Abstract
Non-O1/O139 Vibrio cholerae is naturally present in aquatic ecosystems and has been linked with cholera-like diarrhea and local outbreaks. The distribution of virulence-associated genes and genetic relationships among aquatic isolates from China are largely unknown. In this study, 295 aquatic isolates of V. cholerae non-O1/O139 serogroups from different regions in China were investigated. Only one isolate was positive for ctxB and harbored a rare genotype; 10 (3.4%) isolates carried several types of rstR sequences, eight of which carried rare types of toxin-coregulated pili (tcpA). Furthermore, 16 (5.4%) isolates carried incomplete (with partial open reading frames [ORFs]) vibrio seventh pandemic island I (VSP-I) or VSP-II clusters, which were further classified as 11 novel types. PCR-based analyses revealed remarkable variations in the distribution of putative virulence genes, including mshA (95.6%), hlyA (95.3%), rtxC (89.8%), rtxA (82.7%), IS1004 (52.9%), chxA (30.2%), SXT (15.3%), type III secretion system (18.0%), and NAG-ST (3.7%) genes. There was no correlation between the prevalence of putative virulence genes and that of CTX prophage or TCP genes, whereas there were correlations among the putative virulence genes. Further multilocus sequence typing (MLST) placed selected isolates (n = 70) into 69 unique sequence types (STs), which were different from those of the toxigenic O1 and O139 counterparts, and each isolate occupied a different position in the MLST tree. The V. cholerae non-O1/O139 aquatic isolates predominant in China have high genotypic diversity; these strains constitute a reservoir of potential virulence genes, which may contribute to evolution of pathogenic isolates.
INTRODUCTION
Vibrio cholerae is the causative agent of cholera, a life-threatening diarrheal disease. Of the more than 200 known V. cholerae serogroups, only O1 and O139 are associated with epidemic and pandemic cholera (1). V. cholerae strains in other serogroups (non-O1/O139 V. cholerae) are often nonpathogenic or associated with only mild illness (2). However, depending in part on the virulence factors which they carry, they have been linked with more-severe, cholera-like illness and have been associated with sporadic cases and outbreaks of gastroenteritis and extraintestinal infections in both developing and developed countries (2–7).
Two genetic elements associated with virulence in pathogenic O1 and O139 V. cholerae are a lysogenic filamentous bacteriophage (CTX prophage), which encodes cholera toxin (CT) (8), and the toxin coregulated pilus (TCP) pathogenicity island, which encodes factors involved in intestinal colonization. The CTX prophage uses TCP as a receptor, allowing V. cholerae infection and prophage integration into the bacterial chromosome (8), resulting in the emergence of new toxigenic strains. Non-O1/O139 strains that carry the genes for the CTX prophage and TCP and express CT have been linked with occurrences of severe disease. Other factors that have been associated with virulence include heat-stable toxin (NAG-ST) and hemolysin (Hly) (9). Recently, several novel virulence mechanisms, including a type III secretion system (TTSS) and a type 6 secretion system (T6SS), have been identified in non-O1/O139 isolates (10, 11).
Non-O1/O139 V. cholerae strains are naturally present in aquatic ecosystems, such as rivers, estuaries, and coastal waters (4, 12). We hypothesize that carriage of these and other virulence factors by non-O1/O139 strains creates an environmental reservoir of critical virulence genes, which may contribute to evolution of pathogenic V. cholerae. To explore this hypothesis, we report here results of screening of a collection of 295 environmental non-O1/O139 strains, isolated from 2001 to 2010 from different regions and aquatic environments in China, to determine the frequency of carriage of virulence-associated genes and the underlying phylogenetic relationships on the basis of multilocus sequence typing (MLST) (13).
MATERIALS AND METHODS
Sample location, isolation, and identification of non-O1/O139 V. cholerae.
Samples were procured from estuary environments in eight coastal regions and two inland provinces in China from 2001 to 2010. Non-O1/O139 V. cholerae strains were isolated by previously described methods (14). All isolates were screened for the oxidase reaction and other biochemical tests (bioMérieux, Lyon, France) to identify them as V. cholerae. Non-O1/O139 isolates were identified as positive for species-specific V. cholerae outer membrane protein (ompW) by PCR (15) and negative for agglutination by specific polyvalent antisera against V. cholerae O1 and O139 (S & A Reagents Laboratory, Bangkok, Thailand). A total of 295 V. cholerae non-O1/O139 isolates were used in this study. The sources and times of isolation are shown in Table S1 in the supplemental material.
PCR template preparation.
Genomic DNA from V. cholerae was extracted with a genomic DNA purification kit (Tiangen Biotech, Beijing, China) in accordance with the manufacturer's instructions. DNA was dissolved in Tris-EDTA (TE) buffer and stored at −20°C until PCR assays were performed.
PCR and sequencing analysis.
PCR assays were carried out using conventional PCR amplification. The target genes included cholera toxin B subunit (ctxB), different variants of the rstR repressor gene, including rstRET, rstRclass, rstRcalc, rstR-4**, rstR-5, rstR6, rstR232, and rstRVC06–18 of the CTX prophage, and the classical, El Tor-specific tcpA and tcpI genes of the TCP pathogenicity island. PCR was used to screen for five genes in the VSP-I cluster (VC0175, VC0178, VC0180, VC0183, and VC0185) and eight genes in the VSP-II cluster (VC0490, VC0493, VC0498, VC0502, VC0504, VC0512, VC0514, and VC0516). The putative accessory virulence genes included hemolysin (hlyA), heat-stable enterotoxin (ST), mannose-sensitive hemagglutinin (mshA), RTX toxin (rtxA and rtxC), chxA, SXT, IS1004, and TTSS genes. Table S2 in the supplemental material shows the primer sequences and their origins. Reference isolates for N16961 (O1 El Tor of 7th pandemic) and MO45 (O139 isolate isolated from India in 1993) were used as positive controls for PCR.
PCR products were sequenced commercially (TaKaRa, Dalian, China). Sequences were compared using BioEdit software (Ibis Biosciences, Carlsbad, CA). Clustal-W was used to perform multiple nucleotide alignments. The reference sequences of different types of ctxB, rstR, and open reading frames (ORFs) of VSP-I and VSP-II (VSP-I/II) were accessed from GenBank.
MLST.
MLST was performed as described previously (13). Seven housekeeping genes were targeted for MLST analysis: adk, gyrB, metE, mdh, pntA, purM, and pyrC (see Table S2 in the supplemental material). The PCR products were directly sequenced in both directions (TaKaRa, Dalian, China). Contiguous nucleotide sequences were assembled with MEGA software, and sequence variants were designated allele profiles. Isolates with identical allelic profiles were assigned to the same sequence type (ST). eBURST (16) was used to identify clonal complexes (CCs). A minimum-spanning tree was constructed using the allelic differences between isolates of the seven housekeeping genes and BioNumerics software (Applied Math). The housekeeping genes of 21 reference V. cholerae strains (see Table S1 in the supplemental material) were extracted for MLST analysis. All new housekeeping sequences from this study were deposited in PubMLST and are accessible at http://pubmlst.org/vcholerae/.
Correlation analysis of virulence-associated genes.
The relationships between genes were analyzed using Spearman's correlation with SPSS 17.0 software. Each pair of variables (genes) was compared by correlation measures. The correlation coefficients were obtained, and the correlation was considered significant at the 0.01 or 0.05 level. The gene analysis included CTX prophage elements, TCP genes, and VSP-I/II and putative accessory virulence genes.
Nucleotide sequence accession numbers.
The representative new nucleotide sequences and predicted amino acid sequences for ctxB, rstR, and tcpA genes were deposited in GenBank with accession numbers KJ437653 (ctxB), KJ437633 to KJ437644 (rstR), and KJ437645 to KJ437652 (tcpA).
RESULTS
Distribution of CTX prophage and TCP genes.
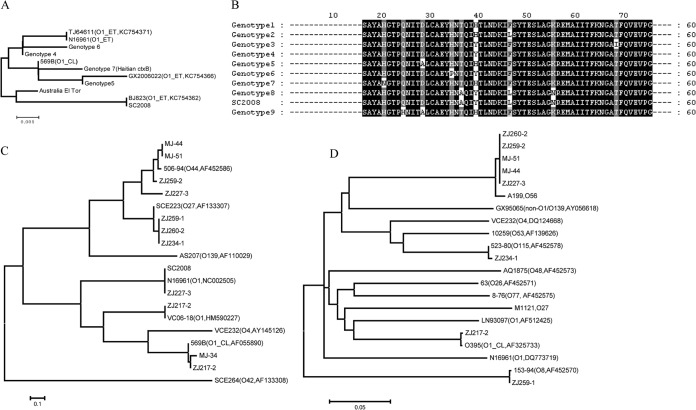
Of the 295 tested non-O1/O139 isolates, only one isolate (SC2008; Table 1) carried ctxB and was thus potentially toxigenic; we did not screen the strain to confirm that it actually expressed CT. Sequencing analysis indicated that SC2008 carried a rare ctxB type (genotype 8) (17) compared with the amino acid substitutions of known types (Fig. 1A and B). In addition, 10 isolates (3.4%) carried several types of rstR (Fig. 1C), some of which are found in environmental O1 and O139 isolates. Two of 10 rstR-positive isolates were positive for two types of rstR, indicating that at least two copies of the CTX prophage were present.
TABLE 1.
Distribution of CTX prophage and TCP genesa
| Strain(s) | Province(s) | ctxB genotype | rstR genotype | tcpA genotype | tcpI |
|---|---|---|---|---|---|
| SC2008 | Sichuan | 8 | ET | − | |
| ZJ217-2 | Guangdong | CL,VC06-18 | CL (O1_CL, AF325733) | + | |
| ZJ227-3 | Guangdong | ET, rstR6 | A199 (O56, EU362122) | + | |
| ZJ234-1 | Guangdong | rstR-4** | 523-80 (O115, AF452578) | + | |
| ZJ259-1 | Guangdong | rstR-4** | 153-94 (O8, AF452570) | + | |
| ZJ260-2 | Guangdong | rstR-4** | A199 (O56, EU362122) | + | |
| ZJ259-2, MJ-44, MJ-51 | Guangdong, Fujian | rstR6 | A199 (O56, EU362122) | + | |
| MJ-34 | Fujian | CL | − |
tcpA genotype data include reference strain identifiers followed by serogroup and GenBank accession numbers in parentheses. ET, El Tor; CL, classical; +, positive; −, negative.
FIG 1.
Phylogenetic trees and sequence alignments of ctxB, rstR, and tcpA. (A, C, and D) Phylogenetic trees of ctxB, rstR, and tcpA, respectively. (B) Alignment of deduced amino acid sequences of ctxB from V. cholerae strains. The serogroups and GenBank accession numbers of the reference sequences for ctxB, rstR, and tcpA are shown in parentheses in panels A, C, and D. Sequence characters in black on a white background represent rare substitutions; sequence characters in white on a black background represent identical sequences in all genotypes; sequence characters in white on a gray background represent common substitutions. El, El Tor biotype; CL, classical biotype.
Because PCR showed that all 295 isolates were negative for El Tor and classical-type-specific tcpA, strains were screened by using a pair of primers that spanned the entire tcpA ORF (tcpA1185 [see Table S2 in the supplemental material]). Eight isolates (2.7%) yielded specific PCR products (1,185 bp), which were sequenced and identified as the environmental types (A199, 153-94 and 523-80) and classical type (Fig. 1D and Table 1) of tcpA. Meanwhile, the eight tcpA-positive isolates were positive for tcpI (Table 1), another ORF within the TCP pathogenicity island. Interestingly, all isolates with the pre-CTX prophage and TCP gene sequences were isolated from only two provinces (Table 1).
Characterization of VSP-I/II clusters and their relationship with elements of CTX prophage and TCP genes.
As previously described (18), we used PCR to detect five ORFs in the VSP-I cluster and eight ORFs in the VSP-II cluster. A total of 16 (5.4%) non-O1/O139 isolates carried several VSP island genes, but none carried a complete VSP island. The isolates were positive for one or more than one ORF of VSP-I and/or VSP-II clusters (see Table S3 in the supplemental material). The positive PCR products were further sequenced and compared to those of V. cholerae O1 El Tor N16961. The VSP-I/II clusters were identified as 11 novel types and divided into three groups (Fig. 2): the partial ORFs of the VSP-I-positive types (group 1), the partial ORFs of the VSP-II-positive types (group 2), and the partial ORFs of the VSP-I- and VSP-II-positive types (group 3). The ORFs were 86% (VC0185) to 99% (VC0490 and VC0502) identical to ORFs from V. cholerae N16961. These results indicated that the isolates carried incomplete VSP-I/II types.
FIG 2.
Characterization of VSP-I/II clusters in non-O1/O139 V. cholerae. PCR primers are shown as blue arrows, and the color gradient from yellow to red indicates the similarity of VSP-I/II clusters in non-O1/O139 V. cholerae and strain N16961 (reference). Green rectangles indicate the ORFs amplified by PCR.
The combination patterns of CTX prophage, TCP genes, and VSP-I/II isolates were as follows: CTX+TCP+(VSP-I)−(VSP-II)+ (n = 3), CTX+TCP+(VSP-I)−(VSP-II)− (n = 5), CTX+TCP−(VSP-I)−(VSP-II)− (n = 2), CTX−TCP−(VSP-I)+(VSP-II)+ (n = 2), CTX−TCP−(VSP-I)+ (VSP-II)− (n = 2), and CTX−TCP−(VSP-I)−(VSP-II)+ (n = 9) (see Table S3 in the supplemental material). Although none carried complete VSP-I and -II islands, there was a correlation between the presence of VSP-I and the presence of VSP-II; only the presence of VSP-II correlated with the presence of TCP and CTX prophage (see Table S4 in the supplemental material).
Prevalence of putative virulence genes and relationship with CTX prophage/TCP genes.
PCR-based analyses revealed remarkable variations in the distribution of putative virulence genes, including mshA (95.6%), hlyA (95.3%), rtxC (89.8%), rtxA (82.7%), IS1004 (52.9%), chxA (30.2%), SXT (15.3%), TTSS (18.0%), and NAG-ST (3.7%) (see Table S5 in the supplemental material). There was no correlation between the prevalence of putative virulence gene(s) and the prevalence of CTX prophage or TCP genes, whereas there were correlations among those nine putative virulence genes (see Table S4 in the supplemental material). rtxA correlated to six other putative virulence genes (mshA, hlyA, rtxC, IS1004, chxA, and TTSS); hlyA correlated to five others (rtxC, chxA, mshA, rtxA, and IS1004). mshA, IS1004, and chxA correlated to four others. rtxC correlated to three others, and TTSS correlated to two others. SXT and ST had no significant correlation to other putative virulence genes.
MLST.
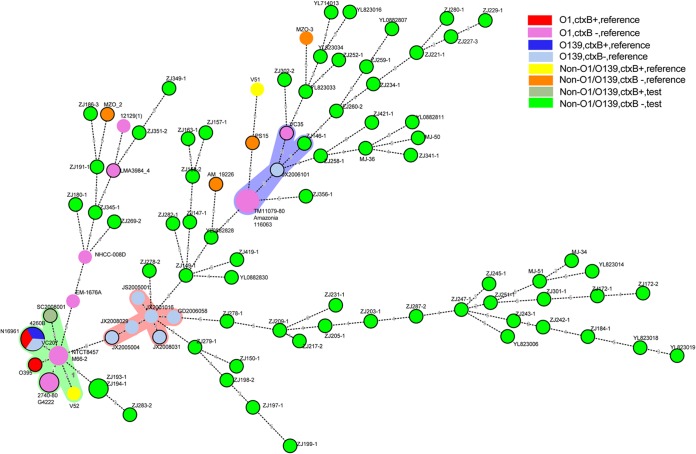
We selected 70 non-O1/O139 isolates for MLST analysis. The isolates were selected based on their different gene characters (types of CTX prophage, TCP genes, and VSP-I/II and putative accessory virulence genes) and sources (different years and provinces or regions). All 70 isolates represented the different types of non-O1/O139 isolates used in this study. The housekeeping genes of 29 reference O1, O139, and non-O1/O139 isolates (see Table S1 in the supplemental material) were also included, resulting in a total of 99 isolates. Excluding two isolates (ZJ193-1 and ZJ194-1), there were 68 non-O1/O139 isolates that had a unique sequence type (ST) and showed high diversity (Fig. 3). Most STs differed from each other by three or more loci. There was no evidence of ST clustering by year or geographic location.
FIG 3.
Genetic relationships between 99 V. cholerae isolates were analyzed by a minimum-spanning tree. Each circle in the tree represents 1 sequence type (ST), and the size of the circle reflects the number of isolates belonging to each ST. The digits on the lines between two circles represent the different numbers of types. The colors of the halos surrounding the ST denote the types that belong to the same clonal complex (CC). The species within different serogroups are represented by different colors, as indicated. Detailed MLST profiles are shown in Table S6 in the supplemental material.
Six ctxB-positive isolates were included in the MLST analysis. O1 El Tor (N16961) and O139 (4260B) isolates were grouped into one ST, while O395 (O1 classical serogroup), V52 (O37), and SC2008 (non-O1/O139 from the current study) and four other ctxB-negative O1 El Tor isolates formed one clonal complex (CC). The only ctxB-positive isolate from the current study, SC2008, had a unique ST. In contrast, V51, an O141 ctxB-positive strain, was far from all ctxB-positive isolates in the MLST tree, forming its own ST.
Unlike the ctxB-positive isolates, all ctxB-negative isolates of the O1, O139, or non-O1/O139 types presented highly diverse single nucleotide polymorphisms (SNPs) in the seven housekeeping genes (see Table S6 in the supplemental material). The nine pre-CTX prophage (ctxB-negative but rstR-positive) isolates and the 16 VSP-I/II-positive isolates were grouped into different positions on the MLST tree and formed STs of their own. Except for two ctxB-negative O139 isolates, the STs of six ctxB-negative O139 isolates formed a CC and also belonged to a different CC with ctxB-positive O139 isolates (Fig. 3). However, the six ctxB-negative O139 isolates were sourced from different years and provinces in China.
DISCUSSION
We found a wide range of putative virulence genes present in our collection of non-O1/O139 strains. Only one strain carried the ctxB gene, which, interestingly, had an unusual genotype, matching ctxB genes previously identified among environmental O1 isolates in China. To date, based on amino acid residue substitutions, nine genotypes of ctxB have been identified (Fig. 1A and B). Genotypes 1, 2, and 3 have been linked with O1 and O139 V. cholerae isolates (19): genotype 3 is responsible for the 7th pandemic, genotype 2 has been found only in El Tor isolates from Australia, and the first report of an atypical El Tor biotype that consisted of classical ctxB (genotype 1) and emerged between 1991 and 1994 in Matlab, Bangladesh, was published in 2002 (20). Other ctxB variants have been reported in association with O139 strains in Bangladesh (21) and El Tor O1 strains in China (17). Here, the ctxB of our SC2008 strain matched that from previously reported Chinese environmental O1 El Tor isolates (17). Factors which select for ctxB gene variants are not well understood. There is a suggestion that the recent emergence of a classical ctxB variant in the predominant global El Tor lineage (22, 23) is associated with increased clinical virulence, linked with a possible increase in CT production (24).
In this study, the pre-CTX prophage and TCP genes identified demonstrated a variety of different rstR and tcpA alleles suggestive of ongoing genetic recombination among non-O1/O139 strains in environmental reservoirs. Non-O1/O139 strains carrying pre-CTX prophage and TCP genes were isolated from only two provinces, consistent with localization of these genes in the available gene pool in a single region. VSP-I and VSP-II are linked with the pandemic potential of seventh pandemic El Tor isolates (25). We found that VSP-I/II gene segments were present in 5.4% of our non-O1/O139 isolates from aquatic environments, although, in all instances, the gene cluster was incomplete, in keeping with studies of environmental non-O1/O139 strains from other areas (26, 27). Environmental non-O1/O139 isolates are also known to carry a variety of other putative accessory virulence genes. Although they were from aquatic environments, the non-O1/O139 isolates analyzed in this study contained roughly the same rates of hlyA and mshA as isolates from diarrheal patients in India (4). The rate of TTSS genes (<20%) was significant lower than in patients from India (4) but a little higher than in environmental isolates from Dhaka, Bangladesh (26). TTSS translocates a number of TTSS effectors, such as VopF and VopE, which interfere with host cell signaling pathways (28): a functional TTSS has been shown to be essential for the pathogenicity of the non-O1/O139 AM-19226 strain (29).
As demonstrated by MLST analysis, the environmental non-O1/O139 population in our sample of aquatic environments in China was phylogenetically heterogeneous. These findings are consistent with a recent report by Octavia et al., in which 77 clinical and environmental non-O1/O139 isolates from a number of different countries were found to form 66 STs (13). The majority of these STs were unique, and only three clonal complexes were formed; as in our study, the clonal complexes centered on epidemic or pandemic, ctx-positive strains, highlighting the close phylogenetic relationship among such strains, in contrast to the diversity seen among non-O1/O139 strains. The nontoxigenic O1 isolates contained STs similar to those seen with the non-O1/O139 isolates, which also scattered into different positions in the MLST tree and showed high diversity. In contrast, the six nontoxigenic O139 isolates from different years and regions formed a CC, consistent with a more recent common origin.
The virulence of non-O1/O139 isolates is multifactorial and combinatorial, with a range of virulence factors involved in disease causation (30). Strains producing cholera toxin have been clearly linked with illness, and, in human volunteer studies, a non-O1/O139 strain that produced NAG-ST (but which did not have the ctx or tcpA gene) caused a major diarrheal purge (30, 31). While other virulence factors have been identified in strains from patients with diarrhea, in the absence of strong epidemiologic data or volunteer studies caution must be taken in assuming that these factors, by themselves, are responsible for illness (31). In this setting, it is difficult to know how to assess the public health risk posed by the environmental strains evaluated in this study. Strain SC2008, with its ctxB gene and position within the same CC as known epidemic strains, is likely to have been pathogenic and may have evolved from an epidemic strain. In contrast, our environmental O1/O139 strains do not appear to share a phylogenetic background with epidemic or pandemic V. cholerae strains (or with each other), suggesting that the virulence genes and gene fragments identified in these strains were acquired through horizontal gene transfer. While we cannot be certain that acquisition of one or more of these putative virulence genes has resulted or will result in human virulence, it remains a theoretical possibility in the appropriate strain background.
In summary, we described the distribution of virulence factors in V. cholerae non-O1/O139 strains from aquatic environments. The findings indicate that strains in aquatic environments are reservoirs for multiple V. cholerae virulence genes, with ongoing recombination events and mutations leading to acquisition and emergence of genes, gene fragments, and new variant genes. Further surveillance and investigation are required to understand the molecular evolution, epidemiology, and pathogenicity of non-O1/O139 isolates of environmental origin.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the NSFC of China (30872260) and the Priority Project on Infectious Disease Control and Prevention (2012ZX10004215) from the Ministry of Health, China.
This publication used the PubMLST website (http://pubmlst.org/), developed by Keith Jolley (32), which is maintained at the University of Oxford and funded by the Wellcome Trust.
We thank Sophie Octavia and Ruiting Lan for obtaining the MLST STs.
Footnotes
Published ahead of print 6 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01021-14.
REFERENCES
- 1.Kaper JB, Morris JG, Jr, Levine MM. 1995. Cholera. Clin. Microbiol. Rev. 8:48–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morris JG., Jr 1990. Non-O group 1 Vibrio cholerae: a look at the epidemiology of an occasional pathogen. Epidemiol. Rev. 12:179–191 [DOI] [PubMed] [Google Scholar]
- 3.Dhar R, Badawi M, Qabazard Z, Albert MJ. 2004. Vibrio cholerae (non-O1, non-O139) sepsis in a child with Fanconi anemia. Diagn. Microbiol. Infect. Dis. 50:287–289. 10.1016/j.diagmicrobio.2004.08.004 [DOI] [PubMed] [Google Scholar]
- 4.Chatterjee S, Ghosh K, Raychoudhuri A, Chowdhury G, Bhattacharya MK, Mukhopadhyay AK, Ramamurthy T, Bhattacharya SK, Klose KE, Nandy RK. 2009. Incidence, virulence factors, and clonality among clinical strains of non-O1, non-O139 Vibrio cholerae isolates from hospitalized diarrheal patients in Kolkata, India. J. Clin. Microbiol. 47:1087–1095. 10.1128/JCM.02026-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kondo S, Ajawatanawong P. 2009. Distribution and sequence analysis of virulence associated genes in Vibrio cholerae O1, O139 and non-O1/non-O139 isolates from Thailand. Southeast Asian J. Trop. Med. Public Health 40:1015–1024 [PubMed] [Google Scholar]
- 6.Luo Y, Ye J, Jin D, Ding G, Zhang Z, Mei L, Octavia S, Lan R. 2013. Molecular analysis of non-O1/non-O139 Vibrio cholerae isolated from hospitalised patients in China. BMC Microbiol. 13:52. 10.1186/1471-2180-13-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma C, Thungapathra M, Ghosh A, Mukhopadhyay AK, Basu A, Mitra R, Basu I, Bhattacharya SK, Shimada T, Ramamurthy T, Takeda T, Yamasaki S, Takeda Y, Nair GB. 1998. Molecular analysis of non-O1, non-O139 Vibrio cholerae associated with an unusual upsurge in the incidence of cholera-like disease in Calcutta, India. J. Clin. Microbiol. 36:756–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waldor MK, Mekalanos JJ. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910–1914. 10.1126/science.272.5270.1910 [DOI] [PubMed] [Google Scholar]
- 9.Bag PK, Bhowmik P, Hajra TK, Ramamurthy T, Sarkar P, Majumder M, Chowdhury G, Das SC. 2008. Putative virulence traits and pathogenicity of Vibrio cholerae non-O1, non-O139 isolates from surface waters in Kolkata, India. Appl. Environ. Microbiol. 74:5635–5644. 10.1128/AEM.00029-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dziejman M, Serruto D, Tam VC, Sturtevant D, Diraphat P, Faruque SM, Rahman MH, Heidelberg JF, Decker J, Li L, Montgomery KT, Grills G, Kucherlapati R, Mekalanos JJ. 2005. Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. Proc. Natl. Acad. Sci. U. S. A. 102:3465–3470. 10.1073/pnas.0409918102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U. S. A. 103:1528–1533. 10.1073/pnas.0510322103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teh CS, Chua KH, Thong KL. 2011. Genetic variation analysis of Vibrio cholerae using multilocus sequencing typing and multi-virulence locus sequencing typing. Infect. Genet. Evol. 11:1121–1128. 10.1016/j.meegid.2011.04.005 [DOI] [PubMed] [Google Scholar]
- 13.Octavia S, Salim A, Kurniawan J, Lam C, Leung Q, Ahsan S, Reeves PR, Nair GB, Lan R. 2013. Population structure and evolution of non-O1/non-O139 Vibrio cholerae by multilocus sequence typing. PLoS One 8:e65342. 10.1371/journal.pone.0065342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faruque SM, Chowdhury N, Kamruzzaman M, Dziejman M, Rahman MH, Sack DA, Nair GB, Mekalanos JJ. 2004. Genetic diversity and virulence potential of environmental Vibrio cholerae population in a cholera-endemic area. Proc. Natl. Acad. Sci. U. S. A. 101:2123–2128. 10.1073/pnas.0308485100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marin MA, Thompson CC, Freitas FS, Fonseca EL, Aboderin AO, Zailani SB, Quartey NK, Okeke IN, Vicente AC. 2013. Cholera outbreaks in Nigeria are associated with multidrug resistant atypical El Tor and non-O1/non-O139 Vibrio cholerae. PLoS Negl. Trop. Dis. 7:e2049. 10.1371/journal.pntd.0002049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518–1530. 10.1128/JB.186.5.1518-1530.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang P, Zhou H, Kan B, Wang D. 2013. Novel ctxB variants of Vibrio cholerae O1 isolates, China. Infect. Genet. Evol. 20:48–53. 10.1016/j.meegid.2013.08.004 [DOI] [PubMed] [Google Scholar]
- 18.O'Shea YA, Finnan S, Reen FJ, Morrissey JP, O'Gara F, Boyd EF. 2004. The Vibrio seventh pandemic island-II is a 26.9 kb genomic island present in Vibrio cholerae El Tor and O139 serogroup isolates that shows homology to a 43.4 kb genomic island in V. vulnificus. Microbiology 150:4053–4063. 10.1099/mic.0.27172-0 [DOI] [PubMed] [Google Scholar]
- 19.Olsvik O, Wahlberg J, Petterson B, Uhlen M, Popovic T, Wachsmuth IK, Fields PI. 1993. Use of automated sequencing of polymerase chain reaction-generated amplicons to identify three types of cholera toxin subunit B in Vibrio cholerae O1 strains. J. Clin. Microbiol. 31:22–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nair GB, Faruque SM, Bhuiyan NA, Kamruzzaman M, Siddique AK, Sack DA. 2002. New variants of Vibrio cholerae O1 biotype El Tor with attributes of the classical biotype from hospitalized patients with acute diarrhea in Bangladesh. J. Clin. Microbiol. 40:3296–3299. 10.1128/JCM.40.9.3296-3299.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhuiyan NA, Nusrin S, Alam M, Morita M, Watanabe H, Ramamurthy T, Cravioto A, Nair GB. 2009. Changing genotypes of cholera toxin (CT) of Vibrio cholerae O139 in Bangladesh and description of three new CT genotypes. FEMS Immunol. Med. Microbiol. 57:136–141. 10.1111/j.1574-695X.2009.00590.x [DOI] [PubMed] [Google Scholar]
- 22.Goel AK, Jain M, Kumar P, Bhadauria S, Kmboj DV, Singh L. 2008. A new variant of Vibrio cholerae O1 El Tor causing cholera in India. J. Infect. 57:280–281. 10.1016/j.jinf.2008.06.015 [DOI] [PubMed] [Google Scholar]
- 23.Morris JG., Jr 2011. Cholera—modern pandemic disease of ancient lineage. Emerg. Infect. Dis. 17:2099–2104. 10.3201/eid1711.111109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh-Banerjee J, Senoh M, Takahashi T, Hamabata T, Barman S, Koley H, Mukhopadhyay AK, Ramamurthy T, Chatterjee S, Asakura M, Yamasaki S, Nair GB, Takeda Y. 2010. Cholera toxin production by the El Tor variant of Vibrio cholerae O1 compared to prototype El Tor and classical biotypes. J. Clin. Microbiol. 48:4283–4286. 10.1128/JCM.00799-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taviani E, Grim CJ, Choi J, Chun J, Haley B, Hasan NA, Huq A, Colwell RR. 2010. Discovery of novel Vibrio cholerae VSP-II genomic islands using comparative genomic analysis. FEMS Microbiol. Lett. 308:130–137. 10.1111/j.1574-6968.2010.02008.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahman MH, Biswas K, Hossain MA, Sack RB, Mekalanos JJ, Faruque SM. 2008. Distribution of genes for virulence and ecological fitness among diverse Vibrio cholerae population in a cholera endemic area: tracking the evolution of pathogenic strains. DNA Cell Biol. 27:347–355. 10.1089/dna.2008.0737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dutta D, Chowdhury G, Pazhani GP, Guin S, Dutta S, Ghosh S, Rajendran K, Nandy RK, Mukhopadhyay AK, Bhattacharya MK, Mitra U, Takeda Y, Nair GB, Ramamurthy T. 2013. Vibrio cholerae non-O1, non-O139 serogroups and cholera-like diarrhea, Kolkata, India. Emerg. Infect. Dis. 19:464–467. 10.3201/eid1903.121156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin OS, Tam VC, Suzuki M, Ritchie JM, Bronson RT, Waldor MK, Mekalanos JJ. 2011. Type III secretion is essential for the rapidly fatal diarrheal disease caused by non-O1, non-O139 Vibrio cholerae. mBio 2:e00106-11. 10.1128/mBio.00106-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tam VC, Serruto D, Dziejman M, Brieher W, Mekalanos JJ. 2007. A type III secretion system in Vibrio cholerae translocates a formin/spire hybrid-like actin nucleator to promote intestinal colonization. Cell Host Microbe 1:95–107. 10.1016/j.chom.2007.03.005 [DOI] [PubMed] [Google Scholar]
- 30.Chen Y, Johnson JA, Pusch GD, Morris JG, Jr, Stine OC. 2007. The genome of non-O1 Vibrio cholerae NRT36S demonstrates the presence of pathogenic mechanisms that are distinct from those of O1 Vibrio cholerae. Infect. Immun. 75:2645–2647. 10.1128/IAI.01317-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris JG, Takeda T, Tall BD, Losonsky GA, Bhattacharya SK, Forrest BD, Kay BA, Nishibuchi M. 1990. Experimental non-O group 1 Vibrio cholerae gastroenteritis in humans. J. Clin. Invest. 85:697–705. 10.1172/JCI114494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. 10.1186/1471-2105-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.