Abstract
When immersed in seawater, substrates are rapidly colonized by both micro- and macroorganisms. This process is responsible for important economic and ecological prejudices, particularly when related to ship hulls or aquaculture nets. Commercial antifouling coatings are supposed to reduce biofouling, i.e., micro- and macrofoulers. In this study, biofilms that primarily settled on seven different coatings (polyvinyl chloride [PVC], a fouling release coating [FRC], and five self-polishing copolymer coatings [SPC], including four commercial ones) were quantitatively studied, after 1 month of immersion in summer in the Toulon Bay (Northwestern Mediterranean Sea, France), by using flow cytometry (FCM), microscopy, and denaturing gradient gel electrophoresis. FCM was used after a pretreatment to separate cells from the biofilm matrix, in order to determine densities of heterotrophic bacteria, picocyanobacteria, and pico- and nanoeukaryotes on these coatings. Among diatoms, the only microphytobenthic class identified by microscopy, Licmophora, Navicula, and Nitzschia were determined to be the dominant taxa. Overall, biocide-free coatings showed higher densities than all other coatings, except for one biocidal coating, whatever the group of microorganisms. Heterotrophic bacteria always showed the highest densities, and diatoms showed the lowest, but the relative abundances of these groups varied depending on the coating. In particular, the copper-free SPC failed to prevent diatom settlement, whereas the pyrithione-free SPC exhibited high picocyanobacterial density. These results highlight the interest in FCM for antifouling coating assessment as well as specific selection among microbial communities by antifouling coatings.
INTRODUCTION
Marine biofouling is now clearly recognized as a source of both economic and environmental hazards. When occurring on ship hulls, the increase of fuel consumption and dry-docking operations leads to significant increases in pollution and economic burden (1), along with the ecological risk of invasive species dispersion (2). Furthermore, biofilms were shown to increase frictional resistance on hulls (3) and to control, at least in some cases, the settlement of macrofouler propagules (4–6). Consequently, knowledge of biofilm communities that colonize antifouling (AF) coatings and how these coatings control microbial communities is of great concern. Unfortunately, the related literature is very scarce, as most studies have dealt with macrofouling.
Most studies of marine biofilm on artificial surfaces have not dealt with AF coatings but with stainless steel, polycarbonate, polystyrene, or glass. Several publications have reported that, whatever the nature of these substrata, prokaryotic communities tend to evolve in a similar pattern with time, depending on the sites and therefore on the environmental conditions (7–10). The Rhodobacterales (Alphaproteobacteria), especially the Roseobacter clade members, have been shown to be the dominant and ubiquitous primary surface colonizers in temperate coastal waters (Pacific and Atlantic coasts) (11–13). The use of PhyloChip (10) but mostly the 454 pyrosequencing approach (14) allowed confirmation of Alphaproteobacteria dominance and showed an important diversity of biofilms on artificial substrata. Quantitative field studies are scarce because of the difficulty in enumeration of bacteria embedded in exopolymeric substances (EPS) within biofilms. Total bacterial abundance has been assessed by using epifluorescence microscopy, especially for early biofilms (e.g., see references 10 and 14–16), or confocal laser scanning microscopy (CLSM), which interestingly allows the three-dimensional (3D) structure of biofilms to be taken into account (17). Fluorescence in situ hybridization (FISH) was used mainly for relative quantification of classes or specific groups of bacterial taxa (7, 9, 12, 18), as was quantitative PCR (qPCR) (19–21). Only very recently were relative abundances of (picocyano)bacteria and pico- and nanoeukaryotes from biofilm on glass determined by using flow cytometry (FCM) (22–24). Microphytobenthos in marine biofilms on artificial surfaces appears to be represented mainly by diatoms and especially pennates. Molecular techniques are rarely used to study eukaryotic microorganisms from biofilms, especially the autotrophic communities of diatoms, due to the lack of a universal molecular target such as 16S rRNA for prokaryotes. Identification to the genus level has generally been carried out by using optical or scanning electron microscopy (e.g., see reference 25). Interestingly, studies of marine microbial biofilm communities mostly have not considered the abundance and diversity of pico-, nano-, and microorganisms, which is the aim of this study.
Antifouling coating can be divided in chemically active self-polishing coatings (SPC), with booster biocides, and silicone- and fluorine-based fouling release coatings (FRC), based on physicochemical and mechanical effects (26). The efficacy of SPC is associated mainly with biocide effects, considering that commercial coatings generally include 2 to 5 different active molecules with different cellular targets. The second major parameter is related to the ability of these coatings to release their biocides over time. The emerging SPC control the release rate through a combination of binder hydrolysis, particle dissolution, and surface polishing in seawater. A “steady-state” constant release rate is achieved as a steady polishing period is established. This steady state continues for as long as antifouling paint film remains. The leaching rates of copper and booster biocides in the self-polishing copolymer are controlled by the degree of polymerization (molecular weight) and the hydrophilicity of the copolymer, which depends on both the ratio of hydrophilic or seawater-hydrolyzable groups and the chemical nature of the macromolecular chains. In addition, antifouling action requires biocides to be available for seawater dissolution and subsequent diffusion toward the surface. However, blended molecules are often entrapped and cannot diffuse and become inactive (26). Considering the biocide-free FRC, the adhesion between foulers and the surface is minimized due to the low surface energy and elastic modulus so that biofouling can be removed by hydrodynamic stress during moving or by a simple mechanical cleaning. Although they inhibit the adhesion of most macrofouling under dynamic conditions, FRC fail to prevent colonization of “slime” layers (i.e., biofilm) (27). Diatoms especially dominate these biofilms, which adhere tenaciously to hydrophobic surfaces and are not released from FRC, even on vessels operating at high speeds (>30 knots) (28). Considering the difficulty in bacterial enumeration when bacteria are embedded in biofilms, their abundance on AF coatings has been reported rarely, and when they have been reported, the analyses were based mostly on microscopy observation (16–30). Successful colonization of all coatings by bacteria soon after immersion has been reported, and significant differences were observed, depending on the coatings and the immersion sites. The diversity of these communities has not been studied, except by Cassé and Swain (30), but the use of a culture-dependent approach represents a significant restriction for the evaluation of bacterial taxon dominance and diversity. Finally, bacterial diversity on AF coating has been addressed for early biofilms only for one copper-based paint and a bromophenol-based surface (using cloning-sequencing), which seems to confirm a high level of diversity, together with Alphaproteobacteria (especially Rhodobacteraceae) dominance and copper-accumulating bacteria (11, 31). Considering diatoms, Amphora (30, 32, 33) or Cylindrotheca, Licmophora, Nitzschia, and Amphora (16, 34, 35) have been reported to be the dominant diatom populations found on all coatings (SPC and FRC) during static trials. Resistance of Amphora species to copper toxicity (32, 33, 36) as well as to the organic biocides Copper Omadine (pyrithione) and Sea Nine 211 (32) was observed. Compared to SPC, FRC exhibited a higher average diatom abundance, without a significant change in diversity (33), or the highest diversity (34).
A previous study conducted at the same site in the Toulon Bay (Northwestern Mediterranean Sea, Southeast France) allowed us to report specific effects of the AF coatings (SPC) on both bacteria and diatoms (35). Nevertheless, the two groups of microorganisms displayed similar community structures on all the coatings, suggesting that biofilm communities did not reach their mature state (“climax”) after 2 weeks. Moreover, bacterial quantification was not achieved. The first aim of this study was to adapt an FCM quantification method for biofilm bacteria but also to extend the study to a more important number of groups to assess the abundances of the main microbial biofilm communities (heterotrophic prokaryotes, pico- and nanoeukaryotes, and diatoms) that are able to colonize AF coatings. The second objective was to show how both commercial and laboratory-based AF coatings impact the biofilms in terms of both the abundance and diversity of the microbial community. Flow cytometry, denaturing gradient gel electrophoresis (DGGE), and microscopy were combined together to address these objectives. Finally, the possible relationship between microorganism abundance and macrofouling was examined. Immersion was performed in the Toulon Bay in July, during the same period as that of the previous study (35). However, this time, the experiment was conducted during 1 month in order to obtain mature biofilms, beyond pioneer stages reported in the previous study. Coated panels were also immersed at the same site during 12 months to determine the long-term efficacy of the same coatings.
MATERIALS AND METHODS
Immersion site.
The immersion site is located in the Toulon Bay (France, Northwestern Mediterranean Sea) in a semienclosed pond in the military harbor (43°06′25″N, 5°55′41″E) (Fig. 1). It is the current immersion site for the assessment of antifouling paints for the MAPIEM laboratory. A static, permanent raft allows the immersion of numerous panels of different sizes at different depths and for long periods of time, if necessary.
FIG 1.
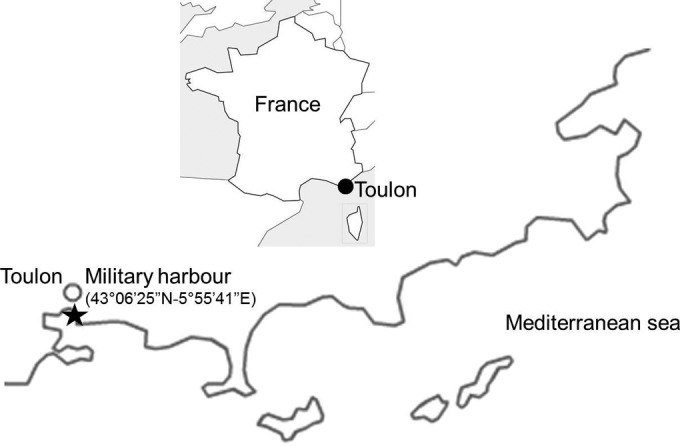
Location of the immersion site on the French Mediterranean coast. (Reprinted from reference 35 [copyright Taylor & Francis 2012].)
Coatings.
Seven different coatings (Table 1) were used, including polyvinyl chloride (PVC) as a reference and Intersleek 757 (IT757) (International Paint Ltd., Le Havre, France), a FRC. The five biocidal SPC AF coatings included four commercial ones: Sea Quantum Classic (SQT) (Jotun France SA, Levallois Perret, France), Intersmooth 360 (IS360) (International Paint Ltd., Le Havre, France), A3S (Nautix, Guidel, France), Nexxium 50 (NEX) (Sigma Coatings, Amsterdam, The Netherlands), and one AF coating synthesized at the MAPIEM laboratory (SL04A) (Ecopaint PACA Program).
TABLE 1.
Reference (PVC) and AF coating characteristics, including measured water contact angles (n = 5)a
| Coating | Manufacturer | Biocide | Mean water contact angle (°) ± SD |
|---|---|---|---|
| PVC | 73.5 ± 1.7 | ||
| IT757 | International Paint Ltd. | 99.6 ± 2.0 | |
| SQT | Jotun | Cu2O-CuPy (ZnO) | 90.9 ± 0.8 |
| IS360 | International Paint Ltd. | Cu2O-ZnPy | 88.2 ± 1.2 |
| NEX | Sigma Coatings | Econea-Sea Nine | 78.5 ± 1.6 |
| SL04A | MAPIEM-USTV | Cu2O-A4S-Sea Nine-Zineb (ZnO) | 81.9 ± 1.8 |
| A3S | Nautix | Cu2O-CuSCN-ZnPy | 83.3 ± 0.9 |
Cu2O, copper oxide; CuPy, copper pyrithione; CuSCN, copper thiocyanate; ZnPy, zinc pyrithione. Zinc oxide (ZnO) is not considered a biocide by regulations but was added if present in the coating formulation. IT757 is an FRC.
For each coating, three panels (two 5- by 5-cm panels for flow cytometry and fingerprinting analyses and one 10- by 10-cm panel for diatom analysis) were prepared in triplicate. The size of the panel was chosen to collect enough biological material for the analyses. AF paints were applied onto sandblasted PVC panels, painted (one side) by using a 300-μm bar coater, and dried for 15 days. PVC panels were sandblasted to obtain a surface as rough as that of the panels coated with current self-polishing antifouling coatings. The average sand grain size is 0.15 to 0.255 mm (Corindon Brown ABC 80; ARENAblast). All the panels were then drilled at the top and bottom in order for them to be fixed to the racks. Experimental facilities were previously described and validated (35, 37). Immersion under static conditions at a 1-m depth was carried out for 30 days, from 10 June to 10 July 2010.
Water contact angle.
The wettability of the surfaces was evaluated by measuring the water contact angle (1.0 μl deionized water) using a Digidrop contact angle meter (GBX Instrument). Surfaces were previously cleaned with deionized water, five water droplets were deposited, and the mean water contact angle was determined for each sample.
Flow cytometry analyses.
One 5- by 5-cm panel was meticulously and totally scraped by using a scalpel, and the collected biofilms were fixed with 4 ml of a 2% formaldehyde–artificial seawater (ASW) (Sigma-Aldrich) sterile solution. The removal was performed identically for all the panels and until the leaching layer of the coatings was partly scraped to ensure that the biofilms were removed totally. Samples were then frozen in liquid N2 and maintained at −80°C until analysis. Prior to the analysis, an experimental procedure for freshwater periphyton was applied to release and separate the microbial cells from the EPS matrix (38).
Flow cytometry is a very powerful technology to perform single-particle (cell) analysis at a high frequency (up to several thousand particles per second) (39). FCM aims to discriminate particles/cells into clusters based on statistical analyses of the set of optical variables (light scatter and various fluorescence intensities) collected by the flow cytometer for each single particle. Cytograms display correlated data from any of the two parameters, and manual gating is performed to define the cell clusters and obtain various types of information (cell numbers and basic statistics such as average intensities and standard deviations, etc.). FCM analyses were performed at the Regional Flow Cytometry Platform for Microbiology (PRECYM) (Aix-Marseille University, Mediterranean Institute of Oceanography) on a MoFlo cell sorter (Beckman-Coulter) equipped with a dual-line, water-cooled argon laser (λex, 351 nm; λex, 488 nm) (Enterprise II; Coherent). All data were collected on a log scale, stored in list mode, and analyzed with the Summit software package (Beckman-Coulter, Miami, FL). Cell concentrations were determined from the flow rate of the instrument calculated by weighing 3 tubes of sampled seawater before and after a 3-min run of the cytometer. Calibration beads were also used as quality controls to monitor the stability of the instrument (further described below).
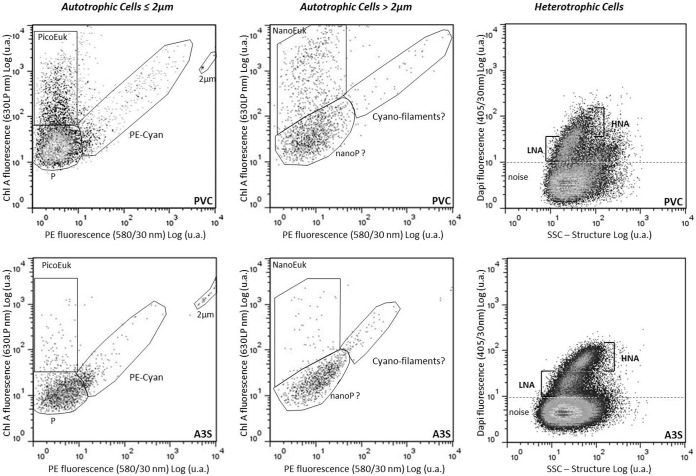
For heterotrophic prokaryote detection by flow cytometry (referred as “bacteria”), 5 μl of a 4′,6-diamidino-2-phenylindole (DAPI) solution (1 mg · ml−1) was added to 995 μl of sample and incubated during 15 min at room temperature in the dark. DAPI is a nucleic acid dye used to induce blue fluorescence when excited by UV (λex = 351 nm). This fluorescence, collected in the range of 405/30 nm (i.e., 405 nm ± 15 nm) and combined with right-angle light scatter (sideward-scatter) intensity, unambiguously distinguishes groups of bacteria, defined by their nucleic acid contents, from inorganic particles, detritus, and free DNA. While green fluorescent dyes such as Sybr green and Picogreen, etc., have been widely used for flow cytometry due to a cheaper light source excitation, DAPI or other UV-excitable dyes, such as Hoechst family dyes, have also been used with success to analyze bacteria by flow cytometry (40, 41). Bacteria were also distinguished from picoautotrophs due to their lack of red fluorescence induced by chlorophyll a or orange fluorescence induced by phycoerythrin. The side-scatter signal related to the size of the bacteria was used as a trigger signal (42). The biological significance of the clusters evidenced by flow cytometry was summarized previously by Bouvier et al. (41). For optical resolution and enumeration of autotrophs, the red fluorescence intensity (630LP ChloroA) related to the chlorophyll a content was used as a trigger signal. Cells were characterized by their scattering signals (with forward scatter related to cell size and side scatter related to cell granularity and shape), and orange fluorescence (580/30 nm) related to phycoerythrin. Before analysis, samples were supplemented with solutions of 2-μm and 6-μm beads (Fluoresbrite; Polysciences). The 2-μm beads were used as an internal standard to discriminate picoautotrophic (<2 μm) and nanoautotrophic (>2 μm) cells in the biofilms. Two groups of autotrophic eukaryotes, characterized by their size and chlorophyll a content, were defined, namely, the picoeukaryotes (PicoEuk) (≤2 to 3 μm) and the nanoeukaryotes (NanoEuk) (>3 μm) (Fig. 2). Although two to three clusters have been separated among the NanoEuk group, in the absence of further characterization, they were considered a single group. Phycoerythrin-rich picocyanobacteria (PE-CYAN) were also discriminated from the previous photosynthetic microorganisms due to their strong orange fluorescence emission. An additional group of picoplankton was discriminated on the basis of its low ChloroA and PE contents.
FIG 2.
Flow cytometry cytograms resolving bacteria, characterized by two populations based on their DAPI fluorescence and structure, discriminating low and high nucleic acid contents (LNA and HNA, respectively) (right) and picoautotrophs (<2 μm), where three populations are distinguished by their red (630LP, chlorophyll A) and orange (580/30 nm, phycoerythrin) fluorescences and named phycoerythrin-rich picocyanobacteria (PE-CYAN), picoeukaryotes (PicoEuk), and picoorganisms (P) (left and middle), isolated from biofilms scraped from two different coatings, PVC and AS3. u.a., arbitrary units; SSC, side scatter.
Biofilm bacterial DNA extraction, amplification, and DGGE analysis.
At the end of immersion, the second small panel (5 by 5 cm) per replicate was washed with sterile artificial seawater. All samples were brought back quickly to the laboratory in a cool box. Biofilm samples were collected by scraping. Water samples were filtered through a 0.2-μm Nuclepore filter. DNA extraction was performed on all samples by using a Power Soil DNA isolation kit (MoBio, Ozyme, St. Quentin, France) according to the manufacturer's instructions. The integrity of the total DNA was checked by agar gel electrophoresis and quantified by determining the absorbance at 260 nm. PCR amplification of the prokaryotic 16S rRNA gene fragments was performed by using 20 ng of template DNA and primers 358f, to which a GC-rich fragment was attached (5′-CGC CCG CCG CGC CCC GCG CCC GTC CCG CCG CCC CCG CCC GCC TAC GGG AGG CAG CAG-3′), and 907rM (5′-CCG TCA ATT C[AC]T TTG AGT TT-3′), which yielded a fragment of ∼590 bp. Both primers targeted the V3-V5 region of the 16S rRNA gene (43, 44).
PCR amplifications were performed in 50-μl volumes containing 5 μl 10× PCR buffer (TaKaRa), 3 μl of a deoxynucleoside triphosphate (dNTP) mixture (2.5 mM each), 1 μl of each primer (12.5 pmol/μl), bovine serum albumin (final concentration, 0.5 mg ml−1; Eurobio), and 1.25 U Taq DNA polymerase (La Taq; TaKaRa). All amplifications were performed by means of the same PCRs using a T-Personal thermal cycler (Biometra) (45). For each set of reactions, a negative control was included, in which the DNA solution was replaced with an equivalent volume of sterile deionized water. DGGE analyses were performed by using the INGENYphorU-2*2 system (Ingeny International). PCR products were separated on a 1-mm-thick vertical gel containing 6% (wt/vol) polyacrylamide, with a linear gradient of the denaturing agents urea and formamide, which increased from 40% to 80% for the 16S rRNA gene amplicons. Individual lines in the gel were loaded with 25 μl of the PCR product. Electrophoresis was performed in 1× Tris-acetate-EDTA (TAE) buffer at 60°C, at a constant voltage of 100 V for 16 h. Nucleic acids were stained for 45 min by immersion in 1× TAE buffer containing Sybr gold (1:5,000 final concentration; Molecular Probes). A digitized image of the DGGE gel was obtained by using a Bio-Rad digital camera and analyzed by using GelCompar II software (Applied Math NV), by determining the position of each nucleic acid band, leading to a matrix based on the presence/absence of bands.
Identification and quantification of fouling microalgae.
At the end of the immersion period, the large panels (10 by 10 cm) were washed with sterile ASW and brushed, and the removed materials were suspended in 10 ml of sterile ASW before Lugol's fixative (final concentration, 0.3% [vol/vol]) was added. Samples were brought back to the laboratory, where identification and cell enumeration were performed. Cells (in 1-ml subsamples) were allowed to settle down for 2 h before being examined under an inverted phase-contrast microscope (magnification, ×400) (Leica DM1 4000 B). At least 100 cells or 30 fields were identified mainly at the genus level according to a French standardized method (NF EN 15-204) (59, 60). In addition to densities, biovolumes were also determined to better estimate the percentage of coverage of the coatings. All of the diatoms were approximated as cylinders (46), except Licmophora, which requires a more complex mathematical model (47). The length and width of 25 organisms per taxa (representing >5% of the total community) were measured.
Procedure for evaluation of the antifouling performance of coatings.
Coated panels were immersed in the same site in the Toulon Bay during 12 months to determine the long-term efficacy of the same coatings studied for microbial colonization. A French standard practice (NF T 34-552) was adapted to assess the AF efficacy of coatings immersed in natural seawater under static conditions. This standard requires reporting of (i) the type of macrofoulers attached to the surface and (ii) the estimated percentage of the surface covered by each type of macrofouler (intensity factor [IF]). To achieve visual determination of macrofouling and to avoid any edge effects on the intensity of colonization, larger panels (29.7 by 21 cm, in duplicate) are recommended by this standard. The inspection was performed 1 cm from the edges of the panel. An efficacy parameter, N, was defined as N = Σ (IF × SF), where SF is the severity factor, which takes into account the frictional drag penalty of ship hulls attributable to increased surface roughness due to foulers (37). SF takes into account composite biofouling, which includes the definition of “hard” and “soft” foulers (1). Soft fouling typically consists of slime layers of bacteria, diatoms, fungi, and protozoa but also green, brown, and red algae; tunicates; hydroids; and anemones. Hard fouling is made of organisms with a calcareous or siliceous structure. The dominant forms of hard fouling are barnacles, tubeworms, and bryozoans. Bivalves such as mussels and oysters are also defined as hard fouling. The latter foulers may dramatically decrease the performance of the coatings.
Statistical analysis.
One-way analysis of variance (ANOVA) and Tukey's posttests were applied to the abundance of the different periphytic communities (GraphPad Prism 5). Values were previously converted to logarithms to become normally distributed for the statistical analysis. Statistical significance was accepted at a P value of <0.05. Spearman nonparametric correlations (r) between the densities of the different groups of organisms, N, and contact angles were also determined.
The DGGE presence/absence matrix was analyzed by using the Jaccard dissimilarity index. Relative abundances of diatom species were compared by using the Bray-Curtis dissimilarity index. Dissimilarity matrices were used to perform multidimensional scale (MDS) analyses. In addition, these matrices of communities were compared by using Mantel's matrix randomization test (48) with Pearson's correlation and 10,000 permutations. All these analyses were performed by using XLSTAT.
RESULTS
Water wettability of the coatings.
Static water contact angles displayed widespread values, from 73.5° ± 1.7° to 99.6° ± 2° (Table 1). Surfaces with water contact angles of <90° were considered hydrophilic surfaces, which was the case for most of the coatings, except IT757, which was consequently considered the only hydrophobic surface. However, SQT and IS360 were noted have water contact angles close to 90°. Finally, Tukey's post hoc tests following ANOVA provided the following order for the hydrophilic coatings: PVC < NEX < SL04A/A3S < SQT/IS360.
Microbial community on coatings characterized by FCM.
Cell densities provided by FCM revealed that PVC always exhibited the highest densities whatever the FCM microbial clusters, except for PE-CYAN (Fig. 2 and 3). Among the various groups identified, heterotrophic bacteria displayed the highest densities, between 1.3 × 106 ± 6.0 × 105 and 1.7 × 107 ± 8.5 × 106 cells/cm2 (Fig. 3). Significant differences compared to PVC were observed only for IS360 and more clearly for NEX and A3S. Among the autofluorescent microorganisms, picoplankton exhibited the highest densities (between 4.9 × 104 ± 1.1 × 104 and 7.2 × 106 ± 8.7 × 105 cells/cm2), except on IS360, where PE-CYAN was most abundant. PicoEuk seemed to be the less abundant group (between 6.1 × 103 ± 2.7 × 103 and 5.6 × 105 ± 2.5 × 104 cells/cm2), with no significant difference between coatings, except for PVC (P < 0.001). A similar pattern was observed for NanoEuk, which presented no significant difference between coatings, except for PVC (P < 0.001), but showed higher densities than those of PicoEuk (between 3.0 × 103 ± 1.6 × 103 and 8.6 × 105 ± 1.8 × 105 cells/cm2).
FIG 3.
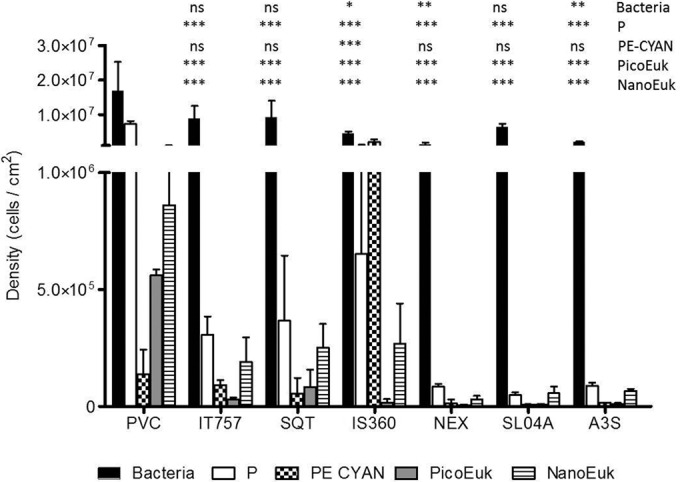
Microorganism density (cells/cm2) on the seven coatings, determined by using flow cytometry. Data for NanoEuk, PicoEuk, low-pigmented picoorganisms (P), PE-CYAN, and bacteria are shown. * (P < 0.05), ** (P < 0.01), and *** (P < 0.001) indicate that the density of microorganisms was significantly different on the coating compared to that on PVC. ns, not significant.
IT757, the only FRC in the study, exhibited no significant difference from the biocidal coatings, whatever the group of organisms, except for PE-CYAN on IS360 (P < 0.001).
A3S appeared to be the less colonized coating. Significant differences from PVC were noted for all groups of organisms, except for PE-CYAN. Conversely, no significant difference appeared for IS360, except with PE-CYAN. Finally, A3S, NEX, and SL04A exhibited no significant differences.
FCM also discriminated several clusters of bacteria based on their nucleic acid contents. Flow cytometry is ataxonomic; however, it makes the discrimination of bacterial clusters possible, as cell fluorescence intensity is induced by DAPI staining. In this study, as the nucleic acid contents varied significantly among bacteria, it was possible to discriminate between up to two clusters: a cluster with a relatively high nucleic acid (HNA) content and a cluster with a relatively low nucleic acid (LNA) content (Fig. 2). Among the coatings, when significant differences were observed (Fig. 4), the LNA cluster was always the dominant group (A3S) (P < 0.05) and was the only cluster detected in two cases (IS360 and SL04A).
FIG 4.
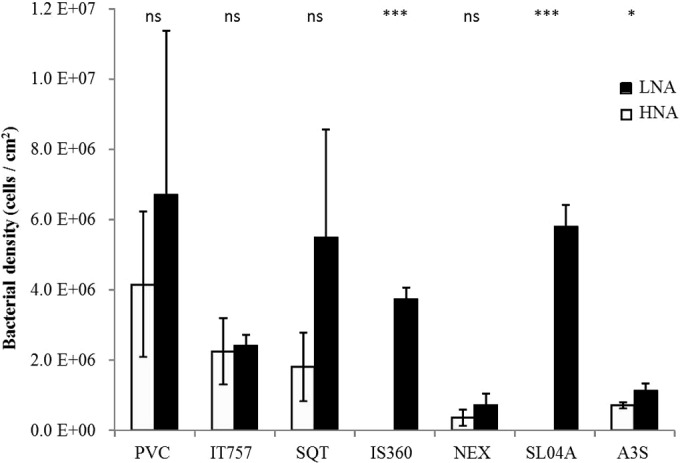
LNA versus HNA bacterial density (cells/cm2), determined by using flow cytometry. * (P < 0.05) or ns (not significant) indicates that the HNA/LNA ratio was significantly different or not on the coating compared to that on PVC.
Diversity of the prokaryotic community on coatings determined by using PCR-DGGE.
All samples (except for one from which the DNA extract was lost) were analyzed in triplicate on the same DGGE gel, and richness was evaluated as DGGE operational taxonomic units (OTUs) (Table 2). DGGE profiles showed that bacteria were present on all the coatings. Up to 53 different OTUs were detected. The richness ranges from 18 ± 2 to 28 ± 2 OTUs, together with a good repeatability for the triplicate biofilm samples. Only NEX and SL04A displayed significantly lower richness than that displayed by PVC. The richness on SQT or A3S was not significantly higher than that on PVC. IS360 appeared to have intermediate richness because no significant difference was observed compared to PVC on the one hand and NEX and SL04A on the other hand.
TABLE 2.
Bacterial richness after DGGE of PCR-amplified 16S rRNA gene fragmentsa
| Coating | Mean richness (no. of OTUs) ± SD | P value |
|---|---|---|
| PVC | 25 ± 2 | |
| IT757 | 23 ± 0 | NS |
| SQT | 28 ± 2 | NS |
| IS360 | 21 ± 2 | NS |
| NEX | 19 ± 3 | <0.05 |
| SL04A | 18 ± 2 | <0.05 |
| A3S | 28 ± 2 | NS |
P values indicate that the richness value was significantly different or not on the coating compared to PVC (n = 3 [n = 2 for IT575]). NS, not significant.
Only 4 OTUs (8%) were common to six among the seven coatings, 8 OTUs were common to <25% of samples, and 4 OTUs were specific for one coating. All these findings suggest that bacterial communities present significant structural differences.
Data from MDS analysis based on DGGE qualitative data are presented in Fig. 5. MDS was used to transform a proximity matrix (similarity in the case of the Jaccard index) between a series of n objects (here 20 16S rRNA gene sequence) to the coordinates of these same objects in a 2-dimensional space so that the objects and their proximity may be visualized easily. All triplicates appeared to be well grouped. Nonbiocidal coatings, i.e., PVC and IT757, appeared separately from biocidal coatings. No clustering between biocidal coatings was observed, which indicates that each of them exhibited a specific prokaryotic community.
FIG 5.
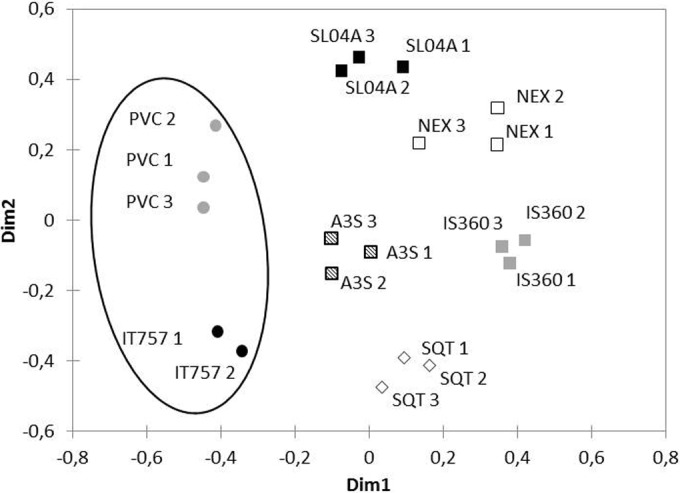
MDS graph showing Jaccard similarity indices of prokaryotic diversity analyzed by DGGE of PCR-amplified 16S rRNA gene fragments for the seven coatings. Dim1 and Dim2, dimensions 1 and 2, respectively.
Structure of the fouling diatom community on coatings.
Microphytobenthos identified in the biofilms on the various coatings belongs exclusively to the class Bacillariophyceae, i.e., diatoms. Only rare taxa (<3%) were centric (Podocystis sp., Delphineis sp., and Coscinodiscus sp.) (data not shown). Dominant to subdominant taxa were all pennates (Table 3 and Fig. 6). Depending on the way in which diatom abundance was expressed (cells/cm2 or cm3/cm2), dissimilar results were observed for NEX, SQT, and, to a lesser extent, IT757.
TABLE 3.
Diatom diversity, richness, and dominant species for biovolumes
| Coating | Mean H′ ± SD |
Mean richness ± SD | Species (% abundance)a |
||
|---|---|---|---|---|---|
| Density | Biovolume | Dominant | Subdominant | ||
| PVC | 1.7 ± 0.2 | 1.2 ± 0.2 | 9 ± 1 | Navicula spp. (59), Licmophora sp. (34) | |
| IT757 | 2.7 ± 0.2 | 2.5 ± 0.4 | 14 ± 5 | Navicula spp. (26) | Nitzschia spp. (21), Cylindrotheca sp. (18), Licmophora spp. (16), Thalassionema sp. (12) |
| SQT | 2.6 ± 0.2 | 1.1 ± 0.4 | 12 ± 1 | Licmophora spp. (75) | Cylindrotheca sp. (18) |
| IS360 | 2.0 ± 0.3 | 2.0 ± 0.5 | 12 ± 5 | Navicula spp. (38), Nitzschia spp. (28) | Thalassionema sp. (14) |
| NEX | 2.0 ± 0.3 | 0.9 ± 0.1 | 11 ± 1 | Thalassionema sp.(85) | |
| SL04A | 2.5 ± 0.1 | 2.0 ± 0.1 | 11 ± 1 | Navicula spp. (36), Licmophora spp. (29) | Nitzschia spp. (18) |
| A3S | 2.3 ± 0.3 | 2.2 ± 0.3 | 10 ± 4 | Licmophora spp. (28) | Nitzschia spp. (24), Cylindrotheca sp. (20), Thalassionema sp. (15) |
Dominant species were present at >25% abundance, and subdominant species were present at abundances between 25 and 10%.
FIG 6.
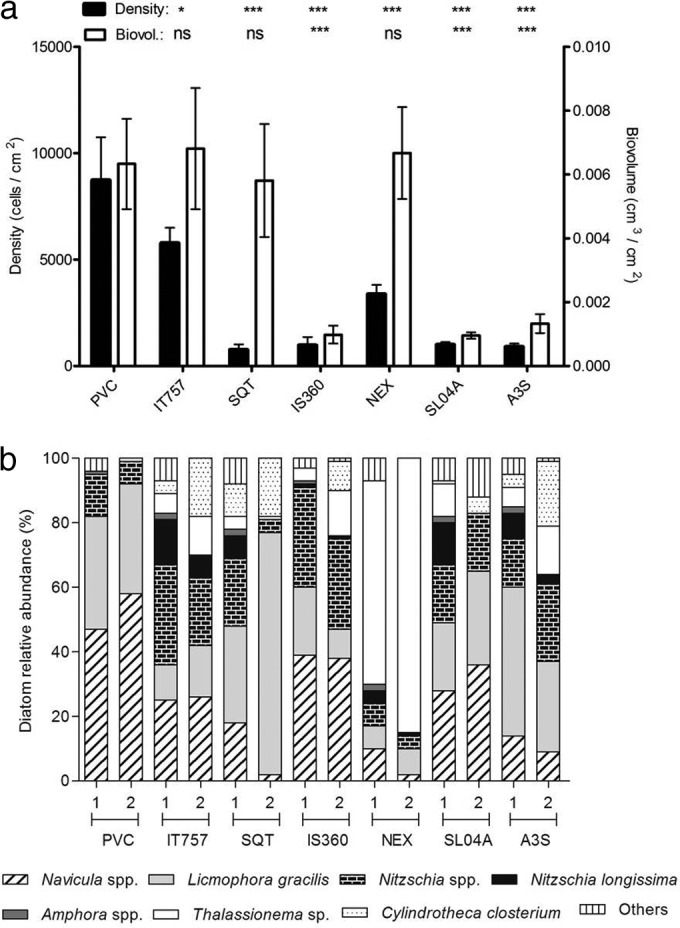
Diatom total (a) and relative (b) abundances on the seven coatings expressed in both cells/cm2 and biovolumes (cm3/cm2). * (P < 0.05) and *** (P < 0.001) indicate that diatom density or biovolume was significantly different on the coating compared to that on PVC. ns (not significant) indicates that diatom density or biovolume on the coating was not significantly different compared to that on PVC.
Total density was the highest on PVC; however, no significant difference were observed with IT757, SQT, and NEX when biovolume was used (Fig. 6a). In both cases, A3S, SL04A, and IS360 exhibited similarly low abundances.
Up to 40 diatom taxa were identified at the genus or species level. Navicula spp. were dominant (>25%) for PVC, IT757, IS360, and SL04A (Fig. 6b and Table 3). Licmophora spp., including L. gracilis, were dominant for PVC, SQT, SL04A, and A3S. Nitzschia spp. (excluding N. longissima) appeared among the dominant taxa only for IS360. Thalassionema sp. was substantially dominant on NEX (85%), especially when based on biovolume, like Licmophora spp. on SQT (75%). Richness values were similar for all coatings (P > 0.05) (Table 3), while significant differences were observed among the Shannon index (H′) values. Based on density, H′ was significantly lower for PVC than for IT757 (P < 0.01), SQT (P < 0.05), and SL04A (P < 0.05). Based on biovolume, more differences in terms of diversity were observed. PVC, SQT, and NEX displayed similarly low values (P > 0.05), significantly lower than those for IT757 (P < 0.01) and A3S (P < 0.05). H′ was also lower for SQT and NEX than for IS360 and SL04A (P < 0.05).
MDS analysis based on both qualitative and quantitative data expressed as biovolumes (Fig. 7) evidenced one cluster where the IS360, SL04A, and A3S replicates are mixed, whereas the four other coatings (PVC, IT757, SQT, and NEX) were separated.
FIG 7.
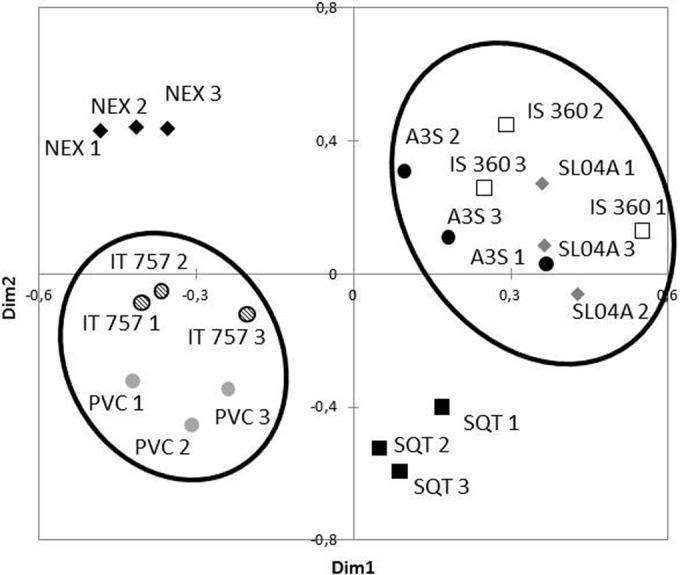
MDS graph showing Bray-Curtis dissimilarity indices of diatom abundance and diversity based on biovolumes for the seven coatings.
Mantel's test showed significant correlations between the similarity matrices of the prokaryotic communities (DGGE) and the diatom communities expressed in cells/cm2 (r = 0.49; P < 0.0001; n = 20) as well as in biovolumes (r = 0.53; P < 0.0001; n = 20).
Antifouling performance of the coatings.
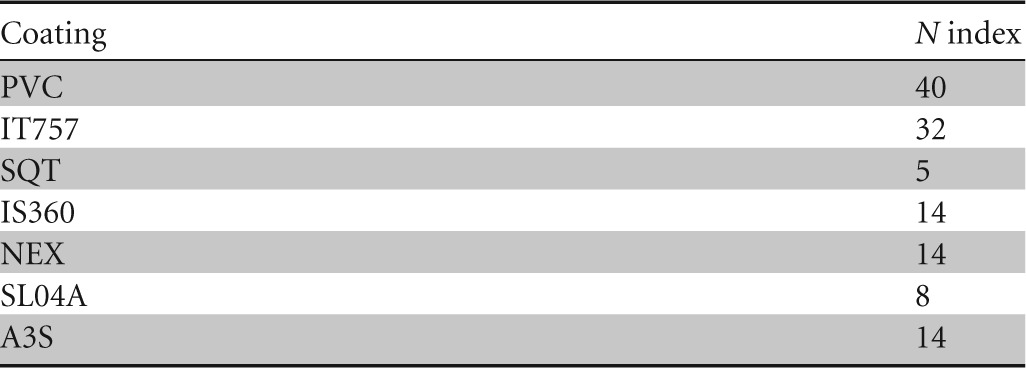
N indices were widespread, ranging from 40 for PVC to 5 for SQT after 12 months (Table 4). Bryozoans and tubeworms could more easily settle down on IT757 panels, whereas brown algae, sponges, ascidians, and barnacles were additionally found on PVC panels (data not shown). Most SPC coatings exhibited only slime and small amounts of algae or hydroids on their surface for samples with N values of >5.
TABLE 4.
N index (macrofouling) after 12 months of exposure in the Toulon Bay for the seven coatings after 1 year of immersion
DISCUSSION
Biofilm microorganisms detected on AF coatings.
Only few studies on biofilms in marine ecosystems have been dedicated to artificial surfaces, especially AF coatings. Pioneer stages of biofouling on artificial surfaces without AF properties are related to the ability of some pioneer bacteria to adhere and to produce extracellular polymeric substances (EPS). These EPS may consolidate the three-dimensional structure of the biofilm, participating in further colonization by other microorganisms, including diatoms, which may themselves produce EPS. Biofilm communities, especially diatoms, are selected from planktonic taxa but exhibit dissimilar community structures and generally lower diversity (e.g., see references 7, 19, 35, and 49). However, bacteria that showed the greatest ability to colonize surfaces seemed to belong to dominant planktonic taxa, at least at a high taxonomic level in some marine studies (see references 6 and 11). However, colonization should not be considered a random process but rather a competition and also a collaboration between microorganisms, influenced by the environmental conditions (bottom-up control by nutrient availability, irradiance, or streams as well as top-down control by grazing).
In this study, flow cytometry was used to quantify pico- and nanoorganisms. While this technique is extensively used for marine planktonic communities, especially for picoplankton enumeration, only three recent articles have reported its use for marine biofilms (22–24), and none of them are related to AF coatings. FCM has the advantage of providing accurate cell abundances for the different subcommunities evidenced by their optical properties, which is not possible with either fingerprinting or deep-sequencing approaches. Depending on the pretreatment used to separate the biofilm matrix, aggregates may influence the determination of cell numbers. However, when aggregates occur, they are usually detected by FCM, as they provide different signals than single particles (the area of the signals collected for doublets is larger than that for single cells). In this study, no cluster of doublets was optically resolved, meaning that there were not enough doublets to bias the cell counts. As far as filaments are concerned, when they are analyzed, in general, they produce particular signals as well (50). In addition to heterotrophic bacterial counts performed after nucleic acid staining, two photosynthetic subcommunities of picobacteria, including PE-rich picocyanobacteria (Synechococcus) and what were recently called “Prochlorococcus-like” picobacteria (22), but also photosynthetic pico- and nanoeukaryotes have been detected and quantified. In this study, Prochlorococcus-like cells are described as the “picoplankton” population. A noticeable observation is the dominance of picoplankton compared to PE-CYAN in the biofilms, which is contradictory to reports for coastal waters if picoplankton were considered Prochlorococcus cells (e.g., see references 51 and 52). Due to the small fluorescence and light scatter intensities of the “events” forming the picoplankton population, one could argue that some debris could also be responsible for the detection of this group. However, staining with DAPI provides a fluorescent signal proving the presence of nucleic acids and therefore the presence of cells. Some bacteria in the Roseobacter clade are aerobic anoxygenic phototrophic bacteria possessing bacteriochlorophyll. This pigment can be excited by a UV laser beam, and its emission is located in the near-infrared region. Unfortunately, the near-infrared photons produced by bacteriochlorophyll are not very energetic compared to photons with shorter wavelengths, and the photomultiplier tubes available at present (including those of the MoFlo cell sorter used in this study) are not efficient enough in catching and converting enough photons to photoelectrons as the cells cross the laser beam (in a few microseconds only). Special dedicated photodetectors should be specifically developed to be implemented on a flow cytometer. A sorting procedure followed by a sequencing approach should provide better identification of this group, which was not possible in this study due to the lack of samples.
In this study, significant differences were evidenced between heterotrophic bacteria, picocyanobacteria, and pico- and nanoeukaryotes from biofilms on AF coatings, as assessed by flow cytometry. Until now, FCM was probably rarely used because it requires single cells in suspension, which involves pretreatment of the biofilm in order to separate the cells (38). This preliminary step could also partly explain the high standard deviations between replicates. Indeed, photosynthetic cells are delicate structures that may be degraded during treatment. Cyanobacteria are especially known to quickly release their pigments (phycoerythrin or phycocyanin) when they start to degrade. It is interesting that in analyses of biofilms that displayed overall low densities after short immersion times (3 days [22] and 8 days [24]) or of biofilms developed in 3 weeks but in a cold immersion site (23), no pretreatment step was performed. It should be noted that in previous studies by Bouchez et al. (24) and Toupoint et al. (23), only one group of picocyanobacteria was mentioned.
Among heterotrophic bacteria, HNA/LNA status, as determined by DAPI staining, varies from no significant difference to dramatic dominance for members of the LNA cluster. Previous studies observed mainly HNA dominance on glass surfaces (e.g., see reference 23). The meaning of these populations remains uncertain and leads to several interpretations, as described previously by Bouvier et al. (41) (various bacterial strains and different physiological states). In addition to the size of bacteria, the active state of the cells was proposed to explain the low DNA content. Nevertheless, Wang et al. (53) showed that during growth after a sorting procedure, characteristics of LNA bacteria (low induced fluorescence intensity and sideward scatter) remained distinct from those of typical HNA bacteria. In addition, as revealed in the literature, several authors have been able to define multiple regions within heterotrophic bacterial cytograms, which indicates a more complex situation in terms of nucleic acid content diversity (e.g., see references 54 and 55).
In the experiment reported here, the prokaryotic richness estimated by DGGE was similar to the richness of marine biofilms obtained by using the same DGGE methodology (18) or of stream biofilm communities obtained by using other fingerprinting methods such as terminal restriction fragment length polymorphism (T-RFLP) analysis (49). Compared to our previous study at the same site in Toulon Bay (35), richness appeared to be similar for PVC but higher for AF coatings (SQT, IS360, and SL04A). The similar richness on PVC after 15 days or 1 month of immersion may indicate that a “stable” state was already reached. Also, considering that most of the OTUs on the coatings (including PVC) were different after 1 month of immersion, unlike the assemblages observed during the short-immersion-time study, where about 25% of the OTUs were shared, our results suggest that biofilm communities diversified faster on PVC than on AF coatings, probably due to biocide diffusion time. Indeed, biocide release induces the selection of resistant bacteria that could delay the stabilization of the richness level of the community; furthermore, SPC included several biocides with different release rates and modes of action. Fingerprinting methods target the most abundant taxa but show reliable community composition patterns. However, obviously, these approaches fail to generate a realistic pattern of the entire diversity compared to high-throughput sequencing methods such as 454 pyrosequencing (49). In addition, rare taxa represented a significant part of the diversity, about 50% (56), especially in biofilms compared to planktonic communities (49). Until now, prokaryotic diversity in marine biofilms on artificial surfaces has been addressed only by using fingerprinting or fluorescence in situ hybridization (FISH) (e.g., see references 9, 19, and 24), cloning-sequencing (11), or PhyloChip (10) studies, and AF coatings were investigated only by using culture-dependent (30, 57) or fingerprinting (19, 35) approaches.
As far as eukaryote diversity is concerned, Bouchez et al. (24) reported various community patterns according to nutrient availability by using a 18S rRNA gene-based fingerprinting approach. Only Toupoint et al. (23), using cloning-sequencing approaches, reported organisms other than diatoms (mainly ciliates, dinoflagellates, and Chlorophyceae). However, those authors failed to detect diatoms by using this molecular approach (but diatoms were identified by scanning electron microscopy). This highlights the issue of the need for up-to-date universal, or at least generalist, primers for eukaryotes adapted to next-generation sequencing (NGS) techniques, as underlined by Taib et al. (58), together with enrichment of international databases with microbial eukaryote sequences. Consequently, literature on the study of the eukaryotic community, especially for diatoms in marine biofilms, remains limited.
Concerning microphytobenthos, only Bacillariophyceae (diatoms) were observed by microscopy. Identified diatom taxa corresponded to the ones already reported as being dominant on AF coatings (Licmophora, Navicula, and Nitzschia), except for Thalassionema on NEX. The issue with the classic expression of density in cells/cm2 is related to the fact that diatom cell size can vary from a few to hundreds of micrometers. Consequently, considering the same density, the coverage rate of the coating could actually be different depending on the species. Thus, the density could be estimated more accurately by using biovolumes. In contrast with our previous study, which reported pioneer stages of colonization (35), and in accordance with DGGE patterns, diatom dominance but also diversity patterns appeared to vary significantly from one coating to another. We assume that climax could be reached after 1 month of immersion, depending on the environmental conditions. Overall, richness and abundance increased, whereas diversity (H′) did not clearly change with increasing time of immersion in the Toulon Bay. For the coatings addressed in this study, H′ values stayed in the same range (1.7 < H′ <2.7), rather low compared to the values obtained in the only reported study on AF coatings where H′ was determined (33). The only FRC showed the highest diversity but the same dominant taxa as those on PVC or SPC. The increase of the immersion time allowed us to clearly see that AF coatings select the microbial communities at their surfaces.
Biofilm densities controlled by AF coatings.
Whatever the type of microorganism, PVC clearly exhibits the highest densities, and A3S exhibits the lowest (Fig. 3 and 6). Overall, about 20 million microorganisms appeared to colonize PVC panels (5 by 5 cm), compared to 2 million for A3S, including in both cases >70% of bacteria. Heterotrophic bacteria are always the most abundant microorganisms whatever the coatings, generally followed by picoplankton populations, which highlights the need for identification of this group of pigmented microorganisms (“Prochlorococcus-like”). Overall, among all the coatings tested, two groups and two single coatings emerged in terms of microbial communities. Surprisingly, the only nonbiocidal coating, IT757 (FRC), did not appear to be frankly different from SQT, with high bacterial densities and diatom biovolumes not significantly different from those obtained on PVC. In fact, relatively speaking, all the microorganisms displayed high densities on these coatings, which constituted the first group of highly colonized coatings. Considering that FRC are known to be efficient essentially under dynamic conditions, this is not surprising for IT757. The determination of the type of microbial community capable of colonizing this coating under such dynamic conditions would be of great interest. As FRC have been reported to fail to inhibit diatom colonization (e.g., see references 26 and 29), the determination of the structure of such diatom communities would be of great interest. IS360 exhibited a singular profile, with especially high levels of PE-CYAN, picoplankton, and NanoEuk together with low diatom densities (and biovolumes). This apparent contradictory result concerning photosynthetic microorganisms was also noted for NEX. Overall, PE-CYAN, picoplankton, and NanoEuk were correlated (P < 0.05) (Table 5). This finding confirmed that photosynthetic microorganisms cannot be considered a single homogenous box regarding their sensitivity to biocides. The second group included two efficient coatings, NEX and SL04A. However, these coatings failed to prevent colonization by diatoms and heterotrophic bacteria, respectively. Finally, A3S is considered the only coating able to control the whole microbial community in this study, performed in the Toulon Bay in July.
TABLE 5.
Spearman nonparametric correlations between densities of the different groups of organisms, N, and contact angles on the coatings (n = 7)a
| Parameter |
r value for correlate |
|||||||
|---|---|---|---|---|---|---|---|---|
| Bacteria | P | PE-CYAN | PicoEuk | NanoEuk | Diatoms (cells/cm2) | Diatoms (cm3/cm2) | Water contact angle (°) | |
| Bacteria | ||||||||
| P | 0.607 | |||||||
| PE-CYAN | 0.429 | 0.929** | ||||||
| PicoEuk | 0.893* | 0.857* | 0.714 | |||||
| NanoEuk | 0.714 | 0.964** | 0.893* | 0.893* | ||||
| Diatoms (cells/cm2) | 0.214 | 0.321 | 0.393 | 0.214 | 0.250 | |||
| Diatoms (cm3/cm2) | 0.143 | 0.179 | 0.179 | 0.250 | 0.000 | 0.571 | ||
| Water contact angle (°) | 0.107 | 0.071 | 0.250 | 0.214 | 0.107 | −0.357 | 0.107 | |
| N | 0.185 | 0.445 | 0.519 | 0.334 | 0.371 | 0.889* | 0.556 | −0.259 |
Significant correlations are shown in boldface type, with P values of <0.05 (*) and <0.01 (**).
Considering diatoms, the FRC IT757 showed a high abundance with a high level of diversity (biovolumes), whereas PVC and two biocidal coatings (SQT and NEX) displayed a similarly high abundance but with a low level of diversity, and the three other biocidal coatings (A3S, SL04A, and IS360) showed opposite characteristics (low abundance with a high level of diversity). In similar static immersion studies, FRC were reported to display a higher level of diversity than biocidal coatings (26), whereas similar diversity levels have also been observed (33). It is notable that relative abundances for the different taxa clearly varied from one coating to another.
The ability of coatings to control microbial communities was clearly not related to their wettabilities, as exhibited in Table 5. The relative role of each biocide remains difficult to determine because the coatings are composed of biocide mixtures, up to five in the same coating (Table 1). Nevertheless, copper thiocyanate (CuSCN), which appeared to be the only difference between IS360 and A3S, seemed to significantly affect PE-CYAN, picoplankton, and NanoEuk. Considering IS360 and SQT, copper pyrithione (CuPy) seemed to control PE-CYAN and picoplankton populations, whereas zinc pyrithione (ZnPy) was more efficient on heterotrophic bacteria and diatoms. Finally, NEX, the only biocidal coating that did not possess copper, was particularly colonized by one diatom species, Thalassionema sp. (85%), which indicates a high sensitivity to copper for this species.
Relationships between populations.
Spearman nonparametric correlations showed that PE-CYAN, picoplankton, and NanoEuk densities were altogether correlated (P < 0.05) (Table 5), which confirms similar sensitivities to biocides. On the contrary, heterotrophic bacterial densities were correlated only with PicoEuk densities. However, a Mantel test showed a significant correlation between the dissimilarity matrices of the prokaryotic communities (DGGE) and the diatom communities (in cells/cm2 [r = 0.49; P < 0.0001; n = 20] as well as in biovolumes [r = 0.53; P < 0.0001; n = 20]). This finding indicates that the ways in which these communities were controlled by coatings are probably more qualitatively than quantitatively related. In this study, the global abundance of prokaryotes on coatings appeared to be influenced to a lesser extent than the structure of the related communities. For subsequent settlement of diatoms, which showed patterns more significantly different, what is the relative importance of these prokaryotic communities versus the coating characteristics? This relationship remains to be understood, as already noted by several studies (16, 29). A more powerful methodology such as NGS would help to better characterize prokaryotic and eukaryotic communities and understand their control by AF coatings.
An additional aim of this study was to investigate the potential for biofilms to serve as a proxy for antifouling coating efficacy. Macrofouling observed on the same coatings at the same site after 12 months of immersion, assessed by the N factor, exhibited no quantitative correlation with bacterial or picoorganism abundances. Conversely, a significant correlation with diatom abundance (cells/cm2) was noted. This result, which remains to be confirmed with a larger number of coatings, could be of interest for the use of biofilms, and especially diatoms, to anticipate the macrofouling efficacy of AF coatings. The communication between biofilms and algae or invertebrate propagules has been clearly established (e.g., see references 5 and 6), but the main biofilm characteristics that control macrofouling settlement remain to be determined.
ACKNOWLEDGMENTS
We acknowledge the financial support of the Direction Générale des Armées, which is part of the French Ministry of Defense, in the framework of the European Defense Agency program Antifouling Coatings for War Ships.
We also thank S. Lafond for paint formulation (SL04A) and her help in surface painting and contact angle measurements.
Footnotes
Published ahead of print 6 June 2014
REFERENCES
- 1.Schultz MP, Bendick JA, Holm ER, Hertel WM. 2011. Economic impact of biofouling on a naval surface ship. Biofouling 27:87–98. 10.1080/08927014.2010.542809 [DOI] [PubMed] [Google Scholar]
- 2.Piola RF, Dafforn KA, Johnston EL. 2009. The influence of antifouling practices on marine invasions. Biofouling 25:633–644. 10.1080/08927010903063065 [DOI] [PubMed] [Google Scholar]
- 3.Schultz MP, Swain GW. 2000. The influence of biofilms on skin friction drag. Biofouling 15:129–139. 10.1080/08927010009386304 [DOI] [PubMed] [Google Scholar]
- 4.Qian P-Y, Lau SCK, Dahms H-U, Dobretsov S, Harder T. 2007. Marine biofilms as mediators of colonization by marine macroorganisms: implications for antifouling and aquaculture. Mar. Biotechnol. 9:399–410. 10.1007/s10126-007-9001-9 [DOI] [PubMed] [Google Scholar]
- 5.Hadfield MG. 2011. Biofilms and marine invertebrate larvae: what bacteria produce that larvae use to choose settlement sites. Annu. Rev. Mar. Sci. 3:453–470. 10.1146/annurev-marine-120709-142753 [DOI] [PubMed] [Google Scholar]
- 6.Salta M, Wharton J, Blache Y, Stokes K, Briand J-F. 2013. Marine biofilms on man-made surfaces: structure and dynamics. Environ. Microbiol. 15:2879–2893. 10.1111/1462-2920.12186 [DOI] [PubMed] [Google Scholar]
- 7.Jones P, Cottrell M, Kirchman D, Dexter S. 2007. Bacterial community structure of biofilms on artificial surfaces in an estuary. Microb. Ecol. 53:153–162. 10.1007/s00248-006-9154-5 [DOI] [PubMed] [Google Scholar]
- 8.Hung OS, Thiyagarajan V, Qian PY. 2008. Preferential attachment of barnacle larvae to natural multi-species biofilms: does surface wettability matter? J. Exp. Mar. Biol. Ecol. 361:36–41. 10.1016/j.jembe.2008.04.011 [DOI] [Google Scholar]
- 9.Huggett MJ, Nedved BT, Hadfield MG. 2009. Effects of initial surface wettability on biofilm formation and subsequent settlement of Hydroides elegans. Biofouling 25:387–399. 10.1080/08927010902823238 [DOI] [PubMed] [Google Scholar]
- 10.Chung HC, Lee OO, Huang YL, Mok SY, Kolter R, Qian PY. 2010. Bacterial community succession and chemical profiles of subtidal biofilms in relation to larval settlement of the polychaete Hydroides elegans. ISME J. 4:817–828. 10.1038/ismej.2009.157 [DOI] [PubMed] [Google Scholar]
- 11.Dang H, Lovell C. 2000. Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes. Appl. Environ. Microbiol. 66:467–475. 10.1128/AEM.66.2.467-475.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dang H, Lovell C. 2002. Numerical dominance and phylotype diversity of marine Rhodobacter species during early colonization of submerged surfaces in coastal marine waters as determined by 16S ribosomal DNA sequence analysis and fluorescence in situ hybridization. Appl. Environ. Microbiol. 68:496–504. 10.1128/AEM.68.2.496-504.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dang H, Li T, Chen M, Huang G. 2008. Cross-ocean distribution of Rhodobacterales bacteria as primary surface colonizers in temperate coastal marine waters. Appl. Environ. Microbiol. 74:52–60. 10.1128/AEM.01400-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobretsov S, Abed RMM, Voolstra CR. 2013. The effect of surface colour on the formation of marine micro and macrofouling communities. Biofouling 29:617–627. 10.1080/08927014.2013.784279 [DOI] [PubMed] [Google Scholar]
- 15.Molino PJ, Childs S, Hubbard MRE, Carey JM, Burgman MA, Wetherbee R. 2009. Development of the primary bacterial microfouling layer on antifouling and fouling release coatings in temperate and tropical environments in Eastern Australia. Biofouling 25:149–162. 10.1080/08927010802592917 [DOI] [PubMed] [Google Scholar]
- 16.Dobretsov S, Thomason JC. 2011. The development of marine biofilms on two commercial non-biocidal coatings: a comparison between silicone and fluoropolymer technologies. Biofouling 27:869–880. 10.1080/08927014.2011.607233 [DOI] [PubMed] [Google Scholar]
- 17.Faÿ F, Carteau D, Linossier I, Vallée-Réhel K. 2011. Evaluation of anti-microfouling activity of marine paints by microscopical techniques. Prog. Org. Coat. 72:579–585. 10.1016/j.porgcoat.2011.04.002 [DOI] [Google Scholar]
- 18.Webster NS, Negri AP. 2006. Site-specific variation in Antarctic marine biofilms established on artificial surfaces. Environ. Microbiol. 8:1177–1190. 10.1111/j.1462-2920.2006.01007.x [DOI] [PubMed] [Google Scholar]
- 19.Shikuma NJ, Hadfield MG. 2010. Marine biofilms on submerged surfaces are a reservoir for Escherichia coli and Vibrio cholerae. Biofouling 26:39–46. 10.1080/08927010903282814 [DOI] [PubMed] [Google Scholar]
- 20.Dang H, Chen R, Wang L, Shao S, Dai L, Ye Y, Guo L, Huang G, Klotz MG. 2011. Molecular characterization of putative biocorroding microbiota with a novel niche detection of Epsilon- and Zetaproteobacteria in Pacific Ocean coastal seawaters. Environ. Microbiol. 13:3059–3074. 10.1111/j.1462-2920.2011.02583.x [DOI] [PubMed] [Google Scholar]
- 21.Bacchetti De Gregoris T, Khandeparker L, Anil AC, Mesbahi E, Burgess JG, Clare AS. 2012. Characterisation of the bacteria associated with barnacle, Balanus amphitrite, shell and their role in gregarious settlement of cypris larvae. J. Exp. Mar. Biol. Ecol. 413:7–12. 10.1016/j.jembe.2011.11.014 [DOI] [Google Scholar]
- 22.Mitbavkar S, Raghu C, Rajaneesh KM, Pavan D. 2012. Picophytoplankton community from tropical marine biofilms. J. Exp. Mar. Biol. Ecol. 426-427:88–96. 10.1016/j.jembe.2012.05.022 [DOI] [Google Scholar]
- 23.Toupoint N, Mohit V, Linossier I, Bourgougnon N, Myrand B, Olivier F, Lovejoy C, Tremblay R. 2012. Effect of biofilm age on settlement of Mytilus edulis. Biofouling 28:985–1001. 10.1080/08927014.2012.725202 [DOI] [PubMed] [Google Scholar]
- 24.Bouchez A, Pascault N, Chardon C, Bouvy M, Cecchi P, Lambs L, Herteman M, Fromard F, Got P, Leboulanger C. 2013. Mangrove microbial diversity and the impact of trophic contamination. Mar. Pollut. Bull. 66:39–46. 10.1016/j.marpolbul.2012.11.015 [DOI] [PubMed] [Google Scholar]
- 25.Patil JS, Anil AC. 2005. Biofilm diatom community structure: influence of temporal and substratum variability. Biofouling 21:189–206. 10.1080/08927010500256757 [DOI] [PubMed] [Google Scholar]
- 26.Bressy C, Margaillan A, Faÿ F, Linossier I, Vallée-Réhel K. 2009. Tin-free self-polishing marine antifouling coatings, p 445–491 In Hellio C, Yebra DM. (ed), Advances in marine antifouling coatings and technologies. Woodhead Publishing Ltd, Cambridge, United Kingdom [Google Scholar]
- 27.Lejars M, Margaillan A, Bressy C. 2012. Fouling release coatings: a nontoxic alternative to biocidal antifouling coatings. Chem. Rev. 112:4347–4390. 10.1021/cr200350v [DOI] [PubMed] [Google Scholar]
- 28.Anderson C, Atlar M, Callow M, Candries M, Milne A, Townsin R. 2003. The development of foul-release coatings for seagoing vessels. J. Mar. Des. Oper. B 4:11–23 [Google Scholar]
- 29.Molino PJ, Wetherbee R. 2008. The biology of biofouling diatoms and their role in the development of microbial slimes. Biofouling 24:365–379. 10.1080/08927010802254583 [DOI] [PubMed] [Google Scholar]
- 30.Cassé F, Swain GW. 2006. The development of microfouling on four commercial antifouling coatings under static and dynamic immersion. Int. Biodeterior. Biodegradation 57:179–185. 10.1016/j.ibiod.2006.02.008 [DOI] [Google Scholar]
- 31.Chen C-L, Maki JS, Rittschof D, Teo SL. 2013. Early marine bacterial biofilm on a copper-based antifouling paint. Int. Biodeterior. Biodegradation 83:71–76. 10.1016/j.ibiod.2013.04.012 [DOI] [Google Scholar]
- 32.Pelletier E, Bonnet C, Lemarchand K. 2009. Biofouling growth in cold estuarine waters and evaluation of some chitosan and copper anti-fouling paints. Int. J. Mol. Sci. 10:3209–3223. 10.3390/ijms10073209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zargiel KA, Coogan JS, Swain GW. 2011. Diatom community structure on commercially available ship hull coatings. Biofouling 27:955–965. 10.1080/08927014.2011.618268 [DOI] [PubMed] [Google Scholar]
- 34.Molino PJ, Campbell E, Wetherbee R. 2009. Development of the initial diatom microfouling layer on antifouling and fouling-release surfaces in temperate and tropical Australia. Biofouling 25:685–694. 10.1080/08927010903089912 [DOI] [PubMed] [Google Scholar]
- 35.Briand J-F, Djeridi I, Jamet D, Coupé S, Bressy C, Molmeret M, Le Berre B, Rimet F, Bouchez A, Blache Y. 2012. Pioneer marine biofilms on artificial surfaces including antifouling coatings immersed in two contrasting French Mediterranean coast sites. Biofouling 28:453–463. 10.1080/08927014.2012.688957 [DOI] [PubMed] [Google Scholar]
- 36.Callow ME. 1986. Fouling algae from ‘in-service' ships. Bot. Mar. 29:351–358 [Google Scholar]
- 37.Bressy C, Briand J-F, Compère C, Réhel K. 2014. Laboratory assays and field test evaluation method, p 332–345 In Williams D, Thomason J, Dobretsov S. (ed), Biofouling methods. Wiley-Blackwell, Oxford, United Kingdom [Google Scholar]
- 38.Villeneuve A, Montuelle B, Bouchez A. 2010. Influence of slight differences in environmental conditions (light, hydrodynamics) on the structure and function of periphyton. Aquat. Sci. 72:33–44. 10.1007/s00027-009-0108-0 [DOI] [Google Scholar]
- 39.Shapiro HM. 2003. Practical flow cytometry, 4th ed, p 681 John Wiley & Sons, Inc, Hoboken, NJ [Google Scholar]
- 40.Robertson BR, Button DK. 1989. Characterizing aquatic bacteria according to population, cell size, and apparent DNA content by flow cytometry. Cytometry 10:70–76. 10.1002/cyto.990100112 [DOI] [PubMed] [Google Scholar]
- 41.Bouvier T, del Giorgio PA, Gasol JM. 2007. A comparative study of the cytometric characteristics of high and low nucleic-acid bacterioplankton cells from different aquatic ecosystems. Environ. Microbiol. 9:2050–2066. 10.1111/j.1462-2920.2007.01321.x [DOI] [PubMed] [Google Scholar]
- 42.Troussellier M, Courties C, Lebaron P, Servais P. 1999. Flow cytometric discrimination of bacterial populations in seawater based on SYTO13 staining of nucleic acids. FEMS Microbiol. Ecol. 29:319–330. 10.1111/j.1574-6941.1999.tb00623.x [DOI] [Google Scholar]
- 43.Schauer M, Massana R, Pedros-Alio C. 2000. Spatial differences in bacterioplankton composition along the Catalan coast (NW Mediterranean) assessed by molecular fingerprinting. FEMS Microbiol. Ecol. 33:51–59. 10.1111/j.1574-6941.2000.tb00726.x [DOI] [PubMed] [Google Scholar]
- 44.Sanchez O, Gasol JM, Massana R, Mas J, Pedros-Alio C. 2007. Comparison of different denaturing gradient gel electrophoresis primer sets for the study of marine bacterioplankton communities. Appl. Environ. Microbiol. 73:5962–5967. 10.1128/AEM.00817-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dorigo U, Leboulanger C, Bérard A, Bouchez A, Humbert JF, Montuelle B. 2007. Lotic biofilm community structure and pesticide tolerance along a contamination gradient in a vineyard area. Aquat. Microb. Ecol. 50:91–102. 10.3354/ame01133 [DOI] [Google Scholar]
- 46.Smayda TJ. 1978. From phytoplankton to biomass, p 273–279 In Sournia A. (ed), Phytoplankton manual. Monographs on oceanographic methodology 6. UNESCO, Paris, France [Google Scholar]
- 47.Hillebrand H, Dürselen C-D, Kirschtel D, Pollingher U, Zohary T. 1999. Biovolume calculation for pelagic and benthic microalgae. J. Phycol. 35:403–424. 10.1046/j.1529-8817.1999.3520403.x [DOI] [Google Scholar]
- 48.Mantel N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res. 27:209–220 [PubMed] [Google Scholar]
- 49.Besemer K, Peter H, Logue JB, Langenheder S, Lindstrom ES, Tranvik LJ, Battin TJ. 2012. Unraveling assembly of stream biofilm communities. ISME J. 6:1459–1468. 10.1038/ismej.2011.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Dijk MA, Grégori G, Hoogveld HL, Rijkeboer M, Denis M, Malkassian M, Gons HJ. 2010. Optimizing the setup of a flow cytometric cell sorter for efficient quantitative sorting of long filamentous cyanobacteria. Cytometry A 77:911–924. 10.1002/cyto.a.20946 [DOI] [PubMed] [Google Scholar]
- 51.Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR, Post AF, Hagemann M, Paulsen I, Partensky F. 2009. Ecological genomics of marine picocyanobacteria. Microbiol. Mol. Biol. Rev. 73:249–299. 10.1128/MMBR.00035-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bec B, Collos Y, Souchu P, Vaquer A, Lautier J, Fiandrino A, Benau L, Orsoni V, Laugier T. 2011. Distribution of picophytoplankton and nanophytoplankton along an anthropogenic eutrophication gradient in French Mediterranean coastal lagoons. Aquat. Microb. Ecol. 63:29–45. 10.3354/ame01480 [DOI] [Google Scholar]
- 53.Wang Y, Hammes F, Boon N, Chami M, Egli T. 2009. Isolation and characterization of low nucleic acid (LNA)-content bacteria. ISME J. 3:889–902. 10.1038/ismej.2009.46 [DOI] [PubMed] [Google Scholar]
- 54.Müller S, Nebe-von-Caron G. 2010. Functional single-cell analyses: flow cytometry and cell sorting of microbial populations and communities. FEMS Microbiol. Rev. 34:554–587. 10.1111/j.1574-6976.2010.00214.x [DOI] [PubMed] [Google Scholar]
- 55.Günther S, Koch C, Hübschmann T, Röske I, Müller RA, Bley T, Harms H, Müller S. 2012. Correlation of community dynamics and process parameters as a tool for the prediction of the stability of wastewater treatment. Environ. Sci. Technol. 46:84–92. 10.1021/es2010682 [DOI] [PubMed] [Google Scholar]
- 56.Pommier T, Neal PR, Gasol JM, Coll M, Acinas SG, Pedrós-Alió C. 2010. Spatial patterns of bacterial richness and evenness in the NW Mediterranean Sea explored by pyrosequencing of the 16S rRNA. Aquat. Microb. Ecol. 61:221–233. 10.3354/ame01484 [DOI] [Google Scholar]
- 57.Inbakandan D, Murthy PS, Venkatesan R, Khan SA. 2010. 16S rDNA sequence analysis of culturable marine biofilm forming bacteria from a ship's hull. Biofouling 26:893–899. 10.1080/08927014.2010.530347 [DOI] [PubMed] [Google Scholar]
- 58.Taib N, Mangot J-F, Domaizon I, Bronner G, Debroas D. 2013. Phylogenetic affiliation of SSU rRNA genes generated by massively parallel sequencing: new insights into the freshwater protist diversity. PLoS One 8:e58950. 10.1371/journal.pone.0058950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.AFNOR. 2006. NF EN 15204. Qualité de l'eau: norme guide pour le dénombrement du phytoplancton par microscopie inversée (methode Utermohl). AFNOR, Paris, France [Google Scholar]
- 60.AFNOR. 1996. NF T 34-552. Peintures et vernis—systèmes de peintures pour la protection des ouvrages en acier—essai d'immersion au radeau en eau de mer vive. AFNOR, Paris, France [Google Scholar]