis and V. harveyi Crl homologs are quite divergent, they can complement E. coli Crl function.
FIG 2.
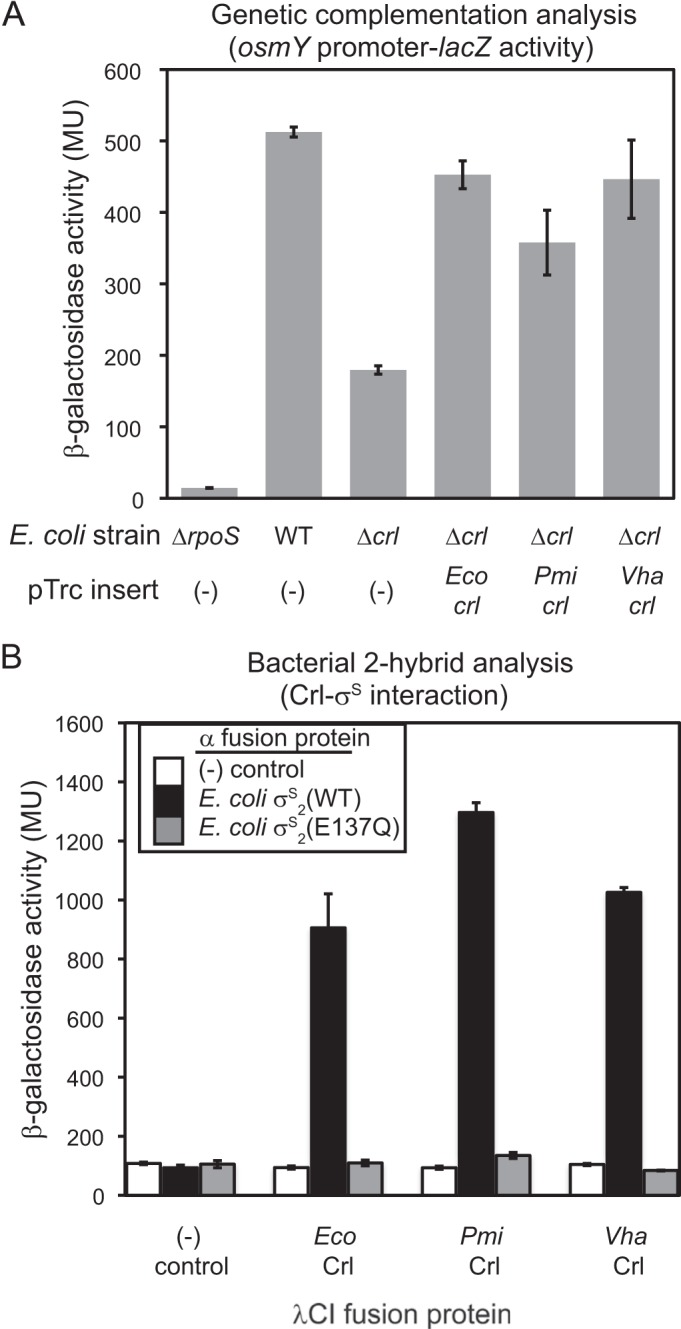
P. mirabilis and V. harveyi Crl homologs interact with E. coli σS and function in E. coli. (A) β-Galactosidase assay of an osmY promoter-lacZ reporter in E. coli strains with E. coli (Eco), V. harveyi (Vha), or P. mirabilis (Pmi) Crl homologs expressed from a plasmid in a strain with a deletion of crl (Δcrl). (B) BTH analysis to determine interaction between the Eco, Vha, or Pmi Crl homolog expressed as a fusion to λCI and E. coli σS2(WT), an E. coli σS2(E137Q) binding variant, or an unfused control expressed as a fusion to α in a promoter-lacZ reporter strain. β-Galactosidase activity is expressed in Miller units (MU), and the error bars show standard deviations (SD) between 2 separate experiments with 2 independent cultures each.
We next examined the interaction of E. coli σS with the P. mirabilis and V. harveyi Crl homologs by BTH analysis to confirm that their complementation reflects an interaction with σS similar to that of E. coli Crl (Fig. 2B). P. mirabilis and V. harveyi Crl interacted with wild-type (WT) E. coli σS2 as well as did E. coli Crl (Fig. 2B, black bars). We showed previously that an E137Q substitution in E. coli σS2 eliminated the interaction with Crl but retained its interaction with the β′ clamp helices of core RNAP (β′CH), indicating that the loss of interaction with Crl did not result from instability of the σS2 peptide (7). Consistent with the hypothesis that the Vibrio and Proteus complexes depend on the same σS2 surface as E. coli Crl, there was no interaction with σS2 E137Q. These results confirmed that the Crl homologs acted like E. coli Crl; therefore, we used the P. mirabilis Crl structure as a model for subsequent studies of the Crl-σS interaction.
The central cleft of Crl contains conserved residues critical for the interaction with σS.
We constructed a homology model of E. coli Crl based on the P. mirabilis Crl structure. We used this model, in combination with data from three BTH data sets, to identify surface-exposed residues necessary for the Crl-σS2 interaction. Solvent exposure was estimated using ESPript 3.0 (40, 41) (Fig. 1B) (see Materials and Methods). Because several of the residues in σS2 that were necessary for interaction with Crl are acidic, D or E (7), we reasoned that conserved basic amino acids in Crl with basic side chains might mediate an electrostatic/salt bridge-mediated interaction with σS. Therefore, as a first step for identifying the surface in Crl that interacts with σS, we tested charge switch and alanine substitutions for seven conserved surface-exposed basic amino acids in Crl (K9, R11, K14, R24, K45, R51, and K78) by BTH analysis. Only substitutions for R51 had a significant defect in the interaction with σS2 (Fig. 3A, teal asterisks, and B, teal).
We next performed a less biased screen for loss of interaction with σS2 by creating a Crl mutant library by error-prone PCR. After performing BTH analysis, we selected 172 colonies with reduced β-galactosidase activities on indicator plates, analyzed their crl sequences, and identified 32 Crl variants with unique single amino acid substitutions at 21 positions. Proline substitutions might disrupt the protein fold, leading to a loss-of-function phenotype without helping to define a surface that interacts with σS2. Therefore, we eliminated the proline variants, leaving 25 unique single amino acid substitutions at 18 positions for which we determined the corresponding β-galactosidase activities in liquid culture (Fig. 3C). Since substitutions of residues on the surface of Crl would be most likely to disrupt the interaction directly (rather than affecting β-galactosidase activity indirectly by causing protein misfolding and/or instability), we also used an additional computational approach, the program Vadar (43), to analyze the Crl homology model in order to determine the residues in Crl with the most surface exposure. Residues are grouped into classes according to the calculated fractional ASA (Fig. 3C). Three substitutions resulted in reduced activities in the BTH assay (<70% of the WT value) and were on the surface of Crl (Vadar ASA > 1), namely, C37Y, L38Q, and E52G (Fig. 3C, teal asterisks; C37 and L38 are in teal in Fig. 3B; E52 is between R51 and F53 but is not visible in the view shown). These evolutionarily conserved surface positions are adjacent to each other and to the residue that was identified as crucial for Crl-σS2 interaction in the BTH screen of basic residues, R51 (Fig. 3A).
We also used our homology model to evaluate an S. enterica Crl BTH data set from a previous study in which residues that were 100% conserved were replaced with alanines (48). Four of the substitutions in that screen met our analysis criteria, namely, Y22A, R51A, E52A, and F53A. Although we did not find Y22 or F53 substitutions in our loss-of-interaction screens in E. coli, we found adjacent residues, I23, R51, E52, and W54, further supporting the importance of this area for Crl interaction.
Since our random-mutagenesis screen was not saturating and our criteria were very stringent, we probably have not identified all the residues that contribute to the Crl-σS2 interaction. Nevertheless, together, the 3 data sets identified a patch of residues in the evolutionarily conserved central cleft of Crl that is necessary for interaction with σS2 (Fig. 3B, teal).
The R51 patch is required for activation of transcription in vivo and in vitro in the context of full-length σS.
The BTH analyses identified a patch on Crl that interacts with σS, consistent with its surface exposure, positive charge (which made it a candidate to interact directly with the critical acidic patch on σS), and evolutionary conservation. We next tested the importance of residues within the proposed σS interaction patch in Crl in a more physiologically relevant context, i.e., when there were native levels of the full-length interacting partners in vivo. Crl R51 was chosen from among the other residues in the patch because of its strong phenotype in the BTH assays, its conservation, and its positive charge, but we also analyzed a few nearby residues in the proposed interaction patch. Substitutions coding for alanine substitutions (which truncate the side chain beyond the C-beta atom), as well as less disruptive conservative substitutions, were constructed and introduced into the crl gene in its native chromosomal context by λ Red-mediated recombination. Relative to a strain lacking crl, wild-type Crl increased the activity of the rpoS-dependent osmY promoter-lacZ fusion ∼3-fold during the transition to stationary phase, when both Crl and σS concentrations increase (Fig. 4A [cf. Fig. 2A]) (12, 49). Mutant strains encoding Crl R51A or even the more conservative variant R51K failed to increase osmY-lacZ activity. The Crl variant F53Y was only slightly defective in activation, but the F53A substitution was inactive, suggesting that maintaining the hydrophobicity of the F53 side chain is necessary for function. E52A did not affect osmY-lacZ activity significantly, suggesting that its side chain is unlikely to contribute directly to the interaction with σS.
FIG 4.
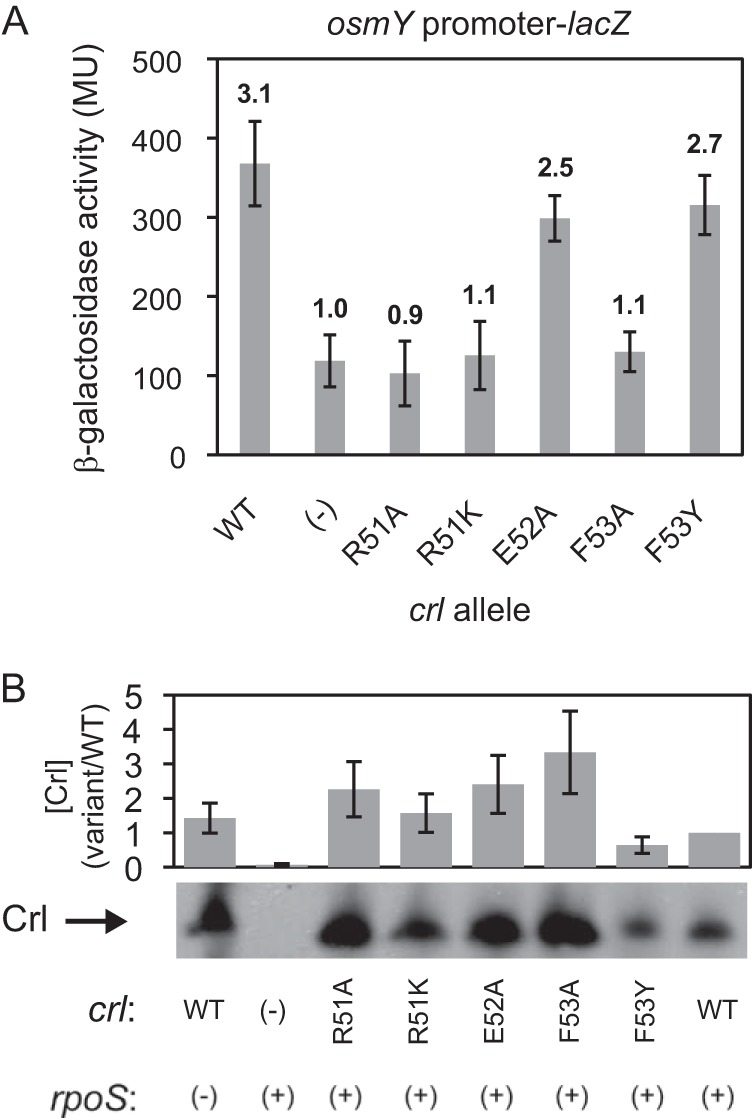
Crl conserved residue R51 is necessary for activation of transcription in vivo. (A) osmY promoter-lacZ expression in early stationary phase in E. coli strains with different chromosomal crl alleles. Fold activation (with or without crl) is shown above the bar for each allele. The error bars show SD for 3 cultures each with 3 replicates each. (B) Western analysis of crl WT and mutant strains. A representative gel is shown. The graph shows the ratios of Crl variants compared to the WT for 3 experiments.
Because loss of function in vivo could result from a reduction in protein stability rather than from loss of an interaction with σS, we analyzed Crl levels in the wild-type and mutant strains using Western blots with a polyclonal anti-Crl antibody (Fig. 4B). Crl levels were not reduced in the activation-defective crl mutants relative to the level of Crl in the wild-type strain. These results indicate that the loss of transcription activation did not result from degradation of the mutant proteins. Interestingly, the levels of some of the Crl variants were greater than the wild-type level (see Discussion).
To ensure that the failure of the crl mutant strains to activate transcription in vivo was not indirect, we overexpressed and purified the most defective Crl variant proteins, R51A and R51K, and tested their effects on transcription by EσS in vitro from the osmY promoter. A representative transcription gel is shown in Fig. 5A, and quantitation of the results with both R51A and R51K, as well as wild-type Crl, is shown in Fig. 5B. Consistent with their effects in vivo, wild-type Crl increased transcription >2-fold in vitro, whereas the R51A and R51K Crl variants failed to increase transcription (Fig. 5A). Interestingly, the activation-defective Crl variants actually inhibited transcription in vitro (Fig. 5B) (see Discussion). Further studies will be needed to define the identities of the complete set of residues in the patch that interacts directly with σS.
FIG 5.
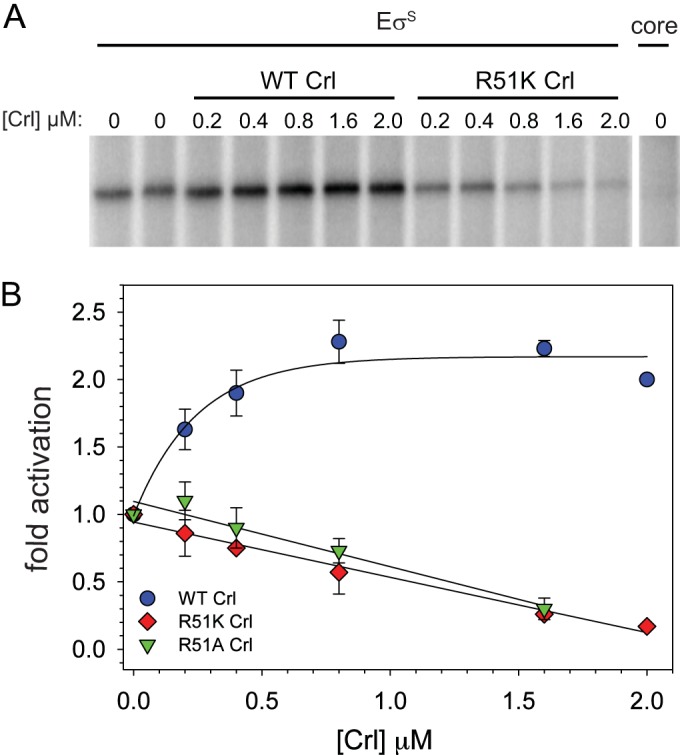
Crl conserved residue R51 is necessary for activation of transcription in vitro. (A) Multiple-round in vitro transcription from a plasmid-borne osmY promoter (pRLG8941) by EσS in the presence of WT Crl and the R51K variant in vitro. A representative experiment is shown. (B) Quantitation of activation by WT Crl and Crl variants over a range of Crl concentrations.
BPA-mediated cross-linking supports the identification of the Crl surface that interacts with σS.
To demonstrate a physical interaction of σS near the proposed Crl interaction patch, we used a UV-cross-linking approach. BPA residues were substituted individually into Crl at positions surrounding the proposed site of interaction with σS using an orthologous tRNA/tRNA synthetase system (23). The positions in Crl for incorporation of BPA were chosen to be near but not within the interaction patch so that they would not interfere with σS binding or perturb the Crl structure. Seventeen Crl variants were purified, each containing a single BPA substitution (for residues S10, A18, E25, G26, K27, V44, A47, V50, E59, F76, A79, R95, E97, H98, E105, T113, or K117).
Although we tried to avoid creating destabilizing substitutions, it was nonetheless possible that some mutant proteins have been inactivated by BPA incorporation, either because the substitution interfered with the interaction with σS or because the proteins did not fold correctly. To identify and eliminate these proteins from further consideration, we utilized σS(F79-BPA), a σS variant that cross-links to wild-type Crl (7). Eleven of the Crl-BPA variants cross-linked to σS(F79-BPA): G26, V44, A47, V50, E59, F76, A79, R95, E97, H98, and E105 [because the σS(F79-BPA) residue cross-linked to Crl and/or because the Crl-BPA residue cross-linked to σS] (Fig. 6A), demonstrating that these BPA substitutions did not interfere with the Crl-σS interaction.
The Crl-BPA proteins were then examined for cross-linking to wild-type σS (i.e., without F79-BPA) (Fig. 6B). Crl proteins containing BPA at position G26, V44, A47, V50, A79, or H98 cross-linked strongly to wild-type σS (Fig. 6C, orange), forming a ring around the proposed interaction patch (Fig. 3B and 6C, teal), which includes the critical residue R51. R95-BPA and E105-BPA cross-linked very weakly (the bands are barely visible in Fig. 6B), and 2 residues on the other surface of Crl, E97-BPA and E59-BPA, failed to cross-link to σS at all (Fig. 6B and D, gray), consistent with the model in which the interaction is located on one surface of the protein in an area we refer to as the central cleft. Despite being near the central cleft, F76-BPA failed to cross-link to wild-type σS, suggesting that proximity is necessary but not sufficient for cross-linking; the reactive group must also be oriented properly. The fact that only a subset of the BPA-containing Crl proteins that cross-linked to σS(F79-BPA) also cross-linked to wild-type σS suggests that complex formation was specific.
DISCUSSION
Identification of the σS binding site on Crl.
Taken together, the structure of Crl and the identification of its evolutionarily conserved regions (Fig. 1; see Fig. S2 in the supplemental material), the complementation of E. coli crl mutant strains by diverse Crl proteins (Fig. 2), the BTH analysis of Crl variants with σS2 (Fig. 3), the transcriptional analyses of crl mutants in vivo (Fig. 4) and in vitro (Fig. 5), and the cross-linking of Crl to σS (Fig. 6) support the model shown in Fig. 6C, in which σS interacts with a highly conserved cleft on one face of Crl. The arginine 51 side chain is critical for this interaction.
In a previous study, Monteil and colleagues introduced alanine substitutions at selected conserved positions in S. enterica Crl and analyzed their interaction with σS2 using the BTH assay (48). Without structural information, it was not possible to evaluate whether defects in interaction with σS2 resulted from substitutions that disrupted protein structure in general or specific defects in the interaction with σS. Nevertheless, three of the substitutions in Crl that eliminated its interaction with σS in that study, Y22A, R51A, and F53A, are within/adjacent to the patch identified here.
Our BTH analysis, combined with that by Monteil and colleagues (48), along with the results from our BPA-mediated cross-linking and functional analyses, indicate that σS interacts with a highly conserved central cleft in Crl. Additional conserved surface-exposed residues in and around this cleft, including several proline and cysteine residues, as well as some nearby residues with charged side chains, suggest that the interaction is complex. Additional residues not identified in our analyses could also contribute to the interaction.
Some of the substitutions in the proposed σS binding interface of Crl increased the concentration of the Crl variant relative to that of wild-type Crl in vivo (Fig. 4B). Potential explanations are that the σS binding interface on Crl is a protease target and that the substitution stabilizes the protein against protease attack or that Crl somehow inhibits its own expression through feedback by a mechanism requiring an intact σS interface.
Mechanism of transcriptional regulation by Crl.
The interaction between Crl and σS is consistent with several possible mechanisms for stimulation of holoenzyme assembly. Crl could help σS interact with core RNAP by acting as a molecular tether, increasing the stability of the holoenzyme. This mechanism could involve repositioning σS within the EσS complex or an increase in the stability of the σS-core RNAP complex. A potential effect of Crl on stabilization of the holoenzyme complex does not rule out the possibility that Crl could also alter the conformation of σS prior to binding to core, for example, by helping unfold free σS and thereby unmasking key core binding determinants in σS. Crl could have effects on holoenzyme assembly in addition to these molecular-tethering-like or chaperone-unfolding-like roles. For example, Crl could remain associated with the holoenzyme and play roles in promoter binding or promoter escape. Because the residues in σS2 that bind Crl are very close to those that bind to the −10 element (7), if Crl were to remain bound to the holoenzyme, it is reasonable to expect that it could affect subsequent steps in transcription initiation.
We observed inhibition of transcription at high concentrations of the R51A and R51K Crl variants in vitro (Fig. 5B). Whatever the mechanism of inhibition, its significance is unclear, since we did not observe inhibition of the same promoter by the same mutant proteins when expressed from the native locus in vivo (Fig. 4A).
We note that a few other transcriptional regulators unrelated to Crl have been identified recently that associate with σ2 in other bacterial species and act positively on transcription initiation (50–52). Although their mechanism(s) of action is not yet understood, we speculate that factors that bind σ2 and influence sigma factor competition could be widespread in nature.
Role of Crl in regulation of transcription initiation.
Our data suggest that Crl homologs from diverse species can interact with σS and stimulate transcription from σS-dependent promoters. Some Crl homologs may be difficult to identify based on sequence similarity alone, and conditions that lead to increases in the concentration of Crl (and therefore to increased σS-dependent transcription) could vary among bacterial species. In this context, we note that residues near the N and C termini of Crl may impact recognition by ClpXP and thus Crl protein stability (53), potentially tying σS-dependent expression to the regulation of ClpXP. It was also reported recently that crl gene expression is regulated by nitrogen stress (36).
Future work may uncover conditions under which Crl stimulates transcription in other bacterial species that are quite different from the conditions under which Crl is needed in E. coli. Conversely, Crl may not be required in organisms that do not experience frequent perturbations in environmental conditions. Even in organisms containing the crl gene, its deletion may not have a prominent phenotype (54). It is possible that Crl recognition sequences are present in different sigma factors in different bacterial species. This would allow Crl to stimulate the expression of different regulons depending on the organism. In this context, we note that we were able to create a Crl-binding version of σ702 (7). Interestingly, when we imported the recognition determinants for Crl binding into the chromosomal (and only) copy of σ70, however, there were severe defects in bacterial growth (55).
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Wollenberg and M. Copeland for bacterial strains. We thank J. Peters for discussions and critical reading of the manuscript, K. Forest for discussions and help with construction of a homology model, R. Burgess and N. Thompson for discussions and σS, and M. Mandel and T. Silhavy for the anti-Crl antibody.
This work was supported by National Institutes of Health (NIH) grant R37 GM37048 (to R.L.G.), by NIH Predoctoral Training Grant T32 GM07215 (to A.B.B. and A.R.M.), and by the Protein Structure Initiative of the National Institutes of Health (Midwest Center for Structural Genomics; GM074942 and GM094585 [to A.J.]). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Sciences, and the use of Structural Biology Center beamline 19-ID was supported by the Office of Biological and Environmental Research under contract DE-AC02-06CH11357.
Footnotes
Published ahead of print 7 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01910-14.
REFERENCES
- 1.Lee DJ, Minchin SD, Busby SJW. 2012. Activating transcription in bacteria. Annu. Rev. Microbiol. 66:125–152. 10.1146/annurev-micro-092611-150012 [DOI] [PubMed] [Google Scholar]
- 2.Österberg S, del Peso-Santos T, Shingler V. 2011. Regulation of alternative sigma factor use. Annu. Rev. Microbiol. 65:37–55. 10.1146/annurev.micro.112408.134219 [DOI] [PubMed] [Google Scholar]
- 3.Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 65:189–213. 10.1146/annurev-micro-090110-102946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hengge R. 2009. Proteolysis of σS (RpoS) and the general stress response in Escherichia coli. Res. Microbiol. 160:667–676. 10.1016/j.resmic.2009.08.014 [DOI] [PubMed] [Google Scholar]
- 5.Maeda H, Fujita N, Ishihama A. 2000. Competition among seven Escherichia coli σ subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res. 28:3497–3503. 10.1093/nar/28.18.3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colland F, Fujita N, Ishihama A, Kolb A. 2002. The interaction between σS, the stationary phase σ factor, and the core enzyme of Escherichia coli RNA polymerase. Genes Cells 7:233–247. 10.1046/j.1365-2443.2002.00517.x [DOI] [PubMed] [Google Scholar]
- 7.Banta AB, Chumanov RS, Yuan AH, Lin H, Campbell EA, Burgess RR, Gourse RL. 2013. Key features of σS required for specific recognition by Crl, a transcription factor promoting assembly of RNA polymerase holoenzyme. Proc. Natl. Acad. Sci. U. S. A. 110:15955–15960. 10.1073/pnas.1311642110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pratt LA, Silhavy TJ. 1998. Crl stimulates RpoS activity during stationary phase. Mol. Microbiol. 29:1225–1236. 10.1046/j.1365-2958.1998.01007.x [DOI] [PubMed] [Google Scholar]
- 9.Gaal T, Mandel MJ, Silhavy TJ, Gourse RL. 2006. Crl facilitates RNA polymerase holoenzyme formation. J. Bacteriol. 188:7966–7970. 10.1128/JB.01266-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monteil V, Kolb A, Mayer C, Hoos S, England P, Norel F. 2010. Crl binds to domain 2 of σS and confers a competitive advantage on a natural rpoS mutant of Salmonella enterica serovar Typhi. J. Bacteriol. 192:6401–6410. 10.1128/JB.00801-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haugen SP, Ross W, Gourse RL. 2008. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat. Rev. Microbiol. 6:507–519. 10.1038/nrmicro1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bougdour A, Lelong C, Geiselmann J. 2004. Crl, a low temperature-induced protein in Escherichia coli that binds directly to the stationary phase σ subunit of RNA polymerase. J. Biol. Chem. 279:19540–19550. 10.1074/jbc.M314145200 [DOI] [PubMed] [Google Scholar]
- 13.Typas A, Barembruch C, Possling A, Hengge R. 2007. Stationary phase reorganisation of the Escherichia coli transcription machinery by Crl protein, a fine-tuner of σS activity and levels. EMBO J. 26:1569–1578. 10.1038/sj.emboj.7601629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.England P, Westblade LF, Karimova G, Robbe-Saule V, Norel F, Kolb A. 2008. Binding of the unorthodox transcription activator, Crl, to the components of the transcription machinery. J. Biol. Chem. 283:33455–33464. 10.1074/jbc.M807380200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feklistov A, Darst SA. 2011. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase σ subunit. Cell 147:1257–1269. 10.1016/j.cell.2011.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cadwell RC, Joyce GF. 1994. Mutagenic PCR. PCR Methods Appl. 3:S136–S140. 10.1101/gr.3.6.S136 [DOI] [PubMed] [Google Scholar]
- 17.Thomason LC, Sawitzke JA, Li X, Costantino N, Court DL. 2014. Recombineering: genetic engineering in bacteria using homologous recombination. Curr. Protoc. Mol. Biol. 106:1.16.1–1.16.39. 10.1002/0471142727.mb0116s106 [DOI] [PubMed] [Google Scholar]
- 18.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomason LC, Costantino N, Court DL. 2007. E. coli genome manipulation by P1 transduction. Curr. Protoc. Mol. Biol. Chapter 1:Unit 1.17. 10.1002/0471142727.mb0117s79 [DOI] [PubMed] [Google Scholar]
- 20.Burgess RR, Jendrisak JJ. 1975. A procedure for the rapid, large-scale purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry 14:4634–4638. 10.1021/bi00692a011 [DOI] [PubMed] [Google Scholar]
- 21.Anthony LC, Foley KM, Thompson NE, Burgess RR. 2003. Expression, purification of, and monoclonal antibodies to sigma factors from Escherichia coli. Methods Enzymol. 370:181–192. 10.1016/S0076-6879(03)70016-0 [DOI] [PubMed] [Google Scholar]
- 22.Kim Y, Babnigg G, Jedrzejczak R, Eschenfeldt WH, Li H, Maltseva N, Hatzos-Skintges C, Gu M, Makowska-Grzyska M, Wu R, An H, Chhor G, Joachimiak A. 2011. High-throughput protein purification and quality assessment for crystallization. Methods 55:12–28. 10.1016/j.ymeth.2011.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryu Y, Schultz PG. 2006. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nat. Methods 3:263–265. 10.1038/nmeth864 [DOI] [PubMed] [Google Scholar]
- 24.Lennon CW, Ross W, Martin-Tumasz S, Toulokhonov I, Vrentas CE, Rutherford ST, Lee J-H, Butcher SE, Gourse RL. 2012. Direct interactions between the coiled-coil tip of DksA and the trigger loop of RNA polymerase mediate transcriptional regulation. Genes Dev. 26:2634–2646. 10.1101/gad.204693.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. 2006. HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62:859–866. 10.1107/S0907444906019949 [DOI] [PubMed] [Google Scholar]
- 26.Langer G, Cohen SX, Lamzin VS, Perrakis A. 2008. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3:1171–1179. 10.1038/nprot.2008.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 28.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255. 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
- 29.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67:235–242. 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66:12–21. 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arthur TM, Burgess RR. 1998. Localization of a σ70 binding site on the N terminus of the Escherichia coli RNA polymerase beta' subunit. J. Biol. Chem. 273:31381–31387. 10.1074/jbc.273.47.31381 [DOI] [PubMed] [Google Scholar]
- 32.Sharp MM, Chan CL, Lu CZ, Marr MT, Nechaev S, Merritt EW, Severinov K, Roberts JW, Gross CA. 1999. The interface of σ with core RNA polymerase is extensive, conserved, and functionally specialized. Genes Dev. 13:3015–3026. 10.1101/gad.13.22.3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dove SL, Joung JK, Hochschild A. 1997. Activation of prokaryotic transcription through arbitrary protein-protein contacts. Nature 386:627–630. 10.1038/386627a0 [DOI] [PubMed] [Google Scholar]
- 34.Nickels BE. 2009. Genetic assays to define and characterize protein-protein interactions involved in gene regulation. Methods 47:53–62. 10.1016/j.ymeth.2008.10.011 [DOI] [PubMed] [Google Scholar]
- 35.Thibodeau SA, Fang R, Joung JK. 2004. High-throughput beta-galactosidase assay for bacterial cell-based reporter systems. Biotechniques 36:410–415 [DOI] [PubMed] [Google Scholar]
- 36.Zafar MA, Carabetta VJ, Mandel MJ, Silhavy TJ. 2014. Transcriptional occlusion caused by overlapping promoters. Proc. Natl. Acad. Sci. U. S. A. 111:1557–1561. 10.1073/pnas.1323413111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Pillay M, Ratner A, Huang J, Woyke T, Huntemann M, Anderson I, Billis K, Varghese N, Mavromatis K, Pati A, Ivanova NN, Kyrpides NC. 2014. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 42:D560–D567. 10.1093/nar/gkt963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 39.Corpet F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881–10890. 10.1093/nar/16.22.10881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gouet P, Robert X, Courcelle E. 2003. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31:3320–3323. 10.1093/nar/gkg556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kabsch W, Sander C. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577–2637. 10.1002/bip.360221211 [DOI] [PubMed] [Google Scholar]
- 42.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. 2010. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38:W529–W533. 10.1093/nar/gkq399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willard L, Ranjan A, Zhang H, Monzavi H, Boyko RF, Sykes BD, Wishart DS. 2003. VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Res. 31:3316–3319. 10.1093/nar/gkg565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kelley LA, Sternberg MJE. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371. 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- 45.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42:W252–W258. 10.1093/nar/gku340. 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benkert P, Biasini M, Schwede T. 2011. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27:343–350. 10.1093/bioinformatics/btq662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krissinel E, Henrick K. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60:2256–2268. 10.1107/S0907444904026460 [DOI] [PubMed] [Google Scholar]
- 48.Monteil V, Kolb A, D'Alayer J, Beguin P, Norel F. 2010. Identification of conserved amino acid residues of the Salmonella σS chaperone Crl involved in Crl-σS interactions. J. Bacteriol. 192:1075–1087. 10.1128/JB.01197-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robbe-Saule V, Jaumouillé V, Prévost M-C, Guadagnini S, Talhouarne C, Mathout H, Kolb A, Norel F. 2006. Crl activates transcription initiation of RpoS-regulated genes involved in the multicellular behavior of Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:3983–3994. 10.1128/JB.00033-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bao X, Nickels BE, Fan H. 2012. Chlamydia trachomatis protein GrgA activates transcription by contacting the nonconserved region of σ66. Proc. Natl. Acad. Sci. U. S. A. 109:16870–16875. 10.1073/pnas.1207300109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tabib-Salazar A, Liu B, Doughty P, Lewis RA, Ghosh S, Parsy M-L, Simpson PJ, O'Dwyer K, Matthews SJ, Paget MS. 2013. The actinobacterial transcription factor RbpA binds to the principal sigma subunit of RNA polymerase. Nucleic Acids Res. 41:5679–5691. 10.1093/nar/gkt277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bortoluzzi A, Muskett FW, Waters LC, Addis PW, Rieck B, Munder T, Schleier S, Forti F, Ghisotti D, Carr MD, O'Hare HM. 2013. Mycobacterium tuberculosis RNA polymerase-binding protein A (RbpA) and its interactions with sigma factors. J. Biol. Chem. 288:14438–14450. 10.1074/jbc.M113.459883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. 2003. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell 11:671–683. 10.1016/S1097-2765(03)00060-1 [DOI] [PubMed] [Google Scholar]
- 54.Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, Chaba R, Lee S, Kazmierczak KM, Lee KJ, Wong A, Shales M, Lovett S, Winkler ME, Krogan NJ, Typas A, Gross CA. 2011. Phenotypic landscape of a bacterial cell. Cell 144:143–156. 10.1016/j.cell.2010.11.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Banta AB. 2013. Molecular interactions between the transcription factor Crl and Sigma S RNA polymerase holoenzyme in Escherichia coli. PhD thesis University of Wisconsin—Madison, Madison, WI [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.