Abstract
Siderophores, which are produced to overcome iron deficiency, are believed to be closely related to the adaptability of bacteria. The high-siderophore-yielding Pseudomonas sp. strain HYS simultaneously secretes the fluorescent siderophore pyoverdine and another nonfluorescent siderophore that is a major contributor to the high siderophore yield. Transposon mutagenesis revealed siderophore-related genes, including the two-component regulators GacS/GacA and a special cluster containing four open reading frames (the nfs cluster). Deletion mutations of these genes abolished nonfluorescent-siderophore production, and expression of the nfs cluster depended on gacA, indicating that gacS-gacA may control the nonfluorescent siderophore through regulation of the nfs cluster. Furthermore, regulation of the nonfluorescent siderophore by GacS/GacA involved the Gac/Rsm pathway. In contrast, inactivation of GacS/GacA led to upregulation of the fluorescent pyoverdine. The two siderophores were secreted under different iron conditions, probably because of differential effects of GacS/GacA. The global GacS/GacA regulatory system may control iron uptake by modulating siderophore production and may enable bacteria to adapt to changing iron environments.
INTRODUCTION
Iron is crucial for almost all organisms to maintain normal life, as it functions as a cofactor for many metabolic enzymes mediating redox reactions and electron transfer (1, 2). However, the iron available in the environment is insufficient because of its low solubility under physiological pH and aerobic conditions, which has led to the development of elaborate high-affinity iron uptake systems in many organisms (3). However, excess free iron can be deleterious due to the formation of oxygen radicals (4). Consequently, iron uptake must be tuned carefully to avoid both starvation and toxicity (5). Microorganisms can synthesize and secrete siderophores to overcome iron deficiency (3, 6, 7). Siderophores are high-affinity iron chelators that can combine Fe(III) from the environment and transport it into cells effectively via membrane-associated transport systems (1, 8). Many bacteria produce more than one siderophore: one primary high-affinity siderophore and one or several lower-affinity siderophores. By employing and regulating these siderophores, bacteria can adjust to changing iron conditions in the environment and maintain intracellular iron homeostasis.
Pseudomonads are widely distributed in diverse niches and are extremely versatile and adaptable. This adaptability is thought to be associated with their diverse and hierarchical siderophore systems (9, 10). Among the various siderophores of pseudomonads, the fluorescent high-affinity peptidic pyoverdines are usually produced as the primary siderophore and have been studied intensively (10–12). Diverse secondary siderophores, such as pyochelin (13), pseudomonine (14), quinolobactin (15), and thioquinolobactin (16), are less well understood because their production and functions are usually masked by pyoverdines. However, these secondary siderophores could play essential roles in the growth of pyoverdine-negative mutants under iron-limited conditions and sometimes could provide additional functions, such as pathogenicity, biocontrol, and transport of other metals (16–18). Because of these interesting functions and the greater diversity of secondary siderophores than of pyoverdines, they may better reflect the competition between strains and the fitness of pseudomonads in diverse environments.
In addition to siderophores, the extreme adaptability of pseudomonads is also strongly related to their exquisite regulatory systems. The intensively investigated GacS/GacA two-component system is a global signal transduction system that is highly conserved in Gram-negative bacteria (19). GacS is a sensing transmembrane protein that can respond to yet-to-be-identified environmental signals and activate the cognate response regulator GacA through a phosphorelay mechanism (20, 21). GacA in turn triggers the expression of small regulatory RNA molecules (e.g., RsmY/RsmZ), which titrate translational repressors (e.g., RsmA/RsmE) and relieve the translational inhibition of target mRNAs (22–24). The GacS/GacA two-component system has been demonstrated to be widely required for the production of many secondary metabolites, including extracellular enzymes, virulence factors, and biocontrol factors, which are closely related to adaptability to the environment (19, 24).
Despite the global control of secondary metabolism by GacS/GacA, the relationship between GacS/GacA and siderophores, ubiquitous secondary metabolites, is still unclear. In Pseudomonas fluorescens and Pseudomonas chlororaphis, a defect in gacS-gacA causes increased production of siderophores, including pyoverdine and pyochelin (25–28), together with an increase in the expression of siderophore-related genes, as shown by transcriptome analysis of P. fluorescens (28, 29). In contrast, gacA mutants of Pseudomonas aeruginosa M18 and Pseudomonas marginalis exhibit reduced pyoverdine production (30, 31). Recently, a study showed that the Gac/Rsm signaling network positively controls pyoverdine and pyochelin production in P. aeruginosa PAO1 (32). In summary, previous studies have focused only on the common siderophores pyoverdine and pyochelin, and the effects are elusive and even discordant. Further exploration of the correlation between this global signaling system and more diverse siderophores may better explain bacterial mechanisms of adaptation to the environment.
In this study, we used the high-siderophore-yielding Pseudomonas strain HYS to investigate the correlation between GacS/GacA and siderophores. We found that this strain can simultaneously secrete the fluorescent siderophore pyoverdine and another nonfluorescent siderophore, and they tend to be produced under different iron conditions. Furthermore, the two types of siderophores are differentially regulated by the GacS/GacA system: GacS/GacA positively controls the nonfluorescent siderophore through the Gac/Rsm signaling pathway and a special nfs (nonfluorescent siderophore) cluster, but a defect in gacS-gacA results in increased pyoverdine. The different regulatory effects may allow HYS to adjust siderophore production through this two-component system to adapt to changing environments.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Table 1 lists the bacterial strains and plasmids used in this study, and the oligonucleotides used are given in Table S1 in the supplemental material. Escherichia coli strains were routinely grown in Luria-Bertani (LB) medium at 37°C. Pseudomonas sp. strain HYS and its derivative strains were grown in iron-poor modified King's B (MKB) medium (15 ml/liter glycerol, 2.5 g/liter K2HPO4, pH 7.2, subsequently supplemented with 5 g/liter Casamino Acids and 2.5 g/liter MgSO4 · 7H2O) and LB medium at 30°C. When required, FeSO4 or 8-hydroxyquinoline was added at the indicated concentrations to the MKB medium. Antibiotics, when necessary, were added at the following concentrations: for Pseudomonas sp. HYS and its derivative strains, 50 μg/ml gentamicin, 25 μg/ml chloramphenicol, 50 μg/ml kanamycin, and 50 μg/ml tetracycline; for E. coli, 10 μg/ml gentamicin, 50 μg/ml kanamycin, 10 μg/ml tetracycline, and 100 μg/ml ampicillin.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | λ− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | Laboratory collection |
| MG1655 | F− λ− ilvG rfb-50 rph-1; enterobactin siderophore producer | Laboratory collection |
| S17-1 λpir | thi pro hsdR recA; chromosomal RP4–2; (Tc::Mu) (Km::Tn7) Tpr Spr | 62 |
| Pseudomonas strains | ||
| P. fluorescens CHA0 | Wild-type P. fluorescens CHA0 | 63 |
| P. fluorescens AB92001 | Wild-type P. fluorescens CCTCC AB92001 | CCTCCb |
| P. putida ATCC 12633 | Wild-type P. putida ATCC 12633 | ATCCc |
| P. aeruginosa PAO1 | Wild-type P. aeruginosa ATCC 15692 | 64 |
| Pseudomonas sp. | ||
| HYS | Wild type; Cmr | Laboratory collection |
| HYS1 | ΔpvdA | This study |
| HYS2 | ΔgacS | This study |
| HYS3 | ΔgacA | This study |
| HYS4 | ΔgacS ΔpvdA | This study |
| HYS5 | ΔgacA ΔpvdA | This study |
| HYS6 | ΔgacS complemented with pBBR2-gacS | This study |
| HYS7 | ΔgacS complemented with pBBR1MCS-2 | This study |
| HYS8 | ΔgacA complemented with pBBR2-gacA | This study |
| HYS9 | ΔgacA complemented with pBBR1MCS-2 | This study |
| HYS18 | Δorf1 | This study |
| HYS17 | Δorf2 | This study |
| HYS16 | Δorf3 | This study |
| HYS15 | Δorf4 | This study |
| HYS22 | ΔrsmY | This study |
| HYS23 | ΔrsmZ | This study |
| HYS24 | ΔrsmY ΔrsmZ | This study |
| HYS33 | ΔgacA ΔrsmA ΔrsmE | This study |
| HYS39 | HYS complemented with pBBR1MCS-2 | This study |
| HYS40 | HYS complemented with pBBR2-rsmA | This study |
| HYS41 | HYS complemented with pBBR2-rsmE | This study |
| HYS45 | HYS containing pBBR5Z-Pnfs | This study |
| HYS46 | ΔgacA containing pBBR5Z-Pnfs | This study |
| Plasmids | ||
| pUC18 | Cloning and sequencing vector; Apr | Commercially available |
| pBT20 | Mariner transposon mutagenesis vector; Apr Gmr | 40 |
| pEX18Tc | Gene replacement vector with MCS from pUC18; Tcr oriT+ sacB+ | 45 |
| pEX18Gm | Gene replacement vector with MCS from pUC18; Gmr oriT+ sacB+ | 45 |
| pEX18Tc-pvdA | Gene replacement vector for pvdA | This study |
| pEX18Tc-gacS | Gene replacement vector for gacS | This study |
| pEX18Tc-gacA | Gene replacement vector for gacA | This study |
| pEX18Gm-orf1 | Gene replacement vector for orf1 | This study |
| pEX18Gm-orf2 | Gene replacement vector for orf2 | This study |
| pEX18Gm-orf3 | Gene replacement vector for orf3 | This study |
| pEX18Gm-orf4 | Gene replacement vector for orf4 | This study |
| pEX18Gm-rsmY | Gene replacement vector for rsmY | This study |
| pEX18Gm-rsmZ | Gene replacement vector for rsmZ | This study |
| pEX18Tc-rsmA | Gene replacement vector for rsmA | This study |
| pEX18Gm-rsmE | Gene replacement vector for rsmE | This study |
| pBBR1MCS-2 | Mobilizable broad-host-range cloning vector; Kmr | 34 |
| pBBR2-gacS | 2,783-bp EcoRI-BamHI fragment containing the gacS gene of HYS cloned into pBBR1MCS-2 | This study |
| pBBR2-gacA | 688-bp EcoRI-BamHI fragment containing the gacA gene of HYS cloned into pBBR1MCS-2 | This study |
| pBBR2-rsmA | 214-bp SalI-EcoRI fragment containing the rsmA gene of HYS cloned into pBBR1MCS-2 | This study |
| pBBR2-rsmE | 223-bp SalI-EcoRI fragment containing the rsmE gene of HYS cloned into pBBR1MCS-2 | This study |
| pBBR1MCS-5 | Mobilizable broad-host-range cloning vector; Gmr | 34 |
| pBBR5-T7 | SacI-PvuI fragment containing the T7 promoter removed from pBBR1MCS-5 | This study |
| pBBR5Z | Promoterless lacZ reporter vector; Gmr | This study |
| pBBR5Z-Pnfs | nfs′-′lacZ transcriptional fusion cloned in pBBR5Z; Gmr | This study |
Ap, ampicillin; Gm, gentamicin; Cm, chloramphenicol; Km, kanamycin; Tc, tetracycline; Sp, spectinomycin; Tp, trimethoprim; MCS, multiple-cloning site.
CCTCC, China Center for Type Culture Collection.
ATCC, American Type Culture Collection.
DNA manipulations and plasmid construction.
Standard molecular methods were used for PCR, agarose gel electrophoresis, restriction enzyme digestion, and transformation (33). All of the reaction enzymes were purchased from Fermentas. Plasmids for laboratory procedures were purified using a Tianprep Mini Plasmid Kit (Tiangen). DNA fragments were purified from agarose gels with a gel extraction kit (Omega). Chromosomal DNA from Pseudomonas spp. was prepared with a Genomic DNA Purification Kit (Promega).
The promoterless lacZ reporter vector pBBR5Z was constructed from the broad-host-range vector pBBR1MCS-5 (34). The plasmid pBBR1MCS-5 was digested with SacI and PvuI to remove the T7 promoter. The resulting products were treated with T4 DNA polymerase and blunt-end ligated to form pBBR5-T7. Then, the 3-kb fragment containing the lacZ gene was amplified by PCR using the primers lacZ-1 and lacZ-2 and cloned into the XhoI-ApaI sites of pBBR5-T7 to generate pBBR5Z.
Fragments containing the promoters and several amino acids of the genes studied were amplified and ligated into pBBR5Z, yielding translational-fusion vectors. Complementation and overexpression plasmids were constructed by cloning the Shine-Dalgarno (SD) sequences and open reading frames (ORFs) of target genes into the broad-host-range vector pBBR1MCS-2 (34).
Siderophore measurements.
The quantities of siderophores were measured using the following methods. On universal chrome azurol S (CAS) agar plates (35), the siderophores secreted by the bacteria can chelate iron from the medium, resulting in a color shift from blue to orange. In our study, 10 μl of overnight MKB culture (optical density at 600 nm [OD600] = 1.0) was dropped onto a CAS agar plate, and siderophore production was assessed by the formation of chelated halos after incubation for 24 h at 30°C. For liquid MKB medium, the appropriate dilution of the filtered culture supernatant was mixed with an equal volume of CAS assay solution (35) using double-distilled water (ddH2O) as a reference. After 1 h of incubation at room temperature, the reference and sample absorbances at 630 nm were measured. Siderophore units were calculated using the difference in absorbance between the reference and the sample according to a previously used equation (36).
The siderophore pyoverdine was detected by its characteristic absorbance at 405 nm in liquid culture supernatant and was observed under UV light by the formation of a fluorescent zone around the colony. Pyoverdine production in liquid MKB medium was estimated using a fluorescence spectrometer (emission at 460 nm after excitation at 405 nm) (37) and normalized to the OD600 of the bacterial culture. The presence of catechol-type and/or hydroxamate-type siderophores in cell-free supernatants of MKB cultures was determined using the Arnow (38) and modified Csáky (39) colorimetric assays, respectively. Cell-free culture supernatants of E. coli MG1655 (which produces enterobactin) and P. aeruginosa PAO1 (which produces pyoverdine) were used as positive controls in the Arnow and modified Csáky tests, respectively. To obtain the characteristic peaks of the siderophores, filtered supernatants of 24-h MKB cultures were normalized to an OD600 of 0.5. Then, the absorption spectra were measured every 0.5 nm using a UV-2550 spectrophotometer (Shimadzu).
Transposon mutagenesis, screening of siderophore-reduced mutants, and insertion site determination.
Bacterial conjugation was performed to introduce the minitransposon vector pBT20 into Pseudomonas sp. HYS according to the method of Kulasekara et al. (40). Then, the siderophore-reduced mutants were screened according to the orange halos produced on CAS agar plates. Transposon-flanking sequences of these mutants were obtained by thermal asymmetric interlaced (TAIL)-PCR (41), arbitrarily primed PCR (42), and rescue cloning (43) methods. TAIL-PCR was carried out as described by Liu and Whittier (41) using U1, U2, and U3 as the specific primers for three sequential PCRs and AD-1, AD-2, AD-3, and AD-4 as the arbitrary degenerate primers. Arbitrarily primed PCR was conducted in two amplification steps using a previously described method (42). The primer pairs U1 plus ARB1 and U2 plus ARB2 were used for the first and second reactions, respectively. For rescue cloning, genomic DNA of the mutant was digested with SalI, ligated into pUC18, and transformed into E. coli DH5α. To isolate clones harboring the transposon, transformants were selected on LB agar containing gentamicin. The products obtained using the three methods were all sequenced using the transposon-derived primer U3. Each sequence was aligned to the HYS whole-genome shotgun contigs in the NCBI database (accession no. NZ_AJJP00000000) (44), and the interrupted genes were identified. Sequence analysis and homology searches were performed using the BLAST tools at NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). For protein motif searching, the Conserved Domain Search at NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and SMART (http://smart.embl-heidelberg.de/) programs were used.
Construction of in-frame deletion mutants.
To construct deletion plasmids, PCR products containing 500- to 600-bp regions upstream and downstream of target genes, and several nucleotides of the ORFs, were digested with primer-specific restriction enzymes and ligated into the pEX18Tc or pEX18Gm gene replacement vector (45). The correct recombinant plasmids were verified by sequencing and introduced into HYS and its derivative strains via conjugation from E. coli S17-1 (λpir). Single recombinants were selected based on resistance to two antibiotics. These transformants were further cultured overnight in 5 ml of liquid LB medium without antibiotics, allowing a second allelic exchange to occur. Appropriately diluted culture was spread onto LB agar plates supplemented with 5% sucrose and incubated at 30°C. The correct gene deletion mutants were selected from these isolates through further verification by PCR and sequencing.
RNA isolation and reverse transcription (RT)-PCR.
Total RNA was extracted from the culture of Pseudomonas sp. HYS grown in MKB at an OD600 of 0.8 (exponential phase) using TRIzol reagent (Ambion) according to the manufacturer's instructions. Residual DNA was removed, and the RNA was then reverse transcribed to generate cDNA using a PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa). To determine the transcriptional relationships of the gene cluster, primer pairs connecting neighboring ORFs were used. As a control, chromosomal DNA and total RNA with genomic DNA removed were used in the same PCRs.
β-Galactosidase assays.
Pseudomonas cells were grown with shaking in MKB medium or MKB medium supplemented with FeSO4 at 30°C. β-Galactosidase activities were determined by the Miller method (46), using cells permeabilized with 0.1% sodium dodecyl sulfate and chloroform, and normalized to the OD600 of the bacterial culture before assays.
Accession numbers.
The GenBank accession numbers for the proteins GacS, GacA, PvdA, RsmA, RsmE, ORF1, ORF2, ORF3, and ORF4 from HYS are WP_010220373, WP_010221877, WP_010222460, WP_010226667, WP_010223936, WP_010220692, WP_010220691, WP_010220690, and WP_010220689, respectively. The small RNAs (sRNAs) rsmY and rsmZ are located in AJJP01000026 (31812 to 31931) and AJJP01000134 (28554 to 28682), respectively.
RESULTS
Characteristics of siderophores of Pseudomonas sp. strain HYS.
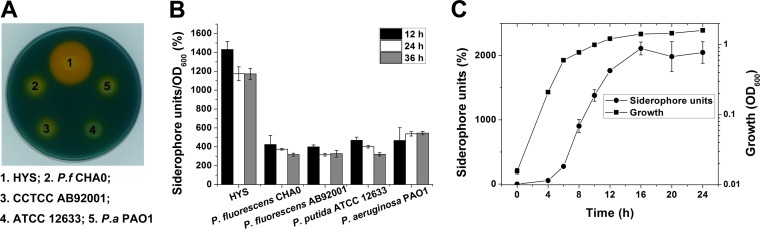
HYS, isolated from the water of East Lake in Wuhan, China, is a siderophore-producing Pseudomonas strain. As shown in Fig. 1A, compared with four common Pseudomonas strains, HYS produced significantly larger chelated halos on CAS agar plates. Consistent with this, the amount of siderophores in the liquid culture supernatant of HYS was approximately two to three times higher than in those of other Pseudomonas strains assayed (Fig. 1B). In conclusion, HYS has a greater ability to secrete siderophores. From the siderophore secretion curve, we found that siderophores of HYS accumulate from the early exponential phase and reach a peak during the late exponential phase (Fig. 1C).
FIG 1.
Comparison of siderophore production in Pseudomonas sp. HYS and other pseudomonads. (A) Siderophore production in the five strains was detected on CAS agar plates. (B) Siderophore production in the five strains in liquid MKB medium was determined as siderophore units (percent) by the CAS liquid assay. (C) Curves of growth and siderophore accumulation of HYS. The error bars indicate standard deviations.
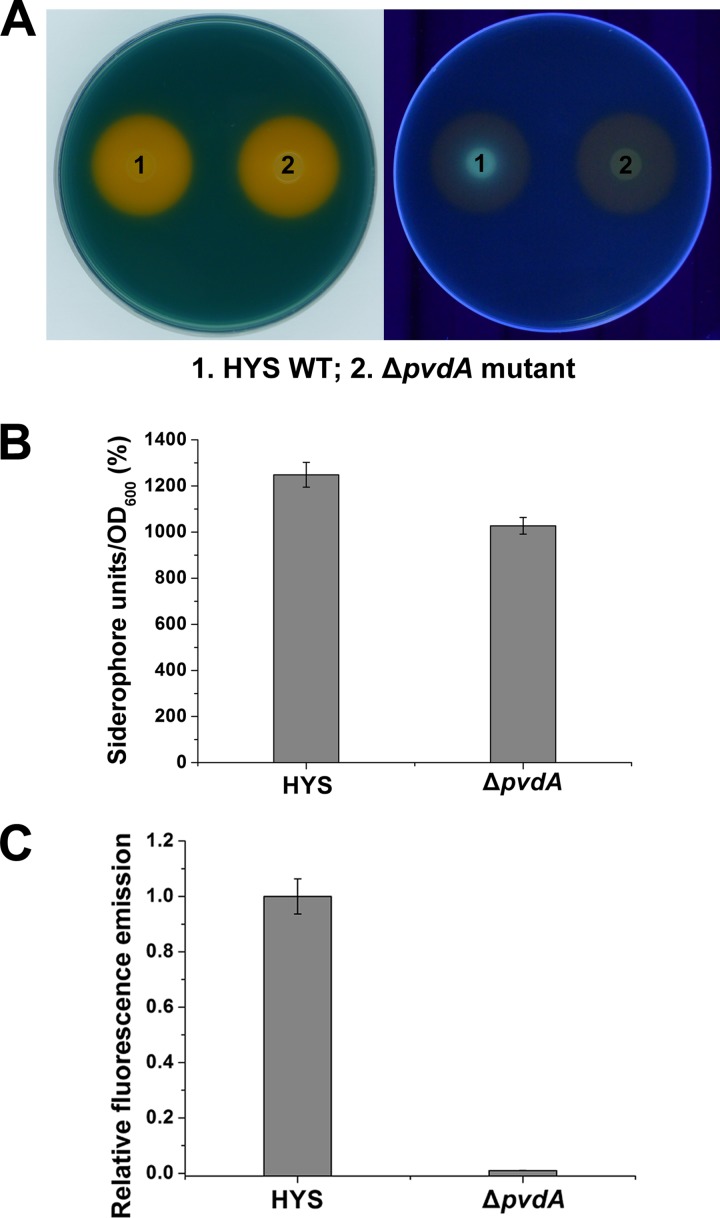
The types of siderophores produced by HYS were further analyzed. In pseudomonads, the fluorescent pyoverdine is the most common siderophore. By sequence analysis, we found the biosynthetic locus of pyoverdine in the HYS genome (44), which implied the production of fluorescent pyoverdine in HYS. Under UV light, the siderophores produced by HYS indeed exhibited yellow-green fluorescence (Fig. 2A), in accordance with that characteristic of pyoverdine (12). To confirm the production of pyoverdine in HYS, we constructed a deletion mutant of pvdA in which pyoverdine production should be abolished. As expected, the yellow-green fluorescence completely disappeared, and pyoverdine was not detected in the liquid culture supernatant of the pvdA mutant (Fig. 2A and C). These results demonstrate that HYS indeed produces pyoverdine.
FIG 2.
Effects of pvdA on siderophore production in Pseudomonas sp. HYS. (A) Siderophore production in wild-type (WT) HYS and a ΔpvdA mutant was detected on CAS agar plates under normal light (left) and under UV light (right). (B) Siderophore production in wild-type HYS and a ΔpvdA mutant in liquid MKB medium was determined as siderophore units (percent) by the CAS liquid assay. (C) Pyoverdine production by 24-h MKB cultures of wild-type HYS and the ΔpvdA mutant. The error bars indicate standard deviations.
It should also be noted that the fluorescence of the chelated halo produced by HYS was faint, and the yield of pyoverdine was low compared with the total siderophore level (Fig. 2A). Furthermore, overall siderophore production in the pvdA mutant was similar to that of the wild type (Fig. 2A and B). Taken together, these results suggest that HYS simultaneously secretes the fluorescent pyoverdine and a nonfluorescent siderophore(s) and that the latter is a major contributor to the high siderophore yield. This situation is quite different from that of many other pseudomonads, in which pyoverdines account for the majority of the total siderophores, suggesting special siderophore production in HYS.
Isolation and characterization of siderophore-reduced transposon mutants.
To identify the siderophore-related genes in HYS, transposon mutagenesis was carried out using the vector pBT20 (40). By screening transconjugants based on the universal CAS method (35), 14 mutants that exhibited strongly decreased siderophore yields were obtained (listed in Table 2).
TABLE 2.
Characteristics of the 14 transposon mutants of HYS
| Mutanta | Insertion site | Insertion gene functionb | Relative siderophore productionc |
|
|---|---|---|---|---|
| CAS plate | Liquid medium | |||
| M32 | Scaffold1 91950–91951 | Pyruvate decarboxylase | 0.31 ± 0.01 | 0.10 ± 0.00 |
| M58 | Scaffold1 93834–93835 | Phenylacetate-CoA ligase | 0.20 ± 0.00 | 0.16 ± 0.01 |
| M30 | Scaffold1 94178–94179 | Phenylacetate-CoA ligase | 0.16 ± 0.01 | 0.21 ± 0.00 |
| M38 | Scaffold1 94422–94423 | Phenylacetate-CoA ligase | 0.17 ± 0.00 | 0.17 ± 0.01 |
| M16 | Scaffold1 94637–94638 | Phenylacetate-CoA ligase | 0.15 ± 0.01 | 0.17 ± 0.00 |
| M52 | Scaffold1 94637–94638 | Phenylacetate-CoA ligase | 0.14 ± 0.01 | 0.16 ± 0.00 |
| M12 | Scaffold1 95250–95251 | Acyl-CoA dehydrogenase | 0.17 ± 0.00 | 0.17 ± 0.01 |
| M55 | Scaffold1 95252–95253 | Acyl-CoA dehydrogenase | 0.17 ± 0.00 | 0.16 ± 0.01 |
| M48 | Scaffold1 95451–95452 | Acyl-CoA dehydrogenase | 0.16 ± 0.02 | 0.12 ± 0.00 |
| M5 | Scaffold1 96196–96197 | Thioesterase | 0.37 ± 0.01 | 0.17 ± 0.01 |
| M13 | Scaffold1 96357–96358 | Thioesterase | 0.36 ± 0.00 | 0.16 ± 0.00 |
| M2 | Scaffold4 115281–115282 | GacA | 0.02 ± 0.00 | 0.17 ± 0.01 |
| M35 | Scaffold33 33826–33827 | GacS | 0.02 ± 0.00 | 0.16 ± 0.00 |
| M11 | Scaffold33 33887–33888 | GacS | 0.02 ± 0.00 | 0.13 ± 0.00 |
The mutants are arranged according to their relative positions in the genome.
Gene designation and proposed functions are based on the annotated genome sequence from Pseudomonas sp. HYS.
Relative siderophore production is expressed as the ratio of mutants to the wild-type HYS. On CAS plates, it was determined as the semidiameter of chelated halos; in liquid medium, it was determined as siderophore units (%) and normalized to the OD600 of the bacterial culture in MKB medium (see Materials and Methods for details). Each value is the average of three different cultures ± standard deviation.
Combining TAIL-PCR, arbitrarily primed PCR, and rescue cloning approaches, we obtained the transposon-flanking sequences of all 14 mutants. Comparison with the draft genome sequence of HYS (44) showed that the insertion sites represent six different genes. Three of these mutants (M35, M11, and M2) showed insertions within gacS or gacA, and the other 11 mutants had interruptions in four ORFs arranged sequentially in the genome (Table 2 shows the details). gacS and gacA form the global two-component transduction system that is responsible for the production of many secondary metabolites (19, 24). The four neighboring ORFs appear to constitute an operon, as inferred from their arrangement and the distances between them.
GacS/GacA has different effects on nonfluorescent and fluorescent siderophores.
It has been proposed that the two-component regulatory system GacS/GacA is related to the production of siderophores, but its effect is debatable and the mechanism remains unclear. In the case of HYS, the gacS-gacA transposon mutants produced markedly decreased total siderophore levels (Table 2). As HYS produces the fluorescent pyoverdine and the nonfluorescent siderophore(s), we constructed gacS-gacA in-frame deletion mutants to investigate the correlation between the two genes and the two types of siderophores in HYS.
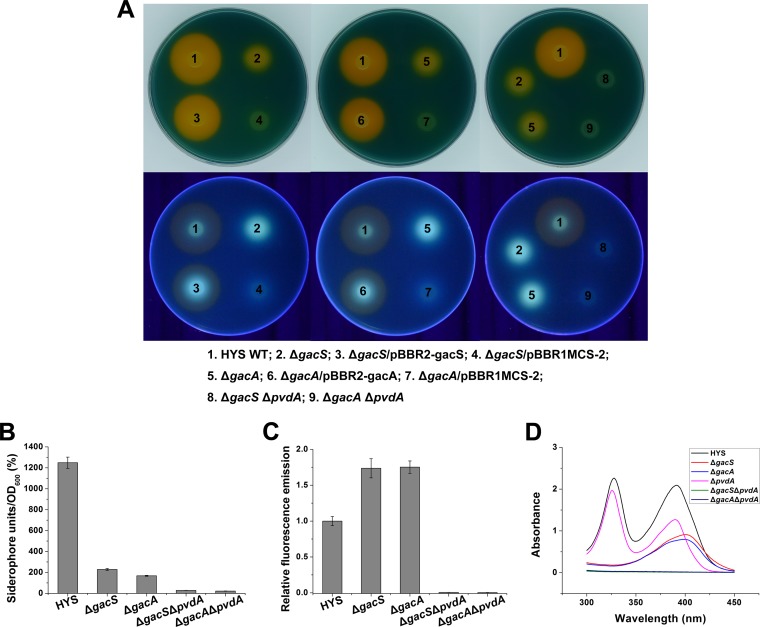
As shown in Fig. 3A, the gacS-gacA mutants showed significantly reduced total amounts of siderophores, which the complemented strains could restore to wild-type levels. Furthermore, the reduced total siderophore levels may be attributable to a marked reduction in the level of the nonfluorescent siderophore; fluorescent-siderophore production did not decrease but increased slightly compared with the wild type. Consistent with these results, the total amount of siderophores in gacS-gacA mutants drastically decreased to ∼15%, but pyoverdine production increased by ∼75% compared with the wild type in the liquid MKB medium (Fig. 3B and C).
FIG 3.
Effects of gacS-gacA on siderophore production in Pseudomonas sp. HYS. (A) Siderophore production in wild-type HYS and its derivative strains was detected on CAS agar plates under normal light (top) and under UV light (bottom). (B) Siderophore production in wild-type HYS and the ΔgacS, ΔgacA, ΔgacS ΔpvdA, and ΔgacA ΔpvdA mutants in liquid MKB medium was determined as siderophore units (percent) by the CAS liquid assay. (C) Pyoverdine production by 24-h MKB cultures of wild-type HYS and ΔgacS, ΔgacA, ΔgacS ΔpvdA, and ΔgacA ΔpvdA mutants. (D) Absorption spectra of filtered supernatants of 24-h MKB cultures of wild-type HYS and ΔgacS, ΔgacA, ΔpvdA, ΔgacS ΔpvdA, and ΔgacA ΔpvdA mutants. The error bars indicate standard deviations.
To further explore the effect of gacS-gacA on the two siderophores, the absorption spectra of filtered culture supernatants of HYS and its mutants were determined. HYS and the pvdA mutant showed two characteristic peaks at 330 nm and 392 nm (Fig. 3D). As the pvdA mutant secretes only the nonfluorescent siderophore, the peaks at 330 nm and 392 nm are the characteristic peaks of the nonfluorescent siderophore. Based on these characteristic peaks, the effect of gacS-gacA on the nonfluorescent siderophore was examined. For the gacS-gacA mutants, the two characteristic peaks of the nonfluorescent siderophore disappeared, and a 405-nm peak, which is reported to be the characteristic peak of pyoverdine (37), emerged instead. These results suggest that the gacS-gacA mutants produce only pyoverdine and not the nonfluorescent siderophore. To confirm this hypothesis, the pyoverdine-related gene pvdA was deleted based on gacS and gacA mutations. Double gacS pvdA and gacA pvdA deletion mutants did not produce any siderophores either on CAS agar plates or in liquid medium (Fig. 3A to C), and all absorbance peaks above 300 nm disappeared (Fig. 3D). All of these results demonstrate that gacS-gacA positively regulates the nonfluorescent siderophore, designated NFS, but has a negative effect on pyoverdine in HYS.
The two types of siderophores respond to external iron conditions differently.
Siderophores are produced under iron-depleted conditions and can be repressed by high iron levels (47). We examined the responses of the two siderophores to iron conditions by adding iron and the iron chelator 8-hydroxyquinoline to MKB medium to create iron-replete and extremely iron-depleted conditions, respectively.
As shown in Fig. 4A, the supernatant of HYS showed strong siderophore absorption peaks in MKB medium, suggesting the production of siderophores. With increased iron concentrations, the absorption peaks of the siderophores declined significantly and finally disappeared when the supplemented iron concentration reached 10 μM, indicating the suppression of siderophore production under high iron concentrations. The responses of the two siderophores to high iron levels were further assessed. No pyoverdine could be detected with a final FeSO4 concentration of 1 μM (Fig. 4B). However, NFS production was completely repressed only when the supplemented iron concentration was increased to 10 μM (Fig. 4B). We conclude from these data that HYS produces NFS under relatively high-iron conditions.
FIG 4.
Regulation of the production of the two siderophores by iron. (A and C) Influence of FeSO4 (A) and 8-hydroxyquinoline (C) on the absorption spectra of filtered supernatants of 24-h MKB cultures of Pseudomonas sp. HYS. (B) Effects of FeSO4 on the production of pyoverdine (■) and NFS (□). (D) Effects of 8-hydroxyquinoline on the production of pyoverdine (■) and NFS (□). The error bars indicate standard deviations.
In the case of iron limitation, with an increasing 8-hydroxyquinoline level, the peak at 330 nm gradually declined (Fig. 4C), showing that levels of NFS decreased when iron availability decreased (Fig. 4D). In contrast, pyoverdine levels increased slightly under the same conditions (Fig. 4D). When the final concentration of 8-hydroxyquinoline was increased to 10 μg/ml, the characteristic absorption peak changed to 405 nm (Fig. 4C), suggesting the secretion of only pyoverdine. Collectively, these results indicate that HYS produces NFS under mildly iron-limited conditions and fluorescent pyoverdine under extremely iron-restricted conditions.
The nfs cluster is essential for NFS production and is under the control of GacS/GacA.
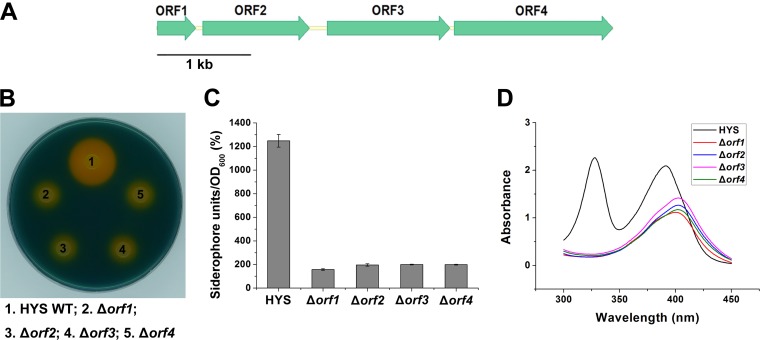
In addition to gacS and gacA, the four genes identified in our transposon mutagenesis also had remarkable effects on siderophore production (Table 2). The four ORFs are arranged sequentially in a 5-kb region of the HYS genome and display the same transcription polarity. Figure 5A shows the arrangement of the four ORFs, which we named ORF1 to ORF4 according to the transcriptional direction.
FIG 5.
Effects of the nfs cluster on siderophore production in Pseudomonas sp. HYS. (A) Representation of the nfs cluster. The arrows and their direction indicate the locations and direction of transcription of predicted genes, respectively. (B) Siderophore production in wild-type HYS and the nfs mutants was detected on CAS agar plates. (C) Siderophore production in wild-type HYS and the nfs mutants in liquid MKB medium was determined as siderophore units (percent) by the CAS liquid assay. (D) Absorption spectra of filtered supernatants of 24-h MKB cultures of wild-type HYS and the nfs mutants. The error bars indicate standard deviations.
To interpret their properties, we performed homology searches and conserved-domain analyses of the four predicted translational products (the results are shown in Table 3). The deduced amino acid sequence of ORF1 showed high homology with thioesterase superfamily proteins and was in the hot_dog superfamily. The ORF2 protein shared remarkable similarity with acyl coenzyme A (CoA) dehydrogenases, containing the N-terminal, middle, and C-terminal domains. The protein encoded by ORF3 possessed the AMP-binding domain and was highly similar to phenylacetate-CoA ligases. The putative product of ORF4 displayed homology with pyruvate decarboxylases, with thiamine pyrophosphate (TPP) binding domains in the N- and C-terminal parts of the sequence. We further compared the entire region containing the four ORFs to the nucleotide database collection at NCBI using BLAST. Unexpectedly, such a string of genes was not found in other sequenced bacteria.
TABLE 3.
Characteristics of genes in the nfs clustera
| Gene | No. of amino acid residues | Conserved domain(s) (position) | Superfamily | Function(s) | Organism | % similarity/% identity | % coverage | Significance (E value) |
|---|---|---|---|---|---|---|---|---|
| orf1 | 137 | 4HBT (20–102) | Hot_dog | Thioesterase superfamily protein TdaD | Phaeobacter gallaeciensis | 83/73 | 99 | 6e−68 |
| orf2 | 380 | Acyl-CoA_dh_N (6–116) | ACAD | Butyryl-CoA dehydrogenase | Burkholderia vietnamiensis | 74/61 | 98 | 2e−161 |
| Acyl-CoA_dh_M (119–170) | ||||||||
| Acyl-CoA_dh_ 1 (225–374) | ||||||||
| orf3 | 437 | AMP-binding domain (67–354) | AFD_class_I | Phenylacetate-CoA ligase | Pseudomonas sp. strain GM49 | 85/72 | 99 | 0 |
| orf4 | 563 | TPP_enzyme_N (3–178) | TPP_enzymes | Putative pyruvate decarboxylase | Burkholderia xenovorans | 69/55 | 98 | 0 |
| TPP_enzyme_M (197–332) | ||||||||
| TPP_enzyme_C (382–528) |
Similarity values are for the most similar proteins, determined by BLASTP analysis, whose functions and source organisms are also shown.
To investigate the effect of this region on siderophores of HYS, we performed gene deletions for each ORF. Similar to their transposon mutants, the individual deletions of ORFs 1 to 4 resulted in significantly reduced siderophore production both on CAS agar plates and in liquid medium (Fig. 5B and C). Moreover, these mutants showed a 405-nm absorption peak (Fig. 5D), suggesting the presence of pyoverdine and the absence of NFS. These results indicate that the region containing ORF1 to ORF4 is essential for NFS production. We therefore named it the nfs cluster.
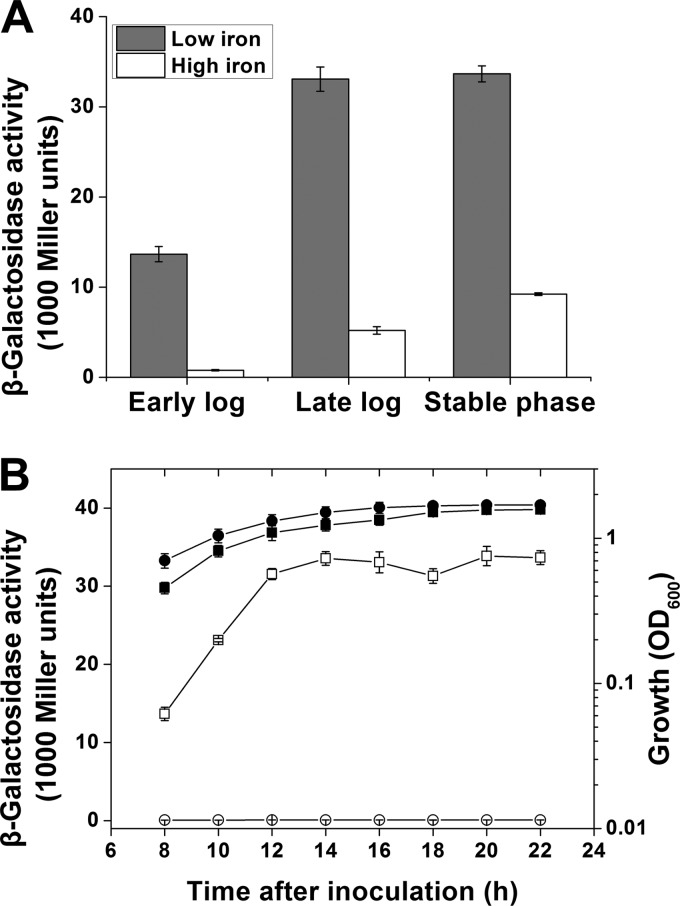
We further examined the correlation between expression of the nfs cluster and the iron concentration. Because of the tandem arrangement and short intergenic regions, we first confirmed the transcriptional relationship among the four ORFs by RT-PCR. The results showed that HYS cDNA generated products similar to those obtained using genomic DNA as the template for all three primer pairs (Fig. 6), suggesting cotranscription of the four ORFs. Furthermore, when we searched for and analyzed potential promoters in this region, we found only a predicted promoter in front of ORF1. This promoter sequence was used to construct an nfs′-′lacZ translational fusion to assess expression of the nfs cluster. We observed significantly decreased levels of β-galactosidase under high-iron conditions (30 μM) relative to those under low-iron conditions (Fig. 7A), indicating that expression of the nfs cluster is suppressed by iron. This suggests that HYS controls NFS production by regulating expression of the nfs cluster under different iron conditions.
FIG 6.
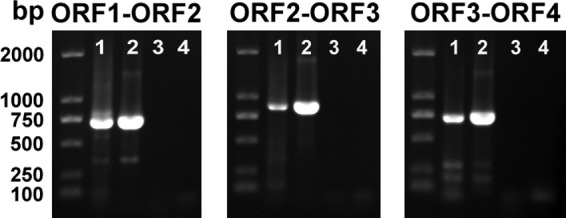
RT-PCR of RNA extracted from Pseudomonas sp. HYS with ORF connecting primer pairs (orf1-orf2RT-1 plus orf1-orf2RT-2, orf2-orf3RT-1 plus orf2-orf3RT-2, and orf3-orf4RT-1 plus orf3-orf4RT-2). The different lanes represent different templates: lanes 1, cDNA; lanes 2, genomic DNA as a positive control; lanes 3, RNA without reverse transcription as a negative control; lanes 4, ddH2O as a blank control.
FIG 7.
Regulation of the nfs cluster expression in Pseudomonas sp. HYS. (A) Expression of a plasmid-encoded nfs′-′lacZ translational fusion in Pseudomonas sp. HYS was determined under low-iron (MKB medium) and high-iron (MKB supplemented with 30 μM FeSO4) conditions. (B) Expression of a plasmid-encoded nfs′-′lacZ translational fusion and growth were determined in wild-type HYS (squares) (HYS/pBBR5Z-Pnfs) and in a ΔgacA mutant (circles) (ΔgacA/pBBR5Z-Pnfs). Open symbols, β-galactosidase activity; solid symbols, OD600. Each value is the average from three different cultures ± standard deviation.
As gacS-gacA and the nfs cluster are both essential for NFS production in HYS, we further tested whether expression of the nfs cluster is regulated by gacS-gacA. To address this question, expression of the nfs cluster in the wild type and the gacA mutant was measured using the nfs′-′lacZ translational fusion. We found that expression of the nfs′-′lacZ fusion throughout the growth cycle was about 167- to 374-fold higher in wild-type HYS than in the gacA mutant (Fig. 7B). Moreover, its expression remained at a constant low level in the gacA mutant, indicating effective inhibition of the nfs cluster because of the gacA defect. We conclude from the above-mentioned results that gacS-gacA positively controls NFS through the nfs cluster in HYS.
GacS/GacA positively regulates NFS through the Gac/Rsm signaling cascade.
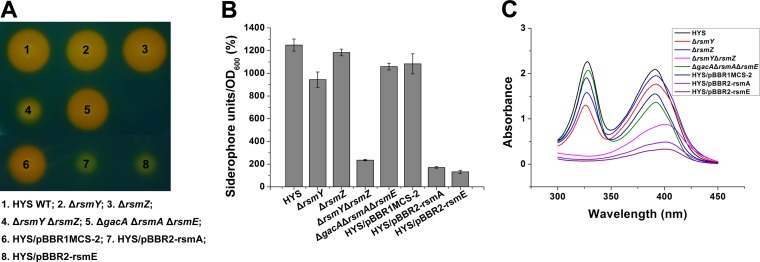
It has been documented that the conserved signal transduction system GacS/GacA regulates many target genes by activating noncoding small RNAs, such as RsmY and RsmZ, which can combine the posttranscriptional repressors RsmA/RsmE to negate their inhibition of target genes (24, 48, 49). To reveal whether gacS-gacA regulates NFS of HYS by the Gac/Rsm pathway, a series of mutant strains was constructed. As expected, the rsmY rsmZ double mutant showed a notable reduction in the production of siderophores both on CAS agar plates and in liquid MKB medium (Fig. 8A and B). Moreover, the absorption maximum of the double mutant was at 405 nm (Fig. 8C), in line with the properties of gacS-gacA mutants. The siderophore production and absorption spectra of the rsmY and rsmZ single mutants were both similar to those of the wild type (Fig. 8). These data convincingly demonstrate that the effect of gacS-gacA on NFS is mediated by two small RNAs (RsmY and RsmZ), whose roles appear to be redundant (22).
FIG 8.
The regulation of NFS by gacS-gacA is through the Gac/Rsm signaling cascade. (A) Siderophore production in wild-type HYS and the genetically modified strains was detected on CAS agar plates. (B) Siderophore production in wild-type HYS and the derivative strains in liquid MKB medium was determined as siderophore units (percent) by the CAS liquid assay. (C) Absorption spectra of filtered supernatants of 24-h MKB cultures of wild-type HYS and the derivative strains. The error bars indicate standard deviations.
HYS also has two posttranscriptional repressors: RsmA and RsmE. In the gacS-gacA mutants, in which no RsmY or RsmZ existed to inhibit their functions, RsmA and RsmE repressed the expression of downstream genes, while in the gacA rsmA rsmE triple mutant, siderophore production was restored to the wild-type level (Fig. 8A and B). The characteristic peaks of the triple-deletion mutant were at 330 nm and 392 nm, indicating restoration of NFS production (Fig. 8C). Furthermore, overexpression of rsmA or rsmE in the wild type greatly reduced the siderophore level, and the characteristic peak was changed to 405 nm (Fig. 8). These results suggest that rsmA and rsmE inhibit NFS production.
Taken together, the roles of RsmY/RsmZ/RsmA/RsmE in HYS indicate that regulation of NFS by gacS-gacA agrees with the Gac/Rsm model.
DISCUSSION
Microorganisms secrete a great variety of siderophores to acquire the iron that is critical for their survival. In changing environments, accurate regulation of siderophore production is very important for bacteria to maintain intracellular iron homeostasis and enhance their adaptability and competitiveness (3, 10, 50). In this study, we investigated the relationship between the global regulatory system GacS/GacA and siderophore production in the high-siderophore-yielding Pseudomonas sp. HYS. For the two siderophores secreted under different iron conditions, GacS/GacA positively regulates the production of the nonfluorescent siderophore NFS through the Gac/Rsm signaling cascade but has a negative impact on the fluorescent siderophore pyoverdine. Through this regulatory strategy, HYS can adapt to diverse environments by modulating the production of the two siderophores.
Pseudomonas sp. HYS has a markedly greater ability to produce siderophores than some common pseudomonads. Like many other Pseudomonas species, HYS produces the fluorescent siderophore pyoverdine. Notably, it secretes a significant amount of NFS, which accounts for the majority of the high siderophore yield. Moreover, siderophores of HYS are secreted during the exponential phase. This secretion at a relatively early stage of growth, in combination with the significantly higher yield of siderophores, may facilitate iron acquisition by HYS in the presence of other bacteria. We preliminarily investigated the chemical nature of NFS. The cell-free supernatant of the pvdA mutant did not give positive reactions in either the Arnow or modified Csáky assay (see Fig. S1 in the supplemental material), indicating that NFS is not a classical catechol-type or hydroxamate-type siderophore. Its characteristic peaks at 330 nm and 392 nm indicate the presence of a relatively large conjugated system (51), implying that NFS might have a unique structure.
Through the genetics experiments, a four-gene iron-regulated cluster (the nfs cluster) was determined to be related to NFS production. To our knowledge, phenylacetate-CoA ligase and pyruvate decarboxylase encoded by ORF3 and ORF4 have not been reported to be related to siderophore biosynthesis. The ORF2 product, acyl-CoA dehydrogenase, has only been demonstrated to be implicated in mycobactin synthesis in Mycobacterium tuberculosis (52). Regarding the ORF1 product thioesterase, many nonribosomal peptide synthetases (NRPSs) required for siderophore synthesis possess a C-terminal thioesterase domain to release the peptide from the enzyme by cyclization or hydrolysis (53). Because of the special gene arrangement and enzyme functions of the nfs cluster, it is probably involved in a previously unreported mode of siderophore biosynthesis.
Further illustrating the roles of the two siderophores produced by HYS, we found that in addition to providing considerable iron-scavenging capacity, they also have individual divisions. NFS tends to be produced under less severely iron-depleted conditions, while pyoverdine is produced under extremely iron-depleted conditions (Fig. 4). This is consistent with the fact that a large amount of NFS is secreted at an early growth stage, when iron limitation is usually not serious. A similar situation has been observed in Erwinia chrysanthemi, which produces two siderophores with different affinities for iron, achromobactin and chrysobactin, in sequential growth phases reflecting different iron conditions (54). The coexistence and complementation of the two different siderophores may enable bacteria to adapt to a wider and more variable external iron environment. In addition, the expression of siderophore systems is believed to require a set of synthesis, assembly, and transport machineries to be triggered and thus consumes a lot of energy (53). The fact that HYS produces different siderophores under different iron conditions indicates that HYS may produce the siderophore that is most suitable for the iron status and thereby avoid wasting energy.
Since two types of siderophores exist, achieving their precise regulation seems to be especially important for HYS. As a master regulatory system, GacS/GacA has been proposed to affect siderophores in other Pseudomonas species, but the effects are discordant. However, in the same strain, GacS/GacA exerts a concordant impact on its multiple siderophores (25, 32). The situation is quite special in HYS. GacS/GacA has different effects on the two siderophores of HYS: its inactivation leads to the loss of NFS but to a slight increase in pyoverdine. The differential effects may be connected with changes in the expression of the two siderophores depending on the iron status. By differential regulation of the two siderophores, HYS may be able to adjust their production according to the environmental iron level. In addition, because GacS/GacA is influenced by many environmental signals, in combination with the spontaneous mutation of gacS-gacA observed in P. fluorescens (26), its state seems to be susceptible. In HYS, being differentially regulated by GacS/GacA, the two siderophores may function in a complementary manner according to the activation state of GacS/GacA, which guarantees stable iron uptake capacity.
In summary, because of the higher siderophore yield and the differential effects of GacS/GacA on the two different siderophores, HYS can survive under a wider range of conditions and adapt to a changing environment. Together with the results for some other Pseudomonas species, our data suggest that GacS/GacA is likely to have universal regulatory roles for siderophores. However, the effects differ between strains because of diverse ecological habitats and genetic backgrounds. Additionally, GacS/GacA and siderophores both have cross talk with many other pathways, such as quorum sensing (55, 56), biofilm formation (57–59), and stress response (19, 60, 61). Further studies on the relationships among GacS/GacA, siderophores, and other factors in diverse strains will provide more insights into the strategies through which bacteria adapt to a changing environment.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kangmin Duan for kindly providing the plasmid.
This work was supported by the National Basic Research Program of China (973 Program, no. 2013CB933904), the National Natural Science Foundation of China (21272182), the National Fund for Fostering Talents of Basic Sciences (J1103513), and the Laboratory (Innovative) Research Fund of Wuhan University.
Footnotes
Published ahead of print 30 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01756-14.
REFERENCES
- 1.Miethke M, Marahiel MA. 2007. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71:413–451. 10.1128/MMBR.00012-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cassat JE, Skaar EP. 2013. Iron in infection and immunity. Cell Host Microbe 13:509–519. 10.1016/j.chom.2013.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews SC, Robinson AK, Rodríguez-Quiñones F. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215–237. 10.1016/S0168-6445(03)00055-X [DOI] [PubMed] [Google Scholar]
- 4.Imlay JA. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77:755–776. 10.1146/annurev.biochem.77.061606.161055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hantke K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172–177. 10.1016/S1369-5274(00)00184-3 [DOI] [PubMed] [Google Scholar]
- 6.Braun V, Killmann H. 1999. Bacterial solutions to the iron-supply problem. Trends Biochem. Sci. 24:104–109. 10.1016/S0968-0004(99)01359-6 [DOI] [PubMed] [Google Scholar]
- 7.Braun V, Hantke K. 2011. Recent insights into iron import by bacteria. Curr. Opin. Chem. Biol. 15:328–334. 10.1016/j.cbpa.2011.01.005 [DOI] [PubMed] [Google Scholar]
- 8.Neilands JB. 1995. Siderophores: structure and function of microbial iron transport compounds. J. Biol. Chem. 270:26723–26726. 10.1074/jbc.270.45.26723 [DOI] [PubMed] [Google Scholar]
- 9.Cornelis P, Matthijs S. 2002. Diversity of siderophore-mediated iron uptake systems in fluorescent pseudomonads: not only pyoverdines. Environ. Microbiol. 4:787–798. 10.1046/j.1462-2920.2002.00369.x [DOI] [PubMed] [Google Scholar]
- 10.Cornelis P. 2010. Iron uptake and metabolism in pseudomonads. Appl. Microbiol. Biotechnol. 86:1637–1645. 10.1007/s00253-010-2550-2 [DOI] [PubMed] [Google Scholar]
- 11.Visca P, Imperi F, Lamont IL. 2007. Pyoverdine siderophores: from biogenesis to biosignificance. Trends Microbiol. 15:22–30. 10.1016/j.tim.2006.11.004 [DOI] [PubMed] [Google Scholar]
- 12.Meyer J-M. 2000. Pyoverdines: pigments, siderophores and potential taxonomic markers of fluorescent Pseudomonas species. Arch. Microbiol. 174:135–142. 10.1007/s002030000188 [DOI] [PubMed] [Google Scholar]
- 13.Cox CD, Rinehart KL, Moore ML, Cook JC. 1981. Pyochelin: novel structure of an iron-chelating growth promoter for Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 78:4256–4260. 10.1073/pnas.78.7.4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mercado-Blanco J, Van der Drift KM, Olsson PE, Thomas-Oates JE, Van Loon LC, Bakker PA. 2001. Analysis of the pmsCEAB gene cluster involved in biosynthesis of salicylic acid and the siderophore pseudomonine in the biocontrol strain Pseudomonas fluorescens WCS374. J. Bacteriol. 183:1909–1920. 10.1128/JB.183.6.1909-1920.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mossialos D, Meyer J-M, Budzikiewicz H, Wolff U, Koedam N, Baysse C, Anjaiah V, Cornelis P. 2000. Quinolobactin, a new siderophore of Pseudomonas fluorescens ATCC 17400, the production of which is repressed by the cognate pyoverdine. Appl. Environ. Microbiol. 66:487–492. 10.1128/AEM.66.2.487-492.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matthijs S, Tehrani KA, Laus G, Jackson RW, Cooper RM, Cornelis P. 2007. Thioquinolobactin, a Pseudomonas siderophore with antifungal and anti-Pythium activity. Environ. Microbiol. 9:425–434. 10.1111/j.1462-2920.2006.01154.x [DOI] [PubMed] [Google Scholar]
- 17.Ratledge C, Dover LG. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881–941. 10.1146/annurev.micro.54.1.881 [DOI] [PubMed] [Google Scholar]
- 18.Braud A, Hannauer M, Mislin GL, Schalk IJ. 2009. The Pseudomonas aeruginosa pyochelin-iron uptake pathway and its metal specificity. J. Bacteriol. 191:3517–3525. 10.1128/JB.00010-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heeb S, Haas D. 2001. Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant Microbe Interact. 14:1351–1363. 10.1094/MPMI.2001.14.12.1351 [DOI] [PubMed] [Google Scholar]
- 20.Rich JJ, Kinscherf TG, Kitten T, Willis DK. 1994. Genetic evidence that the gacA gene encodes the cognate response regulator for the lemA sensor in Pseudomonas syringae. J. Bacteriol. 176:7468–7475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pernestig A-K, Melefors Ö, Georgellis D. 2001. Identification of UvrY as the cognate response regulator for the BarA sensor kinase in Escherichia coli. J. Biol. Chem. 276:225–231. 10.1074/jbc.M001550200 [DOI] [PubMed] [Google Scholar]
- 22.Valverde C, Heeb S, Keel C, Haas D. 2003. RsmY, a small regulatory RNA, is required in concert with RsmZ for GacA-dependent expression of biocontrol traits in Pseudomonas fluorescens CHA0. Mol. Microbiol. 50:1361–1379. 10.1046/j.1365-2958.2003.03774.x [DOI] [PubMed] [Google Scholar]
- 23.Reimmann C, Valverde C, Kay E, Haas D. 2005. Posttranscriptional repression of GacS/GacA-controlled genes by the RNA-binding protein RsmE acting together with RsmA in the biocontrol strain Pseudomonas fluorescens CHA0. J. Bacteriol. 187:276–285. 10.1128/JB.187.1.276-285.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lapouge K, Schubert M, Allain FHT, Haas D. 2008. Gac/Rsm signal transduction pathway of γ-proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol. 67:241–253. 10.1111/j.1365-2958.2007.06042.x [DOI] [PubMed] [Google Scholar]
- 25.Schmidli-Sacherer P, Keel C, Défago G. 1997. The global regulator GacA of Pseudomonas fluorescens CHA0 is required for suppression of root diseases in dicotyledons but not in Gramineae. Plant Pathol. 46:80–90. 10.1046/j.1365-3059.1997.d01-213.x [DOI] [Google Scholar]
- 26.Duffy BK, Défago G. 2000. Controlling instability in gacS-gacA regulatory genes during inoculant production of Pseudomonas fluorescens biocontrol strains. Appl. Environ. Microbiol. 66:3142–3150. 10.1128/AEM.66.8.3142-3150.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poritsanos N, Selin C, Fernando WGD, Nakkeeran S, Kievit TRD. 2006. A GacS deficiency does not affect Pseudomonas chlororaphis PA23 fitness when growing on canola, in aged batch culture or as a biofilm. Can. J. Microbiol. 52:1177–1188. 10.1139/w06-079 [DOI] [PubMed] [Google Scholar]
- 28.Cheng X, Bruijn I, Voort M, Loper JE, Raaijmakers JM. 2013. The Gac regulon of Pseudomonas fluorescens SBW25. Environ. Microbiol. Rep. 5:608–619. 10.1111/1758-2229.12061 [DOI] [PubMed] [Google Scholar]
- 29.Hassan KA, Johnson A, Shaffer BT, Ren Q, Kidarsa TA, Elbourne LDH, Hartney S, Duboy R, Goebel NC, Zabriskie TM, Paulsen IT, Loper JE. 2010. Inactivation of the GacA response regulator in Pseudomonas fluorescens Pf-5 has far-reaching transcriptomic consequences. Environ. Microbiol. 12:899–915. 10.1111/j.1462-2920.2009.02134.x [DOI] [PubMed] [Google Scholar]
- 30.Liao C-H, McCallus DE, Fett WF, Kang Y-G. 1997. Identification of gene loci controlling pectate lyase production and soft-rot pathogenicity in Pseudomonas marginalis. Can. J. Microbiol. 43:425–431. 10.1139/m97-060 [DOI] [PubMed] [Google Scholar]
- 31.Wei X, Huang X, Tang L, Wu D, Xu Y. 2013. Global control of GacA in secondary metabolism, primary metabolism, secretion systems, and motility in the rhizobacterium Pseudomonas aeruginosa M18. J. Bacteriol. 195:3387–3400. 10.1128/JB.00214-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frangipani E, Visaggio D, Heeb S, Kaever V, Cámara M, Visca P, Imperi F. 2014. The Gac/Rsm and cyclic-di-GMP signaling networks coordinately regulate iron uptake in Pseudomonas aeruginosa. Environ. Microbiol. 16:676–688. 10.1111/1462-2920.12164 [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Russell DW. 2001. Molecular cloning: a Laboratory Manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 34.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. 10.1016/0378-1119(95)00584-1 [DOI] [PubMed] [Google Scholar]
- 35.Schwyn B, Neilands JB. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47–56. 10.1016/0003-2697(87)90612-9 [DOI] [PubMed] [Google Scholar]
- 36.Machuca A, Milagres A. 2003. Use of CAS-agar plate modified to study the effect of different variables on the siderophore production by Aspergillus. Lett. Appl. Microbiol. 36:177–181. 10.1046/j.1472-765X.2003.01290.x [DOI] [PubMed] [Google Scholar]
- 37.Höfte M, Buysens S, Koedam N, Cornelis P. 1993. Zinc affects siderophore-mediated high affinity iron uptake systems in the rhizosphere Pseudomonas aeruginosa 7NSK2. Biometals 6:85–91 [DOI] [PubMed] [Google Scholar]
- 38.Arnow LE. 1937. Colorimetric determination of the components of 3,4-dihydroxyphenylalaninetyrosine mixtures. J. Biol. Chem. 118:531–537 [Google Scholar]
- 39.Andrus CR, Walter M, Crosa JH, Payne SM. 1983. Synthesis of siderophores by pathogenic Vibrio species. Curr. Microbiol. 9:209–214. 10.1007/BF01567583 [DOI] [Google Scholar]
- 40.Kulasekara HD, Ventre I, Kulasekara BR, Lazdunski A, Filloux A, Lory S. 2005. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol. 55:368–380. 10.1111/j.1365-2958.2004.04402.x [DOI] [PubMed] [Google Scholar]
- 41.Liu Y-G, Whittier RF. 1995. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25:674–681. 10.1016/0888-7543(95)80010-J [DOI] [PubMed] [Google Scholar]
- 42.Das S, Noe JC, Paik S, Kitten T. 2005. An improved arbitrary primed PCR method for rapid characterization of transposon insertion sites. J. Microbiol. Methods 63:89–94. 10.1016/j.mimet.2005.02.011 [DOI] [PubMed] [Google Scholar]
- 43.Gutierrez JA, Crowley PJ, Brown DP, Hillman JD, Youngman P, Bleiweis AS. 1996. Insertional mutagenesis and recovery of interrupted genes of Streptococcus mutans by using transposon Tn917: preliminary characterization of mutants displaying acid sensitivity and nutritional requirements. J. Bacteriol. 178:4166–4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao J, Yu X, Xie Z. 2012. Draft genome sequence of high-siderophore-yielding Pseudomonas sp. strain HYS. J. Bacteriol. 194:4121. 10.1128/JB.00688-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. 10.1016/S0378-1119(98)00130-9 [DOI] [PubMed] [Google Scholar]
- 46.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 47.Visca P, Leoni L, Wilson MJ, Lamont IL. 2002. Iron transport and regulation, cell signalling and genomics: lessons from Escherichia coli and Pseudomonas. Mol. Microbiol. 45:1177–1190. 10.1046/j.1365-2958.2002.03088.x [DOI] [PubMed] [Google Scholar]
- 48.Babitzke P, Romeo T. 2007. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 10:156–163. 10.1016/j.mib.2007.03.007 [DOI] [PubMed] [Google Scholar]
- 49.Lapouge K, Sineva E, Lindell M, Starke K, Baker CS, Babitzke P, Haas D. 2007. Mechanism of hcnA mRNA recognition in the Gac/Rsm signal transduction pathway of Pseudomonas fluorescens. Mol. Microbiol. 66:341–356. 10.1111/j.1365-2958.2007.05909.x [DOI] [PubMed] [Google Scholar]
- 50.Poole K, McKay GA. 2003. Iron acquisition and its control in Pseudomonas aeruginosa: many roads lead to Rome. Front. Biosci. 8:661–686. 10.2741/1051 [DOI] [PubMed] [Google Scholar]
- 51.Dyke SF, Floyd A, Sainsbury M, Theobald R. 1978. Organic spectroscopy: an introduction. Longman Group Ltd Press, London, United Kingdom [Google Scholar]
- 52.Krithika R, Marathe U, Saxena P, Ansari MZ, Mohanty D, Gokhale RS. 2006. A genetic locus required for iron acquisition in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 103:2069–2074. 10.1073/pnas.0507924103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crosa JH, Walsh CT. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66:223–249. 10.1128/MMBR.66.2.223-249.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Franza T, Mahé B, Expert D. 2005. Erwinia chrysanthemi requires a second iron transport route dependent of the siderophore achromobactin for extracellular growth and plant infection. Mol. Microbiol. 55:261–275. 10.1111/j.1365-2958.2004.04383.x [DOI] [PubMed] [Google Scholar]
- 55.Ren D, Zuo R, Wood TK. 2005. Quorum-sensing antagonist (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone influences siderophore biosynthesis in Pseudomonas putida and Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 66:689–695. 10.1007/s00253-004-1691-6 [DOI] [PubMed] [Google Scholar]
- 56.Kay E, Humair B, Dénervaud V, Riedel K, Spahr S, Eberl L, Valverde C, Haas D. 2006. Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J. Bacteriol. 188:6026–6033. 10.1128/JB.00409-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Molina MA, Ramos J-L, Espinosa-Urgel M. 2003. Plant-associated biofilms. Rev. Environ. Sci. Biotechnol. 2:99–108. 10.1023/B:RESB.0000040458.35960.25 [DOI] [Google Scholar]
- 58.Yang L, Nilsson M, Gjermansen M, Givskov M, Tolker-Nielsen T. 2009. Pyoverdine and PQS mediated subpopulation interactions involved in Pseudomonas aeruginosa biofilm formation. Mol. Microbiol. 74:1380–1392. 10.1111/j.1365-2958.2009.06934.x [DOI] [PubMed] [Google Scholar]
- 59.Parkins MD, Ceri H, Storey DG. 2001. Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol. Microbiol. 40:1215–1226. 10.1046/j.1365-2958.2001.02469.x [DOI] [PubMed] [Google Scholar]
- 60.Suh S-J, Silo-Suh L, Woods DE, Hassett DJ, West SE, Ohman DE. 1999. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J. Bacteriol. 181:3890–3897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vinckx T, Matthijs S, Cornelis P. 2008. Loss of the oxidative stress regulator OxyR in Pseudomonas aeruginosa PAO1 impairs growth under iron-limited conditions. FEMS Microbiol. Lett. 288:258–265. 10.1111/j.1574-6968.2008.01360.x [DOI] [PubMed] [Google Scholar]
- 62.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1:784–791. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- 63.Voisard C, Bull Keel CTC, Laville J, Maurhofer M, Schnider U, Défago G, Haas D. 1994. Biocontrol of root diseases by Pseudomonas fluorescens CHA0: current concepts and experimental approaches, p 67–89 In O'Gara F, Dowling DN, Boesten B. (ed), Molecular ecology of rhizosphere microorganisms: biotechnology and the release of GMOs. VCH Publishers, Weinheim, Germany [Google Scholar]
- 64.Holloway B, Krishnapillai V, Morgan A. 1979. Chromosomal genetics of Pseudomonas. Microbiol. Rev. 43:73–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.