Abstract
Nitrite reductase (NirK) and nitric oxide reductase (NorB) have long been thought to play an essential role in nitrous oxide (N2O) production by ammonia-oxidizing bacteria. However, essential gaps remain in our understanding of how and when NirK and NorB are active and functional, putting into question their precise roles in N2O production by ammonia oxidizers. The growth phenotypes of the Nitrosomonas europaea ATCC 19718 wild-type and mutant strains deficient in expression of NirK, NorB, and both gene products were compared under atmospheric and reduced O2 tensions. Anoxic resting-cell assays and instantaneous nitrite (NO2−) reduction experiments were done to assess the ability of the wild-type and mutant N. europaea strains to produce N2O through the nitrifier denitrification pathway. Results confirmed the role of NirK for efficient substrate oxidation of N. europaea and showed that NorB is involved in N2O production during growth at both atmospheric and reduced O2 tensions. Anoxic resting-cell assays and measurements of instantaneous NO2− reduction using hydrazine as an electron donor revealed that an alternate nitrite reductase to NirK is present and active. These experiments also clearly demonstrated that NorB was the sole nitric oxide reductase for nitrifier denitrification. The results of this study expand the enzymology for nitrogen metabolism and N2O production by N. europaea and will be useful to interpret pathways in other ammonia oxidizers that lack NirK and/or NorB genes.
INTRODUCTION
Ammonia-oxidizing bacteria (AOB) are obligate chemolithotrophs that oxidize ammonia (NH3) through the intermediate hydroxylamine (NH2OH) to nitrite (NO2−) as their primary energy metabolism. During ammonia oxidation AOB produce gaseous nitrogen oxides, including nitrous oxide (N2O), a greenhouse gas (GHG) with more than 300 times the global-warming potential of CO2 (1), across a wide range of substrate and oxygen concentrations (2–4). Genes that encode nitrogen oxide reductases, including a periplasmic copper-containing nitrite reductase (nirK) and a membrane-bound nitric oxide reductase (norB), are present in many closed AOB genome sequences (5), including that of Nitrosomonas europaea strain ATCC 19718 (6), the model organism for this study. Previous work has identified two N2O-producing pathways in N. europaea, the pathway of hydroxylamine oxidation and the pathway of nitrifier denitrification. Generally, hydroxylamine oxidation is favored at atmospheric O2 tension (7, 8) and nitrifier denitrification is favored at low O2 tension (4, 9, 10). Although previous work has been done to describe the roles NirK and NorB may play in electron flow during substrate oxidation and NO2- reduction to N2O (11, 12), many questions remain about the functionality of these gene products, particularly under reduced O2 tension, at which nitrifier denitrification becomes environmentally relevant (13, 14). Furthermore, screening by low-stringency Southern blotting and PCR to identify DNA sequences with similarity to nirK revealed no hybridization signals from genomic DNA of Nitrosococcus mobilis Nc2, Nitrosomonas cryotolerans Nm55, or Nitrosomonas communis Nm2 (15). In addition, the genome of the recently sequenced Nitrosomonas sp. strain Is79 showed no homologues to the norCBQD gene cluster (16). These observations suggest either that NirK and NorB are nonessential to the ammmonia oxidizer lifestyle or that alternate mechanisms of reducing nitrogen oxides are present in AOB that lack these particular nitrogen oxide reductases.
Previous work on a NirK-deficient strain of N. europaea grown at atmospheric O2 tension showed NirK activity to be important in tolerance of the bacteria to NO2− (17) as well as for their efficient oxidation of NH3 and NH2OH (9, 10). Work on a NorB-deficient strain of N. europaea suggested that NorB is important for reduction of nitric oxide (NO) but not for net N2O production under atmospheric O2 tension (18). However, previous studies present conflicting evidence regarding whether NorB is essential for efficient oxidation of NH3 and NH2OH (10, 18). Conflicting results are also present in work on NirK-deficient N. europaea, particularly the role of NirK in pathways of N2O production. When grown in a chemostat, NirK-deficient N. europaea cells were unable to reduce NO2− as an alternate terminal electron acceptor (10), in contrast to batch growth, in which, at reduced O2 tension, there was no difference in the ability of NirK-deficient cells to reduce NO2− to N2O compared to that of wild-type N. europaea (9).
The functional roles of NirK and NorB in growth, substrate oxidation, and N2O production of N. europaea across a range of O2 tensions have not been fully elucidated. In this study, we compared the phenotypes of N. europaea wild-type, NirK-deficient, NorB-deficient, and NirK- plus NorB-deficient strains to solidify our understanding of the enzymology for N2O production as a function of variable O2 levels and to determine the necessity of NirK and NorB for growth, substrate oxidation, and NO2− reduction to N2O.
MATERIALS AND METHODS
Bacterial strains.
Wild-type Nitrosomonas europaea ATCC 19718 was used as the native strain for this study. The nirK::Kan (nirK gene locus NE 0924) strain of N. europaea was created in a previous study (17) and was received as a gift from H. J. E. Beaumont. Confirmation of the nirK::Kan strain was done by PCR using primers nir10f (5′-GGG CGA CAT ACC CAA GAG TG-3′), nir10r (5′-CAA GCC TAT GGG GGT TTA TAG-3′), and nir26r (5′-GTC ATA GCT GTT TCC TGT GTG AAA TT-3′) as described previously (17).
norB::Gen and nirK::Kan norB::Gen N. europaea strains were created by following a methodology described elsewhere (19). Briefly, the norB::Gen strain was generated by amplifying the norB gene (NE 2004) from N. europaea ATCC 19718 genomic DNA using primers Ne_2004F (5′-ACC CAG AAG CTT GCT TAC CC-3′) and Ne_2004R (5′-TGT TCG GTG ACG ATG ACA CT-3′). The amplified fragment was purified and ligated into the pGEM-T vector (Promega, Madison, WI). The ligation mixture was transformed into competent E. coli cells negative for both dam and dcm (New England BioLabs Inc., Ipswich, MA), and transformants were selected via blue-white screening on LB agar plates containing 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), 80 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), and 100 μg/ml of ampicillin. Plasmids from positive recombinants were purified using a Wizard Plus SV Minipreps DNA purification system kit (Promega) and digested with the KpnI restriction enzyme (New England BioLabs Inc.). The digest was run on a 0.8% agarose gel, and linearized vector was gel purified using the Wizard SV gel and PCR clean-up system kit (Promega). The gentamicin resistance cassette from the pUGM vector was digested with KpnI and gel purified (QIAquick gel extraction kit; Qiagen, Venlo, the Netherlands). The purified gentamicin cassette was then ligated into the previously KpnI-digested pGEM-T vector to disrupt the norB gene at nucleotide position 699 to 1347. The ligation mixture was transformed into E. coli JM109 cells, and positive transformants were selected on LB plates containing 100 μg/ml of ampicillin and 10 μg/ml of gentamicin. Positive recombinants were verified by PCR and Sanger sequencing using the BigDye Terminator cycle sequencing kit (Applied Biosystems, Foster City, CA). The plasmid with the correct construct was electroporated into prepared N. europaea cells (19) using an E. coli Pulser transformation apparatus (Bio-Rad Laboratories, Hercules, CA). Electroporated cells were inoculated into mineral salts medium (MSM) (20) without antibiotics and incubated at 28°C without shaking. After 24 h, 5 μg/ml of gentamicin was added and cultures were monitored until turbidity was evident and approximately 10 mM NO2− was produced. Cell culture (1 ml) was then inoculated onto nitrocellulose membranes overlaying agar-solidified mineral medium to select single recombinant colonies as described previously (19). Cultures were PCR screened to confirm the location and orientation of the gentamicin resistance cassette within the norB gene that had recombined in the chromosome using primers Ne_2004F (as reported above), GenF (5′ TGC CTC GGG CAT CCA AGC AG-3′), and GenR (GAG AGC GCC AAC AAC CGC TTC T-3′). The methods for creation of the nirK::Kan norB::Gen strain were identical to generation of the norB::Gen strain except that the nirK::Kan strain of N. europaea (17) was used as the recipient instead of wild-type N. europaea and 5 μg/ml of gentamicin and 30 μg/ml of kanamycin were added to the MSM of electroporated cells after 24 h of incubation as described above.
All N. europaea strains were grown in 500-ml Erlenmeyer flasks with 250 ml of MSM containing 25 mM (NH4)2SO4 (20). Cultures were incubated at 30°C in the dark with shaking. Inoculation of fresh medium used a 1% volume of culture in stationary phase, which was determined by NO2− concentration (21). Concentrations of NO2− were determined using a standard curve from 1 mM to 20 mM NaNO2, and stationary phase was achieved at 10 mM NO2−.
Growth experiments.
Wild-type and mutant N. europaea cultures (1 ml) were inoculated into MSM (100 ml) in Wheaton bottles (250 ml) sealed with caps inlaid with butyl rubber stoppers. Cultures were initiated at atmospheric (ca. 22%) or hypoxic (ca. 5%) levels of O2. Hypoxia was achieved by aseptically sparging the bottles with nitrogen gas and injecting pure O2 into the headspace. Final headspace O2 levels were confirmed by gas chromatography (GC-thermal conductivity detector [TCD] from Shimadzu and molecular sieve column from Alltech, Deerfield, IL). O2 was measured again at the experimental endpoint (72 h) to determine the amount consumed. N2O was measured in the gas headspace at 24, 48, and 72 h by GC-TCD (Hayesep Q column). Headspace concentrations of O2 and N2O in the cultures were determined by comparison to standard curves using pure gases (Sigma-Aldrich). Total cell counts were done at 0, 24, 48, and 72 h using a Petroff-Hausser counting chamber and contrast light microscopy to follow the cells from exponential into stationary phase of growth. NO2− concentrations were determined at 0, 24, 48, and 72 h by colorimetric assay as described above. NH2OH concentration was measured during growth between 0 h and 72 h in increments of 6 h using a colorimetric assay (22). Statistical differences between measured values among the N. europaea strains and experimental conditions were evaluated using Student's t test at a P value of <0.05.
Resting-cell assays.
The wild-type and mutant strains of N. europaea were grown to stationary phase as described above. For each experiment, culture (1 ml) was transferred to a 12-ml vial sealed with a rubber stopper and aluminum crimp seal. The vial was sparged with nitrogen gas to anoxia. An electron donor (ascorbic acid; 1 mM) and electron shuttle (phenazine methosulfate; 0.1 mM) (23) were added to the culture via Hamilton syringe. The vial was left to sit at 30°C in the dark for 72 h to allow adequate time for reduction of NO2−and accumulation of N2O. Headspace N2O concentration was measured at 0 and 72 h as described above. To confirm consistent anoxia, O2 was measured at 0 and 72 h using gas chromatography (GC-TCD from Shimadzu and molecular sieve column from Alltech).
MR measurements.
In preparation for instantaneous O2 consumption and NO2− reduction experiments, the wild-type and mutant strains of N. europaea were grown in 250-ml Wheaton bottles in 100 ml of MSM to stationary phase. Cells were harvested by filtration on Supor200 0.2-μm filters (Pall, Ann Arbor, WI) and rinsed three times with sodium phosphate buffer (50 mM NaH2PO4, 2 mM MgCl2; pH 8) to wash away remaining NO2− produced during growth. Approximately 5 × 1010 total cells were resuspended into 10 ml of sodium phosphate buffer in a 10-ml two-port microrespiratory (MR) chamber with fitted injection lids (Unisense, Aarhus, Denmark). O2 concentration was measuring using an OX-MR 500-μm-tip-diameter MR oxygen electrode (Unisense, Aarhus, Denmark), and N2O concentration was measuring using an N2O-500 N2O minisensor electrode with a 500-μm tip diameter (Unisense). Hydrazine (N2H4) was added to the chamber as an electron donor for NO2− reduction at the beginning of each experiment at a concentration of 250 μM and again at a concentration of 125 μM after the cells had consumed more than half of the available O2. Once the cells had consumed all available O2 they were left to sit for 5 to 10 min under anoxia. An absence of N2O production confirmed that no endogenous NO2− was present, after which 2 mM NaNO2 was added to the chamber through the injection port. Instantaneous NO2− reduction to N2O was measured for approximately 10 min.
RESULTS
Growth phenotype of N. europaea strains.
N. europaea wild-type and mutant strains were grown under atmospheric (ca. 22%) and reduced (ca. 5%) O2 tensions to evaluate and compare the phenotypes of strains deficient in NirK, NorB, or both gene products. Growth experiments beginning at atmospheric O2 tension revealed that only the double mutant (nirK::Kan norB::Gen) strain had a significantly slower doubling time with respect to the other three strains; however, the amounts of NO2− produced by both the nirK::Kan and double mutant strains were significantly less than that of the wild type (Fig. 1; Table 1). In contrast, the norB::Gen strain showed no significant difference in doubling time or NO2− production relative to those of the wild type under atmospheric O2 tension (Fig. 1; Table 1). None of the cultures initiated at atmospheric O2 tension reduced the O2 headspace level to below 6%; however, the double mutant consumed significantly less O2 than the other strains, in congruence with its slower doubling time (Table 1). Previous studies have shown that batch growth of N. europaea with ample O2 is limited by acidification of the medium, which reduces availability of NH3 to the cells (24). Hence, the cells entered stationary phase prior to consuming all of the available O2 in the present experiments due to medium acidification and not O2 limitation.
FIG 1.
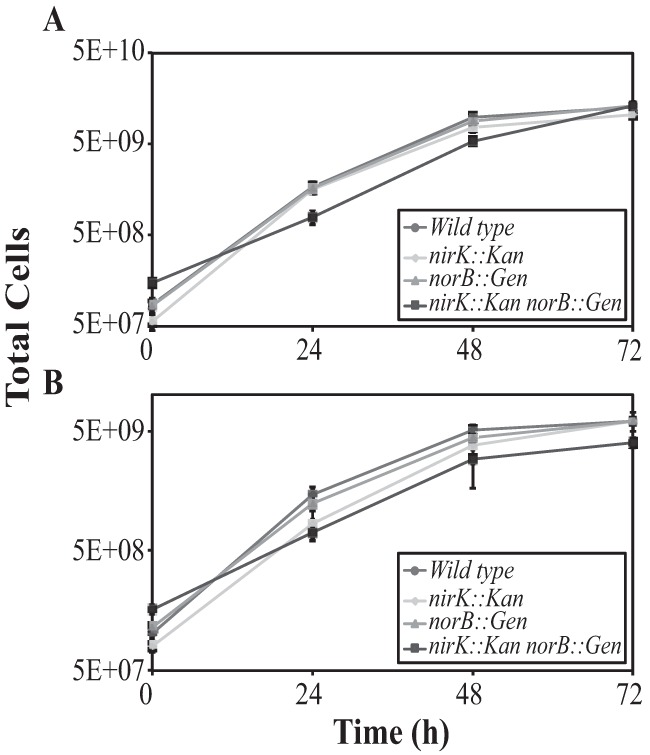
Growth curves for N. europaea strains initiated at 22% O2 (A) and 4.6% O2 (B). Data represent mean values ± SEs (ca. 22% O2, n = 8, ca. 4.6% O2, n = 6).
TABLE 1.
Doubling time, total nitrite production, and percent remaining headspace O2 for wild-type and mutant strains of N. europaea ATCC 19718 cultivated under atmospheric and reduced O2 tensionsa
| Organism description | Value at indicated oxygen tension |
|||||
|---|---|---|---|---|---|---|
| Doubling time (h) |
Total NO2−-N produced (mM × 1010 cells−1) |
Remaining O2 in headspace (%) |
||||
| 22% | 4.6% | 22% | 4.6% | 22% | 4.6% | |
| Wild type | 6.5 d (0.6) | 8.7 d (0.3) | 7.3 bd (0.02) | 4.2 d (0.4) | 6.7 d (9.2e−5) | 1.0 (6.7e−4) |
| nirK::Kan | 6.7 d (0.2) | 9.3 d (0.8) | 5.8 a (0.01) | 4.1 d (0.8) | 6.8 d (1.8e−3) | 1.2 d (2.4e−3) |
| norB::Gen | 7.1 d (0.3) | 9.2 d (0.2) | 6.6 d (0.02) | 4.3 d (0.4) | 6.7 d (1.7e−3) | 1.2 (2.4e−3) |
| nirK::Kan norB::Gen | 9.6 abc (0.4) | 14.3 abc (1.9) | 5.6 ac (0.04) | 8.0 abc (1.1) | 8.0 abc (1.8e−3) | 0.8 b (1.1e−4) |
Doubling times were calculated over the 0- to 48-h period of exponential growth. Total NO2−-N produced and remaining O2 in headspace were determined at 72 h for all cultures. Averages and SEs (in parentheses) were calculated from 8 and 6 replicated experiments for cultures grown under 22 and 4.6% O2, respectively. Significant differences (P < 0.05) are denoted by different letters as follows: “a,” strain versus wild type; “b,” strain versus nirk::Kan strain; “c,” strain vs. norB::Gen strain; and “d,” strain versus nirK::Kan norB::Gen strain.
Growth experiments beginning at reduced O2 tension again revealed a significant reduction in doubling time for the double mutant compared with those of the other strains (Fig. 1; Table 1). The double mutant also showed a significant accumulation of NO2− in comparison to those of the other strains (Table 1). Although NH2OH was assayed from all of the cultures under all O2 tensions, the assay was unable to detect significant differences over time or between strains (data not shown).
Together, the results suggest that regardless of initial O2 levels, NorB alone played no significant role in the growth phenotype or substrate oxidation efficiency of N. europaea; however, NirK was essential for efficient substrate oxidation efficiency, especially during growth initiated at atmospheric O2 tension. The lack of both gene products significantly slowed growth of N. europaea and also allowed for significant accumulation of NO2− during hypoxic growth relative to the other strains.
Effects of NirK and NorB absence on N2O production by N. europaea under variable O2 tensions.
To evaluate the roles of NirK and NorB in the pathways of N2O production for N. europaea, N2O concentration in the gas headspace was measured during growth experiments initiated at both atmospheric and reduced O2 tension and in resting-cell assays in the absence of O2. In confirmation of prior studies, the concentration of N2O in the headspace of the nirK::Kan strain at the endpoint (72 h) of growth in cultures initiated with atmospheric O2 was approximately 15 times that of the wild-type strain (Fig. 2) (9, 17). However, the resulting N2O measured after hypoxic growth and in the anoxic resting-cell assay revealed no difference between wild-type and nirK::Kan strains of N. europaea. In contrast, the norB::Gen strain produced ca. 20% less N2O than did the wild type following growth under atmospheric O2 and approximately 70% less N2O following growth under reduced O2 (Fig. 2). No N2O was detected in the anoxic resting-cell assay of norB::Gen cells. The double mutant produced an amount of N2O similar to that produced by the norB::Gen strain when grown under atmospheric O2 but was unable to produce measurable N2O after 72 h when grown under hypoxia or in the anoxic resting-cell assay.
FIG 2.
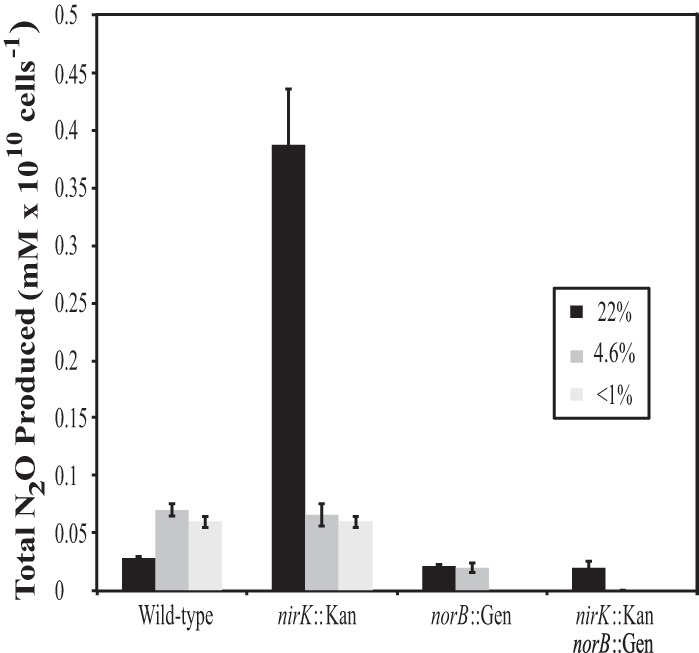
Total N2O produced by all strains after 72 h of growth at high (n = 8) and low (n = 6) oxygen. N2O profiles under anoxia (n = 8) were collected during resting-cell assays. Data are presented as means ± SEs.
Instantaneous O2 consumption and N2O production by wild-type and mutant N. europaea strains.
The anoxic resting-cell assays revealed that N. europaea lacking NorB expression did not produce a measureable quantity of N2O in the gas headspace from the reduction of available NO2− (10 mM) in the medium. Therefore, instantaneous NO2− reduction experiments were conducted to confirm whether NorB is essential to NO2− reduction to N2O by N. europaea and whether an alternative nitrite reductase to NirK was operating in the nirK::Kan strain to produce the same amount of N2O as observed in the wild type (Fig. 2).
Instantaneous NO2− reduction experiments were conducted in microelectrode chambers with the use of a non-nitrite-forming intercellular electron donor, N2H4, and microelectrodes for O2 and N2O. The cells were allowed to consume all of the O2 in the microelectrode chamber via oxidation of N2H4, after which NaNO2− was added. Both the wild type and the nirK::Kan strain produced approximately 8 μmol N2O per liter per min, confirming the presence of an alternate nitrite reductase activity in nirK::Kan cells (Fig. 3A and B). Both the norB::Gen and double mutant strains showed only background levels of N2O production from electrode drift upon the addition of NaNO2, confirming that activity of NorB is essential to this process (Fig. 3C and D).
FIG 3.
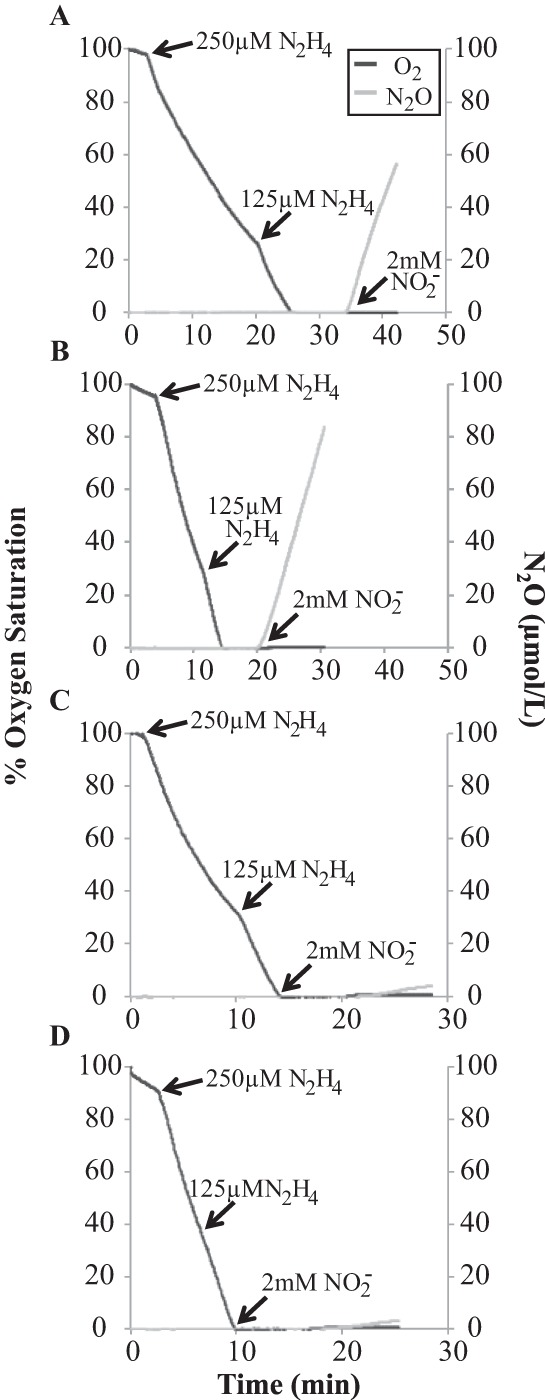
Instantaneous oxygen consumption and nitrite reduction by wild-type (A), nirK::Kan (B), norB::Gen (C), and nirK::Kan norB::Gen (D) strains of N. europaea. Data are single representatives of reproducible results.
DISCUSSION
Function of NirK and NorB in efficient substrate oxidation and growth of N. europaea under variable O2 tensions.
The reduced production of NO2− by both the nirK::kan and double mutant strains of N. europaea when grown at atmospheric O2 tension confirm previous reports of the requirement of NirK for efficient oxidation of NH3 to NO2− during batch (9, 17) and chemostat (10) cultivation. Slowed substrate oxidation in the NirK-deficient strain of N. europaea was previously suggested to be caused by interruption of electron flow from NH2OH to NO2− due to the inability of HAO to pass electrons on to NirK through cytochrome c electron carriers (9). A diminished ability of NirK-deficient N. europaea to oxidize exogenous NH2OH during growth strengthens the hypothesis that NirK functions aerobically to facilitate efficient substrate oxidation (9). Furthermore, extensive accumulation of N2O in the gas headspace of nirK::Kan cultures during growth under atmospheric O2 tension validates previous measurements from growth of this strain in batch (9, 17) and chemostat (10) cultures. A possible explanation for this phenotype is that in the presence of high NH2OH concentrations the HAO enzyme produces NO due to incomplete oxidation of NH2OH, which is then enzymatically reduced to N2O (14). Our data suggest that enzymatic reduction of NO via NorB could lead to production of N2O from NH2OH. During growth under atmospheric O2 tension, the 20% reduction in N2O produced by the norB::Gen strain could be accounted for by the lack of NorB activity, with the remaining N2O being produced from an alternate nitric oxide reductase (Fig. 2).
In the double mutant strain, growth and net NO2− production were likely slowed during growth at atmospheric O2 by both the lack of NirK in speeding substrate oxidation and also from the lack of NorB in preventing toxic accumulation of NO. When cultivated under reduced O2, the alternate nitrite reductase activity could cause the double mutant cultures to accumulate an excess of NO that could not be removed as NorB is more active at low O2. This excess NO could both slow cellular growth and result in net NO2− production due to chemical reactions of nitrogen oxides (NOx) in the culture medium. Future work comparing NO accumulation between the wild type and mutant strains of N. europaea under variable O2 tension would assist in validating these hypotheses.
The growth rate of N. europaea lacking NorB expression alone was not significantly impaired, which is in agreement with data from a previous study (18); however, in contrast to that study, N2O production by norB::Gen cells was significantly lower than that of the wild type (Fig. 2). Schmidt (10) showed that NorB-deficient N. europaea had a significantly lower growth rate and yield than did the wild type, an N2O production profile similar to that of the NirK-deficient strain, and significantly larger amounts of NH2OH released to the growth medium than did the wild type, suggesting similar inefficiency of substrate oxidation by both NirK- and NorB-deficient strains. Under the growth conditions of the present study, however, the results obtained by Schmidt (10) were not validated. Rather, our results suggest that the absence of NorB expression alone in N. europaea had no effect on growth or substrate oxidation rates or on NH2OH accumulation but did result in diminished N2O production in comparison to that of the wild type.
NorB, but not NirK, is required for anoxic reduction of NO2− to N2O in N. europaea.
The inability of N. europaea strains lacking NorB expression to make measurable N2O in anoxic resting-cell assays (Fig. 2) and instantaneous NO2− reduction assays (Fig. 3) pointed to NorB as the essential nitric oxide reductase involved in NO reduction in the absence of O2. The significant accumulation of NO2− only during hypoxic growth of the double mutant strain (Table 1) could be explained by chemical decay of highly reactive NO that may accumulate from activity of the alternative nitrite reductase working in the absence of both NirK and NorB enzymes. However, the lack of similar results with the norB::Gen strain suggests that the activity of NirK in the absence of NorB has an effect on nitrogen oxide metabolism that is substantially different from that of the alternative NO2− reductase. Thus, exploration of alternative nitrogen oxide reductases active in N. europaea with and without expression of NirK and/or NorB will be helpful to elucidate the enzymology behind these phenotypes.
Amended pathways of N2O production in N. europaea and other AOB.
The most important finding of this study is the demonstration that NirK is not essential to the nitrifier denitrification pathway of N. europaea, as has been assumed for many years in the literature. The similar headspace N2O levels produced by both wild-type and nirK::Kan strains of N. europaea during growth under reduced O2 tension, in anoxic resting-cell assays, and in anoxic instantaneous nitrite reduction experiments all revealed that an alternate nitrite reductase to NirK is active in the production of N2O by N. europaea ATCC 19178 (Fig. 4). It was previously suggested that an alternate nitrite reductase may be active in N. europaea (9); however, no other known homologues to nitrite reductase genes have been identified in its closed genome (6). One possible candidate for an alternate nitrite reductase in AOB is C-terminally truncated HAO (HaoA′; NE0962, 2044, 2339) (25). Evolutionary reconstructions showed that HAO evolved from an octaheme cytochrome c nitrite reductase (26), and gene expression of HaoA′ in the methanotrophic strain Methylococcus capsulatus strain Bath was induced in the presence of ammonia (25). M. capsulatus strain Bath can reduce NO2− to N2O in the presence of NH3 and NO2− (27) even though homologues to both nirK and nirS NO-forming nitrite reductases are absent from the closed genome sequence (28). Although nirK genes have been found in the genomes of most AOB (15) and ammonia-oxidizing archaea (29, 30) and nirK has long been used as a marker for denitrification activity in the field of microbial ecology, the present study shows that at least in Nitrosomonas europaea ATCC 19718, nirK is not a marker for denitrification but rather should be considered a marker for ammonia oxidation. It should be noted that due to differences in gene phylogenies and neighborhoods in the Nitrosospira spp. and Nitrosococcus spp. (9), the specific roles of NirK and NorB should be physiologically examined within strains of these genera to determine if the nitrifier denitrification pathways share similar inventories and are similarly regulated among the ammonia-oxidizing bacteria.
FIG 4.
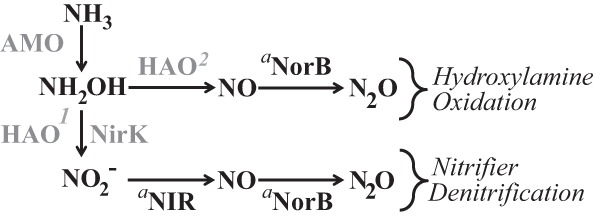
Amended pathways of N2O production by Nitrosomonas europaea ATCC 19718. AMO, ammonia monooxygenase; HAO, hydroxylamine oxidoreductase; NirK, nitrite reductase; NorB, nitric oxide reductase; NIR, unidentified alternate nitrite reductase. The role of enzymes in gray were characterized in previous studies as follows: AMO, reference 32; HAO1, reference 33; HAO2, reference 34; and NirK, reference 9). The roles of enzymes denoted with superscript “a” are from the present study.
In addition to an alternate nitrite reductase, our results also demonstrate that NorB activity plays a role in the hydroxylamine oxidation pathway of N2O production by N. europaea ATCC 19718 (Fig. 4). It is also possible that alternate nitric oxide reductases are active in Nitrosomonas spp. For instance, a complete transcriptome of the nirK::Kan strain showed increased expression of genes for norSY (originally annotated as coxAB2), an alternative nitric oxide reductase, in comparison to the wild-type strain when grown under normal oxic conditions (7). Furthermore, Nitrosomonas eutropha C91 grown under continuous cultivation for 3 months in the presence of nitrogen dioxide (NO2) gas showed increased expression of NorY protein (31). These observations suggest that NorY nitric oxide reductase could potentially contribute to N2O production along with NorB during growth of N. europaea particularly under atmospheric O2 levels (Fig. 2); however, this hypothesis remains to be validated.
Our results showing the inability of the norB::Gen and double mutant strains of N. europaea to reduce NO2− to N2O in instantaneous NO2− reduction experiments (Fig. 3C and D), even with a readily available source of electrons, demonstrate that NorB is the sole enzyme involved in N2O production through the nitrifier denitrification pathway (Fig. 4). While these results are in agreement with those of Cantera and Stein and of Schmidt (9, 10), a discrepancy remains regarding the role of NirK in this pathway.
Conclusions.
This study is unique in its comparison of phenotypes of N. europaea lacking expression of NirK, NorB, and both enzymes together. Furthermore, our assays allowed comparison of phenotypes under O2 initially present at atmospheric, hypoxic, and anoxic levels, each having a different effect on N2O production by the two characterized pathways in N. europaea ATCC 19718 (1–4, 14). The main conclusions from this study are that (i) NirK, but not NorB, plays an essential role in efficient substrate oxidation under atmospheric O2 tension; (ii) an alternate nitrite reductase to NirK is active in N. europaea under both hypoxic and anoxic conditions; (iii) NorB and/or other NOR enzymes are active in N. europaea during growth under atmospheric O2 tension; and (iv) NorB is the only nitric oxide reductase active in the nitrifier denitrification pathway. These results suggest that AOB have diverse enzymology beyond NirK and NorB leading to N2O production that remains to be characterized.
ACKNOWLEDGMENTS
This work was supported by a graduate fellowship awarded to J.A.K. by Alberta Innovates Technology Futures and NSERC Discovery Award 371544-09 to L.Y.S.
Footnotes
Published ahead of print 6 June 2014
REFERENCES
- 1.Stein LY, Yung YL. 2003. Production, isotopic composition, and atmospheric fate of biologically produce nitrous oxide. Annu. Rev. Earth Planet. Sci. 31:329–356. 10.1146/annurev.earth.31.110502.080901 [DOI] [Google Scholar]
- 2.Dundee L, Hopkins DW. 2001. Different sensitivities to oxygen of nitrous oxide production by Nitrosomonas europaea and Nitrosolobus multiformis. Soil Biol. Biochem. 33:1563–1565. 10.1016/S0038-0717(01)00059-1 [DOI] [Google Scholar]
- 3.Shaw LJ, Nicol GW, Smith Z, Fear J, Prosser JI, Baggs EM. 2006. Nitrosospira spp. can produce nitrous oxide via a nitrifier denitrification pathway. Environ. Microbiol. 8:214–222. 10.1111/j.1462-2920.2005.00882.x [DOI] [PubMed] [Google Scholar]
- 4.Poth M, Focht DD. 1985. 15N kinetic analysis of N2O production by Nitrosomonas europaea: an examination of nitrifier denitrification. Appl. Environ. Microbiol. 49:1134–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arp DJ, Chain P, Klotz MG. 2007. The impact of genome analyses on our understanding of ammonia-oxidizing bacteria. Annu. Rev. Microbiol. 61:503–528. 10.1146/annurev.micro.61.080706.093449 [DOI] [PubMed] [Google Scholar]
- 6.Chain P, Lamerdin J, Larimer F, Regala W, Lao V, Land M, Hauser L, Hooper A, Klotz M, Norton J, Sayavedra-Soto L, Arciero D, Hommes N, Whittaker M, Arp D. 2003. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J. Bacteriol. 185:2759–2773. 10.1128/JB.185.9.2759-2773.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho CM-H, Yan T, Liu X, Wu L, Zhou J, Stein LY. 2006. Transcriptome of a Nitrosomonas europaea mutant with a disrupted nitrite reductase gene (nirK). Appl. Environ. Microbiol. 72:4450–4454. 10.1128/AEM.02958-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Law Y, Ni B-J, Lant P, Yuan Z. 2012. N2O production rate of an enriched ammonia-oxidizing bacteria culture exponentially correlates to its ammonia oxidation rate. Water Res. 46:3409–3419. 10.1016/j.watres.2012.03.043 [DOI] [PubMed] [Google Scholar]
- 9.Cantera JJL, Stein LY. 2007. Role of nitrite reductase in the ammonia-oxidizing pathway of Nitrosomonas europaea. Arch. Microbiol. 188:349–354. 10.1007/s00203-007-0255-4 [DOI] [PubMed] [Google Scholar]
- 10.Schmidt I. 2004. Denitrification and ammonia oxidation by Nitrosomonas europaea wild-type, and NirK- and NorB-deficient mutants. Microbiology 150:4107–4114. 10.1099/mic.0.27382-0 [DOI] [PubMed] [Google Scholar]
- 11.Hooper A. 1968. A nitrite-reducing enzyme from Nitrosomonas europaea Preliminary characterization with hydroxylamine as electron donor. Biochim. Biophys. Acta 162:49–65. 10.1016/0005-2728(68)90213-2 [DOI] [PubMed] [Google Scholar]
- 12.Whittaker M, Bergmann D, Arciero D, Hooper AB. 2000. Electron transfer during the oxidation of ammonia by the chemolithotrophic bacterium Nitrosomonas europaea. Biochim. Biophys. Acta 1459:346–355. 10.1016/S0005-2728(00)00171-7 [DOI] [PubMed] [Google Scholar]
- 13.Goreau TJ, Kaplan WA, Wofsy SC, McElroy MB, Valois FW, Watson SW. 1980. Production of NO2- and N2O by nitrifying bacteria at reduced concentrations of oxygen. Appl. Environ. Microbiol. 40:526–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stein LY. 2011. Surveying N2O-producing pathways in bacteria. Methods Enzymol. 486:131–152. 10.1016/B978-0-12-381294-0.00006-7 [DOI] [PubMed] [Google Scholar]
- 15.Cantera JJL, Stein LY. 2007. Molecular diversity of nitrite reductase genes (nirK) in nitrifying bacteria. Environ. Microbiol. 9:765–776. 10.1111/j.1462-2920.2006.01198.x [DOI] [PubMed] [Google Scholar]
- 16.Bollmann A, Sedlacek C, Norton J, Laanbroeck HJ, Suwa Y, Stein LY, Klotz MG, Arp D, Sayavedra-Soto L, Lu M, Bruce D, Detter C, Tapia R, Han J, Woyke T, Lucas S, Pitluck S, Pennacchio L, Nolan M, Land M, Huntemann M, Deshpande S, Han C, Chen A, Kyrpides N, Mavromatis K, Markowitz V, Szeto E, Ivanova N, Mikhailova N, Pagani I, Pati A, Peters L, Ovchinnikova G, Goodwin L. 2013. Complete genome sequence of Nitrosomonas sp. Is79—an ammonia oxidizing bacterium adapted to low ammonium concentrations. Stand. Genomic Sci. 7:469–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beaumont HJE, Hommes NG, Sayavedra-Soto LA, Arp DJ, Arciero DM, Hooper AB, Westerhoff HV, van Spanning RJM. 2002. Nitrite reductase of Nitrosomonas europaea is not essential for production of gaseous nitrogen oxides and confers tolerance to nitrite. J. Bacteriol. 184:2557–2560. 10.1128/JB.184.9.2557-2560.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beaumont HJE, van Schooten B, Lens SI, Westerhoff HV, Van Spanning RJM. 2004. Nitrosomonas europaea expresses a nitric oxide reductase during nitrification. J. Bacteriol. 186:4417–4421. 10.1128/JB.186.13.4417-4421.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sayavedra-Soto LA, Stein LY. 2011. Genetic transformation of ammonia-oxidizing bacteria. Methods Enzymol. 486:389–402. 10.1016/B978-0-12-381294-0.00017-1 [DOI] [PubMed] [Google Scholar]
- 20.Hyman MR, Arp DJ. 1992. 14C2H2- and 14CO2-labeling studies of the de novo synthesis of polypeptides by Nitrosomonas europaea during recovery from acetylene and light inactivation of ammonia monooxygenase. J. Biol. Chem. 267:1543–1545 [PubMed] [Google Scholar]
- 21.Bollmann A, French E, Laanbroek HJ. 2011. Isolation, cultivation, and characterization of ammonia-oxidizing bacteria and archaea adapted to low ammonium concentrations. Methods Enzymol. 486:55–88. 10.1016/B978-0-12-381294-0.00003-1 [DOI] [PubMed] [Google Scholar]
- 22.Frear DS, Burrell RC. 1955. Spectrophotometric method for determining hydroxylamine reductase activity in higher plants. Anal. Chem. 27:1664–1665. 10.1021/ac60106a054 [DOI] [Google Scholar]
- 23.Zumft WG, Vega JM. 1979. Reduction of nitrite to nitrous oxide by a cytoplasmic membrane fraction from the marine denitrifier Pseudomonas perfectomarinus. Biochim. Biophys. Acta 548:484–499 [DOI] [PubMed] [Google Scholar]
- 24.Stein LY, Arp DJ, Hyman MR. 1997. Regulation of the synthesis and activity of ammonia monooxygenase in Nitrosomonas europaea by altering pH to affect NH3 availability. Appl. Environ. Microbiol. 63:4588–4592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poret-Peterson AT, Graham JE, Gulledge J, Klotz MG. 2008. Transcription of nitrification genes by the methane-oxidizing bacterium, Methylococcus capsulatus strain Bath. ISME J. 2:1213–1220. 10.1038/ismej.2008.71 [DOI] [PubMed] [Google Scholar]
- 26.Klotz MG, Stein LY. 2008. Nitrifier genomics and evolution of the nitrogen cycle. FEMS Microbiol. Lett. 278:146–156. 10.1111/j.1574-6968.2007.00970.x [DOI] [PubMed] [Google Scholar]
- 27.Campbell MA, Nyerges G, Kozlowski JA, Poret-Peterson AT, Stein LY, Klotz MG. 2011. Model of the molecular basis for hydroxylamine oxidation and nitrous oxide production in methanotrophic bacteria. FEMS Microbiol. Lett. 322:82–89. 10.1111/j.1574-6968.2011.02340.x [DOI] [PubMed] [Google Scholar]
- 28.Ward N, Larsen Ø, Sakwa J, Bruseth L, Khouri H, Durkin AS, Dimitrov G, Jiang L, Scanlan D, Kang KH, Lewis M, Nelson KE, Methé B, Wu M, Heidelberg JF, Paulsen IT, Fouts D, Ravel J, Tettelin H, Ren Q, Read T, DeBoy RT, Seshadri R, Salzberg SL, Jensen HB, Birkeland NK, Nelson WC, Dodson RJ, Grindhaug SH, Holt I, Eidhammer I, Jonasen I, Vanaken S, Utterback T, Feldblyum TV, Fraser CM, Lillehaug JR, Eisen JA. 2004. Genomic insights into methanotrophy: The complete genome sequence of Methylococcus capsulatus (Bath). PLoS Biol. 2(10):e303. 10.1371/journal.pbio.0020303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartossek R, Nicol GW, Lanzen A, Klenk H-P, Schleper C. 2010. Homologues of nitrite reductases in ammonia-oxidizing archaea: Diversity and genomic context. Environ. Microbiol. 12:1075–1088. 10.1111/j.1462-2920.2010.02153.x [DOI] [PubMed] [Google Scholar]
- 30.Hatzenpichler R. 2012. Diversity, physiology, and niche differentiation of ammonia-oxidizing archaea. Appl. Environ. Microbiol. 78:7501–7510. 10.1128/AEM.01960-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kartal B, Wessels HJCT, van der Biezen E, Francoijs K-J, Jetten MSM, Klotz MG, Stein LY. 2012. Effects of nitrogen dioxide and anoxia on global gene and protein expression in long-term continuous cultures of Nitrosomonas eutropha C91. Appl. Environ. Microbiol. 78:4788–4794. 10.1128/AEM.00668-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hyman MR, Wood PM. 1983. Methane oxidation by Nitrosomonas europaea. Biochem. J. 212:31–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hooper AB, Maxwell PC, Terry KR. 1978. Hydroxylamine oxidoreductase from Nitrosomonas: absorption spectra and content of heme and metal. Biochemistry 17:2984–2989. 10.1021/bi00608a007 [DOI] [PubMed] [Google Scholar]
- 34.Pacheco AA, McGarry J, Kostera J, Corona A. 2011. Techniques for investigating hydroxylamine disproportionation by hydroxylamine oxidoreductases. Methods Enzymol. 486:447–463. 10.1016/B978-0-12-381294-0.00020-1 [DOI] [PubMed] [Google Scholar]


