Abstract
Background
Genome-wide association studies (GWAS) for body mass index (BMI) previously identified a locus near TMEM18. We conducted targeted sequencing of this region to investigate the role of common, low frequency, and rare variation influencing BMI.
Methods and Results
We sequenced TMEM18 and regions downstream of TMEM18 on chromosome 2 in 3976 individuals of European ancestry from three community-based cohorts (Atherosclerosis Risk in Communities, Cardiovascular Health Study and Framingham Heart Study), including 200 adults selected for high BMI. We examined the association between BMI and variants identified in the region from nucleotide position 586,432 to 677,539 (hg18). Rare variants (MAF <1%) were analyzed using a burden test and the Sequence Kernel of Association Test (SKAT). Results from the three cohort studies were meta-analyzed. We estimate that mean BMI is 0.43 kg/m2 higher for each copy of the G allele of SNP rs7596758 (MAF=29%, p=3.46 × 10−4) using a Bonferroni threshold of p <4.6 × 10−4). Analyses conditional on previous GWAS SNPs associated with BMI in the region led to attenuation of this signal and uncovered another independent (r2<0.2), statistically significant association, rs186019316 (p=2.11 × 10−4). Both rs186019316 and rs7596758 or proxies are located in transcription factor binding regions. No significant association with rare variants was found in either the exons of TMEM18 or the 3’ GWAS region.
Conclusions
Targeted sequencing around TMEM18 identified two novel BMI variants with possible regulatory function.
Keywords: body mass index, genetic association, targeted resequencing, TMEM18
Introduction
Body mass index (BMI), an important risk factor for diabetes mellitus and coronary heart disease, is a complex trait with variation attributable to both environmental and genetic factors. The heritability of BMI has been variously estimated between 25–75% overall, indicating a substantial genetic influence1, and both common and rare variants have been associated with BMI. Rare mutations have been identified in families with Mendelian forms of obesity, where family members carrying these highly penetrant rare variants typically demonstrate extreme obesity with early onset in childhood2–4. In the general population, the Genetic Investigation of Anthropometric Traits (GIANT) consortium identified 32 common variants through genome-wide association studies (GWAS), including loci FTO, TMEM18 and MC4R5, associated with BMI.
While the large sample sizes of GWAS have identified loci strongly associated with BMI, together they explain less than 2% of the total variation in BMI5. The majority of variants identified by GWAS are not obviously functional, and associated variants may simply serve as markers for the underlying functional variants. This study evaluated the TMEM18 region (~91kb) including the TMEM18 gene and downstream, using targeted deep resequencing. Study goals were (1) to localize functional variants in this region and (2) to determine whether low frequency and rare functional variants contribute additionally to the genetic signal for BMI in this region. We chose this region for two reasons: first, it contains one of the primary replicated genome-wide signals for BMI5, 6 and second, the GWA signal is intergenic, downstream (3’) of the TMEM18 gene and therefore not covered by ongoing exome sequencing efforts in these populations. In addition, FTO is a well-established locus that has attracted the efforts of other groups such as Almen et al. (2013) 7. Considering the limited resources available for each Phenotype Group in the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE)8 Targeted Sequencing study, we chose TMEM18 and its downstream region to maximize our contribution and impact on the field.
We used data from the CHARGE Targeted Sequencing Study, which completed targeted resequencing and analysis of 3976 individuals with BMI of European ancestry from three community-based cohorts, Atherosclerosis Risk in Communities (ARIC), Cardiovascular Health Study (CHS), and Framingham Heart Study (FHS).
Methods
Study Design and Samples
Briefly, the CHARGE Targeted Sequencing Study implemented a case-cohort study design, in which both a random sample of participants (Cohort Random Sample) and participants with extreme values on 14 traits (14 Phenotype Groups) were selected from each of the three participating cohorts 9. The Phenotype Group for the BMI trait included participants from each cohort who were selected as having the highest BMI levels for their sex and age, 100 from ARIC, 50 from CHS, and 50 from FHS. Our analysis included the 194 participants selected on the basis of high BMI, 1862 participants in other Phenotype Groups, and 1920 participants from the Cohort Random Sample (Table 1). In total, we analyzed 3,974 individuals with available BMI measurements in the Cohort Random Sample and Phenotype Groups (Table 1). To avoid confounding by race, all participants were of European ancestry. In addition, all participants in this study provided informed consent for the use of their genetic and other data to their local institutions. All studies were approved by institutional review committee as well as the informed consent of subjects.
Table 1.
Characteristics of Study Samples in Participanting Cohorts
| Cohort | Sample size |
Age, yrs mean±SD |
Women n (%) | BMI, kg/m2, mean±SD |
BMI, kg/m2, (min, median, 95%-tile, max) |
|---|---|---|---|---|---|
| Sampling Scheme | |||||
| ARIC | 1754 | 54.9±5.7 | 845 (48.2%) | 27.6±6.0 | (14.4, 26.4, 40.2, 56.3) |
| High BMI | 75 | 52.7±5.1 | 35(46.7%) | 45.2±4.4 | (39.0, 45.1, 54.6, 56.3) |
| Random Controls | 810 | 54.5±5.7 | 398(49.1%) | 26.7±4.7 | (15.7, 26.0, 35.2, 51.6) |
| Other phenotype group | 869 | 55.4±5.7 | 412(47.4%) | 27.0±4.8 | (14.4, 26.6, 35.7, 43.0) |
| CHS | 1126 | 72.5±5.5 | 604 (53.6) | 26.7±5.1 | (15.3, 26.0, 35.4, 48.3) |
| High BMI | 59 | 71.6±6.3 | 28 (47.5) | 39.9±4.8 | (32.5, 40.7, 46.7, 48.3) |
| Random Controls | 458 | 72.5±5.4 | 236 (51.5) | 26.3±3.9 | (15.6, 26.1, 33.2, 40.1) |
| Other phenotype group | 609 | 72.5±5.5 | 340 (55.8) | 25.7±4.1 | (15.3, 25.5, 33.2, 39.1) |
| FHS* | 1096 | 37.0±9.7 | 564(51.5) | 26.3±6.4 | (15.7,25.0, 40.5,60.6) |
| High BMI | 58 | 40.6±9.5 | 29(50.0) | 46.8±5.0 | (36.9,46.1,56.5,60.6) |
| Random Controls | 501 | 36.3±9.3 | 249(49.7) | 25.1±4.2 | (15.9,24.7,33.0,40.4) |
| Other phenotype group | 537 | 37.3±9.9 | 286(53.3) | 25.1±4.1 | (15.7, 24.7,32.2,45.2) |
Exam 1 data were used for this analysis.
Sequence Data, QC, and Bioinformatics for Functional Annotation
We used targeted resequencing, covering the region from 586,432 bp to 677,539 bp (hg18), and excluding regions with high GC content or which were highly conserved, for a total sequencing of 71,457 bp. Sequences were custom captured by a NimbleGen Capture array and sequenced using the ABI SOLiD V4.0 platform. Sequence read alignment (mapping) was performed using the BFAST10 algorithm, based on the hg 18 reference genome (NCBI Genome Build 36). SAMtools11 was used to convert the aligned read information into pileup data. The pileup results were filtered producing variant calls and annotated using the ANNOVAR12 software package. Details are fully provided in a separate manuscript 9.
Information for the resequenced region from dbSNP build 135 was retrieved from the NCBI ftp site13, 14. Functional annotations from dbSNP were parsed and RsMergeArch was used to create mappings of all alias SNP identifiers. Regulatory variants were assessed using the current hg19 version of transcription factor binding sites from the ENCODE project, with positions translated to hg1815. These were determined by experiments using chromatin immunoprecipitation combined with next-generation sequencing (ChIP-seq) technologies. Reported variants were confirmed using regulatory information from the HaploReg database16.
Statistical Approach
Details of the statistical approach are available 9.
Single Variant Analysis
We defined common variants as SNPs with at least 50 individuals carrying one or more minor alleles among all participants sequenced; this was approximately equal to 1% allele frequency. For each common variant, we conducted an association analysis, using linear regression for ARIC and CHS samples and a linear mixed effects model for related FHS samples, with BMI as the dependent variable, each single variant as the primary independent variable, and adjustment for sex, age, age-squared, study center and principal components, as appropriate, to account for possible population stratification. Principal components were calculated using genome-wide genotype data within each cohort. We performed unweighted regression analyses to obtain p-values for associations and weighted regression analyses to obtain effect estimates that accounted for the sampling design (Lumley T, Dupuis J, Rice KM, Barbalic M, Bis JC, Cupples LA, et al. http://stattech.wordpress.fos.auckland.ac.nz/files/2012/05/design-paper.pdf). We applied a Bonferroni-corrected threshold to determine statistical significance of association for common variants, defined as 0.05 divided by the effective number of independent SNPs in the targeted region17: 4.6x10−4 =0.05/108, where the effective number of independent SNPs out of 288 common variants is calculated based on the eigenvalues of the genetics correlation matrix proposed by Li and Ji17.
We performed two conditional analyses: each added one of the two regional SNPs (rs6548238 and rs2867125) previously identified by GIANT GWAS into the regression model as a covariate and tested all remaining regional SNPs for association. The interrogated region, near the TMEM18 gene, was first identified by the GIANT consortium in 20096 and the lead SNP, rs6548238, is located 3’ of the gene at 624,905 bp (hg18), a region that contains no other genes. This finding was later confirmed by analysis of data from a larger group of studies in GIANT5 with a different lead SNP, rs2867125 at 612827bp (hg18), which is in complete linkage disequilibrium with the SNP from the original report according to the HapMap CEU 1000G Pilot 1 data (r2 and D’ = 1). Additionally, we conditioned on a SNP identified in the unconditional analyses of these data and tested all remaining SNPs in the region.
Rare Variant Analysis
For variants with minor allele frequency less than 1%, we conducted analyses that collapsed or jointly modeled rare variants within our genomic region. Multiple variant tests have lower power to detect association when they include non-functional variants18, 19. Hence, we conducted rare variant analyses in three ways: the first included all rare SNPs, the second included all ten rare non-synonymous or splice-site SNPs within TMEM18 exons, and the third included all 370 SNPs in the region located in transcription factor binding sites according to ENCODE criteria20. We used a T1 count test by analyzing the association of an aggregated statistic (the number of variants with at least one minor allele present). In addition, we conduct a Sequence Kernel Association Test (SKAT) to detect associations from variants with possibly different directions of effect.
Meta-Analysis
For single-variant analysis of common SNPs and T1 analysis of rare SNPs, we performed fixed-effects, inverse-variance weighted meta-analysis of study-specific association results to combine the association evidence across studies. For SKAT, we combined score and information components of the study-specific statistics to compute a combined SKAT statistic that adjusted for study 9.
Results
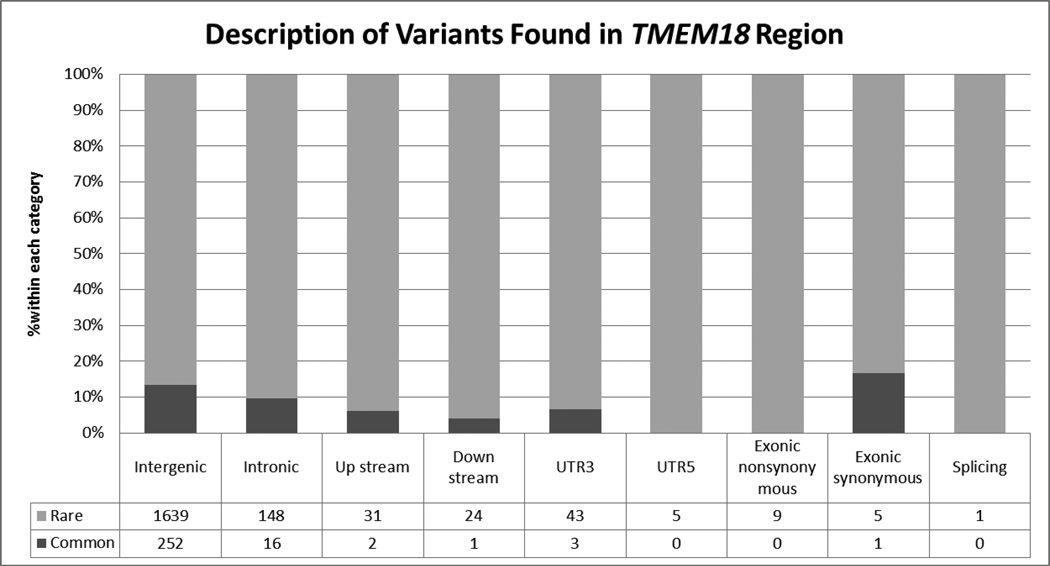
We analyzed a total of 2,180 variants, including 1,629 novel variants not found in the 1000 Genomes Project, in or near TMEM18 with minor allele frequencies ranging from 0.0002 to 0.49. (Figure 1) A total of 288 variants were considered to be common. The vast majority of the variants were intergenic. Of the 2180 investigated variants, 405 (35 common) were located in transcription factor binding regions assayed by ChIP-seq. In the subset of 288 common variants, there were 43 independent (r2<0.2) and nominally significant (p<0.05) SNPs associated to BMI, 8 of them in regulatory regions (Supplementary Table 1).
Figure 1.
Description of Variants Found in TMEM18 Region. There are 2180 variants (288 common variants) in total found in chr2:586432-677539 region. The classification of these variants includes 1891 variants annotated as intergenic, 164 intronic, 33 upstream, 25 downstream, 46 UTR3, 5 UTR5, 9 exonic nonsynonymous, 6 exonic synonymous, and 1 splicing. The figure shows the percent of variants within each category stratified by the allele frequency (rare/common variant). In addition, among 2180 variants, 405 (35 common) are in the transcriptional region.
Single Variant Results
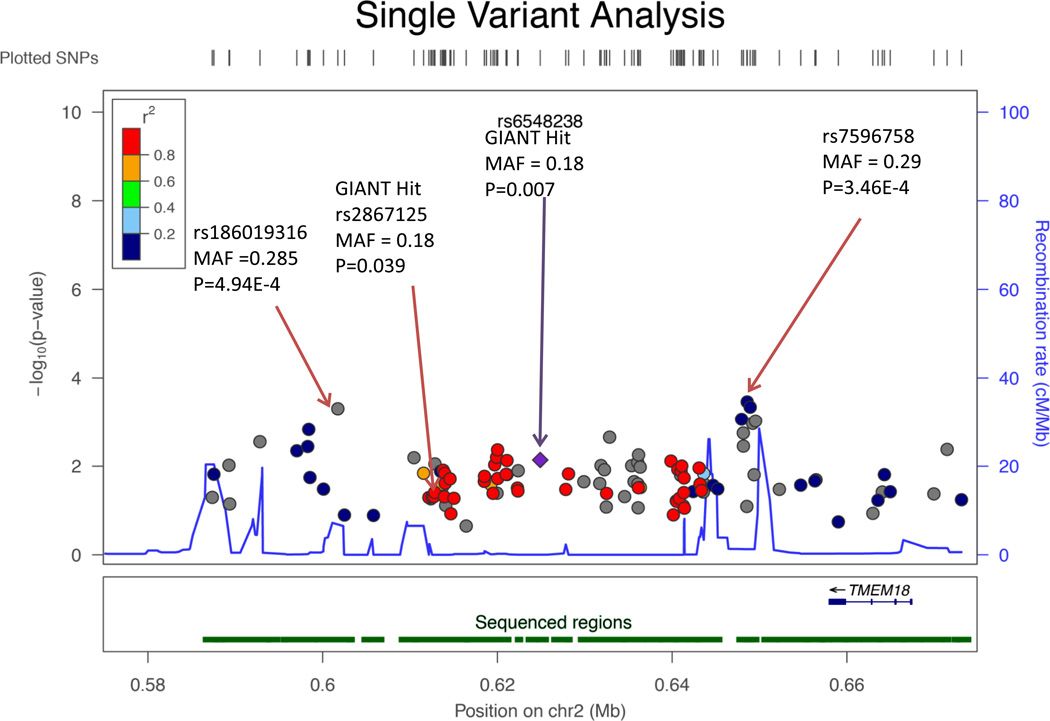
The SNPs previously reported by GIANT GWAS to be associated with BMI5, 6, rs6548238 and rs2867125, displayed nominally significant (p = 0.007 and 0.039, respectively) associations with BMI in our data (Table 2 and Figure 2). Our samples in this study are subsets of GIANT GWAS and our results for these variants are consistent with those of GIANT, with the same direction of effect (C allele of each variant increasing BMI) and a greater magnitude of effect in our data (0.46 vs. 0.26 for rs6548238 and 0.41 vs. 0.31 in GIANT for rs2867125)5, 6. Allele frequencies for these variants (18%) are similar in our data to those of GIANT (17% and 16%).
Table 2.
Single Variant Analysis Results
| rs-number | Position | EA* | NEA* | Effect † | SE † | P | D‡ | I2‡ | Het-P‡ | n | EAF* | HWE-P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Previously Identified Variants | ||||||||||||
| rs6548238§ | 624905 | T | C | −0.462 | 0.167 | 7.27 × 10−3 | --- | 0 | 0.41 | 3963 | 0.175 | 0.076 |
| rs2867125‖ | 612827 | T | C | −0.406 | 0.168 | 3.91 × 10−2 | --- | 0 | 0.59 | 3880 | 0.175 | 0.018 |
| Significant Variants with p-value < 4.6*10−4 | ||||||||||||
| rs7596758 | 648595 | G | T | 0.433 | 0.143 | 3.46 × 10−4 | ++- | 67 | 0.05 | 3927 | 0.288 | 0.104 |
EA: Effect allele; NEA: non-effect allele; EAF: effect allele frequency
Effect and SE were obtained from analysis with sampling weight adjustment
D: direction of effect; I2 % of variation due to heterogeneity; Het-P: heterogeneity test p-value
Variant identified from Willer et al. 20096
Variant identified from Speiliotes et al. 20105
Figure 2.
Regional Association Plot for the Single Variant Analysis. The most significant variant rs7596758 among a cluster of variants which are more significant than the GIANT Hit (rs6548238 and rs2867125 with nominally significant p<0.05) is significant after adjusting for multiple testing. SNP rs186019316 is not significant with p = 4.94E-4 in the single variant analysis but is significant in the analysis conditioning on rs6548238 and rs2867125, respectively.
We identified another significantly associated variant in this region – rs7596758 (MAF = 28.8%, p = 3.46E-4) (Table 2, Figure 2) in a cluster of several variants all showing association. For each additional G allele of this variant, we estimate that mean BMI is higher by 0.43 kg/m2. While this variant is in perfect linkage disequilibrium with both GIANT GWAS SNPs (D’=1), it is not correlated (r2=0.08), according to 1000G Pilot 1. Conditioning on our top SNP, rs7596758, substantially weakened the estimated effect sizes of the GIANT GWAS signals (Table 3) from a magnitude greater than 0.4 to less than 0.1. In contrast, conditioning on either GIANT signal only modestly lowered the effect size of our top signal rs7596758 from 0.43 to ~0.30. Analysis conditional on rs2867125 also identified an additional significant variant rs186019316 (p-value < 4.6E-4, MAF=2.2%), which is only weakly correlated with the GWAS SNPs and our top hit, rs7596758 (r2 < 0.01 in ARIC data).
Table 3.
Association Results of the Top Signals in Un-conditional Analysis, the GIANT Hits and Significant Variants for Analysis Conditional on the Top Signals in Our Data and the GIANT Hits
| rs-number | Position | EA* | NEA* | Effect † | SE † | P | D | I2 | Het-P | n | EAF* | HWE-p |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - Conditional on rs6548238‡: GIANT GWAS Variant | ||||||||||||
| rs186019316 | 601740 | A | G | −1.37 | 0.6 | 1.95 × 10−4 | --+ | 87 | 0.0004 | 3762 | 0.022 | 0.094 |
| rs2867125 | 612827 | T | C | 0.04 | 0.6 | 3.97 × 10−1 | +-+ | 0 | 0.6833 | 3880 | 0.177 | 0.018 |
| rs7596758 | 648595 | G | T | 0.29 | 0.2 | 2.12 × 10−3 | ++- | 69 | 0.0400 | 3927 | 0.288 | 0.104 |
| - Conditional on rs2867125§: GIANT GWAS Variant | ||||||||||||
| rs186019316 | 601740 | A | G | −1.38 | 0.4 | 2.03 × 10−4 | --+ | 88 | 0.0002 | 3635 | 0.022 | 0.094 |
| rs6548238 | 624905 | T | C | −0.39 | 0.6 | 5.40 × 10−2 | --- | 0 | 0.4617 | 3780 | 0.175 | 0.076 |
| rs7596758 | 648595 | G | T | 0.31 | 0.2 | 1.36 × 10−3 | ++- | 74 | 0.0200 | 3765 | 0.288 | 0.104 |
| - Conditional on rs7596758 | ||||||||||||
| rs6548238 | 624905 | T | C | 0.01 | 0.17 | 7.21 × 10−2 | -+- | 22 | 0.2792 | 3780 | 0.175 | 0.076 |
| rs2867125 | 612827 | T | C | −0.06 | 0.17 | 2.26 × 10−1 | -+- | 0 | 0.5230 | 3765 | 0.177 | 0.018 |
None of the 287 additional common or low frequency variants within the TMEM18 region, including coding or functional variants in TMEM18, were significantly associated with BMI.
Analysis of Rare Variants
The results from T1 count tests summarizing all rare SNPs and SKAT-based joint association of all rare SNPs for the TMEM18 region provide no support for an association between rare variants in the region and BMI (p = 0.56 for T1; p=0.44 for SKAT). We also performed T1 and SKAT tests restricting to the 10 exonic functional (nonsynonymous or splice-site) variants or the 370 SNPs localized within transcription factor binding sites in the TMEM18 region; none of these associations were statistically significant (10 exonic SNPs: p=0.50 for T1, p=0.85 for SKAT and 370 transcription factor binding sites: p=0.97 for T1; p=0.72 for SKAT).
Discussion
This study aimed to fine map the previously identified GWAS signal near TMEM18 and to identify any independent variant(s) in the region using the targeted resequencing data. We found a cluster of associated SNPs, one of which, rs7596758, exceeded a Bonferroni-corrected statistical significance threshold of 4.6 × 10 −4. Further analyses conditioning on the previously reported associated SNPs in the region from GWAS identified another low-frequency variant, rs186019316. Both variants lie in transcription factor binding regions.
One possible limitation of our findings is the observed heterogeneity across the cohort samples and attenuated evidence of association in a random effects meta-analysis (Supplementary Table 2). Age differences between the three participating cohorts may have potentially influenced our results. In support of this hypothesis, a growing literature has demonstrated age specific differences in the magnitude of genetic associations 21–25. For example, TMEM18 was reported to have larger BMI effect estimate in adolescents compared with adults23. On average, the participants in the CHS study (mean age of 72.5 in yrs) are older than the two studies (FHS: 37.0 and ARIC: 54.9) and displayed smaller genetic effect sizes.
Advances in genotyping technology and the development of novel, powerful analytical approaches have improved the accuracy and cost-effectiveness of testing variation in large samples, and these facilitated the present study. Here, we detected and verified genetic associations with BMI in a non-exonic region downstream of TMEM18, and extended our investigation to include low frequency variants not represented in typical genome-wide association studies of common haplotypes. This study relies on measurement of low-frequency variants using genetic resequencing. Studies that impute genotypes cannot readily examine low-frequency variant – disease associations, because imputation accuracy tends to be low for rarer variants. Further, while many genetic association studies now concentrate on exonic variation, few focus on association in non-coding regions. Along with the significant progress made following up the 9p21 locus association with myocardial infarction and coronary artery calcification, our study’s dual focus on non-coding variation and sequence variation in the region in and near TMEM18 proved fruitful.
Both newly discovered variants, rs7596758 and rs186019316, reside within regions that show evidence for transcription factor binding in ENCODE’s ChIP-seq results. SNP rs17042288, a proxy for rs7596758 (r2=1, D’=1 in 1000G Pilot 1) with nominally significant results, is in a region annotated for FOXA1 binding. The FOXA (or HNF3) protein family may have a pivotal role in the regulation of metabolism26. FOXA1 has been shown to play an important role in glucose homeostatis27 and glucose-stimulated insulin secretion28. SNP rs186019316 lies in a putative binding region for REST, which represses transcription of neuron-specific genes by binding the neuron-restrictive silencer element (NRSE) and restricting gene expression29, 30, but has not been reported to be related to adiposity biology. However, using Genomatix (Genomatix, Munich, Germany) to query predicted transcription factor binding motif alterations by these variants, we found no evidence that these variants alter the canonical binding motifs for FOXA1 or REST. The functional role of the reported SNPs needs to be further explored using in vitro experiments.
We demonstrate that for BMI, a complex quantitative trait, common variants downstream of TMEM18 may influence trait variation. Although these associations need to be confirmed, our findings add to the growing literature suggesting that GWAS signals tag regions where multiple single-variants may be independently associated with the trait of interest31, 32. These may span the spectrum from low-frequency to common variants, and increase the total variance of the trait that is explained by genetic variation. Indeed, the variants we found associated with BMI in the TMEM18 region differed with respect to their functional significance, effect size, and allele frequency.
This study has several notable strengths. It represents the first attempt to fine-map the BMI associated region near TMEM18 using a comprehensive resequencing approach in a large sample of subjects. Second, TMEM18 represents a region repeatedly shown to be associated with BMI5, 6, 23, 24 (cite a few sources, not just Giant. However, while our sample of nearly 4000 is large for current sequencing studies, even it is insufficient to detect small effects or individual associations with rare or low frequency variants. Specifically, with a total sample size of 3976, we have power of 51.38% to detect an effect size of 2.247, which is 0.1% of the variation in BMI, for a variant with MAF of 1%. Finally, although replication of our results would be desirable, because our focal region is not exonic, we know of no other studies with sequence data available for replication. Despite these limitations, our study does provide evidence for a possible regulatory role of this region with respect to adiposity.
In summary, our targeted resequencing study of the TMEM18 region of chromosome 2 reveals two variants associated with BMI, one of low frequency (2%) and the other common (29%). Our results indicate that conditioning on SNP rs7596758 (MAF 29%) substantially weakens the estimated effects of variants previously identified by GIANT. Further, our data provide no evidence that TMEM18 coding variants influence the phenotype. Instead, the data support a role for a region 3’ of the gene with annotated function that is likely to be regulatory. Expanded studies will be needed to characterize this region and its effects definitively.
Supplementary Material
Acknowledgments
Funding Sources: Support for "Building on GWAS for NHLBI-diseases: the U.S. CHARGE Consortium" was provided by the NIH through the American Recovery and Reinvestment Act of 2009 (ARRA) (5RC2HL102419). Data for "Building on GWAS for NHLBI-diseases: the U.S. CHARGE Consortium" was provided by Eric Boerwinkle on behalf of the Atherosclerosis Risk in Communities (ARIC) Study, L. Adrienne Cupples, principal investigator for the Framingham Heart Study, and Bruce Psaty, principal investigator for the Cardiovascular Health Study. The ARIC Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute (NHLBI) contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN2682011000010C, HHSN2682011000011C, and HHSN2682011000012C), R01HL087641, R01HL59367 and R01HL086694.. The Framingham Heart Study is conducted and supported by the NHLBI in collaboration with Boston University (Contract No. N01-HC-25195), and its contract with Affymetrix, Inc., for genome-wide genotyping services (Contract No. N02-HL-6-4278), for quality control by Framingham Heart Study investigators using genotypes in the SNP Health Association Resource (SHARe) project. A portion of this research was conducted using the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. This CHS research was supported by NHLBI contracts N01-HC-85239, N01-HC-85079, N01-HC-85080, N01-HC-85081, N01-HC-85082, N01-HC-85083, N01-HC-85084, N01-HC-85085, N01-HC-85086, N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, HHSN268201200036C and NHLBI grants HL080295, HL087652, HL105756 with additional contribution from NINDS. Additional support was provided through AG-023629, AG-15928, AG-20098, and AG-027058 from the NIA. See also http://www.chs-nhlbi.org/pi.htm. This work was partially supported by R01-DK-8925601 and R01-DK-075681 (Ingrid B. Borecki, PI) from NIDDK. Baylor Genome Center grant number, U54 HG003273
Footnotes
Conflict of Interest Disclosures: None.
References
- 1.Elks CE, den Hoed M, Zhao JH, Sharp SJ, Wareham NJ, Loos RJ, et al. Variability in the heritability of body mass index: A systematic review and meta-regression. Front Endocrinol (Lausanne) 2012;3:29. doi: 10.3389/fendo.2012.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Montague CT, Farooqi IS, Whitehead JP, Soos MA, Rau H, Wareham NJ, et al. Congenital leptin deficiency is associated with severe early-onset obesity in humans. Nature. 1997;387:903–908. doi: 10.1038/43185. [DOI] [PubMed] [Google Scholar]
- 3.Farooqi IS, Keogh JM, Yeo GS, Lank EJ, Cheetham T, O'Rahilly S. Clinical spectrum of obesity and mutations in the melanocortin 4 receptor gene. N Engl J Med. 2003;348:1085–1095. doi: 10.1056/NEJMoa022050. [DOI] [PubMed] [Google Scholar]
- 4.Krude H, Biebermann H, Luck W, Horn R, Brabant G, Grüters A. Severe early-onset obesity, adrenal insufficiency and red hair pigmentation caused by pomc mutations in humans. Nat Genet. 1998;19:155–157. doi: 10.1038/509. [DOI] [PubMed] [Google Scholar]
- 5.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sällman Almén M, Rask-Andersen M, Jacobsson JA, Ameur A, Kalnina I, Moschonis G, et al. Determination of the obesity-associated gene variants within the entire fto gene by ultra-deep targeted sequencing in obese and lean children. Int J Obes (Lond) 2013;37:424–431. doi: 10.1038/ijo.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Psaty BM, O'Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, et al. Cohorts for heart and aging research in genomic epidemiology (charge) consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin H, Wang M, Brody JA, Bis JC, Dupuis J, Lumley T, et al. Strategies to design and analyze targeted sequencing data: The Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) targeted sequencing study. Circulation Cardiovascular Genetics. 2014 doi: 10.1161/CIRCGENETICS.113.000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Homer N, Merriman B, Nelson SF. Bfast: An alignment tool for large scale genome resequencing. PLoS One. 2009;4:e7767. doi: 10.1371/journal.pone.0007767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and samtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K, Li M, Hakonarson H. Annovar: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. Dbsnp: The ncbi database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, et al. The ucsc genome browser database: Extensions and updates 2011. Nucleic Acids Res. 2012;40:D918–D923. doi: 10.1093/nar/gkr1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenbloom KR, Dreszer TR, Long JC, Malladi VS, Sloan CA, Raney BJ, et al. Encode whole-genome data in the ucsc genome browser: Update 2012. Nucleic Acids Res. 2012;40:D912–D917. doi: 10.1093/nar/gkr1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward LD, Kellis M. Haploreg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity (Edinb) 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 18.Basu S, Pan W. Comparison of statistical tests for disease association with rare variants. Genet Epidemiol. 2011;35:606–619. doi: 10.1002/gepi.20609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. An integrated encyclopedia of dna elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hallman DM, Friedel VC, Eissa MA, Boerwinkle E, Huber JC, Harrist RB, et al. The association of variants in the fto gene with longitudinal body mass index profiles in non-hispanic white children and adolescents. Int J Obes (Lond) 2012;36:61–68. doi: 10.1038/ijo.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hardy R, Wills AK, Wong A, Elks CE, Wareham NJ, Loos RJ, et al. Life course variations in the associations between fto and mc4r gene variants and body size. Hum Mol Genet. 2010;19:545–552. doi: 10.1093/hmg/ddp504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graff M, North KE, Mohlke KL, Lange LA, Luo J, Harris KM, et al. Estimation of genetic effects on bmi during adolescence in an ethnically diverse cohort: The national longitudinal study of adolescent health. Nutr Diabetes. 2012;2:e47. doi: 10.1038/nutd.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graff M, Ngwa JS, Workalemahu T, Homuth G, Schipf S, Teumer A, et al. Genome-wide analysis of bmi in adolescents and young adults reveals additional insight into the effects of genetic loci over the life course. Hum Mol Genet. 2013;22:3597–3607. doi: 10.1093/hmg/ddt205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graff M, Gordon-Larsen P, Lim U, Fowke JH, Love SA, Fesinmeyer M, et al. The influence of obesity-related single nucleotide polymorphisms on bmi across the life course: The page study. Diabetes. 2013;62:1763–1767. doi: 10.2337/db12-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaestner KH. The hepatocyte nuclear factor 3 (hnf3 or foxa) family in metabolism. Trends Endocrinol Metab. 2000;11:281–285. doi: 10.1016/s1043-2760(00)00271-x. [DOI] [PubMed] [Google Scholar]
- 27.Kaestner KH, Katz J, Liu Y, Drucker DJ, Schütz G. Inactivation of the winged helix transcription factor hnf3alpha affects glucose homeostasis and islet glucagon gene expression in vivo. Genes Dev. 1999;13:495–504. doi: 10.1101/gad.13.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao N, Le Lay J, Qin W, Doliba N, Schug J, Fox AJ, et al. Foxa1 and foxa2 maintain the metabolic and secretory features of the mature beta-cell. Mol Endocrinol. 2010;24:1594–1604. doi: 10.1210/me.2009-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (nrsf): A coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 30.Chong JA, Tapia-Ramírez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, et al. Rest: A mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 31.Sanna S, Li B, Mulas A, Sidore C, Kang HM, Jackson AU, et al. Fine mapping of five loci associated with low-density lipoprotein cholesterol detects variants that double the explained heritability. PLoS Genet. 2011;7:e1002198. doi: 10.1371/journal.pgen.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Y, Waite L, Jackson AU, Sheu W, Buyske S, Absher D. Trans-ethnic fine-mapping of lipid loci identifies population-specific signals and extensive allelic heterogeneity that increases the trait variance explained. PLoS Genetics. 2013 Mar;9(3):e1003379. doi: 10.1371/journal.pgen.1003379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.