Abstract
Agrobacterium tumefaciens is a facultative plant pathogen and the causative agent of crown gall disease. The initial stage of infection involves attachment to plant tissues, and subsequently, biofilms may form at these sites. This study focuses on the periplasmic ExoR regulator, which was identified based on the severe biofilm deficiency of A. tumefaciens exoR mutants. Genome-wide expression analysis was performed to elucidate the complete ExoR regulon. Overproduction of the exopolysaccharide succinoglycan is a dramatic phenotype of exoR mutants. Comparative expression analyses revealed that the core ExoR regulon is unaffected by succinoglycan synthesis. Several findings are consistent with previous observations: genes involved in succinoglycan biosynthesis, motility, and type VI secretion are differentially expressed in the ΔexoR mutant. In addition, these studies revealed new functional categories regulated by ExoR, including genes related to virulence, conjugation of the pAtC58 megaplasmid, ABC transporters, and cell envelope architecture. To address how ExoR exerts a broad impact on gene expression from its periplasmic location, a genetic screen was performed to isolate suppressor mutants that mitigate the exoR motility phenotype and identify downstream components of the ExoR regulatory pathway. This suppression analysis identified the acid-sensing two-component system ChvG-ChvI, and the suppressor mutant phenotypes suggest that all or most of the characteristic exoR properties are mediated through ChvG-ChvI. Subsequent analysis indicates that exoR mutants are simulating a response to acidic conditions, even in neutral media. This work expands the model for ExoR regulation in A. tumefaciens and underscores the global role that this regulator plays on gene expression.
INTRODUCTION
Plant-associated and rhizosphere bacteria must sense and respond to many aspects of their local environment, including but not restricted to those cues released from host plants. Some of the best-studied host-responsive signal transduction systems have been identified in members of the family Rhizobiaceae, specifically the symbiotic nitrogen-fixing rhizobia and the plant-pathogenic agrobacteria. These bacteria continuously monitor many features of their environment to control their interactions with host plants and calibrate their physiology to the prevailing conditions. For the rhizobia, leguminous host plants release specific flavonoid derivatives into the environment, to which the rhizobia respond by initiating the complex exchange of morphogenic signals culminating in the formation of symbiotic, nitrogen-fixing nodules (1). In contrast, agrobacteria such as Agrobacterium tumefaciens respond to phenolic lignin precursors released from plant tissues, engaging in an intricate and carefully coordinated transfer of genetic material from the infecting bacterium to plant cell nuclei, one of the only examples of cross-kingdom gene transfer (2–5). In addition to these primary host response pathways, members of the Rhizobiaceae employ a series of other regulatory networks to modulate their interactions with plants and the environmental consequences of host association.
The ExoR regulator represents one of these additional regulatory pathways that can contribute to host interactions (6). The exoR gene was identified originally in Sinorhizobium meliloti through transposon mutant screens for colonies that fluoresced differently from the wild type when grown on medium containing calcofluor white, a reagent that specifically labels several β-linked polysaccharides (7). Subsequent work revealed that the exoR mutation resulted in elevated production of the exopolysaccharide succinoglycan (SCG; also called EPS I in S. meliloti) and that the genes encoding SCG synthesis were derepressed in this mutant (8). The exoR mutant was also shown to be compromised for symbiotic interactions with alfalfa (9, 10). Interestingly, S. meliloti mutants that produced no SCG were also symbiotically deficient, failing to invade nodules during plant colonization (11).
Despite its genetic identification as a repressor of SCG synthesis, ExoR is not a DNA binding protein but rather has an N-terminal secretion signal and in S. meliloti has been shown to be periplasmically localized (12). ExoR also contains tetratricopeptide repeat (TPR) motifs, known to promote protein-protein interactions (13, 14). In a series of elegant genetic and biochemical studies, Long and coworkers found that in S. meliloti the exoR mutant requires the ExoS-ChvI two-component system to drive elevated SCG synthesis (12, 15, 16). ExoR immunoprecipitates with ExoS and is thought to form a physical complex with the periplasmic domain of ExoS, inhibiting its activity (15). ExoS phosphorylates its cognate response regulator ChvI, which is responsible for regulating the expression of a number of target genes. ExoR and ExoS-ChvI also positively regulate flagellar gene expression in S. meliloti (16, 17). Despite being nonmotile, S. meliloti exoR mutants are reported to be hyperadherent and form thicker biofilms than the wild type (12).
A diversity of Alphaproteobacteria have an ExoR homologue, including mammalian pathogens such as Brucella abortus and free-living bacteria such as Nitrobacter hamburgensis, in all cases with a well-supported N-terminal secretion signal and TPRs (18). A. tumefaciens has an ExoR homologue that is highly similar to that from S. meliloti, and A. tumefaciens exoR mutants derepress SCG production and lack flagella, due at least in part by the mutation affecting the transcription of exo and motility genes (18). In contrast to S. meliloti, an A. tumefaciens exoR mutant was isolated by virtue of its inability to form biofilms (18). The exoR mutant is significantly less adherent than an aflagellate A. tumefaciens mutant (lacking flgE) and fails to form biofilms in static cultures, flow cells, and root binding assays (18). Both decreased biofilm formation and loss of motility are independent of SCG, as an exoA exoR double mutant that does not synthesize the polysaccharide retains these phenotypes (18).
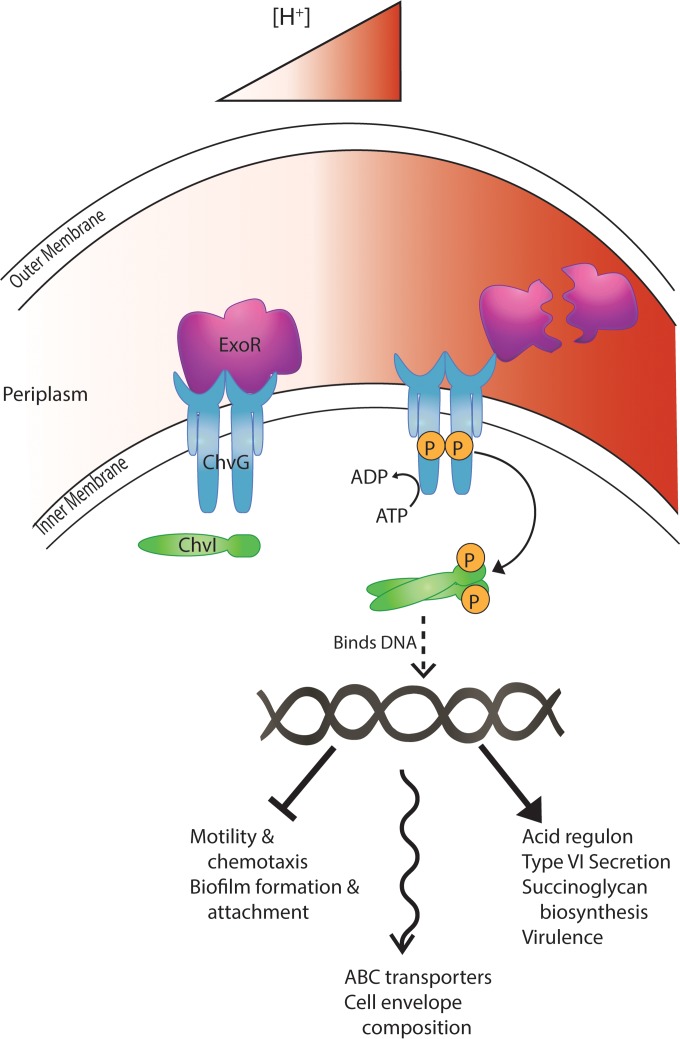
In A. tumefaciens, the ChvG-ChvI two-component system is similar to ExoS-ChvI and is required for plant virulence (19, 20). ChvG-ChvI becomes active under low-pH conditions to elicit their cellular responses. ExoR represses ChvG under neutral conditions, but when A. tumefaciens cells encounter acid, ExoR is proteolyzed to allow the two-component system to become active (21, 22). Known targets of regulation by ChvG-ChvI include motility and chemotaxis genes, SCG biosynthesis genes, a type VI secretion system (T6SS), and various virulence genes (22, 23).
The basis for the A. tumefaciens exoR mutant's inability to attach to surfaces remains unclear, and its effects on SCG synthesis and motility do not provide the complete explanation. In this study, we performed DNA microarray expression analysis comparing the exoR mutant to the wild type. We observed that close to 10% of the A. tumefaciens genome is impacted in an exoR mutant, with many unexpected genes under ExoR control, including virulence functions, type VI and type IV secretion systems, and a wide range of transporter systems. We substantiate the expression results with a range of phenotypic assays that confirm the impact of the exoR mutation on A. tumefaciens physiology. In addition, we demonstrate that approximately 45% of those genes affected for expression in the exoR mutant are responding to the vast overproduction of SCG, which may deplete the nutritional reserves of the bacterium. Genetic analysis also clearly supports the model in which the exoR mutant phenotypes are largely due to unregulated activity of the ChvG-ChvI two-component system.
MATERIALS AND METHODS
Strains, plasmids, reagents, and growth conditions.
All strains and plasmids used in this study are described in Table S1 in the supplemental material. Buffers, antibiotics, and media were obtained from Fisher Scientific (Pittsburgh, PA) and Sigma Chemical Co. (St. Louis, MO). DNA manipulations were performed in accordance with standard protocols (24). DNA sequencing was performed with ABI BigDye Terminator version 3.1 on an ABI 3730 sequencer operated by the Indiana Molecular Biology Institute. Oligonucleotides (see Table S2 in the supplemental material) were obtained from Integrated DNA Technologies (Coralville, IA). Plasmids were electroporated into A. tumefaciens by a standard method (25). A. tumefaciens derivatives were grown in AT minimal salts medium with 15 mM (NH4)2SO4, buffered to pH 7 with KH2PO4 to a final concentration of 79 mM, and either 0.5% (wt/vol) glucose (ATGN) or 0.4% (wt/vol) succinic acid (ATSucN) as the carbon source (26). Acidic medium was prepared with 200 mM MES (morpholineethanesulfonic acid) hydrate buffered to pH 5.5. Virulence-inducing medium is buffered with MES to pH 5.5 and contains 200 μM acetosyringone. Antibiotics were used at the following concentrations (μg/ml): for A. tumefaciens, ampicillin (Ap), 100; gentamicin (Gm), 300; kanamycin (Km), 150; spectinomycin (Sp), 150; streptomycin (Sm), 2,000; and for Escherichia coli, Ap, 100; Gm, 25; Km, 25; Sp, 50. Antibiotic selection was maintained throughout all experiments when using mutants generated by plasmid insertion.
Transposon mutagenesis and isolation of suppressor mutants.
To create a transposon mutant library of A. tumefaciens C58 ΔexoA ΔexoR cells, we used the mariner minitransposon (Himar1) carried by donor strain E. coli SM10/λpir pFD1 (27). Donor and recipient strains were inoculated into 2 ml LB broth and allowed to grow overnight at 37°C and 28°C, respectively. Turbid cultures were diluted 1:10 into 2 ml of LB and incubated for another 4 h. One milliliter of each outgrown culture was removed, and the cells were collected by centrifugation (7,000 × g). The cells were washed in 1 ml of LB and recollected. The supernatant was discarded, and the cell pellets were resuspended in 100 μl of LB. Three independent matings were performed by spotting 25 μl each of the E. coli donor and A. tumefaciens recipient cells onto 0.2-μm cellulose acetate filter disks on LB plates. The remaining 25 μl of each independent suspension was spotted in isolation onto filter disks as controls. Once the spots had dried, the plates were incubated overnight at 28°C to allow mating to occur and transfer of the mariner transposon into A. tumefaciens. The cells were collected off the filter disks after an overnight incubation by placing the filters in Microfuge tubes with ATGN liquid medium and vortexing vigorously. The collected cells were then frozen in 25% glycerol at −80°C for permanent storage. We plated (on ATGN-Km) enough of the libraries to approximate the generation of 20,000 isolated colonies and collected the cells by scraping the colonies into liquid ATGN-Km medium. Collected cells were frozen in 25% glycerol for permanent storage at −80°C. To enrich for suppressor mutants of the parent ΔexoA ΔexoR strain, 5 μl of the collected colonies was inoculated into motility plates (ATGN-Km with 0.3% agar). After 3 days of incubation at room temperature, cells were picked from the outer edge of swim rings formed by motile suppressor mutants. Cells picked from the swim plates were struck for isolation on ATGN-Km plates. Single colonies were reinoculated into motility agar to confirm a motile phenotype. Motile suppressor mutants were then grown in liquid ATGN-Km and frozen in 25% glycerol at −80°C for permanent storage.
Touchdown PCR and sequencing.
To determine the insertion site of the mariner transposon in our mutants of interest, genomic DNA was first isolated from the strains that were determined to have suppressed the motility defect of the parent ΔexoA ΔexoR strain. Genomic DNA was isolated by using a Wizard genomic DNA purification kit (Promega Corp) and quantified using a NanoDrop 1000 spectrophotometer (Thermo Scientific). The insertion site of mariner was mapped using touchdown PCR. The reaction mixture contained a genomic DNA template concentration of 80 to 120 ng; sequence-specific primers MarRSeq and MarLSeq paired with arbitrary primers MarTDL2 and MarTDR1 (see Table S2 in the supplemental material) and Phusion high-fidelity polymerase with HF buffer (New England BioLabs). The reaction was run with a touchdown PCR program as follows: (i) 98°C for 30 s, (ii) 25 cycles of 98°C for 10 s, 60°C for 45 s, decreasing by 0.5°C each cycle, and 72°C for 2 min, (iii) 25 cycles of 98°C for 10 s, 50°C for 45 s, and 72°C for 2 min, and (iv) 72°C for 1 min. A portion of the reactions was subjected to electrophoresis on an agarose gel to confirm amplification products prior to purifying the reaction mixtures via an E.Z.N.A. Cycle Pure kit by Omega Bio-Tek. The purified touchdown PCRs were used as the templates for sequencing with the MarRSeq and MarLSeq primers following the recommended protocol of the Indiana Molecular Biology Institute using BigDye 3.1 sequencing reagent (http://imbi.indiana.edu/index.php?page_id=DNA%20Sequencing%20Facilities&features=1).
Constructing deletion strains.
Creating markerless deletion strains was performed as previously described (28, 29). Fragments upstream and downstream of the genes of interest (0.5 to 1 kb in size) were amplified using primers P1 and P2 (upstream) and P3 and P4 (downstream) (see Table S2 in the supplemental material). These fragments were stitched together via 18-bp complementary sequences by a splicing by overhang extension (SOE) PCR. The deletion construct was first ligated into the pGEM T-Easy (Invitrogen) vector for amplification in E. coli strain Top10F′. Miniprepped pGEM plasmids (collected with the E.Z.N.A. Omega Bio-Tek plasmid minikit) were digested with the appropriate enzymes (see Table S2 in the supplemental material), and the insert was ligated into the suicide vector pNPTS138 (30). This vector was transformed into the E. coli mating strain S17-1λpir in order to conjugate the deletion plasmid into the appropriate A. tumefaciens strain. A. tumefaciens recipient and E. coli donor strains were spotted onto cellulose acetate filter disks as described previously (28, 29). Primary integrants were selected by plating cells collected from filter disks onto ATGN-Km plates. Cells that subsequently lost the integrated plasmid but retained the introduced deletion were selected by plating onto AT medium with 5% sucrose and confirmed by PCR across the deletion junction.
Constructing controlled-expression plasmids.
Complementation analyses were performed by cloning wild-type A. tumefaciens coding sequences into the LacIq-encoding, IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible expression vector with a gentamicin resistance cassette, pSRKGm (31). Coding sequences were amplified using Phusion polymerase (New England BioLabs) from wild-type C58 genomic DNA, using the primers corresponding to each gene, specified in Table S2 in the supplemental material. Amplified fragments were ligated into the cloning vector pGEM-T Easy (Invitrogen), and the inserts were confirmed by sequencing. Coding sequences were excised from pGEM-T by restriction enzyme cleavage and ligated into the appropriately cleaved expression vector using T4 DNA ligase (New England BioLabs). Derived plasmids were verified by PCR amplification across the insert prior to transformation into A. tumefaciens cells via electroporation.
Microarrays.
Custom 60mer oligonucleotide microarrays were purchased from Agilent Technologies. Unique oligonucleotides representing open reading frames were identified using Featurama, a program designed by the Institute for Systems Biology in Seattle, WA. Arrays were produced by Agilent Technologies and consist of 8,455 features that represent 5,419 predicted protein-encoding open reading frames, tRNA- and rRNA-encoding genes, and 2,983 duplicate spots.
To prepare samples, cultures were grown to mid-log phase at 28°C in liquid ATSucN. A culture volume equivalent to 11 ml at an optical density at 600 nm (OD600) of 0.6 was incubated with 2 volumes of RNAProtect reagent (QIAgen) for 15 min at room temperature. Cells were lysed with 10 mg lysozyme, and RNA was extracted using the QIAgen RNEasy Midi kits. Samples were treated with QIAgen on-column DNase. First-strand labeling and reverse transcription were performed using the Invitrogen SuperScript indirect labeling kit, and cDNA was purified on QIAgen QIAQuick columns. cDNA was labeled with Alexa Fluor 542 and 655 dyes using the Invitrogen SuperScript cDNA labeling kit and repurified on QIAQuick columns. cDNA was quantified on a NanoDrop spectrophotometer. Hybridization reactions were performed using the Agilent in situ hybridization kit Plus; the products were boiled for 5 min at 95°C, applied to Agilent custom arrays for A. tumefaciens C58, and hybridized overnight at 65°C. Hybridized arrays were washed with Agilent wash solutions 1 and 2, rinsed with acetonitrile, and incubated in the Agilent stabilization and drying solution immediately prior to scanning the arrays. Four independent biological replicates were performed for each comparison, with dye swaps. Hybridized arrays were scanned on a GenePix Scanner 4200 in the Center for Genomics and Bioinformatics (CGB) at Indiana University. GenePix software was used to define the borders of hybridized spots, subtract background, measure dye intensity at each spot, and calculate the ratio of dye intensities for each spot. Statistical analyses of the raw data as well as processed data were performed in the Indiana University Center for Genomics and Bioinformatics. Genes with significant P values (≤0.05), a false-discovery rate (Q value) of less than or equal to 0.3, and log2 fold change (FC) ratios (M values) of ≥0.5 or less than or equal to −0.5 (representing an FC of ±1.4) are reported here.
Real-time qRT-PCR.
For real-time quantitative reverse transcription-PCR (qRT-PCR), cultures were grown at 28°C in ATGN to mid-log phase. A culture volume equivalent to 1 ml at an OD600 of 0.8 was incubated with 2 volumes of RNAProtect reagent for 15 min at room temperature, lysed with 10 mg lysozyme, and RNA extracted using the QIAgen RNEasy minikit. Samples were treated with DNase (Ambion Inc., Austin, TX) to remove contamination. cDNA was prepared using the qScript cDNA Super Mix kit (Quanta Biosciences, Gaithersburg, MD). qPCR was performed with PerfeCta SYBR green FastMix Low Rox reagent (Quanta Biosciences), on a Stratagene MX3000P instrument. Sample values were normalized using a σ70 primer set (see Table S2 in the supplemental material) and calibrated against wild-type results. Data are representative of two biological replicates, each of which consisted of three technical replicates.
Conjugation assays.
Antibiotic-resistant, genetically marked strains for conjugation assays were utilized. Donor strains were transformed with pSRK-Km to provide a selectable marker of donor cells postconjugation. A Gmr-marked pAtC58 was conjugated into wild-type strain C58 and the different mutant derivatives (ΔexoR mutant, PMM1; ΔexoA mutant, MLL2; ΔexoA ΔexoR mutant, PMM2) already harboring the pSRK-Km plasmid. Transfer of pAtC58Gmr evicted the native pAtC58 to create the corresponding pAtC58Gmr donors, which are Gmr and Kmr. These A. tumefaciens derivatives harboring pAtC58Gmr were conjugated with A. tumefaciens ERM52, lacking both pTiC58 and pAtC58, with a chromosomally integrated Sp/Smr cassette (32). Conjugation assays were performed by mixing suspensions of donor and recipient strains each normalized to an OD600 of 0.8, spotting 100 μl onto cellulose acetate filters on ATGN plates, and incubating the plates for 18 h at 28°C. Sp/Smr Gmr transconjugants and Kmr Gmr donors were selected by plating serial dilutions of the suspended mating mixture on AT medium with appropriate antibiotics, and conjugation efficiency was determined as the ratio of transconjugants per output donor. Genomic DNA of all transconjugants was collected, and PCR was performed to confirm the presence or absence of exoR and exoA, as appropriate.
Calcofluor white staining.
ATGN plates were supplemented with 20 μg/ml of calcofluor white (fluorescent white) dye from a stock solution of 200 μg/ml as previously described (28). A. tumefaciens strains were inoculated into ATGN liquid medium from colonies and grown overnight at 28°C with shaking. Cultures were normalized to an OD of 0.2, and 5 μl was spotted onto each plate. Plates were incubated for 2 days at 28°C and then imaged inside a light cabinet with UV light excitation. All strains were grown and imaged on the same plate.
Acid tolerance/pH sensitivity assays.
All strains were grown to exponential phase and frozen at −80°C in 25% glycerol at an OD600 of 12. AT media of pH 7.0 (AT buffer) or pH 5.5 (MES buffer) were inoculated at an initial density of 0.012 (1:1,000) in 1 ml of medium per well in a 48-well flat-bottomed Falcon plate. Each strain and each condition were represented in quadruplicate; wells 1 to 4 of each row held a strain in neutral medium, and wells 5 to 8 held the same strain in acidic medium. The plate was incubated in a Bio-TEK Synergy HT plate reader for 72 h. Cell culture densities were measured hourly with Bio-TEK Gen5 (version 1.07) software. To calculate the growth yield in acidic medium compared to neutral medium, the growth yield (OD600) of cultures in low-pH medium was divided by the growth yield in neutral medium, with 4 biological replicates under each condition. The average ratio and standard error were calculated from the four growth ratios determined in this way.
Virulence (vir) gene expression measurement with lacZ fusions.
Plasmids carrying vir-lacZ fusions generated by Winans (33) were electroporated into A. tumefaciens cells following a standard procedure (25). Colonies containing plasmids were selected on antibiotic plates. Cells were grown to exponential phase (OD600 between 0.3 and 0.8) and frozen at −80°C in 25% glycerol to a final OD600 of 12. Frozen stocks were inoculated into ATGN minimal medium at pH 7.0, acidic medium of pH 5.5, and vir-inducing medium at pH 5.5 with 200 μM acetosyringone. Cultures were grown to exponential phase, their OD600 was recorded, and they were frozen at −20°C prior to further processing. β-Galactosidase assays were performed as described previously (34), using 100 μl of cell culture per reaction. Reactions were terminated with addition of 600 μl of 1 M NaCO2, intact cells and debris were removed by centrifugation, and free ortho-nitrophenol (ONP) was measured by A420 units using a Bio-TEK Synergy-HT reader with Gen5 (version 1.07) software. Specific activity was reported in Miller units, according to the equation MU = 1,000 [A420/(OD600·t·f)], where t is the reaction time in minutes and f is the ratio of cell volume to total reaction volume.
Starvation assays/carbon limitation.
Frozen stocks (OD600 of 12) of each strain were inoculated into 2 ml of medium at an initial density of 0.012 (1:1,000). Medium was prepared as described previously but with variable concentrations of glucose: 1 μM, 2 μM, and 5 μM glucose as opposed to the 29 μM (0.5%) glucose of ATGN. The OD600 was measured at 13.5, 18.5, 22.5, 40.5, and 71 h postinoculation by spectrophotometry in a Spectronic Genesys (model 5) spectrophotometer.
Biofilm assays.
Biofilm assays were performed as described previously (28, 29). Briefly, polyvinyl chloride (PVC) coverslips were placed upright in 12-well PVC plates. Overnight cultures were diluted to an initial OD600 of 0.05 in ATGN. Each well was inoculated with 3 ml of culture, and the plates were incubated at room temperature for 48 h. Coverslips were stained with 0.1% crystal violet dye. The adherent dye was solubilized with 33% acetic acid. The OD600 of the 48-hour cultures and the A600 of the solubilized crystal violet were measured with a Bio-TEK Synergy HT plate reader using Bio-TEK Gen5 (version 1.07) software.
Swim assays.
Motility medium was prepared as described for ATGN with an agar concentration of 0.3%. Twenty milliliters of medium was added to each petri dish. One microliter of frozen cells (OD600 = 12) was inoculated into the center of the swim plate. Plates were kept at room temperature in a sealed container with an open vial of a saturated solution of K2SO4 to keep the plates hydrated. Swim ring diameters were measured using a standard ruler.
Microarray data accession number.
Microarray data have been deposited into the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) database under accession number GSE59790.
RESULTS
The regulon of ExoR.
To identify all genes directly and indirectly affected by ExoR, we compared the transcriptome of wild-type C58 to a ΔexoR mutant using Agilent custom oligonucleotide microarrays. This analysis identified 491 genes that were differentially expressed between strain C58 and the ΔexoR mutant, with a P value of less than 0.05 and an M value of at least ±0.5, indicating a minimum fold change (FC) of 1.4 (see Fig. S1 in the supplemental material). A subset of these genes are listed in Table 1 (see Table S3 in the supplemental material for the full lists of genes). The distribution of these 491 genes is roughly proportional to the percentage of the genome represented by each replicon: 262 genes (53.4%) are located on the circular chromosome (51.9% of genome), 173 genes (35.2%) on the linear chromosome (34.5% of genome), 40 genes (8.1%) on the At plasmid (9.9% of genome), and 16 genes (3.3%) on the Ti plasmid (3.6% of genome).
TABLE 1.
Gene groups of interest in the core ExoR regulona
| Functional category | No. of genes | Regulation by ExoR | Log2 FC in ΔexoR mutant |
|
|---|---|---|---|---|
| >0 | <0 | |||
| Motility and chemotaxis | 47 (9) | + | 0 | 47 |
| Succinoglycan biosynthesis | 15 | − | 15 | 0 |
| Type VI secretion | 19 (2) | − | 19 | 0 |
| ABC transporters | 20 | ± | 2 | 18 |
| Virulence | 2 | − | 2 | 0 |
| Cell envelope proteins | 19 | ± | 14 | 5 |
| Transcription factors and two-component systems | 8 | ± | 5 | 3 |
| Heat/cold shock proteinsb | 7 | ± | 4b | 3b |
| Plasmid conjugationc | 15 | + | 1c | 14c |
These genes are represented in both “ΔexoR versus wild-type” and “ΔexoA ΔexoR versus ΔexoA” microarrays, except for the last two rows. The functional groups contain annotated genes that correspond to each category, with the number of unannotated genes falling within defined operons in parentheses.
Only in the category “ΔexoA ΔexoR versus ΔexoA.” It is of note that heat shock proteins display increased expression and cold shock proteins display decreased expression in the analysis.
Plasmid conjugation genes—only in the category “ΔexoR versus wild-type.” The only upregulated gene in this category is trbL (atu6035), involved in pTi conjugation; all others are related to pAtC58 conjugation.
The lists of genes that met the cutoff values were characterized using the Blast2GO software. Figure S2 in the supplemental material displays the distribution of the genes at level 2 of the “biological process” category. Many gene sequences were given more than one gene ontology (GO) identification and so are represented more than once in this analysis. Genes not assigned any GO identification were omitted from the analysis.
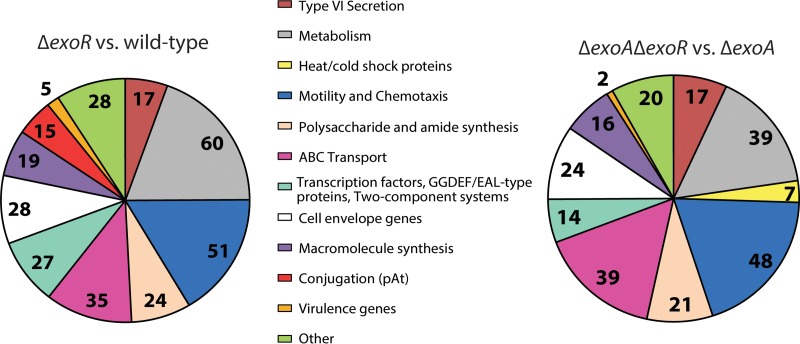
For the bulk of our analyses of the ExoR regulon, we chose to group the genes into functional categories that correspond to the phenotypes that we and others have previously observed for exoR mutants (Fig. 1 and Table 1). There are numerical differences between our functional categories and the GO analysis, as our functional categories include only annotated genes, whereas the Blast2GO software assigns GO IDs to many hypothetical genes. By either analysis, the regulon of ExoR is vast in terms of both breadth and diversity, and many aspects of cell physiology are impacted by this regulator. Notably, the expressions of a large number of regulators are under the control of ExoR (51 in GO analysis and 27 in our analysis in the “transcription factor, GGDEF/EAL protein, and two-component system” category), suggesting that it functions as a global regulator.
FIG 1.
Analysis of the ExoR regulon via microarray. Annotated genes were assigned to functional groups of interest. Microarray analysis of the ΔexoR mutant versus the wild type (182 hypothetical proteins were excluded) (A) and of the ΔexoA ΔexoR double mutant versus the ΔexoA mutant (132 hypothetical proteins were excluded) (B).
The greatest (log2) FC observed in ΔexoR mutants was 5.25 for the unannotated gene atu1221, which is predicted to code for an NlpC/p60 superfamily protein (35–37) by the blastp algorithm on the NCBI website (http://blast.ncbi.nlm.nih.gov/Blast.cgi). While this gene has not been characterized in A. tumefaciens, proteins of the NlpC/p60 family have been studied in other systems and are largely implicated in peptidoglycan maintenance, specifically playing a role in septation (35, 37–39). A small number of annotated genes (6) implicated in cell wall synthesis and maintenance are represented in the microarray and appear as both positively (5) and negatively (1) regulated by ExoR. The most highly upregulated annotated gene in the ΔexoR mutant with empirically demonstrated functions is aopB (Agrobacterium outer-membrane protein), with an FC of 4.99. This gene is highly similar to a family of outer membrane proteins in the Rhizobiaceae that have been shown to influence outer membrane stability, permeability, and topology (40–42). Homologues of aopB are regulated by a variety of environmental conditions, including low pH (23, 40, 41), peptides (42), and calcium (43).
The most dramatic decrease in gene expression in the ΔexoR mutant is for fliN, which encodes the flagellar motor switch protein (log2 FC = −4.58) (see Table S3 in the supplemental material). This is representative of many motility and chemotaxis genes, which make up the largest functional group of genes regulated by ExoR by our characterization (Fig. 1 and Table 1). The genes in this category are not restricted only to the large operons that direct flagellum biosynthesis but also include various genes distributed across the genome involved in chemotaxis, including 10 methyl-accepting chemotaxis proteins.
Relationship to previously identified targets.
Several major groups of genes identified as targets of ExoR regulation were implicated in our previous findings describing the role of exoR in A. tumefaciens (18). These include 16 exo genes, responsible for succinoglycan (SCG) biosynthesis (higher expression in ΔexoR mutants), 51 annotated—and 10 unannotated—motility and chemotaxis genes (lower expression in ΔexoR mutants), and 22 imp or hcp genes, associated with the type VI secretion system (overexpressed in ΔexoR mutants). These trends agree with the established hypermucoid and nonmotile phenotypes of ΔexoR mutants (7, 17, 18, 44) and with the role that ExoR plays in regulating the T6SS presented by Wu et al. (22). Noticeably absent from the motility gene category are the class I flagellar synthesis regulators visN, visR (class Ia), and rem (class Ib). In contrast, it has been observed in S. meliloti that ExoR-ExoS-ChvI affects flagellar synthesis and motility via regulation of the class I genes in response to low pH (45, 46).
These three functional categories of genes—SCG biosynthesis, motility/chemotaxis, and T6S—are known to be regulated by low pH, as described by Yuan et al. (23). Indeed, we noticed a striking overlap between the acid regulon and the ExoR regulon that extends beyond motility, SCG biosynthesis, and T6S gene expression. Furthermore, regulation of T6S genes by ExoR appears to be mediated by ChvG-ChvI (22), and our data demonstrate that many more genes identified as part of the acid regulon (23) are similarly regulated by ExoR. The common genes shared by the acid regulon and the ExoR regulon are noted in Table S3 in the supplemental material.
Novel regulated gene groups.
Four particularly intriguing ExoR-regulated gene categories are ABC transporters, type IV secretion systems, including the At plasmid AvhB conjugation system, virulence genes, and genes related to cell envelope architecture. Thirty-five genes annotated as ABC transporter components were differentially expressed in the ΔexoR mutant compared to the wild type. The majority of these (11) are annotated as amino acid transporters, and all but one are decreased in the exoR mutant. Nine of the ABC transporter genes are annotated as sugar transporters, seven of which are increased in the exoR mutant. This suggests that ExoR can influence the flux of material in and out of the cell by modulating ABC transport gene expression.
Intriguingly, the expression of 15 genes involved in plasmid conjugation was identified as being significantly different in the exoR mutant. Fourteen of these genes are downregulated (log2 FC, between −1.4 and −1.9), and they are all located on the AtC58 plasmid. These genes are involved in synthesis of a type IV conjugal pilus—11 genes belong to the avhB operon (atu5162-5172)—or are otherwise associated with conjugation: 3 genes are annotated as pAtC58 tra genes traG, traD, and traC (atu5108, atu5109, and atu5110, respectively) (47). Additionally, upstream of avhB1 (atu5162), there is an unannotated gene that appears to be in the same operon, and expression of this gene is significantly decreased (log2 FC, −2.1; not included in Fig. 1A due to lack of annotation). A homologue of an activator of conjugal transfer of the Rhizobium etli symbiotic plasmid, rctB (atu5116) (48), is also downregulated in the ΔexoR strain relative to the wild type (log2 FC, −2.43); this gene is grouped with transcriptional regulators and not with pAtC58 conjugation genes. This relationship between exoR and pAtC58 conjugation is of great interest to us because we have observed nearly constitutive conjugation of pAtC58 under laboratory conditions, and this is the first hint of a regulatory mechanism.
Besides virG, virH1, and virH2 (log2 FC, 3.3, 1.1, and 0.9, respectively), other genes related to virulence were upregulated in the ΔexoR mutant, including the agrobacterium chromosomal virulence gene acvB (log2 FC, 0.8) and the trans-zeatin synthase gene tzs (log2 FC, 0.63) (49, 50). The chvI-chvG locus, which responds to low pH and is required for virulence (20), was also upregulated, though we included these genes in the “transcription factor, GGDEF/EAL, and two-component system” group, not the “virulence” group. This locus was also expected to be increased for expression because of the central role of ChvG and ChvI in regulating the acid response (23, 51).
The most highly upregulated annotated gene in our analysis, aopB, falls into another functional category of interest: cell envelope-related genes. There were 28 genes in this category. One-half of the genes in this category are annotated as porins or transporters, which suggests that there is a significant change in the permeability of the cell envelope in exoR mutants. Of the other 14 genes, 3 are annotated as lipoproteins: 2 as rplA, encoding Rare lipoprotein A (atu0290 and atu1500; log2 FC, 2.44 and 1.08, respectively), and 1 as palA (atu3713, log2 FC 0.88), encoding Omp16, which has been characterized as an outer membrane lipoprotein in Brucella spp. (52). Two more genes in the cell envelope category are annotated as modifiers of cell surface-associated factors: lpiA (atu2521; log2 FC, 0.95) is involved in membrane lipid synthesis (53) and low-pH tolerance (45) in rhizobia, and glf (atu3975; log2 FC, −1.54) is required for galactofuranose synthesis, a cell surface component in pathogens (54, 55). These results are consistent with the role that ChvI plays in regulating phospholipid biosynthesis in S. meliloti (56). The appearance of these lipoproteins and cell surface-modifying genes in the expression profile suggests that the architecture of the cell envelope in exoR mutants is distinct from that of wild-type A. tumefaciens.
Overproduction of succinoglycan impacts the transcriptome of exoR mutants.
The role of ExoR in repressing SCG biosynthesis is well established (7, 8, 57). The hypermucoid phenotype of exoR mutants is quite striking: it is clear that cells are allocating significant resources to the production of this exopolysaccharide (18). Therefore, a question of particular interest was whether the overproduction of SCG in an exoR mutant would lead to downstream transcriptional effects. To identify genes that were affected by SCG overproduction, we performed a second microarray comparison with a mutant unable to synthesize SCG due to deletion of the SCG biosynthesis gene exoA (atu4053). We compared the ΔexoA mutant to the ΔexoA ΔexoR double mutant. The genes identified in this comparison represent those that ExoR impacts independently from the effects of SCG overproduction. The annotated genes identified in this comparison that met our cutoff values are represented in Fig. 1B, the entire list can be found in Table S3 in the supplemental material, and the full analysis is displayed in Fig. S1 in the supplemental material.
Most of the genes that fall into the functional categories that we defined above are maintained in the ΔexoA to ΔexoA ΔexoR comparison. All of the SCG biosynthesis genes are still represented in this comparison with the exception of exoA itself. The vast majority of motility and chemotaxis-related genes are maintained, with the exception of three methyl-accepting chemotaxis protein-encoding genes scattered across the genome; one on the circular chromosome (atu0646), one on the At plasmid (atu5442), and one on the Ti plasmid (atu6132). All of the type VI secretion genes identified in the ΔexoR mutant versus wild-type strain analysis are similarly regulated in the ΔexoA mutant analysis.
Despite similar numbers of ABC transporter genes represented in each analysis, there are significant differences in the trends of regulation between the two comparisons. Only one-half of the ABC transport-related genes discovered in the ΔexoA ΔexoR versus ΔexoA comparison are shared with the ΔexoR versus wild-type comparison (20/39 genes). These include the liv operon (atu4515-4519) for transport of branched-chain amino acids (58), which is downregulated in both comparisons, and the genes rbsA (atu4321) and rbsC (atu4322), involved in ribose transport (59), which are upregulated in both comparisons (consistent with ribose utilization patterns in the exoS and chvI mutants of S. meliloti) (60). The pattern of sugar transporters is markedly different between comparisons: 12/14 genes predicted to transport sugars in the ΔexoR versus wild-type comparison are increased in expression, but only 4/24 genes predicted to transport sugars in the ΔexoA ΔexoR versus ΔexoA comparison are increased. This may underlie the carbon-starved condition of ΔexoR mutants as they overproduce SCG, versus the ΔexoA strains, which allocate few if any resources to SCG synthesis.
Another difference between the two microarray experiments is that fewer virulence-related genes are differentially regulated in the ΔexoA ΔexoR versus ΔexoA comparison versus the ΔexoR versus wild-type comparison. Only two of the five virulence genes revealed in the latter comparison appear in the former: acvB (atu2522; log2 FC, 0.74), encoding chromosomal virulence protein, and virG (atu6178; log2 FC, 3.12).
An interesting trend emerges when we analyze the cell envelope gene category for our two microarray experiments: the composition of the membrane itself differs, while the majority of embedded machinery (porins and transporters) remains similar. The gene palA (atu3713; log2 FC, 0.88), encoding the lipoprotein Omp16 (52), is not present in the ΔexoA ΔexoR versus ΔexoA comparison, whereas another annotated lipoprotein, atu1789, is present (log2 FC, −0.83). Additionally, the cell surface-modifying genes lpiA (atu2521; log2 FC,= 0.95) and glf (atu3975, log2 FC, −1.94) are not present in the ΔexoA ΔexoR versus ΔexoA microarray. These differences in gene expression suggest that overproduction of SCG is, in and of itself, contributing to the architecture of the cell membrane in exoR mutants. Of course, there is also no SCG on the outside of the exoA mutant cells, despite the overexpression of most of the SCG biosynthetic genes.
The ΔexoA ΔexoR versus ΔexoA microarray experiment contained a functional group that was absent in the ΔexoR versus wild-type microarray: temperature stress response genes. Four heat shock proteins were upregulated, and three cold shock proteins were downregulated in the ΔexoA ΔexoR strain versus ΔexoA experiment. The presence of these temperature-dependent stress response genes in the exoA comparison may suggest that the overproduction of SCG offers a protective benefit to exoR mutants that is lost when SCG biosynthesis is inhibited. We recognize not only the presence of a unique functional gene group in the ΔexoA ΔexoR versus ΔexoA comparison but also the absence of another gene group: the type IV conjugation machinery of pAtC58.
Succinoglycan overproduction inhibits conjugal transfer of the At plasmid.
An unexpected result of the ΔexoR versus wild-type comparison is that an exoR mutation leads to a decrease in expression of rctB and avhB1 to avhB11, the activator and type IV secretion system associated with pAtC58 conjugation, respectively. However, when SCG production is eliminated, expression of these genes returns to wild-type levels. Curious as to whether these changes in transcription led to measurable differences in conjugation rates, we performed conjugation assays using wild-type C58 and exo gene mutants as donor strains of a gentamicin-resistant copy of pAtC58. These assays demonstrated a wild-type conjugation rate of 1.9 × 10−4 transconjugants per output donor. The ΔexoR mutant was dramatically less efficient, with a conjugation rate decreased by nearly 4 orders of magnitude, to 4.4 × 10−8 transconjugants per donor. The ΔexoA mutant, deficient for SCG production, conjugated at a rate similar to that of the wild type, at 9.5 × 10−3 transconjugants per donor. The ΔexoA ΔexoR double mutant had a conjugation rate of 2.9 × 10−5 transconjugants per donor, much closer to that of the wild type than the ΔexoR mutant. These results are consistent with predictions derived from expression data and suggest a relationship between SCG production and conjugation of pAtC58. The mechanism by which succinoglycan production influences pAtC58 conjugation is currently being investigated.
Overproduction of succinoglycan causes reduced growth yield.
The most striking phenotype of exoR mutants is their hypermucoidy due to SCG overproduction. We also observed that an exoR mutation leads to massive upregulation of sugar transport systems and that many differentially regulated genes in the ΔexoR mutant are specifically affected by the overproduction of SCG (Fig. 1). We hypothesized that this massive overproduction of exopolysaccharide may cause ΔexoR cells to become carbon limited more quickly than the wild type and result in lower overall growth yield. To test this, we compared the growth yield of the ΔexoR and ΔexoA ΔexoR strains to that of wild-type cells in carbon-replete medium (ATGN medium, 29 μM glucose) and in low-carbon media containing 1 μM, 2 μM, and 5 μM glucose. Indeed, we saw an overall reduction in growth yield in the ΔexoR mutant, an effect that was reversed in the ΔexoA ΔexoR double mutant (see Fig. S3 in the supplemental material). However, this trend was consistent across all concentrations of glucose tested. These data indicate that the reduced growth in an exoR mutant is directly related to SCG overproduction but is not sensitive to exogenous carbon availability.
qPCR analysis of predicted regulatory targets.
To verify the validity of our microarray results, we performed quantitative PCR (qPCR) on four genes. These genes were exoY, which encodes an SCG biosynthetic enzyme, flgD, a flagellar hook formation protein, virG, a response regulator activated by VirA, and rctB, an activator of pAtC58 conjugation. The results were consistent with the microarray and phenotypic results that we observed (Table 2). The ΔexoR mutant displayed enhanced expression (FC = 13.5) of exoY, as predicted by its hypermucoid phenotype and the microarray analysis (log2 FC = 3.8). Similarly, we saw a marked reduction in expression of the flgD promoter (FC = 0.02) in the ΔexoR mutant compared to the wild type, corresponding to the reduction in motility gene expression in the microarray (log2 FC = −1.15) and the nonmotile phenotype. The increased expression of virG (log2 FC = 3.35) in the ΔexoR mutant relative to the wild type is mirrored in the enhanced level of transcript measured by qPCR (FC = 10.20). We observed a reduction in rctB transcript in the ΔexoR mutant, as expected from the microarray and phenotypic results of pAtC58 conjugation. Also in agreement with the microarray and phenotypic results, we saw a similar pattern of expression of exoY (FC = 23.59), flgD (FC = 0.04), and virG (FC = 18.51) in the ΔexoA ΔexoR mutant relative to the wild type and an insignificant change in the transcript level of rctB (FC = 1.13). The ΔexoA mutant displayed transcript levels similar to those in the wild type for all the genes tested, with a slight increase in rctB (FC = 4.32), which is consistent with the slight increase in pAtC58 conjugation efficiency.
TABLE 2.
qPCR verification of microarray results
| Strain | Log2 FC (range) of transcript quantity relative to wild typea |
|||
|---|---|---|---|---|
| SCG biosynthesis (exoY) | Motility (flgD) | Virulence (virG) | pAt conjugation (rctB) | |
| C58 | 1.00 (0.81–1.24) | 1.00 (0.84–1.10) | 1.00 (0.79–1.26) | 1.00 (0.84–0.19) |
| ΔexoR mutant | 13.45 (11.06–16.37) | 0.02 (002–0.03) | 10.20 (8.26–12.59) | 0.25 (0.20–0.30) |
| ΔexoA mutant | 2.31 (1.91–2.80) | 3.12 (2.66–3.65) | 1.60 (1.35–1.91) | 4.32 (3.60–5.17) |
| ΔexoA ΔexoR double mutant | 23.59 (19.38–28.70) | 0.04 (0.03–0.05) | 18.51 (15.51–22.08) | 1.13 (0.94–1.38) |
Transcript quantity was calculated by comparing the threshold cycle (CT) of each transcript to the threshold cycle of sigma 70 in each strain. The ΔCT for each transcript was compared to the ΔCT of that transcript in the wild type to yield ΔΔCT, which was used to calculate the fold change (FC) of each transcript in each strain. The fluorescence threshold was set to 3,000.
The ChvG-ChvI two-component system is identified as a target of ExoR regulation in a transposon mutagenesis screen.
To discover downstream components of the ExoR regulatory pathway, a screen was performed on a mariner transposon mutant library of the ΔexoA ΔexoR mutant. The screen was designed to isolate suppressor mutants that restored the mutant's ability to swim in motility agar. We used the ΔexoA ΔexoR double mutant as the parent to avoid complications caused by the hypermucoid phenotype of the ΔexoR mutant. The transposon mutants were plated, and colonies were scraped off selective plates en masse into ATGN broth with glycerol and stored frozen.
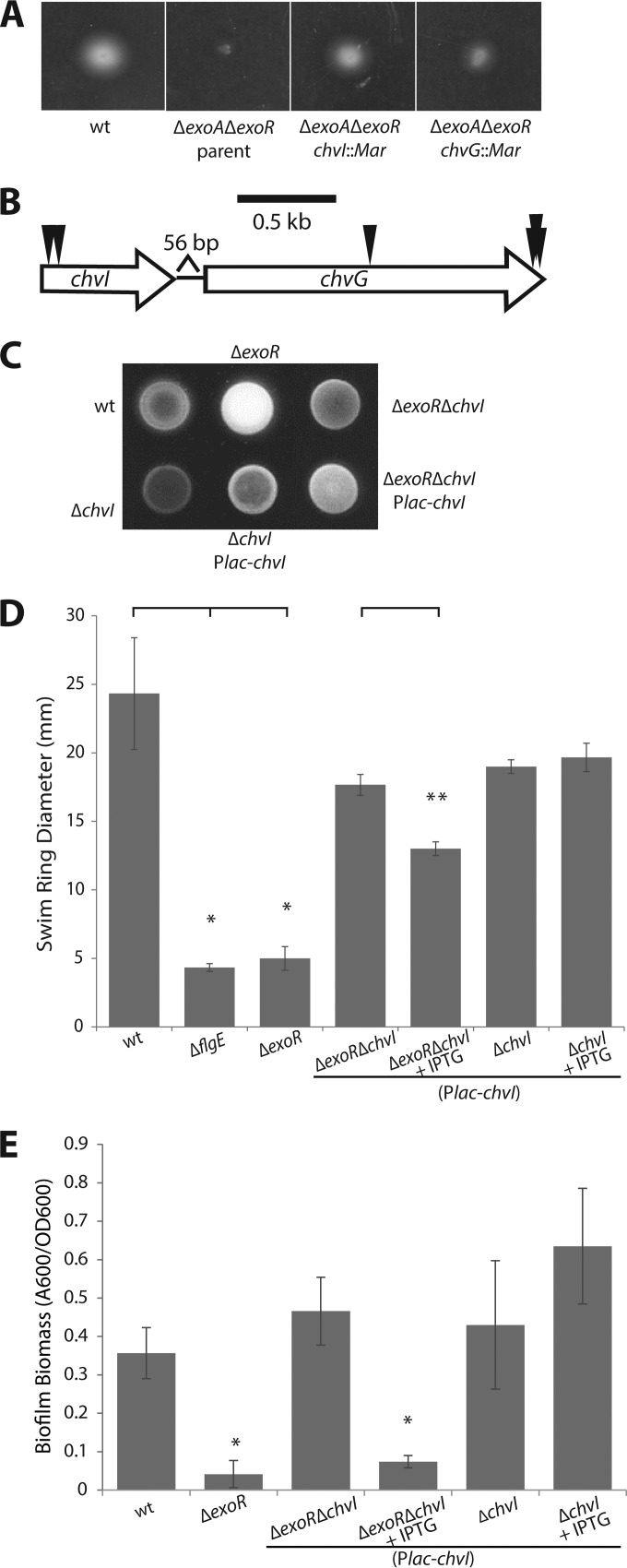
To enrich for transposon mutants that suppressed the nonmotile phenotype of the ΔexoA ΔexoR parent, 5 μl of the mutant library was inoculated into 0.3% swim agar and incubated for 3 days (Fig. 2A). Cells were streaked from the outer edge of the resulting swim ring onto solid media. Isolated colonies were then reinoculated into swim plates to confirm their motility phenotype before the site of the transposon was mapped by touchdown PCR and sequencing.
FIG 2.
Activity of the two-component system ChvG-ChvI directs exoR phenotypes. (A) Image of swim plate 3 days postinoculation of the parent ΔexoA ΔexoR strain and suppressor transposon mutants in chvI and chvG. (B) Genomic organization of the chvI-chvG locus and positions of independent transposon insertions. (C) Calcofluor white fluorescence under UV light exposure as a qualitative representation of the level of SCG biosynthesis. (D) Quantification of flagellar locomotion through swim agar (0.3%) 3 days postinoculation. Single asterisks denote a significant difference relative to the wild type (P < 0.05 with Bonferroni's correction). Double asterisks denote a significant difference between ΔexoR ΔchvI (plac-chvI) strains without and with IPTG (P = 0.009). Error bars represent standard deviations of the means of three biological replicates. (E) Biofilm formation on PVC coverslips in static culture. The A600/OD600 unit represents biofilm biomass as measured by solubilized crystal violet staining of biofilms formed on coverslips normalized to culture density. Asterisks denote a significant difference (P < 0.05 with Bonferroni's correction) compared to the wild type. Error bars represent standard deviations of the means of three biological replicates of a representative experiment.
Twenty-one transposon mutants were initially sequenced, revealing seven independent insertions in the chvI-chvG locus: two insertions in the chvI open reading frame (ORF) and five insertions in the chvG ORF (Fig. 2B). No other genes were identified to be disrupted in more than one mutant; therefore, we focused on chvG-chvI. We continued to characterize the chvI and chvG mutants in ΔexoR mutant backgrounds. It is important to note that transposon mutants with insertions at the far C terminus of chvG could be complemented with a copy of chvG in trans (data not shown), indicating that suppression of the ΔexoA ΔexoR phenotype is not due to polar effects on downstream genes.
Activity of the two-component system ChvG-ChvI causes the characteristic exoR phenotypes.
In-frame deletions were created for both the sensor kinase chvG gene and the response regulator chvI gene in wild-type and ΔexoR backgrounds. The ΔexoR ΔchvI double mutant exhibited full motility compared to the wild-type and the nonmotile ΔexoR parent strains (Fig. 2D). The single ΔchvI mutant displayed motility phenotypes similar to those of the wild type. We observed the same trends for ΔexoR ΔchvG and ΔchvG strains (data not shown). This suggests that the ChvG-ChvI system functions to repress motility in the absence of ExoR.
To determine if the hypermucoidy caused by SCG overproduction and the biofilm defect of the ΔexoR strain were also regulated via ChvG-ChvI, we investigated these phenotypes in the ΔexoR ΔchvI double mutant strain. SCG production can be qualitatively observed by growing cells on solid medium containing calcofluor white. The colony morphology of the ΔexoR ΔchvI strain—as well as that of the ΔexoR ΔchvG strain (data not shown)—is comparable to that of the wild type and distinct from the hypermucoid ΔexoR mutant morphology (Fig. 2C). Additionally, an anthrone assay—which measures the levels of free reducing sugars—revealed a return to wild-type levels of polysaccharide synthesis when either chvG or chvI was deleted in the ΔexoR background (data not shown).
The attachment defect exhibited by the exoR mutant is also suppressed when either chvG (data not shown) or chvI is deleted in an exoR background. ΔexoR ΔchvI double mutants displayed a wild-type biofilm phenotype, in stark contrast to the nonadherent ΔexoR mutant (Fig. 2E). Single-mutant strains of chvI exhibit a biofilm phenotype similar to that of the wild type (Fig. 2E). Expression of chvI in trans in the ΔexoR ΔchvI mutant led to a reduction of swimming motility (Fig. 2D) and biofilm formation (Fig. 2E), recapitulating the phenotypes of the single ΔexoR mutant. All of these phenotypes are consistent between chvI and chvG mutants (data not shown). These observations suggest that ChvG-ChvI plays a substantial role in inhibiting attachment and biofilm formation when active in the exoR mutant.
exoR mutants mimic acid-exposed cells.
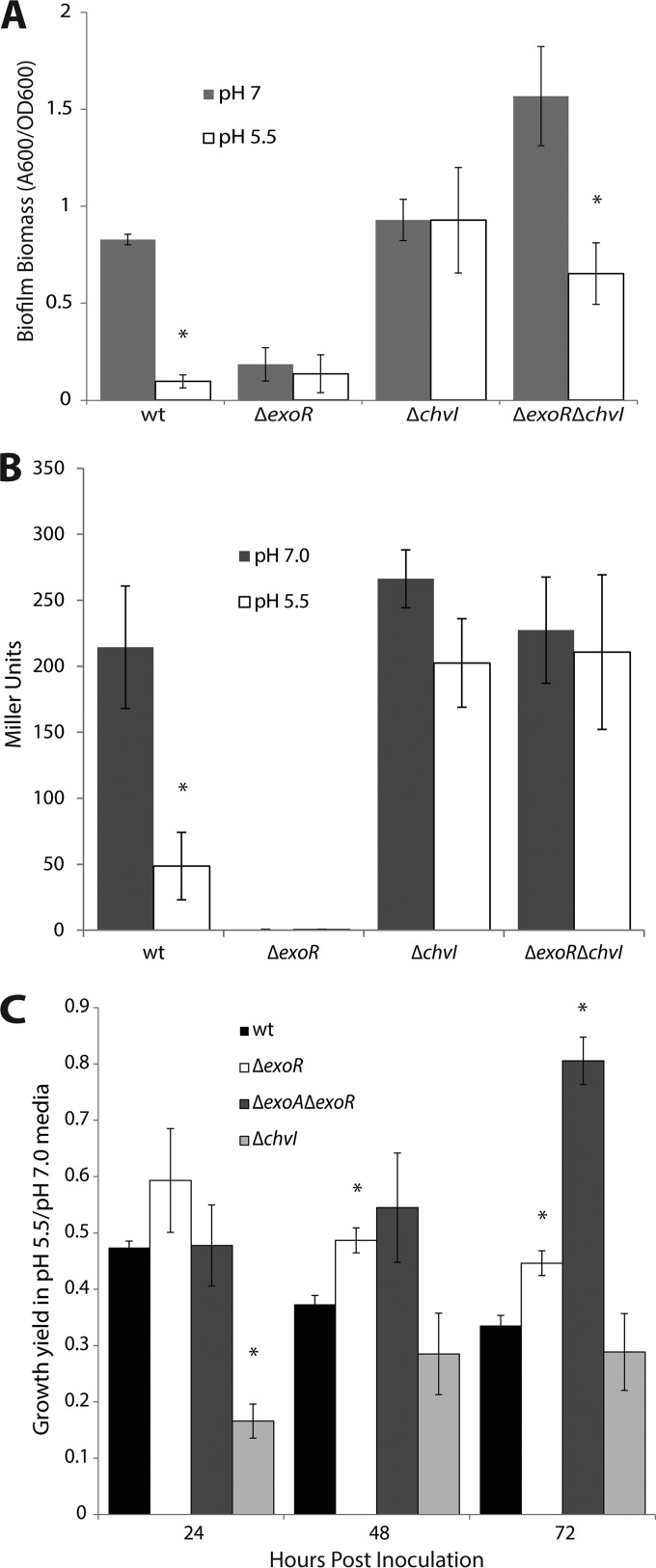
Both the exoR-chvG or -chvI epistasis evidence and the microarray data support the model by which ExoR directly inhibits the activity of ChvG-ChvI and thus suppresses the acid regulon in the absence of an external low-pH signal (22). We hypothesized that when ExoR was absent in A. tumefaciens cells, they would artificially respond to acidic conditions via hyperactivity of ChvG-ChvI. To test this, we examined characteristic exoR phenotypes under neutral and acidic conditions to determine if wild-type cells mimic exoR mutants when exposed to acid. We saw a severe deficiency of biofilm formation in wild-type cells when grown in acidic medium (pH 5.5) compared to neutral medium (Fig. 3A). This mimics the attachment deficiency observed for exoR mutants under neutral conditions and further supports the model that ChvG-ChvI actively represses biofilm formation in the absence of ExoR or in the presence of acid. In a strain lacking chvI, the response to acid is abolished, as predicted (Fig. 3A).
FIG 3.
ExoR mutants mimic acid-exposed cells. (A) Biofilm formation is inhibited under acidic conditions. Asterisks denote a significant difference (P < 0.05) between each strain's biofilm biomass in low pH compared to neutral pH. Error bars represent standard deviations of the means of three biological replicates of a representative experiment. (B) Expression of flgE is downregulated under acidic conditions. Each column represents expression of flgE in a strain grown in medium buffered to pH 5.5 or grown in neutral-pH medium. Asterisks denote a significant difference (P < 0.05) in each strain's expression of flgE in acidic pH compared to neutral pH. Error bars represent standard deviations of the means of three biological replicates. (C) Long-term acid tolerance assay. Each column represents a strain's growth in medium buffered to pH 5.5 relative to its growth in neutral medium. Asterisks denote a significant difference (P < 0.05) in the growth ratio relative to that of the wild type at each time point. Error bars represent standard errors of the means of at least three biological replicates.
To examine the effect of low pH on motility, we measured the promoter activity of the flagellar hook gene flgE in neutral and acidic (pH 5.5) media by a β-galactosidase assay. At low pH, growth is substantially slower, precluding a direct comparison using standard motility agar assays. Based on the nonmotile phenotype of exoR, our microarray data, and the acid regulon data of Yuan et al. (23), we predicted that flagellar gene expression would be reduced under acidic conditions, and this is in fact what we observed (Fig. 3B). Accordingly, we saw a blindness to acidic conditions in chvI mutants (Fig. 3B), confirming that repression of flagellar gene expression depends on ChvG-ChvI activity in exoR mutants or under acidic conditions.
The ability of exoR mutants to proliferate in low-pH media was also tested. We measured the density of wild-type and exoR mutants grown in liquid culture in neutral and acidic media for 72 h. The data are presented as a ratio of growth yield in acidic medium (pH 5.5) relative to growth yield in neutral medium (Fig. 3C). The ΔexoR mutant shows decreased overall growth yield compared to that of the wild type under neutral conditions (see Fig. S3 in the supplemental material). However, the ΔexoR mutant's growth yield in low pH relative to neutral pH is less dramatically affected than that of the wild-type strain, as reflected in a higher ratio (Fig. 3C). The growth yield of the ΔexoA ΔexoR mutant at low pH relative to neutral medium is similar to that of the wild type at 24 h but is substantially greater at 48 and 72 h, suggesting that succinoglycan biosynthesis is not necessary for survival in acid. Altogether, these data support the model whereby exoR mutants artificially experience exposure to low pH through activity of ChvG-ChvI and suggest that an aspect of SCG overproduction in the exoR mutant dampens its ability to tolerate acid pH.
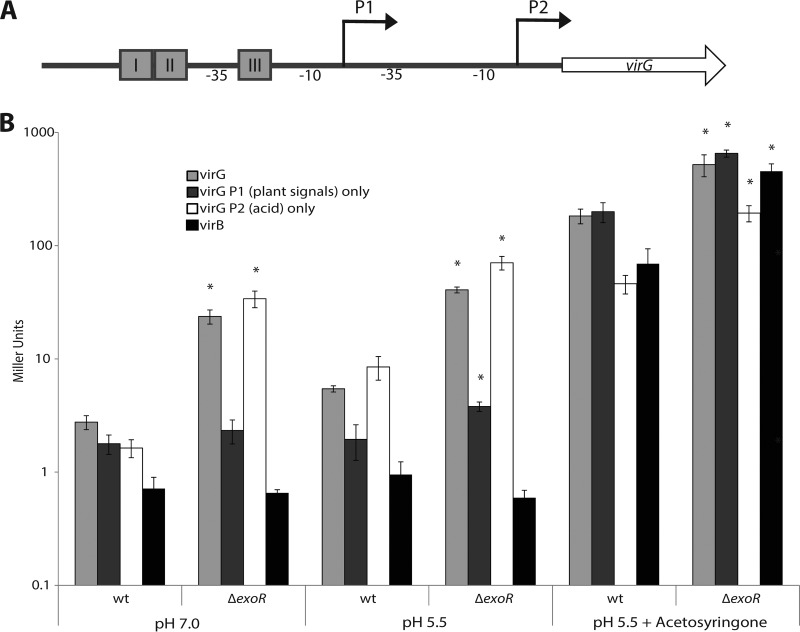
exoR mutants show enhanced expression of virG.
Phosphorylated VirG is limiting for virulence gene induction in A. tumefaciens (61). The virG gene is under the control of two promoters designated P1 and P2 (Fig. 4A); P1 responds to plant signals such as acetosyringone and low phosphate, and P2 responds to low pH (33, 62). Since exoR mutants appear to mimic acid-exposed cells, we hypothesized that expression of virG may be enhanced in exoR mutant strains in a P2-dependent fashion. To test this hypothesis, we used various plasmid-borne virG-lacZ fusions (derived from the octopine-type A. tumefaciens R10 virG gene [Fig. 4A]); one construct contains the entire promoter region of virG, one has a truncated P1 missing the upstream vir boxes (P2 only), and one has a 10-bp deletion between the −10 element and the translation start site of P2 (P1 only) (33). We determined the relative levels of β-galactosidase activity in the wild type and in the ΔexoR mutant in neutral, acidic (pH 5.5), and vir-inducing acidic media. The ΔexoR strain shows enhanced activity of the virG promoter in neutral minimal medium (Fig. 4B). Expression of virG was enhanced with the intact-promoter construct and when only P2 was present but not with the P1-only construct in the ΔexoR mutant (Fig. 4B). This enhanced expression in the ΔexoR mutant was also observed in pH 5.5 medium—when the P2 promoter was normally induced—and in vir-inducing medium when both promoters were induced (Fig. 4B). It appears that the enhanced expression of virG in the ΔexoR mutant is also manifested at the level of vir target gene expression, as the expression of a virB::lacZ fusion is markedly higher in inducing medium in the ΔexoR mutant than in the wild type (Fig. 4B). The same trends were observed in the ΔexoA ΔexoR background, so the effect of ExoR on virG expression is independent of SCG synthesis (data not shown). The enhanced expression of virG in exoR mutants and under acidic conditions is dependent on the activity of ChvG-ChvI, as no virulence gene expression was observed under any condition in a ΔchvI or ΔexoR ΔchvI mutant (data not shown). This confirms the long-standing observation that ChvG-ChvI is required for strong virulence gene expression in A. tumefaciens (20). The identifying feature for chvG and chvI mutants was in fact their avirulent phenotype (20).
FIG 4.
ExoR mutants display enhanced expression of virG through the acid-sensing promoter. (A) Promoter structure of virG showing the relative positions of the vir boxes upstream of P1, which responds to plant signals, and P2, which responds to low pH (33). (B) ΔexoR mutants display enhanced expression of virG through the acid-sensing promoter P2, even under neutral conditions. β-Galactosidase assays were performed on strains harboring plasmid-borne PvirG- and PvirB-lacZ fusions under the conditions indicated. Asterisks denote a significant difference (P < 0.05) in expression in the ΔexoR mutant relative to the wild type for each condition. Error bars represent standard errors of the means of at least three biological replicates.
DISCUSSION
This study reveals the great number and variety of genes regulated through ExoR. We show that the characteristic phenotypes of exoR mutants are caused by hyperactivity of the ChvG-ChvI two-component system. The two-component system is responsive to and active under acidic conditions, and we find that cells lacking ExoR behave similarly to cells exposed to an acidic environment. These results agree well with the study by Wu et al. (22), which showed a direct interaction between ExoR and ChvG that is disrupted when cells are exposed to acidic medium. Accordingly, we see a substantial overlap between the genes defined in the “acid regulon” by Yuan et al. (23) and the ExoR regulon. The exo genes responsible for SCG biosynthesis, the imp type VI secretion system genes, various motility- and chemotaxis-related genes, and virG are common to our defined ExoR regulon and the acid regulon. These commonalities are consistent with a tentative model of ExoR control and low-pH response (Fig. 5). However, there are some noticeable differences that manifest themselves at the defined functional category. For example, none of the acid-repressed genes categorized as “amino acid synthesis and degradation” or “nucleic acid synthesis and metabolism” genes and very few of the acid-repressed “metabolism and cofactor synthesis genes” described by Yuan et al. (23) are represented in our data set. These differences may reflect an integration of other regulatory circuits in low-pH media that are unaffected in our exoR mutants. Additionally, these differences may reflect the different carbon sources used in culture media in the two experiments, i.e., glucose in the Yuan et al. study (23) and succinate in our study.
FIG 5.
Model of ExoR regulation. Under neutral environmental conditions, ExoR represses the sensor kinase ChvG by direct interaction, preventing the phosphorylation cascade to ChvI and the regulation, both repression and activation, of ChvI-regulated genes. Under acidic environmental conditions, ExoR is cleaved by a periplasmic protease, relieving its inhibition of ChvG. This allows ChvG autophosphorylation and transfer of phosphate to ChvI, which then can bind DNA and regulate its target promoters. The T-bar line represents repression, the straight arrow represents activation, and the wavy arrow represents changes in these functional categories in both directions.
A portion of the trends that we have observed for the ΔexoR mutant are consistent with previous studies in S. meliloti (10, 15, 17, 56, 63). However, our results expand and add to these observations, including an analysis of the effects of SCG overproduction. Also, in contrast to S. meliloti, in A. tumefaciens, an exoR mutant is deficient for both motility and biofilm formation, whereas S. meliloti exoR mutants are nonmotile but elevated for biofilms. Furthermore, the A. tumefaciens ΔexoR mutant is unaffected for expression of the class I motility regulators visN-visR and rem. These data are consistent with the regulation of motility genes by ExoR and ExoS in S. meliloti as reported by Yao et al. (17) but inconsistent with trends reported by other groups, who suggest that ExoS and low pH directly influence the expression of class I motility genes (45, 46). Finally, ExoR influences genes on replicons unique to A. tumefaciens, including vir genes on the Ti plasmid and conjugation genes on the At plasmid.
Most of the functional groups that we designated in the “ΔexoR versus wild-type” comparison are maintained in the absence of SCG overproduction, as evidenced by the ΔexoA ΔexoR versus ΔexoA microarray experiment. The functional groups that are common to both analyses comprise the core ExoR regulon (Table 1), and many of these core groups are represented by the phenotypes exhibited by exoR mutants (18). All of the characteristic exoR phenotypes are suppressed when the two-component system ChvG-ChvI is disrupted. However, many genes in our data set are not present in the acid regulon. Since it is clear that the acid regulon is mediated by ChvG-ChvI (22, 23, 51), we suspect that some of the genes that fall outside it may be influenced by additional pathways. Our previous study identified exoR suppressor mutations that were not in chvG or chvI, but these were spontaneous mutants and the site(s) of these mutations was not identified (18). Using transposon mutagenesis and engineered in-frame deletions, it is clear that disruption and loss of function of either chvI or chvG efficiently suppress many of the exoR mutant phenotypes.
We initially characterized ExoR because of its impact on surface adhesion and biofilm formation (18). No obvious regulators of biofilm formation appeared in the ΔexoR versus wild-type analysis, but sinR (atu2394), a fumarate and nitrate reduction regulator (FNR)-type regulator of biofilm formation (64), was greatly decreased in the ΔexoA ΔexoR mutant relative to the ΔexoA mutant (log2 FC = 2.0). We have described SinR's role in biofilm maturation, but it does not appear to be required for attachment, so its reduced gene expression likely does not account for the attachment defect of the ΔexoA ΔexoR mutant, especially considering its absence in the microarray analysis of the equally attachment-deficient ΔexoR mutant (18, 64). We also observed modest downregulation of fnrN (atu1602) in the ΔexoA ΔexoR versus ΔexoA microarray (log2 FC = −0.6), which we have shown regulates sinR expression (64). This finding contributes another potential step in the pathway of oxygen-dependent modulation of biofilm formation that we previously described (64).
Our findings regarding the influence of ExoR on virulence through acid induction of the virG gene now provide a mechanistic link between the response to low pH and potentiation of virG expression (62). Our previous findings revealed that despite its profound attachment defects and its lack of motility, the exoR mutant was as virulent as the wild type in crude tumorigenesis assays. The enhanced level of vir gene expression in the exoR mutant may very well compensate for any deficiencies during surface colonization in these crude assays.
A recent study characterized the role that ntrX, encoding a response regulator, plays in regulating SCG biosynthesis and motility in S. meliloti (65, 66). The study finds that NtrX negatively regulates expression of SCG biosynthesis genes and positively regulates flagellin and the motility regulators visN and visR (65). These authors posit that there is no interaction between NtrY-X and ExoR-ExoS-ChvI because overexpression of chvI or exoR does not rescue the ntrX mutant phenotypes (65). An annotated homologue of this gene (atu1448) appears as part of the core ExoR regulon in our study, its expression being modestly increased in our ΔexoR mutants (log2 FCs, 0.6 in the ΔexoR mutant versus the wild type and 0.59 in the ΔexoA ΔexoR versus the ΔexoA mutants). Though we have not characterized this response regulator in A. tumefaciens C58, it is interesting that it appears as part of the ExoR regulon, given its relationship to described exoR phenotypes in S. meliloti. While the NtrY-X system may not influence the expression of ExoR-ExoS/ChvG-ChvI in S. meliloti, this does not preclude the possibility that ExoR-ChvG-ChvI modulates the expression of ntrX in A. tumefaciens.
The role that the periplasmic regulator ExoR plays in controlling gene expression in A. tumefaciens is substantial. We have elaborated on its intimate control of the acid-responsive two-component system ChvG-ChvI and lent further support to the model that ExoR is required for repressing the acid response under neutral conditions (Fig. 5). The scope of the genes regulated by ExoR extends beyond those included in the acid regulon and, presumably, beyond those directly regulated by ChvG-ChvI. Because ExoR has multiple predicted protein-protein interaction domains—a tetratricopeptide repeat (TPR) domain and two Sel1 domains—there exists the distinct possibility that it has multiple interacting partners in the periplasm and exerts control over other regulatory pathways. The connection between low pH and ExoR regulation is clear, but some genes in the ExoR regulon are influenced by other environmental signals such as low oxygen concentration (sinR [64] and ntrX [66]) and temperature (heat shock and cold shock genes [67–69]). This raises the question, does ExoR control responses to a variety of environmental signals? Another enigma about ExoR is its positive influence on both biofilm formation and motility. Motile bacteria can exist in either a motile or a sessile state, and at some point during interactions with surfaces they make a switch to one lifestyle or the other. ExoR contributes to both these states, and we are now in a position to test whether specific environmental conditions encountered by A. tumefaciens may influence its activity, global gene expression, and the state of the bacterial cell.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by National Institutes of Health (NIH) grant GM080546 (C.F.) and through a grant from the Indiana University META-Cyt program funded in part by a major endowment from the Lilly Foundation (C.F.). B.C.H. and A.D.T. were funded on the Indiana University Genetics, Molecular and Cellular Sciences Training Grant T32-GM007757.
Footnotes
Published ahead of print 30 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01751-14.
REFERENCES
- 1.Peters NK, Frost JW, Long SR. 1986. A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science 233:977–980. 10.1126/science.3738520 [DOI] [PubMed] [Google Scholar]
- 2.Chilton MD, Drummond MH, Merlo DJ, Sciaky D, Montoya AL, Gordon MP, Nester EW. 1977. Stable incorporation of plasmid DNA into higher plant cells: molecular-basis of crown gall tumorigenesis. Cell 11:263–271. 10.1016/0092-8674(77)90043-5 [DOI] [PubMed] [Google Scholar]
- 3.Drummond MH, Gordon MP, Nester EW, Chilton MD. 1977. Foreign DNA of bacterial plasmid origin is transcribed in crown gall tumors. Nature 269:535–536. 10.1038/269535a0 [DOI] [Google Scholar]
- 4.Stachel SE, Messens E, Vanmontagu M, Zambryski P. 1985. Identification of the signal molecules produced by wounded plant cells that activate T-DNA transfer in Agrobacterium tumefaciens. Nature 318:624–629. 10.1038/318624a0 [DOI] [Google Scholar]
- 5.Stachel SE, Nester EW, Zambryski PC. 1986. A plant-cell factor induces Agrobacterium tumefaciens vir gene expression. Proc. Natl. Acad. Sci. U. S. A. 83:379–383. 10.1073/pnas.83.2.379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heindl JE, Wang Y, Heckel BC, Mohari B, Feirer N, Fuqua C. 2014. Mechanisms and regulation of surface interactions and biofilm formation in Agrobacterium. Front. Plant Sci. 5:176. 10.3389/fpls.2014.00176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doherty D, Leigh JA, Glazebrook J, Walker GC. 1988. Rhizobium meliloti mutants that overproduce the R. meliloti acidic calcofluor-binding exopolysaccharide. J. Bacteriol. 170:4249–4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reed JW, Glazebrook J, Walker GC. 1991. The exoR gene of Rhizobium meliloti affects RNA levels of other exo genes but lacks homology to known transcriptional regulators. J. Bacteriol. 173:3789–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozga DA, Lara JC, Leig JA. 1994. The regulation of exopolysaccharide production is important at 2 levels of nodule development in Rhizobium meliloti. Mol. Plant-Microbe Interact. 7:758–765. 10.1094/MPMI-7-0758 [DOI] [PubMed] [Google Scholar]
- 10.Keating DH. 2007. The Sinorhizobium meliloti ExoR protein is required for the downregulation of lpsS transcription and succinoglycan biosynthesis in response to divalent cations. FEMS Microbiol. Lett. 267:23–29. 10.1111/j.1574-6968.2006.00498.x [DOI] [PubMed] [Google Scholar]
- 11.Leigh JA, Signer ER, Walker GC. 1985. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc. Natl. Acad. Sci. U. S. A. 82:6231–6235. 10.1073/pnas.82.18.6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wells DH, Chen EJ, Fisher RF, Long SR. 2007. ExoR is genetically coupled to the ExoS-Chvl two-component system and located in the periplasm of Sinorhizobium meliloti. Mol. Microbiol. 64:647–664. 10.1111/j.1365-2958.2007.05680.x [DOI] [PubMed] [Google Scholar]
- 13.Mittl PRE, Schneider-Brachert W. 2007. Sel1-like repeat proteins in signal transduction. Cell. Signal. 19:20–31. 10.1016/j.cellsig.2006.05.034 [DOI] [PubMed] [Google Scholar]
- 14.D'Andrea LD, Regan L. 2003. TPR proteins: the versatile helix. Trends Biochem. Sci. 28:655–662. 10.1016/j.tibs.2003.10.007 [DOI] [PubMed] [Google Scholar]
- 15.Chen EJ, Sabio EA, Long SR. 2008. The periplasmic regulator ExoR inhibits ExoS/ChvI two-component signalling in Sinorhizobium meliloti. Mol. Microbiol. 69:1290–1303. 10.1111/j.1365-2958.2008.06362.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang CX, Kemp J, Da Fonseca IO, Equi RC, Sheng XY, Charles TC, Sobral BWS. 2010. Sinorhizobium meliloti 1021 loss-of-function deletion mutation in chvI and its phenotypic characteristics. Mol. Plant-Microbe Interact. 23:153–160. 10.1094/MPMI-23-2-0153 [DOI] [PubMed] [Google Scholar]
- 17.Yao SY, Luo L, Har KJ, Becker A, Ruberg S, Yu GQ, Zhu JB, Cheng HP. 2004. Sinorhizobium meliloti ExoR and ExoS proteins regulate both succinoglycan and flagellum production. J. Bacteriol. 186:6042–6049. 10.1128/JB.186.18.6042-6049.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomlinson AD, Ramey-Hartung B, Day TW, Merritt PM, Fuqua C. 2010. Agrobacterium tumefaciens ExoR represses succinoglycan biosynthesis and is required for biofilm formation and motility. Microbiology 156:2670–2681. 10.1099/mic.0.039032-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mantis NJ, Winans SC. 1993. The chromosomal response regulatory gene chvI of Agrobacterium-tumefaciens complements an Escherichia coli phoB mutation and is required for virulence. J. Bacteriol. 175:6626–6636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charles TC, Nester EW. 1993. A chromosomally encoded 2-component sensory transduction system is required for virulence of Agrobacterium tumefaciens. J. Bacteriol. 175:6614–6625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu H-Y, Luo L, Yang M-H, Cheng H-P. 2012. Sinorhizobium meliloti ExoR is the target of periplasmic proteolysis. J. Bacteriol. 194:4029–4040. 10.1128/JB.00313-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu C-F, Lin J-S, Shaw G-C, Lai E-M. 2012. Acid-induced type VI secretion system is regulated by ExoR-ChvG/ChvI signaling cascade in Agrobacterium tumefaciens. PLoS Pathog. 8:e1002938–e1002938. 10.1371/journal.ppat.1002938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan ZC, Liu P, Saenkham P, Kerr K, Nester EW. 2008. Transcriptome profiling and functional analysis of Agrobacterium tumefaciens reveals a general conserved response to acidic conditions (pH 5.5) and a complex acid-mediated signaling involved in Agrobacterium-plant interactions. J. Bacteriol. 190:494–507. 10.1128/JB.01387-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 25.Mersereau M, Pazour GJ, Das A. 1990. Efficient transformation of Agrobacterium tumefaciens by electroporation. Gene 90:149–151. 10.1016/0378-1119(90)90452-W [DOI] [PubMed] [Google Scholar]
- 26.Tempé J, Petit A, Holsters M, Montagu MV, Schell J. 1977. Thermosensitive step associated with transfer of the Ti plasmid during conjugation: possible relation to transformation in crown gall. Proc. Natl. Acad. Sci. U. S. A. 74:2848–2849. 10.1073/pnas.74.7.2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lampe DJ, Akerley BJ, Rubin EJ, Mekalanos JJ, Robertson HM. 1999. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc. Natl. Acad. Sci. U. S. A. 96:11428–11433. 10.1073/pnas.96.20.11428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu J, Kim J, Koestler BJ, Choi J-H, Waters CM, Fuqua C. 2013. Genetic analysis of Agrobacterium tumefaciens unipolar polysaccharide production reveals complex integrated control of the motile-to-sessile switch. Mol. Microbiol. 89:929–948. 10.1111/mmi.12321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merritt PA, Danhorn T, Fuqua C. 2007. Motility and chemotaxis in Agrobacterium tumefaciens surface attachment and biofilm formation. J. Bacteriol. 189:8005–8014. 10.1128/JB.00566-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hibbing ME, Fuqua C. 2011. Antiparallel and interlinked control of cellular iron levels by the Irr and RirA regulators of Agrobacterium tumefaciens. J. Bacteriol. 193:3461–3472. 10.1128/JB.00317-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan SR, Gaines J, Roop RM, Farrand SK. 2008. Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl. Environ. Microbiol. 74:5053–5062. 10.1128/AEM.01098-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morton ER, Platt TG, Fuqua C, Bever JD. 2014. Non-additive costs and interactions alter the competitive dynamics of co-occurring ecologically distinct plasmids. Proc. Roy. Soc. B 281:20132173. 10.1098/rspb.2013.2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winans SC. 1990. Transcriptional induction of an Agrobacterium regulatory gene at tandem promoters by plant-released phenolic compounds, phosphate starvation, and acidic growth media. J. Bacteriol. 172:2433–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 35.Anantharaman V, Aravind L. 2003. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biology 4:R11. 10.1186/gb-2003-4-2-r11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wuenscher MD, Kohler S, Bubert A, Gerike U, Goebel W. 1993. The iap gene of Listeria monocytogenes is essential for cell viability, and its gene product, P60, has bacteriolytic activity. J. Bacteriol. 175:3491–3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tran S-L, Guillemet E, Gohar M, Lereclus D, Ramarao N. 2010. CwpFM (EntFM) is a Bacillus cereus potential cell wall peptidase implicated in adhesion, biofilm formation, and virulence. J. Bacteriol. 192:2638–2642. 10.1128/JB.01315-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh SK, SaiSree L, Amrutha RN, Reddy M. 2012. Three redundant murein endopeptidases catalyse an essential cleavage step in peptidoglycan synthesis of Escherichia coli K12. Mol. Microbiol. 86:1036–1051. 10.1111/mmi.12058 [DOI] [PubMed] [Google Scholar]
- 39.Ishikawa S, Hara Y, Ohnishi R, Sekiguchi J. 1998. Regulation of a new cell wall hydrolase gene, cwlF, which affects cell separation in Bacillus subtilis. J. Bacteriol. 180:2549–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boigegrain RA, Salhi I, Alvarez-Martinez MT, Machold J, Fedon Y, Arpagaus M, Weise CH, Rittig M, Rouot B. 2004. Release of periplasmic proteins of Brucella suis upon acidic shock involves the outer membrane protein Omp25. Infect. Immun. 72:5693–5703. 10.1128/IAI.72.10.5693-5703.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jia YH, Li LP, Hou QM, Pan SQ. 2002. An Agrobacterium gene involved in tumorigenesis encodes an outer membrane protein exposed on the bacterial cell surface. Gene 284:113–124. 10.1016/S0378-1119(02)00385-2 [DOI] [PubMed] [Google Scholar]
- 42.Foreman DL, Vanderlinde EM, Bay DC, Yost CK. 2010. Characterization of a gene family of outer membrane proteins (ropB) in Rhizobium leguminosarum bv. viciae VF39SM and the role of the sensor kinase ChvG in their regulation. J. Bacteriol. 192:975–983. 10.1128/JB.01140-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim WS, Sun-Hyung J, Park RD, Kim KY, Krishnan HB. 2005. Sinorhizobium fredii USDA257 releases a 22-kDa outer membrane protein (Omp22) to the extracellular milieu when grown in calcium-limiting conditions. Mol. Plant-Microbe Interact. 18:808–818. 10.1094/MPMI-18-0808 [DOI] [PubMed] [Google Scholar]
- 44.Cheng HP, Walker GC. 1998. Succinoglycan production by Rhizobium meliloti is regulated through the ExoS-ChvI two-component regulatory system. J. Bacteriol. 180:20–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hellweg C, Puehler A, Weidner S. 2009. The time course of the transcriptomic response of Sinorhizobium meliloti 1021 following a shift to acidic pH. BMC Microbiol. 9:37. 10.1186/1471-2180-9-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoang HH, Gurich N, Gonzalez JE. 2008. Regulation of motility by the ExpR/Sin quorum-sensing system in Sinorhizobium meliloti. J. Bacteriol. 190:861–871. 10.1128/JB.01310-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen LS, Chen YC, Wood DW, Nester EW. 2002. A new type IV secretion system promotes conjugal transfer in Agrobacterium tumefaciens. J. Bacteriol. 184:4838–4845. 10.1128/JB.184.17.4838-4845.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perez-Mendoza D, Sepulveda E, Pando V, Munoz S, Nogales J, Olivares J, Soto MJ, Herrera-Cervera JA, Romero D, Brom S, Sanjuan J. 2005. Identification of the rctA gene, which is required for repression of conjugative transfer of rhizobial symbiotic megaplasmids. J. Bacteriol. 187:7341–7350. 10.1128/JB.187.21.7341-7350.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winans SC. 1992. 2-way chemical signaling in Agrobacterium-plant interactions. Microbiol. Rev. 56:12–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang HW, Wirawan IGP, Kojima M. 1994. Cellular-localization and functional analysis of the protein encoded by the chromosomal virulence gene (acvB) of Agrobacterium tumefaciens. Biosci. Biotechnol. Biochem. 58:2024–2032. 10.1271/bbb.58.2024 [DOI] [PubMed] [Google Scholar]
- 51.Li LP, Jia YH, Hou QM, Charles TC, Nester EW, Pan SQ. 2002. A global pH sensor: Agrobacterium sensor protein ChvG regulates acid-inducible genes on its two chromosomes and Ti plasmid. Proc. Natl. Acad. Sci. U. S. A. 99:12369–12374. 10.1073/pnas.192439499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tibor A, Decelle B, Letesson JJ. 1999. Outer membrane proteins Omp10, Omp16, and Omp19 of Brucella spp. are lipoproteins. Infect. Immun. 67:4960–4962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sohlenkamp C, Galindo-Lagunas KA, Guan Z, Vinuesa P, Robinson S, Thomas-Oates J, Raetz CRH, Geiger O. 2007. The lipid lysyl-phosphatidylglycerol is present in membranes of Rhizobium tropici CIAT899 and confers increased resistance to polymyxin B under acidic growth conditions. Mol. Plant-Microbe Interact. 20:1421–1430. 10.1094/MPMI-20-11-1421 [DOI] [PubMed] [Google Scholar]
- 54.Oppenheimer M, Valenciano AL, Sobrado P. 2011. Isolation and characterization of functional Leishmania major virulence factor UDP-galactopyranose mutase. Biochem. Biophys. Res. Commun. 407:552–556. 10.1016/j.bbrc.2011.03.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gruber TD, Borrok MJ, Westler WM, Forest KT, Kiessling LL. 2009. Ligand binding and substrate discrimination by UDP-galactopyranose mutase. J. Mol. Biol. 391:327–340. 10.1016/j.jmb.2009.05.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bélanger L, Charles TC. 2013. Members of the Sinorhizobium meliloti ChvI regulon identified by a DNA binding screen. BMC Microbiol. 13:132. 10.1186/1471-2180-13-132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tomlinson AD. 2010. PhD thesis Indiana University, Bloomington, IN [Google Scholar]
- 58.Adams MD, Wagner LM, Graddis TJ, Landick R, Antonucci TK, Gibson AL, Oxender DL. 1990. Nucleotide-sequence and genetic characterization reveal 6 essential genes for the LIV-I and LS transport systems of Escherichia coli. J. Biol. Chem. 265:11436–11443 [PubMed] [Google Scholar]
- 59.Bell AW, Buckel SD, Groarke JM, Hope JN, Kingsley DH, Hermodson MA. 1986. The nucleotide-sequences of the RbsD, RbsA, and RbsC genes of Escherichia coli K12. J. Biol. Chem. 261:7652–7658 [PubMed] [Google Scholar]
- 60.Bélanger L, Dimmick KA, Fleming JS, Charles TC. 2009. Null mutations in Sinorhizobium meliloti exoS and chvI demonstrate the importance of this two-component regulatory system for symbiosis. Mol. Microbiol. 74:1223–1237. 10.1111/j.1365-2958.2009.06931.x [DOI] [PubMed] [Google Scholar]
- 61.Jin SG, Komari T, Gordon MP, Nester EW. 1987. Genes responsible for the supervirulence phenotype of Agrobacterium tumefaciens A281. J. Bacteriol. 169:4417–4425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mantis NJ, Winans SC. 1992. The Agrobacterium tumefaciens vir gene transcriptional activator virG is transcriptionally induced by acid pH and other stress stimuli. J. Bacteriol. 174:1189–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen EJ, Fisher RF, Perovich VM, Sabio EA, Long SR. 2009. Identification of direct transcriptional target genes of ExoS/ChvI two-component signaling in Sinorhizobium meliloti. J. Bacteriol. 191:6833–6842. 10.1128/JB.00734-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramey BE, Matthysse AG, Fuqua C. 2004. The FNR-type transcriptional regulator SinR controls maturation of Agrobacterium tumefaciens biofilms. Mol. Microbiol. 52:1495–1511. 10.1111/j.1365-2958.2004.04079.x [DOI] [PubMed] [Google Scholar]
- 65.Wang D, Xue H, Wang Y, Yin R, Xie F, Luo L. 2013. The Sinorhizobium meliloti ntrX gene is involved in succinoglycan production, motility, and symbiotic nodulation on alfalfa. Appl. Environ. Microbiol. 79:7150–7159. 10.1128/AEM.02225-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.del Carmen Carrica M, Fernandez I, Adrian Marti M, Paris G, Alberto Goldbaum F. 2012. The NtrY/X two-component system of Brucella spp. acts as a redox sensor and regulates the expression of nitrogen respiration enzymes. Mol. Microbiol. 85:39–50. 10.1111/j.1365-2958.2012.08095.x [DOI] [PubMed] [Google Scholar]
- 67.Jomaa A, Iwanczyk J, Tran J, Ortega J. 2009. Characterization of the autocleavage process of the Escherichia coli HtrA protein: implications for its physiological role. J. Bacteriol. 191:1924–1932. 10.1128/JB.01187-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lipinska B, Fayet O, Baird L, Georgopoulos C. 1989. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at elevated temperatures. J. Bacteriol. 171:1574–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lipinska B, Sharma S, Georgopoulos C. 1988. Sequence-analysis and regulation of the htrA gene of Escherichia coli: a sigma-32-independent mechanism of heat-inducible transcription. Nucleic Acids Res. 16:10053–10067. 10.1093/nar/16.21.10053 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.