Abstract
In Candida albicans, the transcription factor Upc2 is central to the regulation of ergosterol biosynthesis. UPC2-activating mutations contribute to azole resistance, whereas disruption increases azole susceptibility. In the present study, we investigated the relationship of UPC2 to fluconazole susceptibility, particularly in azole-resistant strains. In addition to the reduced fluconazole MIC previously observed with UPC2 disruption, we observed a lower minimum fungicidal concentration (MFC) for a upc2Δ/Δ mutant than for its azole-susceptible parent, SC5314. Moreover, the upc2Δ/Δ mutant was unable to grow on a solid medium containing 10 μg/ml fluconazole and exhibited increased susceptibility and a clear zone of inhibition by Etest. Time-kill analysis showed higher fungistatic activity against the upc2Δ/Δ mutant than against SC5314. UPC2 disruption in strains carrying specific resistance mutations also resulted in reduced MICs and MFCs. UPC2 disruption in a highly azole resistant clinical isolate containing multiple resistance mechanisms likewise resulted in a reduced MIC and MFC. This mutant was unable to grow on a solid medium containing 10 μg/ml fluconazole and exhibited increased susceptibility and a clear zone of inhibition by Etest. Time-kill analysis showed increased fungistatic activity against the upc2Δ/Δ mutant in the resistant background. Microarray analysis showed attenuated induction by fluconazole of genes involved in sterol biosynthesis, iron transport, or iron homeostasis in the absence of UPC2. Taken together, these data demonstrate that the UPC2 transcriptional network is universally essential for azole resistance in C. albicans and represents an attractive target for enhancing azole antifungal activity.
INTRODUCTION
The increased frequency of invasive fungal infections in recent decades is directly related to an expansion in the immunocompromised patient population, including those with HIV/AIDS, cancer chemotherapy patients, neutropenic patients, recipients of transplants with indwelling catheters, and patients receiving antibiotics (1–3). Candida species collectively are the fourth leading cause of nosocomial infections in the United States, and such infections are associated with unacceptably high rates of mortality (2, 4–6). Candida albicans is the most prevalent opportunistic human fungal pathogen and causes a wide variety of infections, from superficial mucosal infections to invasive disseminated disease, despite the availability of effective antifungal treatment. Moreover, the most common opportunistic infection among AIDS patients is oropharyngeal candidiasis (OPC), and chronic cases continue despite the availability of highly active antiretroviral therapy (HAART), due to poor compliance, lack of access, and failure of HAART (7–11).
The azoles, particularly fluconazole (FLC), are the most widely used class of antifungals for the treatment of Candida infections (2, 12). Fluconazole acts by inhibiting the protein product of ERG11, lanosterol 14α-demethylase, which leads to ergosterol depletion and the accumulation of toxic methylated sterol precursors, resulting in the inhibition of growth (13). In an immunocompromised host, the fungistatic nature of fluconazole limits its efficacy against Candida species (14, 15). C. albicans exhibits inhibited growth in the presence of this fungistatic drug, and it is generally believed that such fungistatic activity facilitates the development of resistance, creating problems for treatment, especially in immunocompromised patients with OPC (16, 17). Currently known mechanisms of resistance to the azoles include alterations in the expression of drug efflux pump (CDR1, CDR2, and MDR1) and ergosterol biosynthesis (ERG) genes and mutations in ERG11 (18). These mechanisms are frequently combined, resulting in a stepwise development of resistance over time.
In C. albicans, UPC2 encodes a zinc cluster transcription factor that regulates the expression of genes involved in ergosterol biosynthesis, including ERG11 (19, 20). Disruption of ERG11 or pharmacologic inhibition of the enzyme it encodes leads to reduced ergosterol production and the accumulation of alternate sterols, including lanosterol, eburicol, obtusifoliol, 14α-methyl fecosterol, and 14α-methylergosta-8,24(28)-dien-3β,6α-diol (21). Biosynthesis of these alternate sterols does not require ERG11 but does require other genes in the ergosterol biosynthesis pathway, some of which appear to be regulated by Upc2. We reasoned that since Upc2 is required for the transcriptional activation of other genes of the sterol biosynthesis pathway, the disruption of UPC2 might result in enhanced activity of the azole antifungals in azole-resistant as well as azole-susceptible isolates. In the present study, we further examined the role of UPC2 in azole antifungal activity against both azole-susceptible and azole-resistant strains of C. albicans, in particular an azole-resistant clinical isolate and mutant strains that represent four of the major azole resistance mechanisms. Taken together, our results indicate that the UPC2 transcriptional network is universally essential for fluconazole resistance in C. albicans.
MATERIALS AND METHODS
Strains and growth conditions.
All C. albicans strains (Table 1) were stored as frozen stocks in 40% glycerol at −80°C. YPD (1% yeast extract, 2% peptone, and 1% dextrose) agar plates and YPD liquid medium were used for routine growth of strains at 30°C. Iron-poor agar plates and medium were prepared by adding 200 μM bathophenanthroline disulfonic acid (BPS) to YPD. Iron-rich agar plates and medium were prepared by adding 100 μM iron chloride (FeCl3) to YPD. For CFU counts during time-kill analysis, PDA (0.4% potato starch, 2% dextrose, and 1.5% agar) plates were used, and cultures grown on PDA were incubated at 35°C.
TABLE 1.
C. albicans strains used in this study
| Strain | Designation | Strain backgrounda | Relevant characteristic(s) or genotypeb | Source or reference |
|---|---|---|---|---|
| SC5314 | SC5314 | N/A | UPC2-1/UPC2-2 | ATCC |
| Clinical isolate 2-79 | 2-79 | N/A | Susceptible isolate | 53 |
| Clinical isolate 12-99 | 12-99 | N/A | Resistant isolate | 53 |
| UPC2M4A | upc2Δ/Δ strain | SC5314 | upc2-1Δ::FRT/upc2-2Δ::FRT | 25 |
| UPC2M2K21A | upc2Δ/Δ+UPC2 strain | UPC2M2A | upc2-1Δ::FRT/UPC2S1-1-caSAT1 | 25 |
| 12-99UPC2A5C1A | 12-99upc2Δ/Δ | 12-99 | upc2Δ::FRT/upc2Δ::FRT | This study |
| 12-99UPC2A5C1A4A | 12-99upc2Δ/Δ+UPC2 | 12-99UPC2A5C1A | upc2Δ::FRT/UPC212-99-caSAT1 | This study |
| 10C1B1M1 | ERG11K143R mutant | SC5314 | ERG11K143R::FRT/ERG11K143R::FRT UPC2-1/UPC2-2 | This study |
| 10C1B1M1UPC2C9H | ERG11upc2Δ/Δ mutant | 10C1B1M1 | ERG11K143R::FRT/ERG11K143R::FRT upc2-1Δ::FRT/upc2-2Δ::FRT | This study |
| SCMRR1R34A | MRR1P683S mutant | SC5314 | MRR1P683S::FRT/MRR1P683S::FRT UPC2-1/UPC2-2 | 20 |
| Δupc2MRR1R34A | MRR1upc2Δ/Δ mutant | SCMRR1R34A | MRR1P683S::FRT/MRR1P683S::FRT upc2-1Δ::FRT/upc2-2Δ::FRT | 20 |
| SCTAC1R34A | TAC1G980E mutant | SC5314 | TAC1G980E::FRT/TAC1G980E::FRT UPC2-1/UPC2-2 | 32 |
| SCupc2TAC1R34A1A14A | TAC1upc2Δ/Δ mutant | SCTAC1R34A | TAC1G980E::FRT/TAC1G980E::FRT upc2-1Δ::FRT/upc2-2Δ::FRT | This study |
N/A, not applicable.
FRT, FLP recombination target.
Drug susceptibility testing.
The MICs of fluconazole (FLC) were determined by using broth microdilution as described by NCCLS (now CLSI) standard M27-A2 (22), modified by using YPD medium (iron replete), iron-poor medium, or iron-rich medium, and were read both visually and spectrophotometrically at 24, 48, and 72 h. Minimum fungicidal concentrations (MFCs) were measured by removing 2 μl from each well of the MIC plate and plating onto YPD agar. Also, serial dilutions from a suspension with an optical density at 600 nm (OD600) of 0.1 were diluted 4-fold, and 2 μl of each dilution was plated onto YPD agar plates or iron-poor agar plates with or without 10 μg/ml fluconazole and was then incubated at 30°C for 24 and 48 h. Fluconazole activity was also assessed by Epsilometer test strips (Etest strips) (bioMérieux) according to the manufacturer's instructions with the following modifications. A standardized cell suspension (a 0.5 McFarland standard) was used to create a confluent lawn across YPD agar plates or iron-poor agar plates prior to Etest strip placement, and the cells were then incubated at 30°C for 24 and 48 h. Time-kill analyses were performed with a cell suspension at a 0.5 McFarland standard, which was 10-fold diluted into YPD medium with or without 10 μg/ml fluconazole and was incubated at 35°C. Aliquots were removed at 0, 6, 12, and 24 h, 10-fold serially diluted, and plated onto PDA. CFU were counted in duplicate after 48 h at 35°C and were plotted on a log-scale curve versus time (23).
Construction of strains with ERG11 mutant alleles.
Candida albicans ERG11 (CaERG11) coding sequences were amplified by PCR (Pfu DNA polymerase; Stratagene) from C. albicans genomic DNA using primers ERG11-A and ERG11-E. Products were cloned into pCR-BLUNTII-TOPO using a Zero Blunt TOPO PCR cloning kit (Invitrogen) and were transferred into Escherichia coli TOP10 cells with selection on LB agar plates containing 50 μg/ml kanamycin. Plasmid DNA was purified (QIAprep; Qiagen, Germantown, MD) and was sequenced on an ABI model 3130XL genetic analyzer using the ERG11 sequencing primers (Table 2), resulting in full-length sequence from both strands of the CaERG11 gene. The sequencing was performed using six sets of clones derived from three independent PCRs for each strain/isolate sequenced.
TABLE 2.
Primers used in this study
| Primer | Sequencea |
|---|---|
| qRT-PCR | |
| ACT1-F | 5′-ACGGTGAAGAAGTTGCTGCTTTAGTT-3′ |
| ACT1-R | 5′-CGTCGTCACCGGCAAAA-3′ |
| BMR1-F | 5′-ACATAAATACTTTGCCCATCCAGAA-3′ |
| BMR1-R | 5′-AAGAGTTGGTTTGTAATCGGCTAAA-3′ |
| CDR1-F | 5′-ATTCTAAGATGTCGTCGCAAGATG-3′ |
| CDR1-R | 5′-AGTTCTGGCTAAATTCGTAATGTTTTC-3′ |
| CDR2-F | 5′-TAGTCCATTCAACGGCAACATT-3′ |
| CDR2-R | 5′-CACCCAGTATTTGGCATTGAAA-3′ |
| CFL4-F | 5′-GCAATGGTTGACAGGTTGGAA-3′ |
| CFL4-R | 5′-GCAATGTGACGATGATAAGTGACAA-3′ |
| ERG11-F | 5′-CCCCTATTAATTTTGTTTTCCCTAATTTAC-3′ |
| ERG11-R | 5′-CACGTTCTCTTCTCAGTTTAATTTCTTTC-3′ |
| FET3-F | 5′-GCCGGTGTCTTAGGTTTAGCC-3′ |
| FET3-R | 5′-CTAGCAACTCTTTCTTCAACATCGG-3′ |
| FRP1-F | 5′-CTTCCAATACCATCCATTCACGAT-3′ |
| FRP1-R | 5′-ATCTCCCCACTTTCAGCAAGAC-3′ |
| FTR1-F | 5′-ATTGTTGTTTCAGTGCTTTTGGC-3′ |
| FTR1-R | 5′-GGTCGGAACTACCACCCATAGA-3′ |
| ERG11 mutant construction | |
| ERG11-A | 5′-GGGCCCGGGTTATTTGAGAACAGCC-3′ |
| ERG11-B | 5′-ATCCGTTCTCGAGCACTAAGGGACAA-3′ |
| ERG11-C | 5′-GTAATCAATTGAGCTCTTTTAACTTT-3′ |
| ERG11-D | 5′-GATTATAGTTCCGCGGTGGTTTTACC-3′ |
| ERG11-E | 5′-TGATGGTTTTTGTCCACTGGCTCGAG-3′ |
| ERG11 sequencing | |
| ERG11 seq B | 5′-TATTTTCACTGCTTCAAGATCT-3′ |
| ERG11 seq C | 5′-CCAAAAGGTCATTATGTTTTAG-3′ |
| ERG11 seq E | 5′-AATGAGGTTTTTCACCTAAATG-3′ |
| ERG11 seq F | 5′-CCCTTTACCGAAAACTGGAGTA-3′ |
| T7 | 5′-TAATACGACTCACTATAGGG-3′ |
| M13R | 5′-CAGGAAACAGCTATGACC-3′ |
| UPC2 mutant construction | |
| UPC2-A | 5′-GGGCCCGAGATCTTGATGTCATTAG-3′ |
| UPC2-B | 5′-CTCGAGCTATATCTTCAATGAACTG-3′ |
| UPC2-C | 5′-CCGCGGACAGGTCAATACCGCGTAG-3′ |
| UPC2-D | 5′-GAGCTCGTTCCTCTAGTATCACTCTT-3′ |
| UPC2-E | 5′-CTCGAGCACCACAGTAACGAATCAC-3′ |
Underlining reflects the introduction of a restriction site sequence.
Sequenced plasmids of the ERG11 open reading frame (ORF) whose predicted translation indicated an amino acid substitution were digested with restriction enzymes ApaI and XhoI, which excised the full-length ORF from the plasmid, and the ERG11 alleles were cloned upstream of the SAT1-flipper cassette into the ApaI and XhoI sites of plasmid pSFS2 (24). The ERG11 downstream segments were amplified with Ex Taq polymerase (TaKaRa) using primers ERG11-C and ERG11-D and were cloned downstream of the SAT1-flipper cassette in pSFS2 using the NotI and SacII sites.
Construction of UPC2 deletion strains.
Plasmid pSFS2 contains the entire SAT1-flipper cassette (24). The SAT1-flipper cassette consists of the SAT1 selectable marker, which confers resistance to nourseothricin, and the FLP flipper recombinase gene, both flanked by FRT sites (flipper recombinase target sequences). The UPC2 deletion cassette (pUPC2M2), where the 5′ upstream sequence from position −373 to +15 was cloned downstream of the SAT1-flipper cassette while the 3′ sequence from position +2097 to +2437 was cloned upstream of the SAT1-flipper cassette, was developed by Dunkel et al. (25) (Table 1). Upon transformation of the parent strain with the gel-purified SacI-ApaI fragment from pUPC2M2, the SAT1-flipper cassette was inserted into the coding region of one allele from position +16 to +2096, and such positive transformants (NouR) were selected on YPD-nourseothricin agar plates containing 200 μg/ml of nourseothricin. The FLP gene was induced by growing the transformants in YPD medium for 24 h without selective pressure. Positive cells (NouS) were selected by replica plating onto YPD plates with or without 200 μg/ml of nourseothricin. Upon induction of the FLP gene, the cassette was excised such that only one copy of the FRT site remained in the locus. Another round was required to disrupt the second allele. Appropriate gene disruption and complementation for two independent strains were confirmed by Southern hybridization (24).
Isolation of genomic DNA and Southern hybridization.
Genomic DNA was isolated as described previously (26). Four micrograms of DNA was digested with an appropriate restriction endonuclease, separated on a 1% agarose gel, and, after staining with ethidium bromide, was transferred by vacuum blotting to a nylon membrane and fixed by UV cross-linking. Southern hybridization with enhanced-chemiluminescence-labeled probes was performed with the Amersham ECL Direct nucleic acid labeling and detection system according to the manufacturer's instructions.
RNA isolation for quantitative reverse transcription-PCR (qRT-PCR).
RNA was isolated using a small-scale hot phenol method of RNA isolation described by Schmitt et al. (27). Briefly, overnight cultures were diluted to an OD600 of 0.2 in 20 ml YPD and were then incubated at 30°C with shaking for 3 h. Cells were collected by centrifugation and were stored at −80°C. Cell pellets were resuspended in 950 μl of AE buffer (50 mM sodium acetate [pH 5.3], 10 mM EDTA [pH 8.0]) and were then transferred to a 2-ml RNase-free microcentrifuge tube containing 950 μl acid phenol (pH 4.3) with 1% SDS. Cells were incubated at 65°C for 10 min; then lysates were clarified by centrifugation. The supernatant was then divided into two new 2-ml microcentrifuge tubes containing 950 μl of chloroform, and the contents of each tube were mixed. The sample was then subjected to centrifugation again, and the top aqueous layer was transferred to a new tube containing 1 ml of isopropanol and 100 μl 2 M sodium acetate. The RNA pellet was subsequently washed with 500 μl of 70% ethanol and was collected by centrifugation. The RNA pellet was resuspended in DNase/RNase-free H2O. Quantity and purity were determined spectrophotometrically by A260 and A280.
Quantitative RT-PCR.
First-strand cDNAs were synthesized from 1 μg of total RNA using the SuperScript first-strand synthesis system for RT-PCR (Invitrogen). Gene-specific primers (Table 2) were designed using Primer Express software (Applied Biosystems) and were synthesized by Integrated DNA Technologies (Coralville, IA). Quantitative PCRs were performed in triplicate using the 7000 sequence detection system (Applied Biosystems), independently amplifying ACT1 (normalizing gene) and the genes of interest (GOI) as described previously (28).
RNA isolation for microarray.
RNA was isolated using a large-scale version of the hot phenol method of RNA isolation described by Schmitt et al. (27). Briefly, overnight cultures were diluted to an OD600 of 0.005 in 100 ml YPD and were then incubated at 30°C with shaking for an additional 8 h to an OD600 of 1.0. Cultures were diluted again to an OD600 of 0.025 in 100 ml fresh YPD, allowed to incubate at 30°C with shaking for one doubling, inoculated with or without 10 μg/ml FLC, and then incubated at 30°C with shaking for 6 h. Cells were collected by centrifugation and were stored at −80°C. Cell pellets were resuspended in 12 ml of AE buffer (50 mM sodium acetate [pH 5.3], 10 mM EDTA [pH 8.0]) and were then transferred to 50-ml Oak Ridge tubes treated with RNase Away (Molecular BioProducts) containing 12 ml acid phenol (pH 4.3) with 1% SDS. Cells were incubated at 65°C for 10 min; then lysates were clarified by centrifugation. The supernatant was then transferred to a new tube containing 15 ml of chloroform, and the contents of the tube were mixed. The sample was then subjected to centrifugation again, and the top aqueous layer was transferred to a new tube containing 1 volume of isopropanol and 0.1 volume of 2 M sodium acetate. The RNA pellet was subsequently washed with 10 ml of 70% ethanol and was collected by centrifugation. The RNA pellet was resuspended in DNase/RNase-free H2O. Quantity and purity were determined spectrophotometrically by A260 and A280.
Transcriptional profiling.
Gene expression profiles were obtained by hybridizing labeled cRNAs generated from C. albicans total RNA onto Affymetrix C. albicans custom expression arrays (CAN07; 49-5241 array format) (25), which have been described previously (29). Microarray hybridization and analysis were performed as described previously (29). Genes were considered to be differentially expressed in response to the drug if their expression changed ≥1.5-fold in two independent experiments. Genes induced by FLC were considered to be UPC2 dependent if the induction in the deletion mutant was ≥2.0-fold (50%) less than that in the wild type.
Microarray data accession number.
All microarray data are available for download from the NCBI under GEO accession number GSE57929.
RESULTS
UPC2 disruption results in enhanced fluconazole activity.
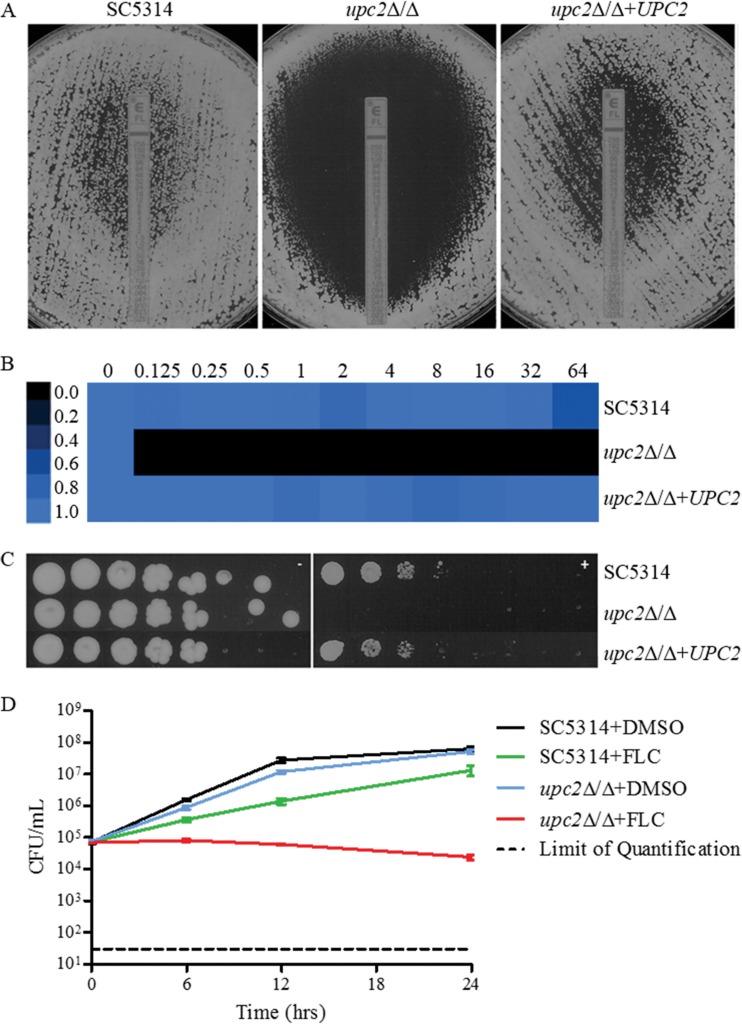
In order to further investigate the requirement of UPC2 for susceptibility to fluconazole, we subjected the upc2Δ/Δ mutant derived from azole-susceptible isolate SC5314 to various azole susceptibility tests, examining both MICs and MFCs by using nutrient-rich YPD medium in order to detect strong phenotypes in an environment that promotes growth. In agreement with previous observations, disruption of UPC2 resulted in marked reductions in fluconazole MICs by broth microdilution (Table 3), Etest (Fig. 1A), 72-h regrowth (Fig. 1B), and spot assays (Fig. 1C). Interestingly, at 24 h in YPD, the fluconazole MFC was >64 μg/ml for SC5314, whereas for the upc2Δ/Δ mutant it was 0.25 μg/ml (Table 3). At 48 h, the MIC for SC5314, as measured by Etest, was 1.0 μg/ml, and a halo of reduced growth (but not a clear zone of inhibition) was observed up to the Etest strip, a result consistent with the fungistatic nature of fluconazole. The 48-h MIC by Etest for the upc2Δ/Δ mutant was 0.032 μg/ml, and a clear zone of inhibition around the Etest strip was observed (Fig. 1A). We also used a 72-h endpoint for a broth microdilution assay in YPD as a way to assess the ability of the organism to resume growth in the presence of fluconazole. SC5314 was able to resume growth in fluconazole at all concentrations tested, whereas the upc2Δ/Δ mutant was not (Table 3; Fig. 1B). When cells were plated on YPD agar plates containing 10 μg/ml fluconazole, the level of growth in the presence of fluconazole was lower for the upc2Δ/Δ mutant than for SC5314 (Fig. 1C). Time-kill analysis showed higher fungistatic activity of 10 μg/ml fluconazole against the upc2Δ/Δ mutant than against its parent strain (Fig. 1D). All phenotypes reverted with reintegration of one allele of the disrupted gene.
TABLE 3.
MICs and MFCs in YPD in the SC5314 background
| Medium and strain | Relevant characteristics or genotype | MIC (μg/ml) |
MFC (μg/ml) |
||||
|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | ||
| YPD | |||||||
| SC5314 | UPC2-1/UPC2-2 | 0.5 | 0.5 | >64 | >64 | >64 | >64 |
| upc2Δ/Δ mutant | upc2-1Δ::FRT/upc2-2Δ::FRT | ≤0.125 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 |
| upc2Δ/Δ+UPC2 strain | upc2-1Δ::FRT/UPC2S1-1-caSAT1 | 0.5 | 0.5 | >64 | >64 | >64 | >64 |
| YPD + BPS | |||||||
| SC5314 | UPC2-1/UPC2-2 | 0.5 | 0.5 | 0.5 | >64 | >64 | >64 |
| upc2Δ/Δ mutant | upc2-1Δ::FRT/upc2-2Δ::FRT | NGa | NG | <0.125 | 0.125 | 0.125 | <0.125 |
| upc2Δ/Δ+UPC2 strain | upc2-1Δ::FRT/UPC2S1-1-caSAT1 | 0.5 | 0.5 | 0.5 | >64 | >64 | >64 |
| YPD + FeCl3 | |||||||
| SC5314 | UPC2-1/UPC2-2 | 1 | 1 | >64 | >64 | >64 | >64 |
| upc2Δ/Δ mutant | upc2-1Δ::FRT/upc2-2Δ::FRT | <0.125 | 0.25 | 0.5 | 0.25 | 0.25 | 1 |
| upc2Δ/Δ+UPC2 strain | upc2-1Δ::FRT/UPC2S1-1-caSAT1 | 1 | 1 | >64 | >64 | >64 | >64 |
NG, no growth.
FIG 1.
(A) Effects of UPC2 on MICs and growth on YPD agar as determined by Etest. A confluent lawn of C. albicans was streaked prior to the addition of Etest strips and was then incubated for 48 h. (B) MIC heat map of SC5314, the UPC2 mutant, and a complemented derivative. Susceptibility was determined by broth microdilution in YPD at 72 h (MICs in µg/ml above heat map). Growth was quantified spectrophotometrically and was assigned to a colorimetric scale. (C) Effect of UPC2 on the ability to grow on a solid medium containing fluconazole. From 4-fold serial dilutions of C. albicans strains, 2-μl aliquots were spotted onto YPD agar with (right) or without (left) 10 μg/ml FLC and were incubated for 48 h. (D) Effect of fluconazole on UPC2 in a time-kill assay. SC5314 or upc2Δ/Δ cells were diluted in YPD medium containing fluconazole (10 μg/ml) or the solvent dimethyl sulfoxide (DMSO). After 0, 6, 12, and 24 h, samples from each strain and medium were diluted and were plated for CFU.
UPC2 disruption in strains containing resistance mutations in MRR1, TAC1, or ERG11 also enhances fluconazole activity.
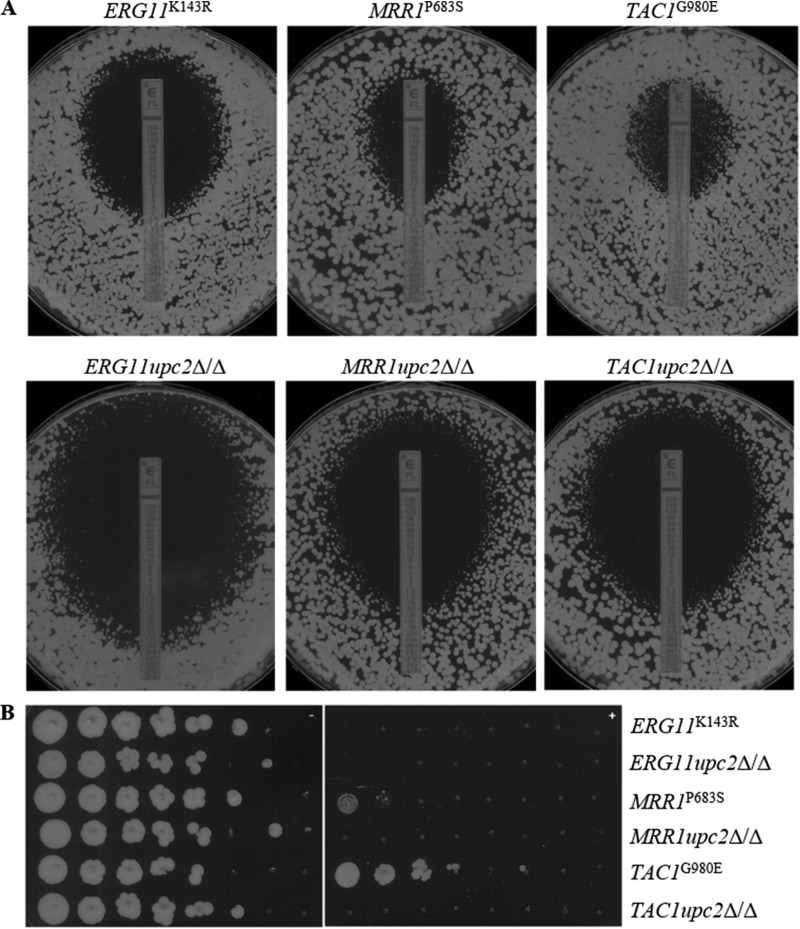
In order to investigate the requirement for UPC2 in the setting of specific mechanisms of azole resistance, independent mutants were constructed in strains containing two copies of a gene conferring reduced susceptibility to fluconazole: the MRR1P683S, TAC1G980E, or ERG11K143R gene. The MRR1P683S and TAC1G980E alleles contain gain-of-function mutations that render the transcription factors they encode constitutively active, resulting in the upregulation of either MDR1 (MRR1P683S) or CDR1 and CDR2 (TAC1G980E), respectively, and decreased fluconazole susceptibility (30–33). The ERG11K143R allele contains a point mutation postulated to be located near the azole access channel, interfering with the entry of fluconazole and thus resulting in decreased fluconazole susceptibility (34). Again, the disruption of UPC2 in each background resulted in marked reductions in MICs and MFCs as determined by all methods, and the trend in susceptibility seen for the MRR1upc2Δ/Δ mutant was consistent with what we have observed previously (20). The MFCs at 24 h in YPD for the ERG11upc2Δ/Δ, MRR1upc2Δ/Δ, and TAC1upc2Δ/Δ mutants were reduced from 8 μg/ml, >64 μg/ml, and >64 μg/ml in their background strains to 1 μg/ml, 4 μg/ml, and 2 μg/ml, respectively (Table 4). At 48 h, the MICs for the ERG11upc2Δ/Δ, MRR1upc2Δ/Δ, and TAC1upc2Δ/Δ mutants by Etest were reduced from 1.5 μg/ml, 4 μg/ml, and 8 μg/ml in their background strains to 0.19 μg/ml, 0.5 μg/ml, and 0.5 μg/ml, respectively (Fig. 2A). The growth of the MRR1upc2Δ/Δ and TAC1upc2Δ/Δ strains when plated on YPD agar plates containing 10 μg/ml fluconazole was also reduced from that of their background strains (Fig. 2B). For the ERG11K143R strain and its upc2Δ/Δ derivative, the results of growth experiments were consistent with the fluconazole MICs: both were unable to grow in the presence of fluconazole at this concentration.
TABLE 4.
MICs and MFCs in YPD in the backgrounds of strains expressing resistance mechanisms
| Strain | Relevant characteristics or genotype | MIC (μg/ml) |
MFC (μg/ml) |
||||
|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | ||
| SC5314 | UPC2-1/UPC2-2 | 0.5 | 0.5 | >64 | >64 | >64 | >64 |
| ERG11K143R mutant | ERG11K143R::FRT/ERG11K143R::FRT UPC2-1/UPC2-2 | 4 | 8 | 8 | 8 | 8 | 32 |
| ERG11upc2Δ/Δ mutant | ERG11K143R::FRT/ERG11K143R::FRT upc2-1Δ::FRT/upc2-2Δ::FRT | 0.5 | 1 | 1 | 1 | 1 | 1 |
| MRR1P683S mutant | MRR1P683S::FRT/MRR1P683S::FRT UPC2-1/UPC2-2 | 16 | >64 | >64 | >64 | >64 | >64 |
| MRR1upc2Δ/Δ mutant | MRR1P683S::FRT/MRR1P683S::FRT upc2-1Δ::FRT/upc2-2Δ::FRT | 2 | 2 | 4 | 4 | 4 | 4 |
| TAC1G980E mutant | TAC1G980E::FRT/TAC1G980E::FRT UPC2-1/UPC2-2 | 16 | >64 | >64 | >64 | >64 | >64 |
| TAC1upc2Δ/Δ mutant | TAC1G980E::FRT/TAC1G980E::FRT upc2-1Δ::FRT/upc2-2Δ::FRT | 2 | 2 | 2 | 2 | 2 | 2 |
FIG 2.
(A) Effects of UPC2 in resistant backgrounds on MICs and growth on YPD agar as determined by Etest. A confluent lawn of C. albicans was streaked prior to the addition of Etest strips and was then incubated for 48 h. (B) Effects of UPC2 in resistant backgrounds on the ability to grow on a solid medium containing fluconazole. From 4-fold serial dilutions of C. albicans strains, 2-μl aliquots were spotted onto YPD agar with (right) or without (left) 10 μg/ml FLC and were incubated for 48 h.
Disruption of Upc2 overrides clinical drug resistance.
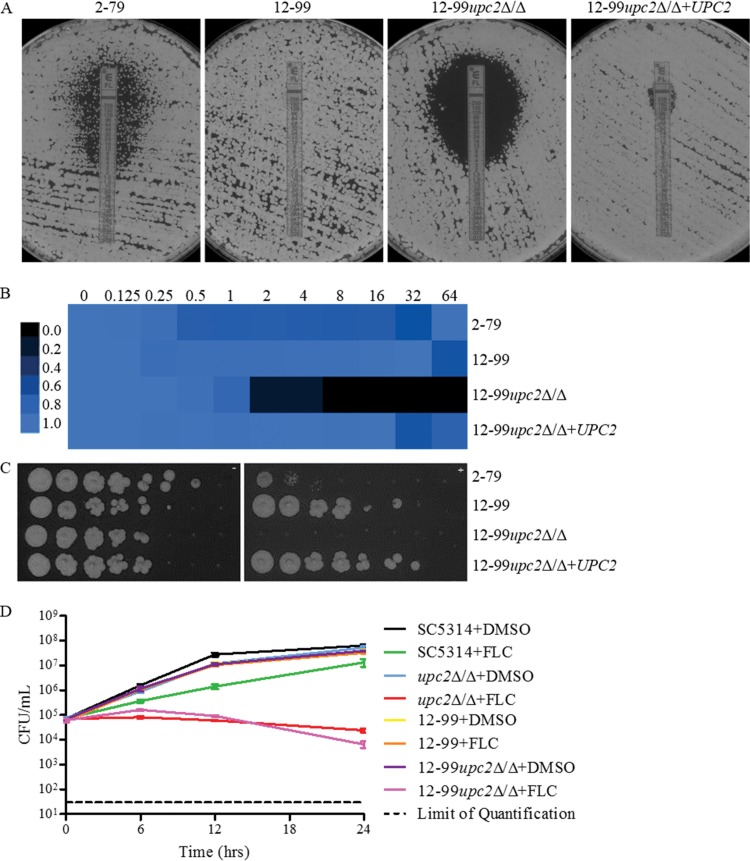
Since UPC2 disruption in strains containing one resistance mechanism resulted in enhanced fluconazole activity, we wanted to examine further the extent to which disruption of UPC2 influences high-level azole resistance in the presence of multiple resistance mechanisms. We constructed upc2Δ/Δ mutant strains in the background of an azole-resistant clinical isolate (isolate 12-99) known to carry four of the most common mechanisms of azole resistance: overexpression of CDR1 and CDR2, overexpression of MDR1, overexpression of ERG11, and a mutation in ERG11 (35). As was observed for isolate SC5314 and the isogenic resistant strains, the disruption of UPC2 resulted in marked reductions in MICs and MFCs as determined by all methods. The MFC at 24 h in YPD was >64 μg/ml for 12-99, whereas it was 4 μg/ml for 12-99upc2Δ/Δ (Table 5). The fluconazole MIC by Etest at 48 h was >256 μg/ml for 12-99, and confluent growth was observed, whereas a MIC of 3 μg/ml and a clear zone of inhibition were observed for 12-99upc2Δ/Δ (Fig. 3A). Likewise, in broth microdilution assays after 72 h, the parent strain was able to grow in the presence of fluconazole at all concentrations tested, whereas 12-99upc2Δ/Δ grew less well (Fig. 3B). The growth of 12-99upc2Δ/Δ was also reduced from that of its parent strain when they were plated on YPD agar plates containing 10 μg/ml fluconazole (Fig. 3C). As was observed in the SC5314 background, time-kill analysis also revealed an increased fungistatic effect for fluconazole at 10 μg/ml against 12-99upc2Δ/Δ (Fig. 3D). All phenotypes reverted with the reintegration of one allele of the disrupted gene.
TABLE 5.
MIC and MFCs in YPD for strains in the 12-99 background
| Medium and strain | Relevant characteristics or genotype | MIC (μg/ml) |
MFC (μg/ml) |
||||
|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | ||
| YPD | |||||||
| 2-79 | Susceptible isolate | 1 | 1 | >64 | >64 | >64 | >64 |
| 12-99 | Resistant isolate | >64 | >64 | >64 | >64 | >64 | >64 |
| 12-99upc2Δ/Δ | upc2Δ::FRT/upc2Δ::FRT | 4 | 4 | 4 | 4 | 4 | 4 |
| 12-99upc2Δ/Δ+UPC2 | upc2Δ::FRT/UPC2-caSAT1 | 64 | >64 | >64 | >64 | >64 | >64 |
| YPD + BPS | |||||||
| 2-79 | Susceptible isolate | 2 | 2 | 4 | >64 | >64 | >64 |
| 12-99 | Resistant isolate | 32 | 32 | 32 | >64 | >64 | >64 |
| 12-99upc2Δ/Δ | upc2Δ::FRT/upc2Δ::FRT | NGa | NG | NG | NG | NG | NG |
| 12-99upc2Δ/Δ+UPC2 | upc2Δ::FRT/UPC2-caSAT1 | 32 | 32 | 32 | >64 | >64 | >64 |
| YPD + FeCl3 | |||||||
| 2-79 | Susceptible isolate | 2 | >64 | >64 | >64 | >64 | >64 |
| 12-99 | Resistant isolate | >64 | >64 | >64 | >64 | >64 | >64 |
| 12-99upc2Δ/Δ | upc2Δ::FRT/upc2Δ::FRT | 4 | 4 | 8 | 8 | 8 | 16 |
| 12-99upc2Δ/Δ+UPC2 | upc2Δ::FRT/UPC2-caSAT1 | 64 | >64 | >64 | >64 | >64 | >64 |
NG, no growth.
FIG 3.
(A) Effects of UPC2 in 12-99 on MICs and growth on YPD agar as determined by Etest. A confluent lawn of C. albicans was streaked prior to the addition of Etest strips and was then incubated for 48 h. (B) MIC heat map of 2-79, 12-99, the UPC2 mutant, and a complemented derivative. Susceptibility was determined by broth microdilution in YPD at 72 h (MICs in µg/ml above heat map). Growth was quantified spectrophotometrically and was assigned to a colorimetric scale. (C) Effect of UPC2 in 12-99 on the ability to grow on a solid medium containing fluconazole. From 4-fold serial dilutions of C. albicans strains, 2-μl aliquots were spotted onto YPD agar with (right) or without (left) 10 μg/ml FLC and were incubated for 48 h. (D) Effect of fluconazole on UPC2 in 12-99 by a time-kill assay. 12-99 or 12-99upc2Δ/Δ cells were diluted in YPD medium containing fluconazole (10 μg/ml) or the solvent dimethyl sulfoxide (DMSO). After 0, 6, 12, and 24 h, samples from each strain and medium were diluted and were plated for CFU.
Expression of ERG11, CDR1, CDR2, and MDR1 when UPC2 is disrupted in resistant backgrounds.
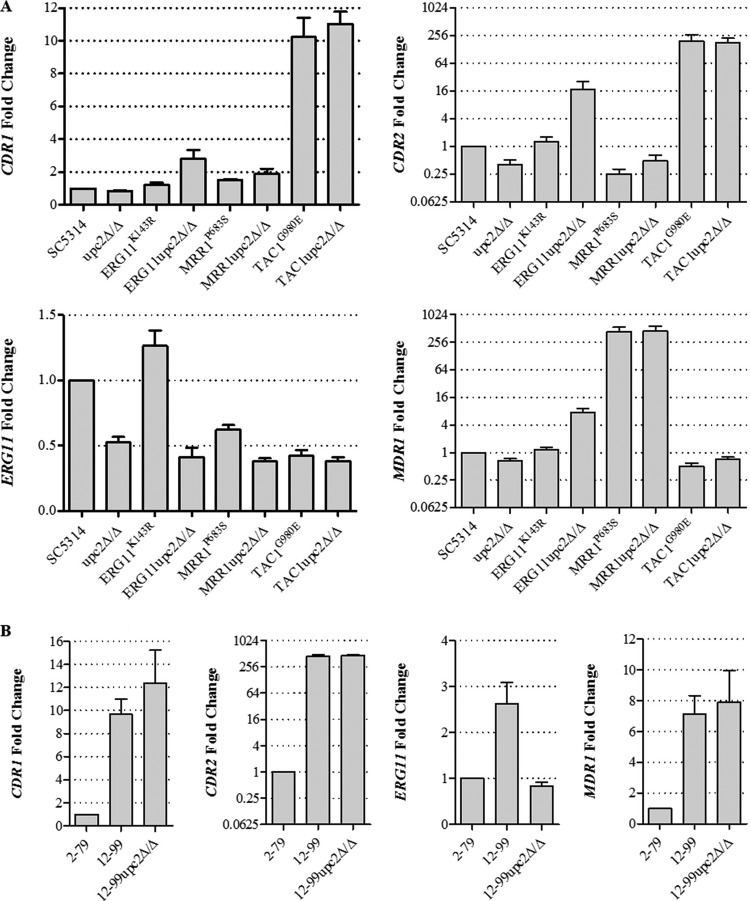
In order to determine if the enhanced fluconazole activity was due to decreased expression of ERG11 or genes encoding efflux pumps, we measured the abundances of ERG11, CDR1, CDR2, and MDR1 mRNAs by qRT-PCR in the strains containing a single resistance mechanism, clinical isolate 12-99, and their respective upc2Δ/Δ mutants (Fig. 4). As expected, the upc2Δ/Δ mutant constructed in the SC5314 background showed a reduction in baseline ERG11 expression from that of its parent strain. This was also the case for the upc2Δ/Δ mutants constructed in the ERG11K143R and 12-99 backgrounds. However, both the TAC1G980E and MRR1P683S strains exhibited levels of ERG11 expression lower than that of SC5314, with no appreciable additional reduction in expression when UPC2 was disrupted. Disruption of UPC2 did not result in decreased expression of CDR1, CDR2, or MDR1 in any background. Interestingly, disruption of UPC2 in the ERG11upc2Δ/Δ mutant resulted in increases in the expression of these transporter genes, the significance of which is unclear. These data suggest that the enhanced activity of fluconazole observed in resistant strains lacking UPC2 is not due to changes in transporter gene expression levels but may be associated with a reduction in the level of expression of ERG genes, particularly ERG11.
FIG 4.
Expression levels of ERG11, CDR1, CDR2, and MDR1. Levels of ERG11, CDR1, CDR2, and MDR1 expression in various strains were measured in triplicate by qRT-PCR and were compared to expression levels in SC5314 (A) and 2-79 (B). Error bars represent the standard errors of the means.
Comparison of the gene expression profiles of wild-type strain SC5314 and the upc2Δ/Δ mutant exposed to fluconazole.
In order to identify genes whose expression in response to fluconazole is influenced by Upc2, we compared the transcriptional profiles of SC5314 and its upc2Δ/Δ derivative after treatment with or without 10 μg/ml fluconazole for 6 h. Genes were considered to be differentially expressed in response to fluconazole if their expression changed by ≥1.5-fold in two independent experiments. Fluconazole-inducible genes were also considered to be UPC2 dependent if their induction was reduced (i.e., the level of expression was ≥2.0-fold [50%] lower than that in SC5314) in the deletion mutant. By use of these criteria, there were 127 genes upregulated by fluconazole whose induction was abrogated in the absence of UPC2 (Table 6; see also Data Set S1 in the supplemental material). The most common biological processes represented by these genes included the lipid metabolic process, iron ion transport and iron homeostasis, transport, responses to stress and chemical stimuli, and the oxidation-reduction process.
TABLE 6.
Genes upregulated at least 1.5-fold by fluconazole that are dependent on Upc2
| Processa | orf19 designation | CGD name | Fold change in expressionb in: |
Ratio (fold change in the upc2Δ/Δ strain/fold change in SC5314) |
||||
|---|---|---|---|---|---|---|---|---|
| SC5314 |
upc2Δ/Δ strain |
|||||||
| Expt 1 | Expt 2 | Expt 1 | Expt 2 | Expt 1 | Expt 2 | |||
| Lipid metabolic process | orf19.1598 | ERG24 | 2 | 1.6 | 0.9 | 0.7 | 0.5 | 0.4 |
| orf19.1631 | ERG6 | 1.8 | 1.7 | 0.4 | 0.5 | 0.2 | 0.3 | |
| orf19.2670 | 1.6 | 1.8 | 0.4 | 0.7 | 0.2 | 0.4 | ||
| orf19.3240 | ERG27 | 2.8 | 2.4 | 1.4 | 1.1 | 0.5 | 0.4 | |
| orf19.4982 | 2.1 | 2.3 | 0.7 | 0.8 | 0.3 | 0.4 | ||
| orf19.7585 | INO1 | 9.2 | 20.5 | 0.9 | 0.8 | 0.1 | 0 | |
| orf19.922 | ERG11 | 1.6 | 1.5 | 0.8 | 0.7 | 0.5 | 0.4 | |
| Iron ion transport | orf19.1415 | FRE10 | 2 | 4.2 | 0.2 | 0.1 | 0.1 | 0 |
| orf19.1932 | CFL4 | 35.6 | 359 | 8 | 22 | 0.2 | 0.1 | |
| orf19.4211 | FET3 | 2.8 | 6.2 | 0.7 | 0.2 | 0.3 | 0 | |
| orf19.4215 | FET34 | 2.1 | 5.2 | 0 | 0.1 | 0 | 0 | |
| orf19.5634 | FRP1 | 8.9 | 8.2 | 0.1 | 0.2 | 0 | 0 | |
| orf19.7219 | FTR1 | 3.2 | 7.8 | 0.1 | 0 | 0 | 0 | |
| Iron ion homeostasis | orf19.1264 | CFL2 | 2 | 6 | 0.2 | 0.8 | 0.1 | 0.1 |
| orf19.1715 | IRO1 | 4.2 | 8.1 | 2.2 | 1.4 | 0.5 | 0.2 | |
| orf19.5636 | RBT5 | 1.7 | 1.6 | 0.2 | 0.2 | 0.1 | 0.2 | |
| orf19.7114 | CSA1 | 1.7 | 3.2 | 0.7 | 0.6 | 0.4 | 0.2 | |
| Transport | orf19.1352 | TIM22 | 3.6 | 4.4 | 1.3 | 2 | 0.4 | 0.4 |
| orf19.2023 | HGT7 | 20.8 | 21.6 | 9.4 | 7.8 | 0.5 | 0.4 | |
| orf19.2292 | OPT4 | 5.2 | 3.7 | 0 | 0 | 0 | 0 | |
| orf19.2350 | 2.5 | 4.1 | 0.9 | 0.6 | 0.4 | 0.1 | ||
| orf19.2785 | ATP7 | 3.1 | 3.1 | 1.3 | 1.5 | 0.4 | 0.5 | |
| orf19.3026 | MAS1 | 1.7 | 1.6 | 0.9 | 0.8 | 0.5 | 0.5 | |
| orf19.3195 | HIP1 | 1.5 | 2.6 | 0.7 | 0.8 | 0.5 | 0.3 | |
| orf19.3232 | 24.6 | 5.3 | 1.6 | 1.6 | 0.1 | 0.3 | ||
| orf19.3668 | HGT2 | 48.3 | 35.5 | 20.1 | 12.3 | 0.4 | 0.3 | |
| orf19.3746 | IFC1 | 2.3 | 2.7 | 0.2 | 0 | 0.1 | 0 | |
| orf19.4335 | TNA1 | 194.9 | 29.8 | 0.4 | 0.2 | 0 | 0 | |
| orf19.4384 | HXT5 | 70.6 | 75.5 | 16.9 | 9.7 | 0.2 | 0.1 | |
| orf19.4682 | HGT17 | 45 | 24.7 | 4.2 | 4.5 | 0.1 | 0.2 | |
| orf19.4690 | 16.3 | 20.2 | 1 | 1.4 | 0.1 | 0.1 | ||
| orf19.5307 | JEN2 | 10.3 | 2.7 | 0.4 | 1.3 | 0 | 0.5 | |
| orf19.5753 | HGT10 | 20.6 | 1.6 | 2.6 | 0.8 | 0.1 | 0.5 | |
| orf19.6148 | 4.4 | 47.3 | 2 | 13.8 | 0.4 | 0.3 | ||
| orf19.6249 | HAK1 | 5.4 | 5.8 | 1.4 | 1.7 | 0.3 | 0.3 | |
| orf19.6993 | GAP2 | 30.6 | 10.6 | 6 | 3 | 0.2 | 0.3 | |
| orf19.7093 | HGT13 | 40.9 | 18.8 | 9.8 | 2.6 | 0.2 | 0.1 | |
| Response to stress | orf19.1434 | 1.7 | 2.7 | 0.8 | 0.9 | 0.5 | 0.3 | |
| orf19.3239 | CTF18 | 2.7 | 2.4 | 1.4 | 0.8 | 0.5 | 0.4 | |
| orf19.3672 | GAL10 | 6 | 7.3 | 2.7 | 3.8 | 0.5 | 0.5 | |
| orf19.4082 | DDR48 | 5.8 | 5.7 | 1.9 | 1.3 | 0.3 | 0.2 | |
| orf19.4093 | PES1 | 2.7 | 3.5 | 0.6 | 1.3 | 0.2 | 0.4 | |
| orf19.4317 | GRE3 | 1.5 | 1.5 | 0.3 | 0.7 | 0.2 | 0.5 | |
| orf19.496 | 2.6 | 2.3 | 1.1 | 0.6 | 0.4 | 0.3 | ||
| orf19.5902 | RAS2 | 7.6 | 6.2 | 2.3 | 1.5 | 0.3 | 0.2 | |
| orf19.7221 | SET3 | 4.6 | 2.9 | 2.1 | 1.4 | 0.5 | 0.5 | |
| orf19.921 | HMS1 | 2.6 | 2.8 | 0.8 | 1.3 | 0.3 | 0.5 | |
| Response to chemical stimulus | orf19.4645 | BEM1 | 1.5 | 1.9 | 0.4 | 0.6 | 0.3 | 0.3 |
| orf19.5591 | ADO1 | 2.4 | 2.9 | 0.7 | 0.8 | 0.3 | 0.3 | |
| orf19.6102 | RCA1 | 2.5 | 3.4 | 1.1 | 1.4 | 0.5 | 0.4 | |
| orf19.7374 | CTA4 | 1.7 | 2.2 | 0.8 | 1 | 0.5 | 0.5 | |
| Oxidation-reduction process | orf19.1411 | 2.7 | 4.8 | 1.4 | 1.1 | 0.5 | 0.2 | |
| orf19.1710 | ALI1 | 2 | 1.9 | 0.7 | 0.8 | 0.4 | 0.4 | |
| orf19.1940 | 1.7 | 1.5 | 0.4 | 0.7 | 0.2 | 0.5 | ||
| orf19.2091 | 2.2 | 2.3 | 0.8 | 0.9 | 0.4 | 0.4 | ||
| orf19.2108 | SOD6 | 7.9 | 10.6 | 1.1 | 1.6 | 0.1 | 0.1 | |
| orf19.4274 | PUT1 | 6.8 | 8.8 | 2.5 | 0.6 | 0.4 | 0.1 | |
| orf19.4747 | HEM14 | 1.7 | 2 | 0.1 | 0.2 | 0 | 0.1 | |
Descriptions are from the Candida Genome Database (CGD) (http://www.candidagenome.org).
Given as the ratio of expression in the presence of FLC to expression in the absence of FLC.
Validation of microarray data by real-time RT-PCR.
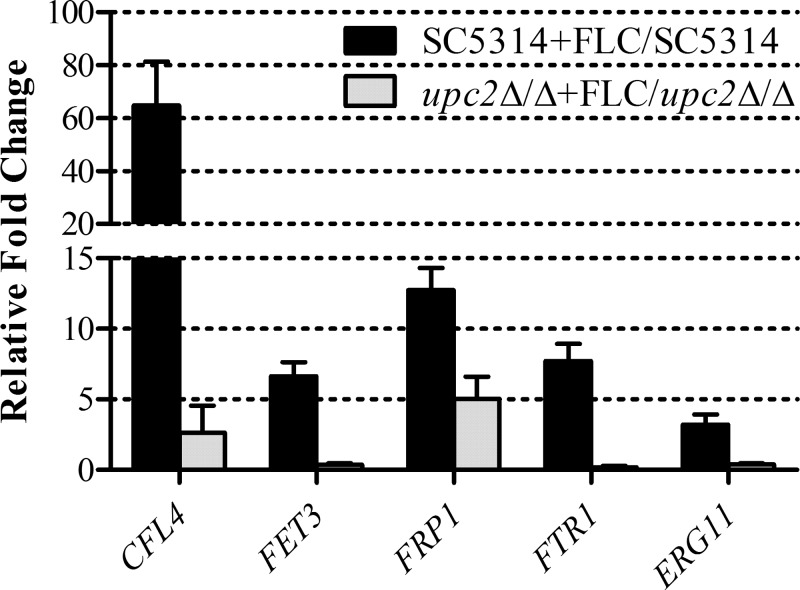
In order to validate the differential expression of genes identified by the microarray, we examined the mRNA abundances for five genes of interest by using the same RNA isolated for the microarray experiments. In addition to ERG11, four other genes were chosen based on their involvement in iron transport and homeostasis. The correlation between the microarray data and those obtained by real-time RT-PCR was good (Fig. 5). The expression of CFL4, FET3, FRP1, and FTR1 was upregulated in the wild-type strain SC5314 when treated with fluconazole but could not respond to the same extent when UPC2 was disrupted. As expected, ERG11 was also shown to respond to fluconazole in a UPC2-dependent fashion. These data suggest that the enhanced activity of fluconazole observed in both susceptible and resistant strains lacking UPC2 may be due to dysregulation of iron homeostasis, in addition to the inability to upregulate genes involved in the ergosterol biosynthesis pathway.
FIG 5.
Validation of fluconazole-inducible and Upc2-dependent iron gene expression. Levels of CFL4, FET3, FRP1, FTR1, and ERG11 expression were measured in triplicate by qRT-PCR and were compared to the expression levels in SC5314. Shown are the relative n-fold changes in gene expression in SC5314 and upc2Δ/Δ cells treated with fluconazole (FLC). Error bars represent the standard errors of the means.
Upc2 is required for growth under iron-poor conditions.
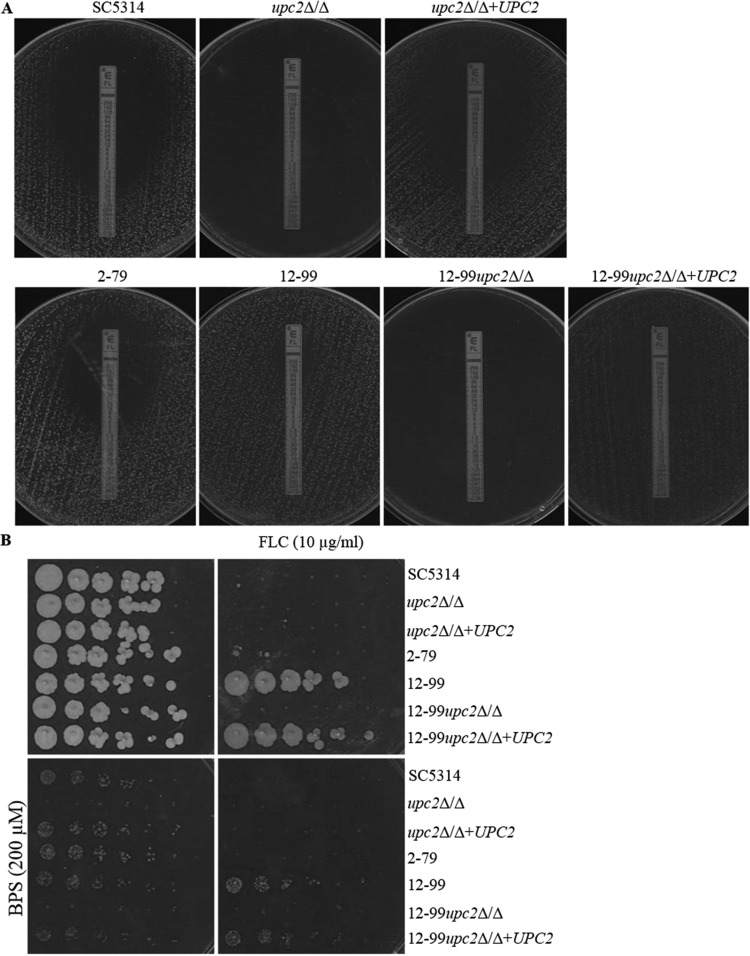
In order to investigate the relationship between Upc2 and iron transport and homeostasis, and to examine the impact of iron on susceptibility to fluconazole, we examined the growth and fluconazole susceptibilities of SC5314, 12-99, and their respective upc2Δ/Δ derivatives in media with varying concentrations of iron. In broth microdilution assays using iron-replete medium (YPD only), isolates SC5314 and 12-99 were able to resume growth in the presence of all concentrations of fluconazole tested after 72 h, whereas in iron-poor medium (YPD plus BPS), these strains were unable to grow at concentrations exceeding their 24-h MICs (Tables 3 and 5). The MICs at all time points were 0.5 μg/ml for SC5314 and 32 μg/ml for 12-99. No change in MFC was observed for these strains at any time point. Meanwhile, both upc2Δ/Δ mutants showed little to no growth at all time points based on both MICs and MFCs. In an iron-poor medium, the fluconazole MIC for SC5314 at 48 h by Etest was 0.38 μg/ml, compared to 1 μg/ml in an iron-replete medium (Fig. 1A); however, a clear zone of inhibition was observed (Fig. 6A). A MIC of <0.016 μg/ml and a clear zone of inhibition were observed for the upc2Δ/Δ mutant in an iron-poor medium (Fig. 6A), compared to 0.032 μg/ml and a clear zone of inhibition in an iron-replete medium (Fig. 1A). A MIC of 64 μg/ml and a small clear zone were observed for 12-99 in an iron-poor medium (Fig. 6A), compared to >256 μg/ml and confluent growth in an iron-replete medium (Fig. 3A), and a MIC of <0.016 μg/ml and a clear zone of inhibition were observed for 12-99upc2Δ/Δ in an iron-poor medium (Fig. 6A), compared to 1.5 μg/ml and a clear zone in an iron-replete medium (Fig. 3A). Growth was also reduced for all strains when plated on YPD agar plates containing BPS compared to YPD alone and was further reduced by 10 μg/ml of fluconazole (Fig. 6B). Only 12-99 and its complemented derivative were able to grow on YPD agar containing both BPS and 10 μg/ml fluconazole. Importantly, disruption of UPC2 in both SC5314 and 12-99 precluded the growth of either strain under iron-poor conditions. Conversely, high-iron conditions (YPD plus FeCl3) only very modestly enhanced the abilities of these isolates to grow in the presence of fluconazole (Tables 3 and 5). These data indicate that UPC2 is required for growth under iron-poor conditions.
FIG 6.
(A) Effect of BPS on MICs and growth on YPD agar with 200 μM BPS as determined by Etest. A confluent lawn of C. albicans was streaked prior to the addition of Etest strips and was then incubated for 48 h. (B) Effect of BPS on the ability to grow on a solid medium containing fluconazole. From 4-fold serial dilutions of C. albicans strains, 2-μl aliquots were spotted onto YPD agar with (right) or without (left) 10 μg/ml FLC and with (bottom) or without (top) 200 μM BPS and were incubated for 48 h.
DISCUSSION
Identifying novel drug targets that improve the efficacy of fluconazole is important in order to develop new therapeutic strategies to preserve the azole class of antifungals and overcome azole resistance. UPC2 has been well characterized with regard to its impact on fluconazole susceptibility and its role in regulating genes of the ergosterol biosynthesis pathway (19, 29, 36, 37). Silver et al. and MacPherson et al. identified Upc2p as the key regulator of ergosterol metabolism in C. albicans, showing that azole-inducible expression of ERG2, ERG7, ERG11, and ERG25 is diminished in the absence of UPC2 (19, 36). We and others have established that in some azole-resistant isolates, specific mutations render UPC2 constitutively active, resulting in increases in the expression of ERG genes (including ERG11), cellular ergosterol levels, and fluconazole resistance (25, 28, 29, 38–41). Moreover, UPC2 disruption results in a reduction in cellular ergosterol content (28). This suggests that UPC2 influences azole susceptibility through the regulation of this pathway.
In the present study, we observed that UPC2 disruption resulted not only in enhanced fluconazole susceptibility as measured by MICs but also in a substantial reduction in fluconazole MFCs at 24, 48, and 72 h. Indeed, UPC2 disruption in an azole-susceptible strain prevented its regrowth in YPD medium in the presence of high fluconazole concentrations after 72 h, resulted in a clear zone of inhibition around a fluconazole Etest strip, and prevented growth on a solid medium containing a therapeutically relevant concentration of fluconazole (10 μg/ml). Time-kill analysis also demonstrated a greater effect of 10 μg/ml fluconazole against the upc2Δ/Δ mutant than against its parent strain. Taken together, these data underscore the contribution of the Upc2 transcriptional activation pathway to azole susceptibility.
We then wanted to determine if disruption of UPC2 might have similar effects on fluconazole-resistant isolates. For this purpose, we chose isogenic strains containing resistance mutations in ERG11, MRR1, or TAC1. For the strain containing the ERG11K143R mutation, the fluconazole MIC was 8 μg/ml at 48 and 72 h in YPD. Accordingly, this strain was unable to grow in the presence of 10 μg/ml fluconazole. Although this background was not as highly resistant as others, its respective UPC2 deletion mutant exhibited a marked drop in both the MIC and the MFC of fluconazole. The MICs and MFCs for the resistant MRR1P683S and TAC1G980E strains were >64 μg/ml at 48 and 72 h in YPD medium, and those for the respective upc2Δ/Δ mutants were reduced markedly at all time points. This trend in susceptibility is consistent with what has been observed previously for this MRR1P683Supc2Δ/Δ mutant (20). However, in contrast to the halo of reduced confluent growth observed around the Etest strip with clinical isolate SC5314 (and generally observed with other C. albicans isolates), such a reduction in growth was not observed with these resistant strains, despite higher MICs and MFCs of >64 μg/ml. It has been shown recently that the constructed MRR1P683S and TAC1G980E mutants exhibit a fitness defect associated with the introduction of these specific resistance mutations, whereas clinical isolates carrying such mutations appear to have regained fitness (42). Such clinical isolates have likely evolved compensatory mutations that mitigate these fitness defects. This may explain the unusual growth pattern of these mutants when their susceptibilities are tested by Etest. Despite the absence of reduced confluent growth, the upc2Δ/Δ mutants in each background showed increased susceptibility to fluconazole by Etest and an inability to grow in the presence of fluconazole at 10 μg/ml. Therefore, the loss of UPC2 in strains containing a single mechanism of resistance also resulted in enhanced susceptibility to fluconazole, to a greater extent than that observed for their susceptible parent strain, SC5314.
The same phenotype was also observed upon disruption of UPC2 in a highly resistant clinical isolate, 12-99, which carries all four of the most common mechanisms of resistance. 12-99upc2Δ/Δ exhibited a substantial reduction in fluconazole resistance from that of isolate 12-99. Although 12-99upc2Δ/Δ was able to resume growth in lower concentrations of fluconazole at 72 h in broth microdilution plates, it was not able to grow to the extent of 12-99 or its matched susceptible isolate, 2-79, both of which were able to grow even in the highest concentration of fluconazole. Similarly to what was observed for the upc2Δ/Δ mutant in the SC5314 background, fluconazole exhibited a greater fungistatic effect against 12-99upc2Δ/Δ than against its azole-resistant parental isolate. Indeed, the loss of UPC2 allowed fluconazole to overcome all four mechanisms of fluconazole resistance operative in isolate 12-99 and resulted in greater fluconazole susceptibility than that seen with its matched susceptible isolate, 2-79. Moreover, the loss of UPC2 resulted in more-pronounced effects by MFC, 72-h regrowth, and time-kill analysis than those observed for the susceptible wild-type strain, SC5314.
One hypothesis for the way in which UPC2 disruption overcomes drug resistance is that it influences the expression of the efflux pump genes CDR1, CDR2, and MDR1 in these strains. Indeed, it has been shown that in C. albicans, Upc2 binds to the promoters of CDR1 and MDR1 and can regulate their expression (29). However, the expression of these genes was not affected by the absence of UPC2 in either the constructed mutant strains or the resistant clinical isolate 12-99. The disruption of UPC2 had such an effect on susceptibility that even with constitutive upregulation of these efflux pumps, the strains were no longer resistant to fluconazole. CDR1 and CDR2 remained upregulated in the TAC1G980E strain and 12-99, as did MDR1 in the MRR1P683S strain and 12-99.
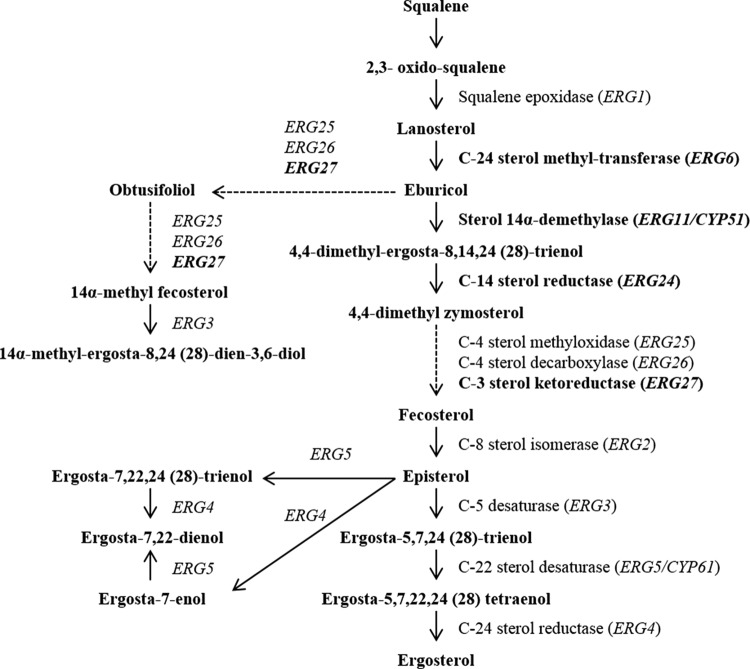
Another possible explanation for the increased susceptibility to fluconazole observed in the upc2Δ/Δ mutants is a reduction in baseline or inducible expression of genes involved in sterol biosynthesis. In the presence of fluconazole, the ERG11 gene product, lanosterol demethylase, is inhibited, leading to the accumulation of toxic sterol precursors and the production of alternate sterols, such as lanosterol, eburicol, obtusifoliol, 14α-methyl fecosterol, and 14α-methylergosta-8,24(28)-dien-3β,6α-diol (21). Integration of these sterols into the plasma membrane disrupts its integrity, resulting in its altered structure and function (12, 43, 44). Under these conditions, growth is inhibited but the organism remains viable (45). Fluconazole treatment also induces the expression of ERG11 and other genes of the ergosterol biosynthesis pathway, leading to increased production not only of lanosterol demethylase but also of enzymes involved in alternate sterol production (46). However, UPC2 disruption would reduce the level of ERG11 expression, which would diminish the amount of ergosterol in the cell. The expression of genes encoding enzymes needed for the biosynthesis of alternate sterols would also be reduced, further compromising the organism in the face of fluconazole exposure. Indeed, the level of ERG11 expression was reduced in the ERG11upc2Δ/Δ mutant as well as in 12-99upc2Δ/Δ as measured by real-time RT-PCR, and inducible expression of ERG6, ERG24, and ERG27 (as well as ERG11) was reduced in the upc2Δ/Δ mutant from that in its parent strain, SC5314, upon exposure to fluconazole as measured by microarray analysis. ERG6 and ERG27 both encode enzymes believed to be involved in the production of alternate sterols (Fig. 7).
FIG 7.
Sterol biosynthesis pathway in C. albicans. Genes whose products are shown in boldface were found to be responsive to fluconazole in a Upc2-dependent manner. Dotted lines denote multiple enzymatic steps.
Of particular interest, another core set of fluconazole-induced genes found to be dependent on UPC2 included those in the Gene Ontology process categories of iron transport (CFL4, FET3, FET34, FRE10, FRP1, FTR1) and iron homeostasis (CFL2, CSA1, IRO1, RBT5). Iron starvation has been shown to enhance the susceptibility of C. albicans to fluconazole (47–50). Prasad et al. observed that an ftr1Δ/Δ mutant, defective in high-affinity iron uptake, also exhibited enhanced susceptibility to fluconazole (47). Examination of the ERG11 mRNA abundance and membrane ergosterol levels in the ftr1Δ/Δ mutant revealed reductions in both from those for wild-type cells, which led to increased membrane fluidity and consequently increased passive diffusion of fluconazole. In contrast, ERG3, which acts in the biosynthesis of 14α-methylergosta-8,24(28)-dien-3β,6α-diol (51), was upregulated. This phenotype reverted upon the addition of iron to the medium. A similar observation has been made for Cryptococcus neoformans, where disruption of the genes encoding the ferroxidase (CFO1) and an iron permease (CFT1) of the high-affinity reductive iron uptake system resulted in increased susceptibility to azole antifungals (52). In the present study, we found that when UPC2 is intact, fluconazole exposure results in the upregulation of four genes associated with high-affinity iron uptake (FTR1, FET3, FET34, and FRP1). When UPC2 is disrupted, fluconazole exposure results in the downregulation of these genes, suggesting that UPC2 may be required for this process. Indeed, we found that disruption of UPC2 in isolates SC5314 and 12-99 precluded their ability to grow under low-iron conditions. Moreover, the fluconazole susceptibility of the upc2Δ/Δ mutants was greatly enhanced in iron-poor medium. Further investigation of the relationship between UPC2, high-affinity iron uptake, and fluconazole resistance is warranted and is under way in our laboratory.
Since many enzymes in the ergosterol biosynthesis pathway require iron, it is tempting to speculate that decreased iron uptake would have a significant impact on the activities of these enzymes. This, combined with decreased expression of the genes encoding enzymes involved in ergosterol biosynthesis (ERG11 in particular), may account for the substantial effect of UPC2 disruption on fluconazole susceptibility observed in these strains. While further investigation is needed to determine which Upc2 targets influence susceptibility to the azole antifungals, our findings suggest that Upc2 and the transcriptional activation pathway it regulates represent potential targets for overcoming azole antifungal resistance in C. albicans.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported in part by a grant from the Children's Foundation Research Center at Le Bonheur Children's Hospital, Memphis, TN (to E.M.V.) and by National Institutes of Health grant R01AI058145 (to P.D.R.).
We are grateful to Joachim Morschhäuser, Spencer Redding, and Ted White for mutant strains and clinical isolates. We thank Qing Zhang for invaluable assistance in the laboratory.
We have no financial or commercial conflicts of interest to declare.
Footnotes
Published ahead of print 21 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00221-13.
REFERENCES
- 1.Pfaller MA, Sheehan DJ, Rex JH. 2004. Determination of fungicidal activities against yeasts and molds: lessons learned from bactericidal testing and the need for standardization. Clin. Microbiol. Rev. 17:268–280. 10.1128/CMR.17.2.268-280.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marie C, White TC. 2009. Genetic basis of antifungal drug resistance. Curr. Fungal Infect. Rep. 3:163–169. 10.1007/s12281-009-0021-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sussman A, Huss K, Chio LC, Heidler S, Shaw M, Ma D, Zhu G, Campbell RM, Park TS, Kulanthaivel P, Scott JE, Carpenter JW, Strege MA, Belvo MD, Swartling JR, Fischl A, Yeh WK, Shih C, Ye XS. 2004. Discovery of cercosporamide, a known antifungal natural product, as a selective Pkc1 kinase inhibitor through high-throughput screening. Eukaryot. Cell 3:932–943. 10.1128/EC.3.4.932-943.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis RE, Rogers PD. 2007. Opportunistic mycoses, p 1600 In Chisholm-Burns M, Wells B, Schwinghammer T, Malone P, Kolesar J, Rotschafer J, DiPiro J. (ed), Pharmacotherapy principles and practice, 1st ed. McGraw-Hill Medical, New York, NY [Google Scholar]
- 5.Jarvis WR. 1995. Epidemiology of nosocomial fungal infections, with emphasis on Candida species. Clin. Infect. Dis. 20:1526–1530. 10.1093/clinids/20.6.1526 [DOI] [PubMed] [Google Scholar]
- 6.Gudlaugsson O, Gillespie S, Lee K, Vande Berg J, Hu J, Messer S, Herwaldt L, Pfaller M, Diekema D. 2003. Attributable mortality of nosocomial candidemia, revisited. Clin. Infect. Dis. 37:1172–1177. 10.1086/378745 [DOI] [PubMed] [Google Scholar]
- 7.Silverman S, Jr, Gallo JW, McKnight ML, Mayer P, deSanz S, Tan MM. 1996. Clinical characteristics and management responses in 85 HIV-infected patients with oral candidiasis. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 82:402–407. 10.1016/S1079-2104(96)80304-0 [DOI] [PubMed] [Google Scholar]
- 8.Feigal DW, Katz MH, Greenspan D, Westenhouse J, Winkelstein W, Jr, Lang W, Samuel M, Buchbinder SP, Hessol NA, Lifson AR, Rutherford GW, Moss A, Osmond D, Shiboski S, Greenspan JS. 1991. The prevalence of oral lesions in HIV-infected homosexual and bisexual men: three San Francisco epidemiological cohorts. AIDS 5:519–525. 10.1097/00002030-199105000-00007 [DOI] [PubMed] [Google Scholar]
- 9.Klein RS, Harris CA, Small CB, Moll B, Lesser M, Friedland GH. 1984. Oral candidiasis in high-risk patients as the initial manifestation of the acquired immunodeficiency syndrome. N. Engl. J. Med. 311:354–358. 10.1056/NEJM198408093110602 [DOI] [PubMed] [Google Scholar]
- 10.Kontoyiannis DP, Lewis RE. 2002. Antifungal drug resistance of pathogenic fungi. Lancet 359:1135–1144. 10.1016/S0140-6736(02)08162-X [DOI] [PubMed] [Google Scholar]
- 11.Traeder C, Kowoll S, Arasteh K. 2008. Candida infection in HIV positive patients 1985–2007. Mycoses 51(Suppl 2):58–61. 10.1111/j.1439-0507.2008.01574.x [DOI] [PubMed] [Google Scholar]
- 12.Ghannoum MA, Rice LB. 1999. Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin. Microbiol. Rev. 12:501–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hitchcock CA, Whittle PJ. 1993. Chemistry and mode of action of fluconazole, p 183–197 In Rippon JW, Fromtling RA. (ed), Cutaneous antifungal agents: selected compounds in clinical practice and development. Marcel Dekker, Inc, New York, NY [Google Scholar]
- 14.Levin DE. 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69:262–291. 10.1128/MMBR.69.2.262-291.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson JB. 2005. Evolution of antifungal-drug resistance: mechanisms and pathogen fitness. Nat. Rev. Microbiol. 3:547–556. 10.1038/nrmicro1179 [DOI] [PubMed] [Google Scholar]
- 16.Sanglard D. 2003. Resistance and tolerance mechanisms to antifungal drugs in fungal pathogens. Mycologist 17(2):74–78. 10.1017/S0269915X03002076 [DOI] [Google Scholar]
- 17.Ruhnke M, Eigler A, Tennagen I, Geiseler B, Engelmann E, Trautmann M. 1994. Emergence of fluconazole-resistant strains of Candida albicans in patients with recurrent oropharyngeal candidosis and human immunodeficiency virus infection. J. Clin. Microbiol. 32:2092–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White TC, Marr KA, Bowden RA. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacPherson S, Akache B, Weber S, De Deken X, Raymond M, Turcotte B. 2005. Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob. Agents Chemother. 49:1745–1752. 10.1128/AAC.49.5.1745-1752.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schubert S, Barker KS, Znaidi S, Schneider S, Dierolf F, Dunkel N, Aid M, Boucher G, Rogers PD, Raymond M, Morschhäuser J. 2011. Regulation of efflux pump expression and drug resistance by the transcription factors Mrr1, Upc2, and Cap1 in Candida albicans. Antimicrob. Agents Chemother. 55:2212–2223. 10.1128/AAC.01343-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly SL, Lamb DC, Corran AJ, Baldwin BC, Kelly DE. 1995. Mode of action and resistance to azole antifungals associated with the formation of 14-α-methylergosta-8,24(28)-dien-3-β,6-α-diol. Biochem. Biophys. Res. Commun. 207:910–915. 10.1006/bbrc.1995.1272 [DOI] [PubMed] [Google Scholar]
- 22.NCCLS. 2002. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard, 2nd ed. M27–A2. NCCLS, Wayne, PA [Google Scholar]
- 23.Klepser ME, Wolfe EJ, Jones RN, Nightingale CH, Pfaller MA. 1997. Antifungal pharmacodynamic characteristics of fluconazole and amphotericin B tested against Candida albicans. Antimicrob. Agents Chemother. 41:1392–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reuss O, Vik A, Kolter R, Morschhäuser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127. 10.1016/j.gene.2004.06.021 [DOI] [PubMed] [Google Scholar]
- 25.Dunkel N, Liu TT, Barker KS, Homayouni R, Morschhäuser J, Rogers PD. 2008. A gain-of-function mutation in the transcription factor Upc2p causes upregulation of ergosterol biosynthesis genes and increased fluconazole resistance in a clinical Candida albicans isolate. Eukaryot. Cell 7:1180–1190. 10.1128/EC.00103-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amberg DC, Burke DJ, Strathern JN. 2006. Isolation of yeast genomic DNA for Southern blot analysis. CSH Protoc. 2006:pii:pdb.prot4149. 10.1101/pdb.prot4149 [DOI] [PubMed] [Google Scholar]
- 27.Schmitt ME, Brown TA, Trumpower BL. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091–3092. 10.1093/nar/18.10.3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flowers SA, Barker KS, Berkow EL, Toner G, Chadwick SG, Gygax SE, Morschhauser J, Rogers PD. 2012. Gain-of-function mutations in UPC2 are a frequent cause of ERG11 upregulation in azole-resistant clinical isolates of Candida albicans. Eukaryot. Cell 11:1289–1299. 10.1128/EC.00215-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Znaidi S, Weber S, Al-Abdin OZ, Bomme P, Saidane S, Drouin S, Lemieux S, De Deken X, Robert F, Raymond M. 2008. Genomewide location analysis of Candida albicans Upc2p, a regulator of sterol metabolism and azole drug resistance. Eukaryot. Cell 7:836–847. 10.1128/EC.00070-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coste AT, Karababa M, Ischer F, Bille J, Sanglard D. 2004. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot. Cell 3:1639–1652. 10.1128/EC.3.6.1639-1652.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morschhäuser J, Barker KS, Liu TT, Blaß-Warmuth J, Homayouni R, Rogers PD. 2007. The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog. 3:e164. 10.1371/journal.ppat.0030164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasse C, Schillig R, Dierolf F, Weyler M, Schneider S, Mogavero S, Rogers PD, Morschhäuser J. 2011. The transcription factor Ndt80 does not contribute to Mrr1-, Tac1-, and Upc2-mediated fluconazole resistance in Candida albicans. PLoS One 6:e25623. 10.1371/journal.pone.0025623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schubert S, Popp C, Rogers PD, Morschhäuser J. 2011. Functional dissection of a Candida albicans zinc cluster transcription factor, the multidrug resistance regulator Mrr1. Eukaryot. Cell 10:1110–1121. 10.1128/EC.05100-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marichal P, Koymans L, Willemsens S, Bellens D, Verhasselt P, Luyten W, Borgers M, Ramaekers FC, Odds FC, Bossche HV. 1999. Contribution of mutations in the cytochrome P450 14α-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology 145(Part 10):2701–2713 [DOI] [PubMed] [Google Scholar]
- 35.White TC, Holleman S, Dy F, Mirels LF, Stevens DA. 2002. Resistance mechanisms in clinical isolates of Candida albicans. Antimicrob. Agents Chemother. 46:1704–1713. 10.1128/AAC.46.6.1704-1713.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silver PM, Oliver BG, White TC. 2004. Role of Candida albicans transcription factor Upc2p in drug resistance and sterol metabolism. Eukaryot. Cell 3:1391–1397. 10.1128/EC.3.6.1391-1397.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliver BG, Song JL, Choiniere JH, White TC. 2007. cis-Acting elements within the Candida albicans ERG11 promoter mediate the azole response through transcription factor Upc2p. Eukaryot. Cell 6:2231–2239. 10.1128/EC.00331-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heilmann CJ, Schneider S, Barker KS, Rogers PD, Morschhäuser J. 2010. An A643T mutation in the transcription factor Upc2p causes constitutive ERG11 upregulation and increased fluconazole resistance in Candida albicans. Antimicrob. Agents Chemother. 54:353–359. 10.1128/AAC.01102-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoot SJ, Smith AR, Brown RP, White TC. 2011. An A643V amino acid substitution in Upc2p contributes to azole resistance in well-characterized clinical isolates of Candida albicans. Antimicrob. Agents Chemother. 55:940–942. 10.1128/AAC.00995-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White TC. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41:1482–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rogers PD, Barker KS. 2003. Genome-wide expression profile analysis reveals coordinately regulated genes associated with stepwise acquisition of azole resistance in Candida albicans clinical isolates. Antimicrob. Agents Chemother. 47:1220–1227. 10.1128/AAC.47.4.1220-1227.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sasse C, Dunkel N, Schafer T, Schneider S, Dierolf F, Ohlsen K, Morschhäuser J. 2012. The stepwise acquisition of fluconazole resistance mutations causes a gradual loss of fitness in Candida albicans. Mol. Microbiol. 86:539–556. 10.1111/j.1365-2958.2012.08210.x [DOI] [PubMed] [Google Scholar]
- 43.Kelly SL, Lamb DC, Kelly DE, Manning NJ, Loeffler J, Hebart H, Schumacher U, Einsele H. 1997. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol Δ5,6-desaturation. FEBS Lett. 400:80–82. 10.1016/S0014-5793(96)01360-9 [DOI] [PubMed] [Google Scholar]
- 44.Klepser ME. 2006. Candida resistance and its clinical relevance. Pharmacotherapy 26:68S–75S. 10.1592/phco.26.6part2.68S [DOI] [PubMed] [Google Scholar]
- 45.Watson PF, Rose ME, Ellis SW, England H, Kelly SL. 1989. Defective sterol C5–6 desaturation and azole resistance: a new hypothesis for the mode of action of azole antifungals. Biochem. Biophys. Res. Commun. 164:1170–1175. 10.1016/0006-291X(89)91792-0 [DOI] [PubMed] [Google Scholar]
- 46.Henry KW, Nickels JT, Edlind TD. 2000. Upregulation of ERG genes in Candida species by azoles and other sterol biosynthesis inhibitors. Antimicrob. Agents Chemother. 44:2693–2700. 10.1128/AAC.44.10.2693-2700.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prasad T, Chandra A, Mukhopadhyay CK, Prasad R. 2006. Unexpected link between iron and drug resistance of Candida spp.: iron depletion enhances membrane fluidity and drug diffusion, leading to drug-susceptible cells. Antimicrob. Agents Chemother. 50:3597–3606. 10.1128/AAC.00653-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuipers ME, Beljaars L, Van Beek N, De Vries HG, Heegsma J, Van Den Berg JJ, Meijer DK, Swart PJ. 2002. Conditions influencing the in vitro antifungal activity of lactoferrin combined with antimycotics against clinical isolates of Candida. Impact on the development of buccal preparations of lactoferrin. APMIS 110:290–298. 10.1034/j.1600-0463.2002.100403.x [DOI] [PubMed] [Google Scholar]
- 49.Kuipers ME, de Vries HG, Eikelboom MC, Meijer DK, Swart PJ. 1999. Synergistic fungistatic effects of lactoferrin in combination with antifungal drugs against clinical Candida isolates. Antimicrob. Agents Chemother. 43:2635–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venkatesh MP, Rong L. 2008. Human recombinant lactoferrin acts synergistically with antimicrobials commonly used in neonatal practice against coagulase-negative staphylococci and Candida albicans causing neonatal sepsis. J. Med. Microbiol. 57:1113–1121. 10.1099/jmm.0.2008/001263-0 [DOI] [PubMed] [Google Scholar]
- 51.Martel CM, Parker JE, Bader O, Weig M, Gross U, Warrilow AG, Rolley N, Kelly DE, Kelly SL. 2010. Identification and characterization of four azole-resistant erg3 mutants of Candida albicans. Antimicrob. Agents Chemother. 54:4527–4533. 10.1128/AAC.00348-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim J, Cho YJ, Do E, Choi J, Hu G, Cadieux B, Chun J, Lee Y, Kronstad JW, Jung WH. 2012. A defect in iron uptake enhances the susceptibility of Cryptococcus neoformans to azole antifungal drugs. Fungal Genet. Biol. 49:955–966. 10.1016/j.fgb.2012.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Redding S, Smith J, Farinacci G, Rinaldi M, Fothergill A, Rhine-Chalberg J, Pfaller M. 1994. Resistance of Candida albicans to fluconazole during treatment of oropharyngeal candidiasis in a patient with AIDS: documentation by in vitro susceptibility testing and DNA subtype analysis. Clin. Infect. Dis. 18:240–242. 10.1093/clinids/18.2.240 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.