Letter to the Editor
Allogeneic hematopoietic stem-cell transplantation (HSCT) in general, and haploidentical HSCT in particular, is a therapeutic approach for leukemias and especially acute myeloid leukemia (AML). Residual malignant disease is mitigated, in part, post-transplant by graft-versus-leukemia (GvL) activity of donor-derived T cells expressing αβ T-cell receptors that recognize major and minor histocompatibility antigens. In situations of human-leukocyte-antigen (HLA) mismatched HSCT the extensive T-cell depletion of the allograft undertaken to prevent graft-versus-host disease (GvHD) results in delayed T-cell engraftment, raises the probability of infection by opportunistic pathogens, and increases the possibility of relapse. However, the infused and engrafted allogeneic natural killer (NK) cells also contribute to controlling infections and the GvL-effect1 albeit using a different activation mechanism.
Donor-derived NK cells engrafting after haploidentical HSCT and adoptive transfer of haploidentical NK cells have demonstrated therapeutic effects. NK-cell alloreactivity resulting from appropriate killer cell Ig-like receptor (KIR)-ligand disparity in HLA-haplotype mismatched HSCT has resulted in improved engraftment, decreased incidence of leukemia relapse, and increased survival rates in patients with AML2 and possibly also in children with acute lymphoblastic leukemia (ALL). 3 Pre-transplant infusion of alloreactive NK cells has been shown to eliminate AML blasts and kill recipient T cells which may allow a reduction in the toxicity of the conditioning regimen. Furthermore, NK-cell mediated eradication of recipient dendritic cells may offer protection against GvHD and support an increased T-cell content in the allograft.1 Infusion of haploidentical NK cells after lymphodepleting chemotherapy has also been used to treat subsets of patients with relapsed AML2 and this observation has formed the basis of clinical trials infusing haploidentical NK cells for patients with relapsed/recurrent neuroblastoma4 and pediatric ALL5.
It is hypothesized that a KIR-ligand-mismatched donor provides the best chance for achieving a clinical effect against an underlying malignancy.2 Yet, not all patient-donor pairs apparently benefit with an allogeneic NK-cell mediated anti-tumor effect based on genotype predictions. The ‘missing self’ recognition hypothesis6 predicts that target cells are vulnerable to NK-cell lysis in the absence or alteration of major histocompatibility complex (MHC) class I molecules and NK-cell attack can be blocked by inhibitory KIR that recognize a subset of classical HLA class I molecules. As a result, HLA mismatches of an altered allelic repertoire, as in haploidentical HSCT, leads to KIR-ligand mismatch and alloreactive NK cell-mediated killing of tumor cells. Though a haplo-identical donor is mismatched to the recipient on a genetic level, observed alloreactivity may not occur in part due to allelic KIR variants that may not allow for full receptor expression1 and that the relative numbers of alloreactive NK cells lacking KIR for self HLA class I molecules may vary dramatically.7 Further, since each KIR has the ability to recognize allotypic determinants that are shared by different HLA class I alleles, an HLA-mismatch between NK cells and target cells does not inevitably lead to NK-cell mediated alloreactivity.8 While a prediction of therapeutic effectiveness may be approached with the combined use of genotyping and profiling of KIR and HLA on donor cells, this remains an approximation of the actual killing potential between donor NK cell and recipient targets as most available antibodies used to detect KIR expression do not distinguish between activating and inhibitory KIRs.7
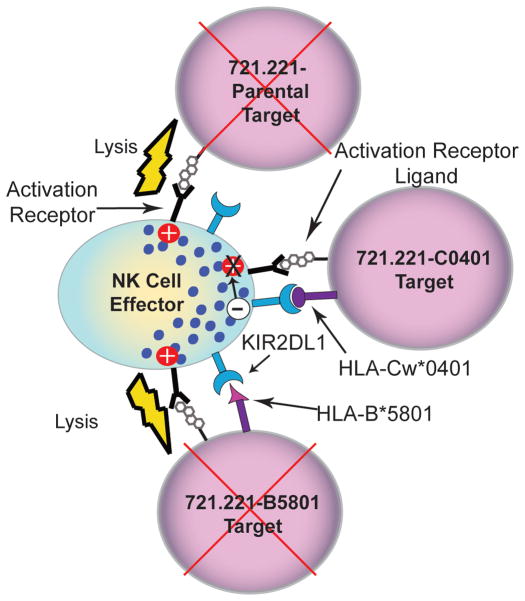
Since we cannot predict the actual frequency of alloreactive NK cells from genotyping or phenotyping of donor-derived KIR haplotypes and recipient HLA, we developed an approach to selecting a donor that exhibits an NK-cell alloreactivity for HLA molecules expressed in the recipient. This was undertaken with a functional assay to directly measure NK-cell killing through KIR-ligand interactions based on a panel of targets with enforced expression of individual HLA class I genes recognized by KIR (Figure 1) and tested using populations of NK cells that were predicted to kill based on KIR and HLA typing genotype.
Figure 1.
Schematic of alloreactivity generated between NK cells that are KIR-ligand mismatched with targets. HLA-B*5801, C*0702 and C*0401 alleles were amplified from cDNA using 5UTCE (5’GGG CGA ATTC GCC CGA GAT GCG GGT CAT GGC GCC CC 3’) and 3UTCH (5’CCG CAA GCT T TC GGG GAG GGA ACA CAG GTC AGT GTG GGG AC 3’) primers. The resulting PCR amplicon which encodes the full length classical HLA class I antigen was cloned into the mammalian expression vector pcDNA3.1- (Invitrogen). A clone was isolated and the fidelity of the HLA class I gene was confirmed by DNA sequencing. DNA for each class I allele was transfected by electroporation into the HLA class I deficient human B-cell line, 721.2219. Antibiotic selection with G418 identified positive transfectants and class I expressing subclones were subsequently screened by flow cytometry using anti-HLA class I clone W6/32 as a primary antibody and PE-conjugated goat anti-mouse IgG as a secondary antibody. Following flow cytometric identification of a population of transfected cells expressing class I HLA, the HLA-expressing 721.221 cells were subjected to single-cell sorting to obtain stable transfectants of a clonal nature. Three stable/clonal HLA class I transfectants (B*5801, Cw*0702 and Cw*0401) were established by this approach. The B*5801 transfectant was produced in a similar manner in the laboratory of Dr. Jennifer Gumperz (University of Wisconsin, Madison, WI) and the other transfectants were produced in the laboratory of Dr. William Hildebrand (University of Oklahoma Health Sciences Center, Oklahoma City, OK).
A panel of putative NK-cell targets was developed using 721.221 parental B cells that do not express HLA class I molecules and 721.221 cells with enforced expression of the individual HLA alleles B*5801, Cw*0401, and Cw*0702 which function as KIR ligands.9 These ligands bind to three main NK-cell inhibitory receptors, KIR3DL1, KIR2DL1, and KIR2DL2/3, respectively. NK-cell alloreactivity is determined by a balance of activating and inhibitory signaling, primarily through activating and inhibitory KIR. Therefore, target cells expressing single HLA alleles were anticipated to identify donors with demonstrable NK-cell mediated alloreactivity, and the magnitude of killing would identify donors with the largest subset of alloreactive NK cells for a particular KIR-ligand mismatch (Figure 1). The observed killing was compared against the predicated alloreactivity based on genotyping. NK-cell cytolysis of 721.221 cell transfectants was expected if: (i) HLA-KIR ligands were absent in the donor and in the target cells or (ii) HLA-KIR ligands were present in the donor and absent in the target cells.
Normal blood samples were obtained from the Gulf Coast Regional Center (GCRC) with peripheral blood mononuclear cells (PBMC) isolated using Ficoll gradient. PBMC were HLA and KIR genotyped (Table 1), and stored frozen until use. Though all donors had a complete KIR repertoire of the receptors tested for, they each had different combinations of HLA-KIR ligands present and thus different patterns of predicted NK-cell inhibition versus activation for cytolysis (Table 1). NK cells were isolated from thawed PBMC from five healthy donors using magnetic negative cell sorting with NK-cell isolation kit (Miltenyi Biotec, Auburn, CA). The cell-surface phenotypes of the NK cells and 721.221 cells were assessed by staining with the following: fluorescein isothiocyanate (FITC)-conjugated or phycoerythrin (PE)-conjugated mAbs specific for HLA-A,B,C (ab33257, Abcam, Cambridge, MA), CD3 (349201, BD Biosciences, San Jose, CA) and CD56 (555518, BD Biosciences). The B*5801-, Cw*0401-, and Cw*0702-modified 721.221 targets were 85%, 71%, and 83%, respectively, positive for expression of class I HLA molecules. HLA-A,B,C expression of parental 721.221 was at a base-line level of 11%. Isolated bulk NK-cell populations averaged 50% CD3−CD56+ and ranged from 37% to 88%. The bulk NK-cell populations were co-cultured with the 721.221 cell panel in a standard four-hour chromium release assay at an effector to target ratio of 20:1 and the results plotted as percent inhibition relative to 721.221 parental lytic inhibition.
Table 1.
NK cells were KIR-typed using two methodologies testing for the presence or absence of KIR genes. This analysis also distinguished two groups of 2DS4 alleles, those carrying a deletion of 22 nucleotides (003, 004, 006, 007), and alleles carrying full length allele. Genomic DNA was extracted from nucleated cells utilizing the QiaGen DNA purification kit EZ1 DNA Tissue Card (Qiagen, Germantown, MD, USA). KIR typing was performed utilizing PCR amplification combined with hybridization with oligonucleotide probes (PCR-SSOP). For SSOP KIR genotyping we used KIR SSO genotyping kits from One Lambda Inc. (Canoga Park, CA, USA). DNA was amplified with specific primers and products were incubated with beads labeled by specific oligonucleotide probes. Beads were collected by the Luminex 100 platform (Luminex Corporation, Austin, TX, USA) and analyzed using HLA Visual 2.2 (One Lambda Inc.) software. KIR typing was also performed by PCR amplification utilizing sequence specific primers (SSP) using the Pel-Freeze Genotyping SSP KIR kit (Invitrogen/Pel-Freeze, Brown Deer, WI, USA) containing 21 locus-specific primer mixes. Donors were typed for alleles at HLA-A, B, C, DRB1, DRB3/4/5/ and DQB1 by PCR amplification and oligonucleotide hybridization by molecular methods utilizing commercial kits (Invitrogen, Carlsbad, CA, USA and One Lambda) that achieve intermediate resolution. The donors were also typed for these loci by high resolution methods utilizing PCR amplification and nucleotide sequencing (Abbott Laboratories, Abbott Park, IL, USA). Additional high resolution tests for selected loci were performed in donors in whom an allele level mismatch could not be ruled out. HLA-KIR ligands were identified by analyzing the intermediate resolution and high resolution results of HLA-B and C loci in the donors. The polymorphism at residue 80 of HLA-B and C loci was analyzed to classify for the presence of the KIR ligands. The alleles of HLA-B having a codon encoding for 'threonine' (T) or isoleucine (I) at codon 80 were classified as 'Bw4-positive' being the ligands of the KIR receptor 3DL1. The alleles of HLA-C having a codon encoding for 'asparagine' (N) at codon 80 were classified belonging to the HLA-C 'Group-1' being the ligands of the KIR receptors 2DL2 and 2DL3. The alleles of HLA-C having a codon encoding for 'lysine' (K) at codon 80 were classified belonging to the HLA-C 'Group-2' being the ligands of the KIR receptor 2DL1. The assignment into KIR ligand groups was made from the interpretation of the high resolution results or from the reactivity of oligonucleotide probes detecting sequence polymorphisms at codon 80. The (+) or (−) symbols denote the presence or absence of KIR ligand on donor-derived NK cells. Observed inhibition was deemed as: Low – less than 25%, Moderate (Mod) – 25% to 60%, and High – greater than 60% relative NK-cell inhibition. Blue boxes indicate disparity between predicted inhibition and observed inhibition.
| KIR | KIR Ligand | Expected Inhibition by Ligand | Observed Inhibition | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||||
| Donor | KIR3DL1 | KIR2DL1 | KIR2DL2/3 | Bw4 | Cw4 | Cw7 | B*5801 | Cw*0401 | Cw*0702 | B*5801 | Cw*0401 | Cw*0702 |
| 1 | + | + | + | + | + | − | Pos. | Pos. | Neg. | Mod | High | Low |
| 2 | + | + | + | + | − | + | Pos. |
|
Pos. | Mod |
|
High |
| 3 | + | + | + | − | − | + | Neg. | Neg. | Pos. | Low | Low | Mod |
| 4 | + | + | + | − | + | − | Neg. | Pos. |
|
Low | High |
|
| 5 | + | + | + | + | + | + | Pos. | Pos. | Pos. | Mod | Mod | Mod |
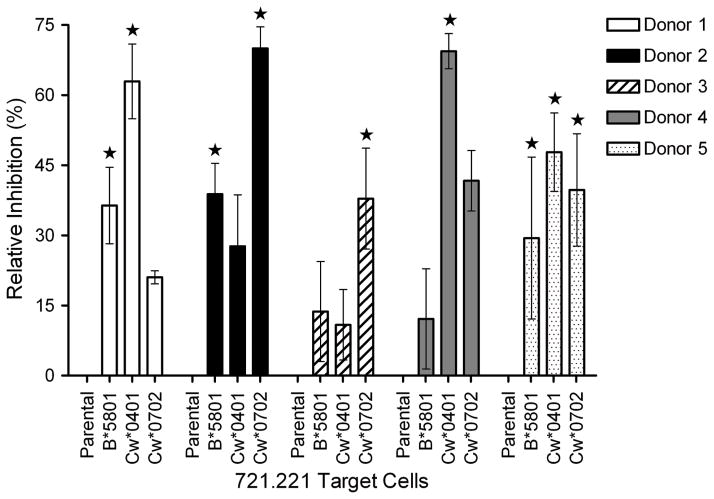
To evaluate the NK-cell mediated killing from one donor to the next we evaluated the ability of the HLA class I-transfected 721.221 cells to resist being lysed. Donors 2 and 4 demonstrated unpredicted NK-cell lytic inhibition of 721.221-Cw*0401 and Cw*0702 targets, respectively (Table 1, blue boxes). We also examined the degree of inhibition between donor NK cells that were predicted to be alloreactive (Figure 2). Donor 2 had predicted alloreactivity in the presence of allele Cw*0401, yet moderate inhibition was observed. Donor 4 was predicted to be alloreactive in the presence of the Cw*0702 allele, but rather displayed moderate inhibition. NK cells from donor 5 were predicted to be inhibited in the presence of all three alleles tested and while this was the case, the magnitude of inhibition was moderate for all three targets. As this donor expresses all three KIR ligands, the moderate inhibition may be interpreted as the donor having three populations of NK cells that are mutually exclusively inhibited by their corresponding ligand, thus since not all of the NK-cell populations are inhibited at once there will be some activity from NK cells within each of these populations. In comparing the alloreactivity of donors 1 and 4, it is suggested that donor 1 has a greater subset of alloreactive NK cells in the presence of the Cw*0702 allele. Like-wise in the comparison of donors 2 and 3, NK cells from donor 3 showed a greater magnitude of alloreactivity than donor 2 in the presence of Cw*0401. In aggregate, the genotype-based prediction of lytic potential for 721.221 cell transfectants failed to fully predict the actual lysis for 2 of 5 donor-”recipient” pairs.
Figure 2.
Inhibition of NK-cell mediated lysis of HLA+ 721.221 transfectants, relative to the HLAneg 721.221 parental target. The star symbol indicates predicted inhibition based on presence/absence of KIR ligand in donor NK cells. Error bars are ± SD. The cytolytic activity of NK-cell populations from each donor was determined in a 4-hour chromium release assay (CRA) using 51Cr-labeled 721.221 parental target and a panel of three 721.221 transfectants expressing HLA-B*5801, HLA-Cw*0401, and HLA-Cw*0702. The isolated NK cells were washed and plated in 96-well V-bottom microtiter plates (Costar, Cambridge, MA) at 37°C for 4 hours in triplicate at 105/well with 5 × 103 target cells. After centrifugation and incubation, 100 μL aliquots of cell-free supernatant were transferred to LumiPlates, dried, and read on a Perkin Elmer Gamma Counter (Perkin Elmer, Waltham, MA). The percent specific cytolysis (SC) was calculated from the release of 51Cr as follows: [(CPSexperimental – CPScontrol)/(CPSmaximal – CPScontrol)]*100. Control wells contained target cells incubated in media. The maximal 51Cr was determined by measuring the 51Cr content released by target cells lysed with 2% SDS. Data are reported as an average ± standard deviation (SD). Percent relative inhibition (RI) to the 721.221 parental target was determined by: RI = 100 – (target cell SL/parental SL)*100.
Overall, we noted marked discrepancies between the potential for alloreactivity based on KIR-ligand mismatches and the observed degree of cytolysis. Moreover, the ability to observe the actual lytic potential of bulk populations of NK cells enabled the magnitude of alloreactivity to be observed and compared between potential donors. Future applications of this NK-cell mediated killing assay will be to correlate survival data with magnitude of observed lysis of 721.221 cells and inhibition of observed lysis based on KIR-HLA binding. Indeed, this type of information may prove a valuable addition to the selection criteria for alloreactive NK-cell donors as the magnitude and proportion of alloreactive NK-cell repertoire are apparently not parameters that can be predicted solely from donor genotyping. Detection of genes encoding for the receptors and ligands do not warrant the definition of a complete repertoire, as allelic variations in the receptors and other structural variations in the ligands as well as other genes (e.g., activating genes) may have an impact in defining the NK repertoire. However, pairing functional and genetic studies may allow for the elucidation of the molecular basis of the NK repertoire. We conclude that when a “KIR-mediated anti-tumor effect” is desired, the donor and recipients should be selected based on the combined information from observed and predicted lysis rather than using genetic studies alone.
Acknowledgments
The project described was supported in part by Award Number TL1RR024147 from the National Center for Research Resources. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health. Additional support is from CCSG Grant (CA16672), RO1 (CA124782, CA120956), R21 (CA129390, CA116127), DOD (PR064229), The Alex Lemonade Stand Foundation, The Burroughs Welcome Fund, The Department of Veterans Affairs Career Development Award, The Gillson Longenbaugh Foundation, The Leukemia and Lymphoma Society, The Lymphoma Research Foundation, The Miller Foundation, The National Foundation for Cancer Research, The Pediatric Cancer Research Foundation, The William Lawrence and Blanche Hughes Foundation.
References
- 1.Ruggeri L, Mancusi A, Burchielli E, Capanni M, Carotti A, Aloisi T, et al. NK cell alloreactivity and allogeneic hematopoietic stem cell transplantation. Blood Cells Mol Dis. 2008 Jan-Feb;40(1):84–90. doi: 10.1016/j.bcmd.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 2.Miller JS, Soignier Y, Panoskaltsis-Mortari A, McNearney SA, Yun GH, Fautsch SK, et al. Successful adoptive transfer and in vivo expansion of human haploidentical NK cells in patients with cancer. Blood. 2005 Apr 15;105(8):3051–7. doi: 10.1182/blood-2004-07-2974. [DOI] [PubMed] [Google Scholar]
- 3.Locatelli F, Pende D, Maccario R, Mingari MC, Moretta A, Moretta L. Haploidentical hemopoietic stem cell transplantation for the treatment of high-risk leukemias: how NK cells make the difference. Clin Immunol. 2009 Nov;133(2):171–8. doi: 10.1016/j.clim.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 4.NCT00698009, http://clinicaltrials.gov
- 5.NCT009419285, http://clinicaltrials.gov
- 6.Ljunggren HG, Kärre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol Today. 1990 Jul;11(7):237–44. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 7.Ljunggren HG, Malmberg KJ. Prospects for the use of NK cells in immunotherapy of human cancer. Nat Rev Immunol. 2007 May;7(5):329–39. doi: 10.1038/nri2073. [DOI] [PubMed] [Google Scholar]
- 8.Velardi A, Ruggeri L, Moretta A, Moretta L. NK cells : a lesson from mismatched hematopoietic transplantation. Trends in Immunology. 2002 Sep;23(9):438–44. doi: 10.1016/s1471-4906(02)02284-6. [DOI] [PubMed] [Google Scholar]
- 9.Shimizu Y, Geraghty DE, Koller BH, Orr HT, DeMars R. Transfer and expression of three cloned human non-HLA-A,B,C class I major histocompatibility complex genes in mutant lymphoblastoid cells. Proc Natl Acad Sci USA. 1988 Jan;85(1):227–31. doi: 10.1073/pnas.85.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]