Abstract
Human intoxication or infection due to bacterial food contamination constitutes an economic challenge and a public health problem. Information on the in situ distribution and expression of pathogens responsible for this risk is to date lacking, largely because of technical bottlenecks in detecting signals from minority bacterial populations within a complex microbial and physicochemical ecosystem. We simulated the contamination of a real high-risk cheese with a natural food isolate of Staphylococcus aureus, an enterotoxin-producing pathogen responsible for food poisoning. To overcome the problem of a detection limit in a solid matrix, we chose to work with a fluorescent reporter (superfolder green fluorescent protein) that would allow spatiotemporal monitoring of S. aureus populations and targeted gene expression. The combination of complementary techniques revealed that S. aureus localizes preferentially on the cheese surface during ripening. Immunochemistry and confocal laser scanning microscopy enabled us to visualize, in a single image, dairy bacteria and pathogen populations, virulence gene expression, and the toxin produced. This procedure is readily applicable to other genes of interest, other bacteria, and different types of food matrices.
INTRODUCTION
Humans are susceptible to numerous food-borne diseases that are transmitted via water and food consumption. There are an estimated 48 million cases of food-borne illnesses in the United States every year, resulting in 128,000 hospitalizations, 3,000 deaths (1), and large economic and productivity losses (2). In Europe, the European Food Safety Authority reported a total of 5,363 outbreaks of food-borne illness in 2012, affecting almost 55,000 people and causing 41 deaths (3). Food-borne infection or intoxication is attributed to the pathogen itself or to toxins released in the food product.
Staphylococcus aureus is a worldwide cause of food-borne infections (4, 5). This bacterium is a leading cause of gastroenteritis resulting from the consumption of foods in which enterotoxigenic staphylococci have grown and produced toxins (6, 7). Thus, even if the bacteria are killed, e.g., by heat treatment, the heat-resistant enterotoxins can persist, leading to staphylococcal food poisoning (SFP) (6, 8). A notable example is staphylococcal enterotoxin D (SED), which is clearly involved in both cheese official controls performed in France (9) according to European Union regulation 1441/2007 (10) and in SFP outbreaks (11–17). The sed gene is carried by a plasmid (6, 18, 19) and controlled by two regulators, the accessory gene regulator (agr) quorum-sensing system, a main regulatory system controlling virulence gene expression in S. aureus (6, 12, 20) and the staphylococcal accessory regulator sarA (21).
In Europe, 777 outbreaks in 2012 were caused by bacterial toxins produced by Bacillus spp., Clostridium spp., and coagulase-positive staphylococci; the latter are the second most common causative agents of food-borne outbreaks (3). Among them, 346 were due to staphylococcal enterotoxins, of which 20% correlated with cheese as the food vehicle. Among all cheese families, soft and uncooked semihard cheeses are most often involved in SFP outbreaks (22, 23).
Cheese is a complex environment, constituted principally by water, proteins, fat, minerals, and a dynamic microbial ecosystem characterized by the presence of a large variety of bacteria, yeasts, and molds. The monitoring of minority bacterial populations of food pathogens and specific genes that they express within such a complex microbial physicochemical ecosystem is a major challenge in food microbiology. Indeed, the detection of microbial pathogens in food is complicated by low bacterial counts, which may not be recovered by using traditional culturing and sampling techniques. Traditional culture-based approaches may be affected by large populations of lactic acid bacteria in the cheese matrix (24). Implementation of DNA microarray technology has proven effective for the profiling of microbial communities (25, 26) and for S. aureus gene expression analysis in pure and mixed cultures with Lactococcus lactis in a simplified model cheese matrix (27). More recently, next-generation sequencing technologies (24, 28) and real-time reverse transcription-quantitative PCR (RT-qPCR) (29) have been used to measure enterotoxin and virulence gene expression and regulation in simulations of environmental conditions (27, 30, 31). While these methods can be sensitive and give both qualitative and quantitative information about the microorganisms tested, there is also a need to evaluate the distribution of microbial populations in situ. Fluorescence in situ hybridization with 16S rRNA provides microbial identification and physical detection of uncultivable microorganisms in fragile matrices like cheese (32), and its use is being expanded to pathogens in different environments (33–35). The spatial distribution of bacterial flora in cheese has also been explored by using scanning electron microscopy, fluorescence and light microscopy, and laser scanning microscopy (36–40). An in situ approach used to investigate the spatial distribution of bacterial colonies of fluorescent Lactococcus lactis in a solid-food matrix with a model system has been recently developed (41), demonstrating that live cells can be visualized in cheese. However, in situ approaches to visualize minority gene expression in food matrices are still lacking.
Green fluorescent protein (GFP) variants have been used to quantify the expression of numerous S. aureus genes under various conditions, including different stages of biofilm maturation and dispersal (42–48). To date, all of these studies have been conducted either with a single bacterial population at high cell concentrations or under laboratory conditions.
The aim of the present study was to monitor the distribution of S. aureus and the spatial and temporal expression of virulence genes during the manufacture and ripening of semihard cheeses. To best approach a real-life situation, a fluorescent reporter was applied for use in S. aureus cheese isolates. The procedures developed for cheese sample preparation, immunochemistry, and confocal laser scanning microscopy (CLSM) enabled the visualization of dairy bacteria and minor pathogen populations, pathogenic gene expression, and the toxin produced, all in a single image.
MATERIALS AND METHODS
Bacterial strains.
S. aureus strain CIM433 (sed, sej, and ser enterotoxin genes), from the ARILAIT collection (La Roche-sur-Foron, France), was used throughout this study. CIM433 is a cheese isolate that produces SED, which can be quantified by the confirmatory method of the European Union Reference Laboratory for coagulase-positive staphylococci (EU RL for CPS) (49).
Industrial starter and ripening cultures (Ets COQUARD, Villefranche sur Saône, France) lyophilized Beta 1 (homofermentative Lactococcus lactis/Lactococcus cremoris strains), Lambda 5/2 (Lactobacillus bulgaricus/Streptococcus thermophilus 25/75), Sigma 63 (Brevibacterium linens), and liquid Sigma 52 (Geotrichum candidum) were used for cheese making. Cultures were stored at −20°C prior to use.
Plasmid construction.
Plasmid construction was performed with Escherichia coli strain TG1. Plasmid pCM11 (47) contains a sarA promoter that drives the expression of a synthetic version of the gene encoding superfolder GFP (sGFP), which gives a strong fluorescent signal in S. aureus (44, 46, 47). pCM11 is derived from pE194, which has an estimated copy number of 55 per cell (50). A promoterless version of sgfp, called pIF1, was constructed as a negative control by digesting pCM11 with HindIII and KpnI, followed by filling in and ligation steps (New England BioLabs, Ipswich, MA). To monitor sed expression, the sed promoter was fused to sgfp by the Gibson assembly method, giving rise to plasmid pIF2 (51). The oligonucleotides used for sed amplification were 5′ GTAAAACGACGGCCAGTGCCAAGCTTGGTACCCCGGCGTAGAGGATCAAATATATTG 3′ and 5′ CATCCTCCTAAGGTACCCGGGGATCCGCCTTTTTTTCAATAAATTTGAGCACC 3′, such that the transcription start site and −35 and −10 promoter elements were amplified. The pCM11 vector, including the sgfp ribosome binding site, was amplified with oligonucleotides 5′ GGTGCTCAAATTTATTGAAAAAAAGGCGGATCCCCGGGTACCTTAGGAGGATG 3′ and 5′ CAATATATTTGATCCTCTACGCCGGGGTACCAAGCTTGGCACTGGCCGTCGTTTTAC 3′. All constructs were verified by DNA sequencing (GATC Biotech, Constance, Germany). The pIF1 and pIF2 plasmids were introduced into S. aureus RN4420 (52), extracted, and used to electrotransform S. aureus CIM433 as described previously (53).
Growth conditions in reconstituted milk.
S. aureus CIM433, CIM433/pIF1 (sgfp without promoter), CIM433/pCM11 (sarA promoter-driven sgfp), and CIM433/pIF2 (sed promoter-driven sgfp) were cultured in brain heart infusion (BHI; Oxoid, Dardilly, France) broth; plasmid-carrying strains were grown in medium with erythromycin (Ery) at 10 μg/ml. Precultures were grown at 37°C with shaking (200 rpm) for 8 h. A preculture containing 106 CFU/ml was used to inoculate 50 ml of sterile reconstituted milk (100 g of semiskim milk powder [Régilait, Saint-Martin-Belle-Roche, France] per liter of distilled water sterilized at 108°C for 10 min). Bacterial growth in shaking cultures was followed for 24 h. Serial dilutions of milk cultures were prepared in sterile 1% (wt/vol) peptone water and plated on BHI solid medium, supplemented or not for antibiotic selection, for differential S. aureus count determination. Plates prepared in duplicate were incubated for 48 h at 37°C before bacterial enumeration.
Inoculum preparation for cheese manufacture.
Strains were cultured in 10 ml of BHI broth with antibiotic (except for CIM433) at 37°C with shaking for 8 h. A 100-μl volume of this preculture was then added to 100 ml of BHI broth without antibiotic, and the culture was incubated for 4 h at 37°C without shaking so that S. aureus was in the exponential growth phase at the time of milk inoculation for cheese manufacture. The lactic acid bacterial starter culture was prepared according to the manufacturer's instructions and suspended, just before use, in pasteurized milk from the cheese vat to ensure homogenization (see below).
Cheese manufacture.
Raw bulk milk (pH 6.5) cooled at 4°C and collected from a local farm (Viltain, Jouy-en-Josas, France) was pasteurized (30 s at 72°C). Cheeses were prepared in four automated 20-liter tanks in a P2-level experimental cheese plant at INRA (Jouy-en-Josas, France). Following the addition of CaCl2 (12.5 g/100 liters of milk), 15 liters of pasteurized milk preheated at the maturation temperature was inoculated with S. aureus (to a final concentration of 104 CFU/ml) and with starter culture (to a final level of 106 CFU/ml). The equivalent of 33 ml of filtered rennet extract (520 mg of chymosin/liter; Berthelot) for 100 liters of milk was added after 1 h of milk maturation at 34°C. Coagulation then proceeded for about 40 min before the curd was cut into small cubes to corn grain size, and after 20 min of slow stirring, 10 min with no stirring, and then 10 min of rapid stirring at 34°C, 33% (5 liters) of the whey was drained. The curd was then poured into molds and pressed for 3 h with 1.5-kg weights. After being taken out of the molds (molding lasted 4 h), cheeses were salted for 1 h in sterile brine (24% NaCl, pH 5.10, 13°C), turned over daily, and dried for 4 days at 16°C. Cheeses were washed and smeared at 12°C with brine solution including B. linens and G. candidum at days 6 and 12 during ripening. Cheeses were approximately 13 to 15 cm in diameter and 2.5 cm thick. The mean levels of moisture on a fat-free basis and fat in dry matter of 1-day-old cheeses were, respectively, 64.7 and 46.2%; the curd pH was around 5.2. The mean moisture on a fat-free basis of 15-day-old cheeses was 52.3%, and the mean NaCl content was 2.2%. The core pH reached values of around 4.8, and the surface pH was around 6.9.
Bacterial enumeration in cheeses.
The absence of S. aureus was checked in all pasteurized milk samples before inoculation. S. aureus and starter bacteria in cheeses were estimated over the 24 h of the cheese-making procedure and after 15 days of ripening (after core-surface separation) by plating as described previously (31).
FCM.
Cell pellets were recovered from 1-ml milk cultures by centrifugation (6,000 × g for 2 min), immediately frozen in liquid nitrogen, and stored at −80°C. For flow cytometry (FCM), cell pellets from cheese were prepared as for RNA extraction (see below). All cell pellets were suspended in 1 ml of sterile 1% (wt/vol) peptone water, and the cell density was adjusted to 106 CFU/ml after filtering with a 50-μm CellTrics filter (Partec, Ste. Geneviève des Bois, France). Fluorescence levels of 20,000 cells were determined with a CyFlow Space cytometer (Partec) equipped with a blue laser (488-nm emission). Fluorescence signals (from the sGFP reporter) were collected with a 527-nm bandpass filter (512 to 542 nm) (FL1 channel). FCM analyses were performed by using logarithmic gains and specific detector settings as adjusted on CIM433, the nonfluorescent parental strain, to correct for autofluorescence. Data were collected and analyzed with FlowMax software (Partec).
Gene expression analysis.
sed gene expression was analyzed by RT-qPCR analysis of total RNA extracted from cheese (same homogenate as for bacterial cell counts) as described previously (31). Three reference genes shown to be stably expressed during cheese manufacturing, pta, gyrB, and rpoB, were used in this study (31). Primers 5′ GATCTCCTGTACTTTTATTTTCTCC 3′ and 5′ AAACGTTAAAGCCAATGAAAAC 3′, designed for sed real-time PCR, were purchased from Eurogentec SA (Seraing, Belgium).
Total RNA extracted from cheese (375 ng) was annealed with Random Nonamers (Eurogentec) for 10 min at 20°C after a denaturation step (5 min at 65°C) to remove RNA secondary structures. cDNA was synthesized by 1 h of RT at 42°C with PrimeScript reverse transcriptase and Ultrapure deoxynucleoside triphosphate (Clontech-TaKaRa Bio Europe, Saint Germain en Laye, France) in a 30-μl final volume, followed by enzyme inactivation (15 min at 70°C). cDNA levels were analyzed by qPCR with the ABI Prism 7900 HT Sequence detection system (SDS; Applied Biosystems, Foster City, CA). Each sample was tested in duplicate in a 96-well plate (Applied Biosystems). The reaction mixture (20 μl, final volume) consisted of 10 μl of SYBR green PCR master mix (Applied Biosystems), 1.2 μl of each primer (300 nM, final concentration), 2.6 μl ultrapure H2O, and 5 μl of a 2.5-fold dilution of the relevant cDNA as the template. Absence of genomic DNA in RNA samples was checked by real-time PCR before cDNA synthesis (minus RT control). A blank (no-template control) was incorporated into each assay. The thermocycling program was as follows: 50°C for 2 min, 95°C for 10 min, and 40 cycles of 15 s at 95°C and 1 min at 60°C. Melting-curve data were then collected to check PCR specificity, contamination, and the absence of primer dimers. To minimize interrun variations, a calibrator sample (RNA extracted from a pure culture of strain CIM433) was used to determine and fix the fluorescence threshold. The CT values calculated by the SDS software were exported to Excel for relative quantification analysis as described previously (31).
Enterotoxin determination.
Cheese samples (25 g) were tested for the presence of SED after molding for 4 h and after 1 and 15 (core and surface separately analyzed) days of ripening, according to the European Screening Method of the EU RL for CPS, consisting of extraction followed by a dialysis concentration step coupled to detection with the Vidas SET2 kit, a qualitative detection test (bioMérieux, Marcy l'Etoile, France). Staphylococcal enterotoxin-positive samples were further analyzed by quantitative double-sandwich enzyme-linked immunosorbent assay, the confirmatory method of the EU RL for CPS (49), to quantify the amount of enterotoxin produced.
Microstructural analysis.
Cheese pieces of approximately 1 by 1 by 0.5 cm were cut from fresh samples with a scalpel and fixed in 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) in 0.1 M phosphate buffer (pH 7.4) for 2 h at room temperature. Samples were rinsed and immersed in 30% sucrose in 0.1 M phosphate buffer (pH 7.4) for 20 h at 4°C as described previously (54). After direct freezing with liquid nitrogen vapor, 15-μm-thick embedded cheese sections were cut at −20°C with a Cryostat (Leica CM 1950) and stored at −80°C for immunohistochemistry. Each section was placed on a SuperFrost ULTRA PLUS slide (Thermo Scientific Menzel, Illkirch, France), blocked in 0.5% Triton X-100 (Sigma-Aldrich, St. Louis, MO) and 2% bovine serum albumin (Sigma-Aldrich) for 1 h at 4°C, rinsed, and incubated overnight at 4°C with S. aureus-specific rabbit antibody (1:200) (Biodesign International, Saco, ME) and sheep antibody specific to SED (1:120; Toxin Tech, Sarasota, FL). After rinsing, slides were incubated with the following secondary polyclonal antibodies: diluted (1:200) goat anti-rabbit IgG antibody coupled with cy5 (Rockland Immunochemicals Inc., Gilbertsville, PA) and donkey anti-sheep IgG antibody (1:200) coupled with tetramethyl rhodamine isocyanate (TRITC; Jackson ImmunoResearch, West Grove, PA). Nucleic acids were stained with a 4′,6-diamidino-2-phenylindole (DAPI; Molecular Probes, Eugene, OR) solution at 2 μg/ml for 30 min at 4°C. Microstructural analysis was performed with Zen 2011 software and a Zeiss confocal laser scanning microscope 700 (MIMA2 Platform; INRA, Jouy-en-Josas) with an immersion 63× objective (numerical aperture 1.40, oil M27) at zoom 2.0. GFP fluorescence was excited with a 488-nm laser diode (at 5% intensity), cy5 was excited with a 639-nm laser diode (at 2% intensity), TRITC was excited with a 555-nm laser diode (at 5% intensity), and DAPI was excited with a 405-nm laser diode (at 5% intensity). Fluorescence was detected with a 490- to 555-nm band-pass filter for GFP, a 560- to 630-nm band-pass filter for TRITC, a 640-nm-and-higher filter for Cy5, and a 410-nm-and-higher filter for DAPI. Sequential tracking was performed, i.e., each fluorescent marker was detected sequentially, to minimalize potential spectral overlap.
RESULTS AND DISCUSSION
Fitness and stability of genetically modified food isolates in milk.
The main objective of this work was to monitor targeted gene expression with the use of the fluorescent reporter sGFP. We chose the S. aureus sarA-sgfp fluorescent reporter fusion in plasmid pCM11 to initiate this project, as the sarA promoter region is a well-studied transcriptional element and is frequently used as a constitutive promoter in biofilm and host labeling experiments (42, 44, 45, 47, 55). Using this plasmid, we constructed pIF1, which lacks the sarA promoter, to evaluate basal fluorescence and the background and pIF2, in which the sed promoter is fused to sgfp, to monitor enterotoxin expression under simulated food contamination conditions. The three plasmids were successfully introduced into S. aureus food isolate CIM433, demonstrating that this strain is transformable.
We tested the transformed isolates in milk without antibiotic to mimic the conditions encountered in cheese, in which dairy bacteria are antibiotic sensitive and antibiotic addition is forbidden. The growth kinetics of the parental CIM433 strain and transformed strains in aerated reconstituted milk without antibiotic over a 24 h-period show that the fitness of the S. aureus food isolate is not affected by plasmid transformation (Table 1). However, after 24 h in milk, about 45, 14, and 7% of the cultivable CIM433 bacteria carrying pCM11, pIF1, and pIF2, respectively, lost their plasmids (Table 2). This loss of stability will be considered in further analyses. While plasmid loss would clearly lower signal detection, the fact that strain fitness was essentially unaffected in milk led us to use these strains and evaluate their behavior in the cheese matrix.
TABLE 1.
Cell counts of S. aureus CIM433 (parental strain without sgfp plasmid), CIM433/pIF1 (sgfp without promoter), CIM433/pCM11 (sarA promoter-driven sgfp), and CIM433/pIF2 (sed promoter-driven sgfp) in shaking reconstituted milk without antibiotic for 24 ha
| Time (h) | Cell count on BHI (CFU/ml) |
|||
|---|---|---|---|---|
| CIM433 | CIM433/pIF1 | CIM433/pCM11 | CIM433/pIF2 | |
| 0 | 1.2 × 106 | 1.2 × 106 | 1.1 × 106 | 1.3 × 106 |
| 2 | 3.0 × 107 | 3.5 × 107 | 2.0 × 107 | 3.2 × 107 |
| 4 | 3.5 × 108 | 2.3 × 108 | 1.4 × 108 | 1.9 × 108 |
| 6 | 2.3 × 108 | 3.1 × 108 | 1.7 × 108 | 2.5 × 108 |
| 24 | 3.2 × 108 | 4.1 × 108 | 2.2 × 108 | 3.4 × 108 |
Bacterial cultures were plated on BHI solid medium. Data are expressed as the means of two biological replicates.
TABLE 2.
Stability of plasmids pIF1 (sgfp without promoter), pCM11 (sarA promoter-driven sgfp), and pIF2 (sed promoter-driven sgfp) carried by S. aureus CIM433 in milk for 24 ha
| Time (h) | Cell count on BHI + Ery as % of cell count on BHI alone |
||
|---|---|---|---|
| pIF1 | pCM11 | pIF2 | |
| 0 | 103 | 105 | 94 |
| 2 | 101 | 117 | 96 |
| 4 | 105 | 105 | 103 |
| 6 | 101 | 116 | 118 |
| 24 | 86 | 55 | 93 |
Bacterial cultures were plated in duplicate on BHI and on BHI supplemented with 10 μg/ml erythromycin. Cell counts on BHI with antibiotic selection are expressed as percentages of cell counts on BHI alone.
Detection of sGFP fluorescence in milk.
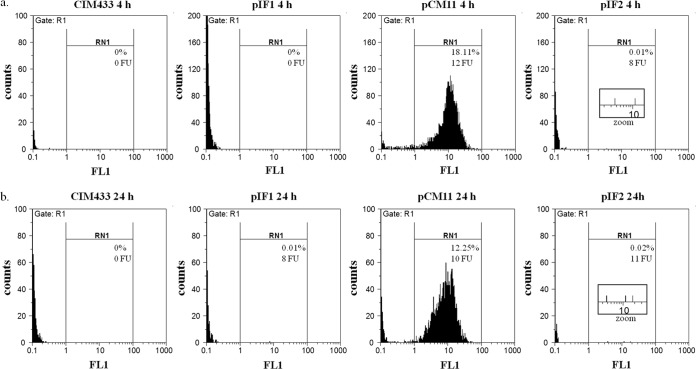
The fluorescence levels of each strain in milk were measured by FCM after 4 or 24 h. CIM433 and CIM433/pIF1 served as a negative gating control to gauge the green fluorescence intensity shifts (Fig. 1). In comparison with control strains (Fig. 1a and b), the presence of pCM11 in CIM433 conferred robust fluorescence in the late logarithmic growth (4 h) and stationary phases (24 h), as already observed for the sarA promoter-sgfp fusion under laboratory conditions (44). These observations demonstrate that pCM11 can be used to monitor S. aureus in milk even if the fluorescence levels are underestimated because of plasmid loss. A very weak fluorescent signal was detected for CIM433 carrying the sed reporter plasmid (pIF2) (8 fluorescence units [FU] at 4 h and 11 FU at 24 h) relative to the GFP background level of pIF1 (0 FU at 4 h and 8 FU at 24 h) (Fig. 1a and b, insets), indicating that sed promoter activity is very low under the conditions tested.
FIG 1.
Cell flow sorting of S. aureus labeled with a fluorescent reporter under growth conditions in milk. CIM433 served as a negative gating control. CIM433 carrying plasmid pIF1, pCM11, or pIF2 was used for the FCM test. Each strain was grown for 24 h in reconstituted milk, the cell density was adjusted to 106 CFU/ml after filtering, and populations of 20,000 cells were separated by FCM after 4 (a) and 24 (b) h of growth (insets show zoom on cytographs). The percentage of fluorescent cells collected with a 527-nm bandpass filter (FL1 channel) and the fluorescence intensity of each signal are indicated.
Fitness and stability of genetically modified food isolates in cheese.
S. aureus fitness and plasmid stability were evaluated all along the cheese-making process. Bacterial cell counts in cheese showed <5.5-fold differences between parental and plasmid-carrying strains after 15 days, indicating that plasmid maintenance has a minor impact on bacterial fitness (Table 3).
TABLE 3.
Cell counts of S. aureus CIM433, CIM433/pIF1, CIM433/pCM11, and CIM433/pIF2 in cheese for 15 daysa
| Stage and location | Cell count (CFU/g of cheese) on BHI |
|||
|---|---|---|---|---|
| CIM 433 | CIM433/pIF1 | CIM433/pCM11 | CIM433/pIF2 | |
| Molding for 4 h | 3.0 × 107 | 2.3 × 107 | 1.9 × 107 | 2.4 × 107 |
| Day 1 | 1.1 × 108 | 4.5 × 107 | 4.3 × 107 | 5.2 × 107 |
| Day 15 | ||||
| Core | 2.8 × 107 | 2.5 × 107 | 1.9 × 107 | 8.3 × 106 |
| Surface | 5.9 × 108 | 1.1 × 108 | 1.2 × 108 | 1.6 × 108 |
After 15 days of ripening, the core had separated from the surface. Bacterial cultures were plated on BHI solid medium. Data are expressed as the means of two biological replicates.
Independently of the presence of a plasmid, we observed that S. aureus counts were about 10-fold higher on the cheese surface than in the core, which can be attributed to more favorable aerobic conditions and higher pH values on the cheese surface than in the core (56, 57). This difference was also observed in soft cheeses (58) and in other semihard cheese studies in our laboratory (data not shown).
In 1-day-old cheese, plasmids pCM11 and pIF2 were lost by about half of the S. aureus CIM433 populations (Table 4). However, the levels of all three plasmids were stable for up to 15 days (46 to 67% according to plasmid and core versus surface sampling; Table 4). This stability likely reflects the absence of significant bacterial growth (i.e., cell divisions) in the core and little growth on the cheese surface, during which plasmid loss would occur. We concluded that the presence of reporter plasmids in at least half of the bacterial population is sufficient to address the question of S. aureus localization and sed expression in the cheese matrix.
TABLE 4.
Stability of plasmids pIF1, pCM11, and pIF2 carried by S. aureus CIM433 during cheese manufacturinga
| Stage and location | Cell count on BHI + Ery as % of cell count on BHI alone |
||
|---|---|---|---|
| CIM433/pIF1 | CIM433/pCM11 | CIM433/pIF2 | |
| Molding for 4 h | 114 | 94 | 88 |
| Day 1 | 95 | 41 | 55 |
| Day 15 | |||
| Core | 61 | 63 | 46 |
| Surface | 67 | 61 | 60 |
Bacterial cultures were plated in duplicate on BHI and on BHI supplemented with 10 μg/ml erythromycin. Cell counts on BHI with antibiotic selection are expressed as percentages of cell counts on BHI alone.
Detection of sGFP fluorescence in cheese.
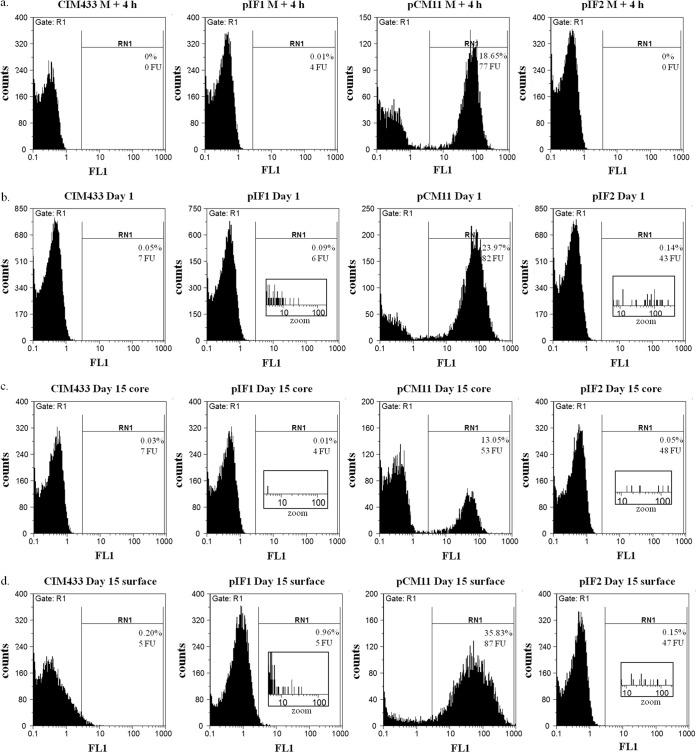
The fluorescence levels of the CIM433 strain and plasmid-carrying derivatives in cheeses were monitored by FCM after molding for 4 h and after 1 and 15 days. CIM433 and CIM433/pIF1 served as negative gating controls (Fig. 2). Compared to CIM433 or CIM433/pIF1, CIM433/pCM11 displayed robust fluorescence after molding for 4 h (77 FU) (Fig. 2a), at day 1 (82 FU) (Fig. 2b), and during ripening (Fig. 2c and d), with a lower fluorescence level in the core (53 FU) (Fig. 2c) than on the cheese surface (87 FU) (Fig. 2d). We observed a very slight but significant fluorescence level (43 FU) of the CIM433 strain carrying the sed-sgfp reporter fusion (pIF2) relative to the GFP background of pIF1 (6 FU) in 1-day-old cheese (Fig. 2b, insets). The same low but significant level of sed expression as measured by GFP was maintained in 15-day-old cheese in both core and surface samples (48 FU versus 4 FU in the core and 47 FU versus 5 FU on the surface) (Fig. 2c and d, insets). We note that the fluorescence intensities were likely underestimated by at least 2-fold because of plasmid loss (Table 4). Weak fluorescence from pIF2 may reflect low sed promoter activity under our conditions. The expected lower plasmid copy number in the absence of selection may further account for the weak signal. The above factors may also explain the differences between the FCM (Fig. 2) and culture counts (Table 3) of CIM433/pIF2. Our results reveal that S. aureus cell counts were about 10-fold higher on the cheese surface than in the core (Table 3). As FCM is corrected for a given cell number, we project that the cheese surface would contain about 10-fold more SED, accordingly.
FIG 2.
Cell flow sorting of S. aureus labeled with a fluorescent reporter in cheese. CIM433 served as a negative gating control. CIM433 carrying plasmid pIF1, pCM11, or pIF2 was used for the FCM test. The cell density was adjusted to 106 CFU/ml after filtering, and populations of 20,000 cells were separated by FCM after molding for 4 h (a) and after 1 (b) and 15 (after core [c]-surface [d] separation) days (insets show zoom on cytographs). The percentage of fluorescent cells collected with a 527-nm bandpass filter (FL1 channel) and the fluorescence intensity of each signal are indicated.
Gene expression correlated with enterotoxin production in cheese.
To validate the measurements above of sed expression with pIF2, we also quantified endogenous S. aureus sed transcripts and protein production in cheese. First, gene expression in the food matrix was quantified by RT-qPCR as described previously (27, 30, 31). Transcript levels were comparable in the four CIM433 strains, indicating that the introduced plasmids do not significantly impact sed expression (Fig. 3). A slight variation in sed pCM11 expression at day 1 (∼1.6-fold) compared to that of the other strains was not considered significant. A more marked difference in sed expression between the core and surface observed was observed for all four strains in 15-day-old cheeses, with ∼5-fold greater expression on the surface (Fig. 3). Importantly, the results obtained by direct measurement of sed expression (Fig. 3) are consistent with those obtained with the gfp reporter fusions (Fig. 2), i.e., weak expression that increased from day 1 (Fig. 2b) and a fluorescent subpopulation that was slightly more numerous on the surface (0.15%) than in the core (0.05%) (Fig. 2c and d). These results allowed us to correlate fluorescence intensities with gene expression.
FIG 3.
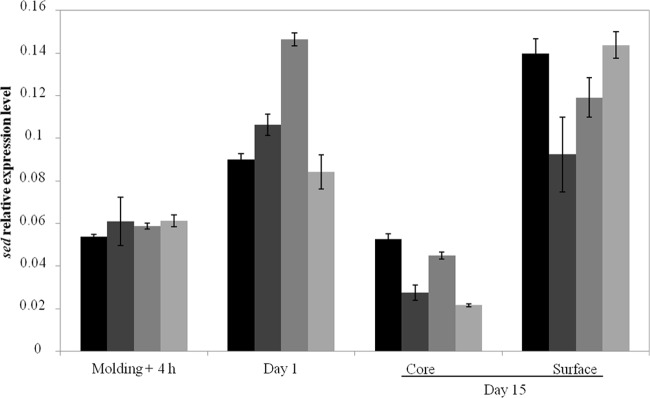
sed expression levels quantified by RT-qPCR during growth of S. aureus CIM433 (black), CIM433/pIF1 (dark gray), CIM433/pCM11(medium gray), and CIM433/pIF2 (light gray) in cheese. Vertical bars indicate standard deviations.
SED protein was measured after molding for 4 h and after 1 and 15 days (Fig. 4) (49, 59). At day 1, low toxin production did not correlate with sed gene expression. Importantly, the four strains tested behaved similarly, suggesting that SED is produced late in the cheese production process. Amounts of SED expectedly represent its accumulation since the start of the cheese-making process. The presence of ∼5-fold larger amounts of SED on the surface than in the cheese core may indicate that SED production occurs during ripening, when bacteria are compartmentalized. Greater sed expression and SED production correlate with higher counts of CIM433 CFU on the surface (Table 3). In keeping with these results, staphylococcal enterotoxin production is often positively correlated with the growth of S. aureus (17, 60), and we also observed a correlation between sed mRNA levels and SED production during semihard cheese manufacturing (31).
FIG 4.
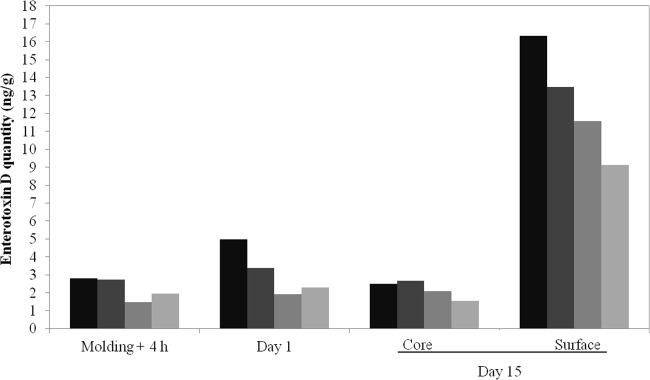
SED produced by S. aureus CIM433 (black), CIM433/pIF1 (dark gray), CIM433/pCM11(medium gray), and CIM433/pIF2 (light gray) in cheese was quantified by the confirmatory method.
Combined visualization of bacterial localization, gene expression, and toxin production.
In this part of the work, our goal was to visualize in situ S. aureus localization, gene expression, and toxin production. Nondestructive techniques using viability staining in conjunction with CLSM have been used for direct observation of bacteria in foods (61, 62) and detection of bacterial microcolonies in cheese (40, 41). Despite this progress, none of these reports aimed at discriminating a minority pathogen among a highly dense population, i.e., of dairy bacteria. Our additional goal was to detect SED production in this system, which required thin-slice sample preparation and multiple compatible immunohistochemistry markers.
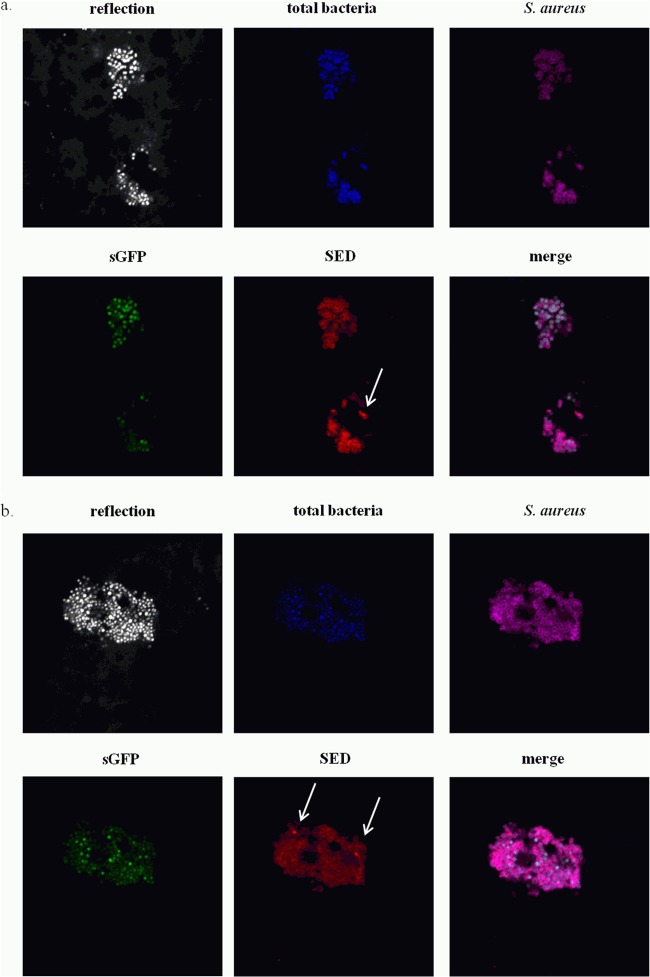
We first verified that there was no inherent autofluorescence in the matrix in the absence of bacteria by using cheese models obtained by chemical acidification. The model cheese was prepared with glucono δ-lactone (food additive E575), an organic acid authorized for use as an acidity regulator in the dairy industry (CODEX STAN 283-1978). No fluorescent background was observed in cheeses at any stage when they were contaminated with parental strain CIM433 or CIM433/pIF1 (data not shown). The cheese sample structure was visualized by the reflection of a 405-nm laser diode in a grayscale image (Fig. 5). DAPI staining was used to visualize the total bacteria (i.e., dairy and pathogen bacteria) (stained blue in Fig. 5). At day 15, S. aureus clusters were numerous on the surfaces of all of the cheeses contaminated with CIM433, CIM433/pIF1, CIM433/pIF2 (data not shown), and CIM433/pCM11 (Fig. 5b). In contrast, S. aureus clusters in the core (Fig. 5a) or in cheeses at day 1 and after molding for 4 h were rarer and more scattered (data not shown). These observations support the enumeration results (Table 3) and suggest that S. aureus imprisoned in the curd mass continued to multiply on the aerated surface, which is not the case in the core.
FIG 5.
Confocal laser scanning micrographs of S. aureus CIM433/pCM11 in the cheese core (a) and on the cheese surface (b) at day 15. The cheese sample structure is visualized by the reflection of the 405-nm laser diode in a grayscale image. Dairy bacteria and pathogens are blue, S. aureus strains are magenta, sgfp expression is green, and SED is red. Representative images of merged channels are also shown. SED-positive bacteria are indicated by arrows.
The strong sarA promoter in pCM11 allowed GFP visualization in real cheese during manufacturing (data not shown) and ripening (Fig. 5). However, the low fluorescence of pIF2 (carrying the sed-sgfp fusion) in cheeses at days 1 and 15 was insufficient for detection by CLSM (data not shown).
We also employed an immunohistochemistry-based method to detect SED protein. Fluorescence appeared in scattered spots (Fig. 5), probably reflecting the weak subpopulation expressing sed (Fig. 2) and producing toxin. This approach allowed us to obtain a qualitative image of bacterial distribution with a distinction among dairy bacteria, S. aureus, and the toxin produced (Fig. 5).
In conclusion, the present study demonstrated that a combined set of tools can be used to monitor the in situ behavior, including population distribution and gene expression, of a pathogen within a complex ecosystem and food matrix. The simulated contamination involved a real high-risk cheese produced with a complete starter culture and an enterotoxin-producing S. aureus food isolate. While the hazard of this pathogen is attributed to enterotoxin production, our results suggest that ∼10-fold higher bacterial density on the cheese surface is correlated with higher SED production, indicating that cell density is nevertheless an important factor in predicting intoxication.
To our knowledge, this is the first time that a single image reveals dairy bacteria and pathogen populations, pathogenic gene expression, and the toxin produced. This procedure could be easily transferred to other genes of interest and other bacteria within different types of matrices, such as contaminated meat or infected tissues.
ACKNOWLEDGMENTS
We thank the MIMA2 Imaging Facility (INRA, Jouy-en-Josas) for the fluorescence microscopy images and the ICE platform (INRA, Jouy-en-Josas) for use of the ABI Prism SDS 7900 HT. We are grateful to A. Horswill for providing plasmid pCM11, B. Cesselin for helpful discussions about the Gibson assembly method, G. Champeil-Potokar for the idea of glucose cheese embedding, and C. Boulesteix for use of the Cryostat.
The P2-level experimental cheese plant was constructed thanks to regional funding from the Ile-de-France.
Footnotes
Published ahead of print 13 June 2014
REFERENCES
- 1.Scallan E, Griffin PM, Angulo FJ, Tauxe RV, Hoekstra RM. 2011. Foodborne illness acquired in the United States—unspecified agents. Emerg. Infect. Dis. 17:16–22. 10.3201/eid1701.P21101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scharff RL. 2012. Economic burden from health losses due to foodborne illness in the United States. J. Food Prot. 75:123–131. 10.4315/0362-028X.JFP-11-058 [DOI] [PubMed] [Google Scholar]
- 3.European Food Safety Authority. 2014. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2012. EFSA J. 12:3547. 10.2903/j.efsa.2014.3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergdoll MS. 1989. Staphylococcus aureus, p 463–523 In Doyle MP. (ed), Foodborne bacterial pathogens. Marcel Dekker, New York, NY [Google Scholar]
- 5.Zahoor S, Bhatia A. 2007. Bacteria: silent killers in food. Sci. Rep. 2007:33–34 [Google Scholar]
- 6.Le Loir Y, Baron F, Gautier M. 2003. Staphylococcus aureus and food poisoning. Genet. Mol. Res. 2:63–76 http://www.funpecrp.com.br/gmr/year2003/vol1-2/sim0009_full_text.htm [PubMed] [Google Scholar]
- 7.Bhatia A, Zahoor S. 2007. Staphylococcus aureus enterotoxins: a review. J. Clin. Diagn. Res. 1:188–197 http://www.jcdr.net/article_fulltext.asp?issn=0973-709x&year=2007&month=June&volume=1&issue=3&page=188&id=85 [Google Scholar]
- 8.Schelin J, Wallin-Carlquist N, Cohn MT, Lindqvist R, Barker GC, Radstrom P. 2011. The formation of Staphylococcus aureus enterotoxin in food environments and advances in risk assessment. Virulence 2:580–592. 10.4161/viru.2.6.18122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ostyn A, De Buyser ML, Guillier F, Krys S, Hennekinne JA. 2011. Benefits of the combined use of immunological and PCR-based methods for determination of staphylococcal enterotoxins food safety criteria in cheeses. Food Anal. Methods 5:173–178. 10.1007/s12161-011-9244-y [DOI] [Google Scholar]
- 10.Anonymous. 2007. Commission regulation (EC) no. 1441/2007 of 5 December 2007 amending regulation (EC) no. 2073/2005 on microbiological criteria for foodstuffs. Off. J. Eur. Union L322:12–29 http://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2007:322:0012:0029:EN:PDF [Google Scholar]
- 11.Kérouanton A, Hennekinne JA, Letertre C, Petit L, Chesneau O, Brisabois A, De Buyser ML. 2007. Characterization of Staphylococcus aureus strains associated with food poisoning outbreaks in France. Int. J. Food Microbiol. 115:369–375. 10.1016/j.ijfoodmicro.2006.10.050 [DOI] [PubMed] [Google Scholar]
- 12.Hennekinne JA, De Buyser ML, Dragacci S. 2012. Staphylococcus aureus and its food poisoning toxins: characterization and outbreak investigation. FEMS Microbiol. Rev. 36:815–836. 10.1111/j.1574-6976.2011.00311.x [DOI] [PubMed] [Google Scholar]
- 13.Gücükoğlu A, Kevenk TO, Uyanik T, Cadirci O, Terzi G, Alisarli M. 2012. Detection of enterotoxigenic Staphylococcus aureus in raw milk and dairy products by multiplex PCR. J. Food Sci. 77:M620–M623. 10.1111/j.1750-3841.2012.02954.x [DOI] [PubMed] [Google Scholar]
- 14.Dambrosio A, Quaglia NC, Saracino M, Malcangi M, Montagna C, Quinto M, Lorusso V, Normanno G. 2013. Microbiological quality of Burrata cheese produced in Puglia region: southern Italy. J. Food Prot. 76:1981–1984. 10.4315/0362-028X.JFP-13-067 [DOI] [PubMed] [Google Scholar]
- 15.Pexara A, Solomakos N, Sergelidis D, Govaris A. 2012. Fate of enterotoxigenic Staphylococcus aureus and staphylococcal enterotoxins in Feta and Galotyri cheeses. J. Dairy Res. 79:405–413. 10.1017/S0022029912000325 [DOI] [PubMed] [Google Scholar]
- 16.Lamprell H. 2003. La production des entérotoxines dans les fromages en fonction de la diversité phénotypique et génétique des souches de Staphylococcus aureus. Ph.D. thesis. Université de Bourgogne, Dijon, France [Google Scholar]
- 17.Gómez-Lucía E, Goyache J, Orden JA, Domenech A, Javier Hernandez F, Ruiz-Santa Quiteria JA, Lopez B, Blanco JL, Suarez G. 1992. Growth of Staphylococcus aureus and synthesis of enterotoxin during ripening of experimental Manchego-type cheese. J. Dairy Sci. 75:19–26. 10.3168/jds.S0022-0302(92)77733-9 [DOI] [PubMed] [Google Scholar]
- 18.Bayles KW, Iandolo JJ. 1989. Genetic and molecular analyses of the gene encoding staphylococcal enterotoxin D. J. Bacteriol. 171:4799–4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang S, Iandolo JJ, Stewart GC. 1998. The enterotoxin D plasmid of Staphylococcus aureus encodes a second enterotoxin determinant (sej). FEMS Microbiol. Lett. 168:227–233. 10.1111/j.1574-6968.1998.tb13278.x [DOI] [PubMed] [Google Scholar]
- 20.Balaban N, Rasooly A. 2000. Staphylococcal enterotoxins. Int. J. Food Microbiol. 61:1–10. 10.1016/S0168-1605(00)00377-9 [DOI] [PubMed] [Google Scholar]
- 21.Cheung AL, Bayer AS, Zhang G, Gresham H, Xiong YQ. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 40:1–9. 10.1016/S0928-8244(03)00309-2 [DOI] [PubMed] [Google Scholar]
- 22.Meyrand A, Vernozy-Rozand C. 1999. Croissance et entérotoxinogenèse de Staphylococcus aureus dans différents fromages. Rev. Med. Vet. 150:601–616 [Google Scholar]
- 23.De Buyser ML, Dufour B, Maire M, Lafarge V. 2001. Implication of milk and milk products in food-borne diseases in France and in different industrialised countries. Int. J. Food Microbiol. 67:1–17. 10.1016/S0168-1605(01)00443-3 [DOI] [PubMed] [Google Scholar]
- 24.Ndoye B, Rasolofo EA, Lapointe G, Roy D. 2011. A review of the molecular approaches to investigate the diversity and activity of cheese microbiota. Dairy Sci. Technol. 91:495–524. 10.1007/s13594-011-0031-8 [DOI] [Google Scholar]
- 25.Zhou XX, Pan YJ, Wang YB, Li WF. 2008. Optimization of medium composition for nisin fermentation with response surface methodology. J. Food Sci. 73:M245–M249. 10.1111/j.1750-3841.2008.00836.x [DOI] [PubMed] [Google Scholar]
- 26.Liu-Stratton Y, Roy S, Sen CK. 2004. DNA microarray technology in nutraceutical and food safety. Toxicol. Lett. 150:29–42. 10.1016/j.toxlet.2003.08.009 [DOI] [PubMed] [Google Scholar]
- 27.Cretenet M, Nouaille S, Thouin J, Rault L, Stenz L, Francois P, Hennekinne JA, Piot M, Maillard MB, Fauquant J, Loubiere P, Loir YL, Even S. 2011. Staphylococcus aureus virulence and metabolism are dramatically affected by Lactococcus lactis in cheese matrix. Environ. Microbiol. Rep. 3:340–351. 10.1111/j.1758-2229.2010.00230.x [DOI] [PubMed] [Google Scholar]
- 28.Jany JL, Barbier G. 2008. Culture-independent methods for identifying microbial communities in cheese. Food Microbiol. 25:839–848. 10.1016/j.fm.2008.06.003 [DOI] [PubMed] [Google Scholar]
- 29.Graber HU, Casey MG, Naskova J, Steiner A, Schaeren W. 2007. Development of a highly sensitive and specific assay to detect Staphylococcus aureus in bovine mastitic milk. J. Dairy Sci. 90:4661–4669. 10.3168/jds.2006-902 [DOI] [PubMed] [Google Scholar]
- 30.Ulve VM, Monnet C, Valence F, Fauquant J, Falentin H, Lortal S. 2008. RNA extraction from cheese for analysis of in situ gene expression of Lactococcus lactis. J. Appl. Microbiol. 105:1327–1333. 10.1111/j.1365-2672.2008.03869.x [DOI] [PubMed] [Google Scholar]
- 31.Duquenne M, Fleurot I, Aigle M, Darrigo C, Borezee-Durant E, Derzelle S, Bouix M, Deperrois-Lafarge V, Delacroix-Buchet A. 2010. Tool for quantification of staphylococcal enterotoxin gene expression in cheese. Appl. Environ. Microbiol. 76:1367–1374. 10.1128/AEM.01736-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ercolini D, Hill PJ, Dodd CE. 2003. Development of a fluorescence in situ hybridization method for cheese using a 16S rRNA probe. J. Microbiol. Methods 52:267–271. 10.1016/S0167-7012(02)00162-8 [DOI] [PubMed] [Google Scholar]
- 33.Malic S, Hill KE, Hayes A, Percival SL, Thomas DW, Williams DW. 2009. Detection and identification of specific bacteria in wound biofilms using peptide nucleic acid fluorescent in situ hybridization (PNA FISH). Microbiology 155:2603–2611. 10.1099/mic.0.028712-0 [DOI] [PubMed] [Google Scholar]
- 34.Lawson TS, Connally RE, Vemulpad S, Piper JA. 2011. Express fluorescence in situ hybridization methods for the detection of Staphylococcus aureus. Clin. Lab. 57:789–794 [PubMed] [Google Scholar]
- 35.Gey A, Werckenthin C, Poppert S, Straubinger RK. 2013. Identification of pathogens in mastitis milk samples with fluorescent in situ hybridization. J. Vet. Diagn. Invest. 25:386–394. 10.1177/1040638713486113 [DOI] [PubMed] [Google Scholar]
- 36.Yiu SH. 1985. A fluorescence microscopic study of cheese. Food Microstruct. 4:99–106 [Google Scholar]
- 37.Marcellino SN, Benson DR. 1992. Scanning electron and light microscopic study of microbial succession on Bethlehem St. Nectaire cheese. Appl. Environ. Microbiol. 58:3448–3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parker ML, Gunning PA, Macedo AC, Malcata FX, Brocklehurst TF. 1998. The microstructure and distribution of micro-organisms within mature Serra cheese. J. Appl. Microbiol. 84:523–530. 10.1046/j.1365-2672.1998.00375.x [DOI] [PubMed] [Google Scholar]
- 39.De Freitas I, Pinon N, Lopez C, Thierry A, Maubois JL, Lortal S. 2005. Microstructure, physicochemistry, microbial populations and aroma compounds of ripened Cantal cheeses. Lait 85:453–468. 10.1051/lait:2005033 [DOI] [Google Scholar]
- 40.Lopez C, Maillard MB, Briard-Bion V, Camier B, Hannon JA. 2006. Lipolysis during ripening of Emmental cheese considering organization of fat and preferential localization of bacteria. J. Agric. Food Chem. 54:5855–5867. 10.1021/jf060214l [DOI] [PubMed] [Google Scholar]
- 41.Jeanson S, Chadoeuf J, Madec MN, Aly S, Floury J, Brocklehurst TF, Lortal S. 2011. Spatial distribution of bacterial colonies in a model cheese. Appl. Environ. Microbiol. 77:1493–1500. 10.1128/AEM.02233-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheung AL, Nast CC, Bayer AS. 1998. Selective activation of sar promoters with the use of green fluorescent protein transcriptional fusions as the detection system in the rabbit endocarditis model. Infect. Immun. 66:5988–5993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kahl BC, Goulian M, van Wamel W, Herrmann M, Simon SM, Kaplan G, Peters G, Cheung AL. 2000. Staphylococcus aureus RN6390 replicates and induces apoptosis in a pulmonary epithelial cell line. Infect. Immun. 68:5385–5392. 10.1128/IAI.68.9.5385-5392.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malone CL, Boles BR, Lauderdale KJ, Thoendel M, Kavanaugh JS, Horswill AR. 2009. Fluorescent reporters for Staphylococcus aureus. J. Microbiol. Methods 77:251–260. 10.1016/j.mimet.2009.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yarwood JM, Bartels DJ, Volper EM, Greenberg EP. 2004. Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 186:1838–1850. 10.1128/JB.186.6.1838-1850.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boles BR, Horswill AR. 2008. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4(4):e1000052. 10.1371/journal.ppat.1000052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lauderdale KJ, Malone CL, Boles BR, Morcuende J, Horswill AR. 2010. Biofilm dispersal of community-associated methicillin-resistant Staphylococcus aureus on orthopedic implant material. J. Orthop. Res. 28:55–61. 10.1002/jor.20943 [DOI] [PubMed] [Google Scholar]
- 48.Moormeier DE, Endres JL, Mann EE, Sadykov MR, Horswill AR, Rice KC, Fey PD, Bayles KW. 2013. Use of microfluidic technology to analyze gene expression during Staphylococcus aureus biofilm formation reveals distinct physiological niches. Appl. Environ. Microbiol. 79:3413–3424. 10.1128/AEM.00395-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hennekinne JA, Guillier F, Perelle S, De Buyser ML, Dragacci S, Krys S, Lombard B. 2007. Intralaboratory validation according to the EN ISO 16 140 standard of the Vidas SET2 detection kit for use in official controls of staphylococcal enterotoxins in milk products. J. Appl. Microbiol. 102:1261–1272. 10.1111/j.1365-2672.2006.03183.x [DOI] [PubMed] [Google Scholar]
- 50.Novick RP. 1989. Staphylococcal plasmids and their replication. Annu. Rev. Microbiol. 43:537–565. 10.1146/annurev.mi.43.100189.002541 [DOI] [PubMed] [Google Scholar]
- 51.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6:343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- 52.Kreiswirth BN, Lofdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712. 10.1038/305709a0 [DOI] [PubMed] [Google Scholar]
- 53.Schenk S, Laddaga RA. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73:133–138 [DOI] [PubMed] [Google Scholar]
- 54.Hajjar T, Yong Meng G, Rajion MA, Vidyadaran S, Ai Li T, Ebrahimi M. 2013. Alterations in neuronal morphology and synaptophysin expression in the rat brain as a result of changes in dietary n-6:n-3 fatty acid ratios. Lipids Health Dis. 12:113. 10.1186/1476-511X-12-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Femling JK, Nauseef WM, Weiss JP. 2005. Synergy between extracellular group IIA phospholipase A2 and phagocyte NADPH oxidase in digestion of phospholipids of Staphylococcus aureus ingested by human neutrophils. J. Immunol. 175:4653–4661. 10.4049/jimmunol.175.7.4653 [DOI] [PubMed] [Google Scholar]
- 56.Cretenet M, Even S, Le Loir Y. 2011. Unveiling Staphylococcus aureus enterotoxin production in dairy products: a review of recent advances to face new challenges. Dairy Sci. Technol. 91:127–150. 10.1007/s13594-011-0014-9 [DOI] [Google Scholar]
- 57.Maijala R, Becker H, Hennekinne JA, Mantis A. 2010. Opinion of the scientific committee on veterinary measures relating to public health on staphylococcal enterotoxins in milk products, particularly cheeses. European Commission Health & Consumer Protection Directorate-General, Brussels, Belgium [Google Scholar]
- 58.Vernozy-Rozand C, Meyrand A, Mazuy C, Delignette-Muller ML, Jaubert G, Perrin G, Lapeyre C, Richard Y. 1998. Behaviour and enterotoxin production by Staphylococcus aureus during the manufacture and ripening of raw goats' milk lactic cheeses. J. Dairy Res. 65:273–281. 10.1017/S0022029997002781 [DOI] [PubMed] [Google Scholar]
- 59.Macaluso L, Lapeyre C, Dragacci S. 1998. Determination of influential factors during sample preparation for staphylococcal enterotoxin detection in dairy products. Analusis 26:300–304. 10.1051/analusis:1998177 [DOI] [Google Scholar]
- 60.Consentino S, Palmas F. 1997. Hygienic conditions and microbial contamination in six ewe's-milk-processing plants in Sardinia, Italy. J. Food Prot. 60:283–287 [DOI] [PubMed] [Google Scholar]
- 61.Auty MA, Gardiner GE, McBrearty SJ, O'Sullivan EO, Mulvihill DM, Collins JK, Fitzgerald GF, Stanton C, Ross RP. 2001. Direct in situ viability assessment of bacteria in probiotic dairy products using viability staining in conjunction with confocal scanning laser microscopy. Appl. Environ. Microbiol. 67:420–425. 10.1128/AEM.67.1.420-425.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takeuchi K, Frank JF. 2001. Confocal microscopy and microbial viability detection for food research. J. Food Prot. 64:2088–2102 [DOI] [PubMed] [Google Scholar]