Abstract
Carrion decomposition is an ecologically important natural phenomenon influenced by a complex set of factors, including temperature, moisture, and the activity of microorganisms, invertebrates, and scavengers. The role of soil microbes as decomposers in this process is essential but not well understood and represents a knowledge gap in carrion ecology. To better define the role and sources of microbes in carrion decomposition, lab-reared mice were decomposed on either (i) soil with an intact microbial community or (ii) soil that was sterilized. We characterized the microbial community (16S rRNA gene for bacteria and archaea, and the 18S rRNA gene for fungi and microbial eukaryotes) for three body sites along with the underlying soil (i.e., gravesoils) at time intervals coinciding with visible changes in carrion morphology. Our results indicate that mice placed on soil with intact microbial communities reach advanced stages of decomposition 2 to 3 times faster than those placed on sterile soil. Microbial communities associated with skin and gravesoils of carrion in stages of active and advanced decay were significantly different between soil types (sterile versus untreated), suggesting that substrates on which carrion decompose may partially determine the microbial decomposer community. However, the source of the decomposer community (soil- versus carcass-associated microbes) was not clear in our data set, suggesting that greater sequencing depth needs to be employed to identify the origin of the decomposer communities in carrion decomposition. Overall, our data show that soil microbial communities have a significant impact on the rate at which carrion decomposes and have important implications for understanding carrion ecology.
INTRODUCTION
Understanding the ecology of carrion decomposition is important for ecosystem ecology and has implications for forensic science. When an animal dies, it becomes an ephemeral nutrient pulse, which stimulates a rapid response from organisms across the tree of life (1), including bacteria, archaea, and microbial eukaryotes. As this microbial decomposer community recycles nutrients, the corpse progresses through six recognized stages of decomposition (fresh, bloated, active decay, advanced decay, dry, and remains) reported by Payne (2) and further described by Carter and colleagues (1). Soil microbial communities are likely important contributors to the decomposer community in terrestrial ecosystems. It is well known that soil microbes play a critical role in mineralizing much of the annual primary production of terrestrial ecosystems (3, 4). For instance, soil fungi produce cellulolytic and oxidative enzymes capable of breaking down the different chemical constituents of plant litter (5). It is likely that similar soilborne microbial processes are important for carrion breakdown. However, carrion decomposition is complicated by the fact that both the soil and the host are replete with microbes, many of which are capable of mineralizing the macromolecules present (nucleic acids, proteins, etc.) in vertebrate flesh.
Several recent studies have begun to reveal the complex dynamics associated with decomposing mammals, including mice and swine carrion, as well as human cadavers (6–8). Although these studies each involved different mammalian taxa decomposing on different soil substrates, they each discovered a significant change in microbial communities during decomposition. For example, Pechal et al. (8) discovered a significant and succession-like change in bacterial communities on skin sites of swine carrion that were decomposing in an outdoor scenario on native soil of temperate forest soils in Ohio. Moreover, Metcalf et al. (7) demonstrated a similar succession-like trend in abdominal, skin, and gravesoil bacterial and microbial eukaryotic communities associated with mouse cadavers decomposing in a lab setting using soils collected from a riparian shrubland in the foothills of Colorado. Importantly, Metcalf et al. (7) discovered that as decomposition progressed, similar microbial taxa became abundant in and on the mouse cadavers as well as the associated gravesoils. For example, in in the abdominal cavity and on the skin, Gammaproteobacteria become highly abundant after rupture (7). Furthermore, the nematode Oscheius tipulae dominated microbial eukaryotic communities during late-stage decomposition at all sites, including the gravesoils.
The extent to which the presence of an endogenous soil microbial community influences the decomposer community dynamics of carrion decomposition is not well understood. Therefore, we investigated microbial communities of decomposing mice in the presence and in the absence of an endogenous soil microbial community. By comparing the mouse decomposition rate on untreated soil (data originally published by Metcalf et al. [7]) to that on soils subjected to three rounds of sterilization, we were able to assess the influence of endogenous soil communities on decomposition rates and decomposer microbial community composition. Our major questions were as follows. (i) Do we see a difference in decomposition rates of mice under the different soil treatments? (ii) Are the microbial communities different in late decomposition rates between the treatment groups (both in composition and in measures of diversity)? (iii) Can we determine from where abundant active and advanced decay decomposer taxa originate (soil, skin, or abdominal cavity)?
MATERIALS AND METHODS
Soil collection, characterization, and treatment.
As previously described by Metcalf et al. (7), we collected soils from the top 10 cm of the soil profile in Eldorado Canyon, CO, sieved through 4-mm mesh to remove rocks, and transported the soil samples to the lab at ambient temperature. The soil is part of the Juget series and classified as sandy-skeletal, mixed, frigid lithic Haplustolls (https://soilseries.sc.egov.usda.gov/OSD_Docs/J/JUGET.html). Once in the lab, water-holding capacity (WHC) was determined by saturating soil in a filter apparatus that was allowed to drain for 30 min. The water remaining in the soil was assumed to represent 100% WHC and was used to calculate the amount of water needed to maintain ∼55 to 60% WHC throughout the decomposition experiment. Half of the collected soil was sterilized by subjecting it to heat and pressure (121°C, 100 kPa for 30 min) in an autoclave 3 times over 4 days to destroy microbes, fungi, and their spores (to the extent possible).
Experimental setup and sampling.
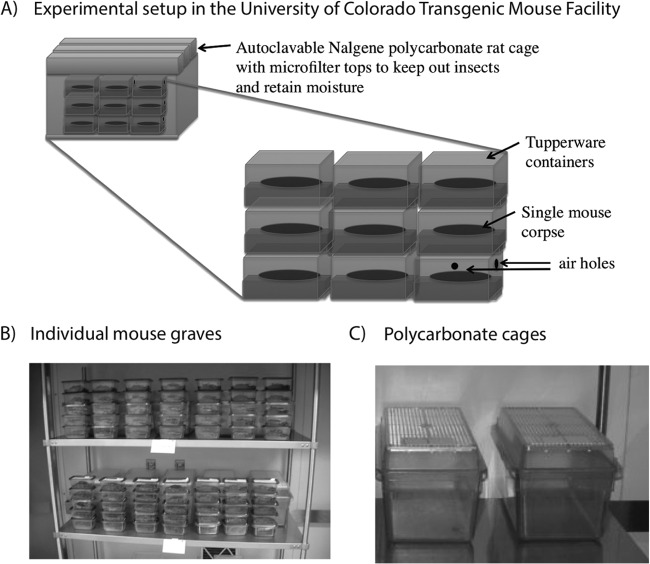
Graves for individual mice were constructed by placing ∼200 to 300 g of untreated or sterile soil in clean polypropylene containers (∼500-ml capacity) with one small hole (∼2 mm) drilled into each side (4 holes per grave) of the container above soil level to allow gas exchange (Fig. 1). We humanely sacrificed (University of Colorado IACUC protocol 1101.02) lab-reared mice (Mus musculus, strain B6C3F1) ranging in age from 36 to 109 days and randomly assigned each carcass (n = 80) to one of eight graves for destructive sampling across time to avoid biases related to age. Each mouse was placed on its right side on top of the soil. We monitored WHC of soils to ensure it remained at ∼55 to 60% throughout the experiment. WHC was maintained by keeping humidity high in the rat cage secondary containers (Fig. 1). We lined cages with soaked lab diaper paper. We also weighed each grave throughout the experiment to monitor water loss. (It was a closed system.) In cases when we detected water loss, we added sterile water to the soil with a spray bottle to bring moisture levels of soil back up to 55 to 60% WHC. We distributed water evenly around the mouse cadaver as best as possible.
FIG 1.
Experimental setup. Each mouse carcass was placed on either sterile or untreated soil in a polypropylene container “grave” with air holes to prevent anaerobic conditions. Tupperware containers were grouped by treatment (sterilized soil versus untreated soil) in secondary filter-top polycarbonate cages.
For each treatment, five replicates were sampled at eight time points over 48 days following the experimental setup (n = 80 total samples). We sampled skin on the head, skin on the belly, and the abdominal cavity of each mouse as well as the soil underneath the mouse while still in an aboveground position on the gravesoil using sterile swabs (BD BBL CultureSwab; Becton, Dickinson). An additional sample (>1 g) of soil was collected for pH measurements. Details on sampling of mouse skin and abdominal cavity can be found in reference 7. At each time point before sampling of the 10 mouse carcasses, we recorded a visual decomposition score for the head and torso following Megyesi et al. (9) (see Table S1 in the supplemental material).
Soil pH measurements.
Soil pH for each grave was estimated by mixing 1 g of soil with 5 ml deionized water followed by vortexing for 1 min. Large particulates were allowed to settle before measurement of the pH with an Orion 3 benchtop pH meter. The average of three replicates was used to estimate the pH for each gravesoil.
DNA extraction and next-generation sequencing.
We followed the extraction and amplification protocols described by Metcalf et al. (7) and the Earth Microbiome project (http://www.earthmicrobiome.org/emp-standard-protocols/) to produce 18S and 16S rRNA amplicons for high-throughput sequencing on the Illumina platform. Briefly, DNA extractions were performed using the 96-well high-throughput PowerSoil DNA isolation kit (MoBio Laboratories, Carlsbad, CA) at the University of Colorado, Boulder, and stored at −20°C until needed. Triplicate PCRs for each sample were performed using primers specific for 18S rRNA (Euk1391-EukBr) (10) or 16S rRNA (515f-806r) (11). Additionally we used a blocking primer (GCCCGTCGCTACTACCGATTGG/ideoxyI//ideoxyI//ideoxyI//ideoxyI//ideoxyI/TTAGTGAGGCCCT/3SpC3/) (12) in conjunction with the 18S primers to minimize the amplification of host ribosomal DNA. The blocking primers reduce the amplification of vertebrate 18S rRNA, which may dominate in pools of genomic DNA derived from host-associated samples. Triplicate PCR mixtures were pooled, quantified (13), and sequenced on the Illumina HiSeq 2000 platform at the BioFrontiers Institute at the University of Colorado.
18S rRNA sequence processing.
Processing of 18S rRNA data was performed in QIIME (14) using the curated SILVA database (version 108) as a reference (15) for open reference operational taxonomic unit (OUT) picking. (The curated version can be found at qiime.org/home_static/dataFiles.html) Sequences with little similarity to the SILVA tree (e.g., less than 70%) were discarded, while the remaining sequences were aligned with PyNAST (16) and used to construct a phylogenetic tree using RAxML (17). Additionally sequences that were similar to 16S rRNA or appeared to be host ribosomal sequences were filtered from the data. Taxonomy assignment was done using the RDP classifier (18) implemented in QIIME. Sequences were rarefied to 1,000 sequences per sample for phylogenetic and taxonomic analyses.
16S rRNA amplicon sequence processing.
We followed the approach outlined by Metcalf et al. (7), in which data for the untreated soil treatment group were originally published, for quality filtering and open reference OTU picking using workflow pipelines available in the software package QIIME (14). We used Greengenes 2012 as a reference data set. The resulting OTUs were aligned and used to construct a phylogenetic tree using PyNAST (16) and FastTree (19), respectively, for downstream analysis. Taxonomy was assigned using the RDP classifier (18) in QIIME. Low-abundance OTUs (those with abundance of <0.005% in the total data set) were discarded (20) prior to analysis. Samples were rarefied to 3,000 sequences per sample for all downstream analyses.
Statistical analysis.
For statistical analysis, duplicates of samples (e.g., abdominal swab and abdominal liquid/syringe) were removed. We explored β diversity patterns by performing principal coordinate analysis (PCoA) with phylogeny-based (UniFrac) unweighted distances (21). We tested whether different soil treatments resulted in significantly different microbial communities by comparing unweighted UniFrac distances with a permutational multivariate analysis of variance (PERMANOVA) test between treatments at each sample site during active/advanced decomposition. α diversity was estimated using the phylogenetic diversity metric Faith's phylogenetic diversity (PD) (22) and tested for significance using the nonparametric option in the compare_alpha_diversity.py command, while differences in taxon abundances were performed using the bootstrapped Mann-Whitney U test in QIIME. We utilized Bayesian source tracking software (23) to better understand which sample sites may have hosted bacterial communities that contributed to the active and advanced decay decomposer communities. Therefore, we used day 0 communities from each sample site as a potential source for all active and advanced decay decomposition communities. Since we sampled the abdominal cavity in three different specific sites at day 0 (fecal, cecum, and abdominal cavity swab/liquid), we used each of these sites as a separate potential source. We tested that each of our potential sources was distinct (untreated soil, sterile soil, abdominal cavity, cecum, fecal, skin of torso, or skin of head).
Nucleotide sequence accession numbers.
DNA sequences have been deposited in the QIIME database as studies 714 and 1889 (http://www.microbio.me/emp) under EBI accession no. ERP003930 and ERP003931.
RESULTS
The effect of the sterilization treatment on soil microbial communities.
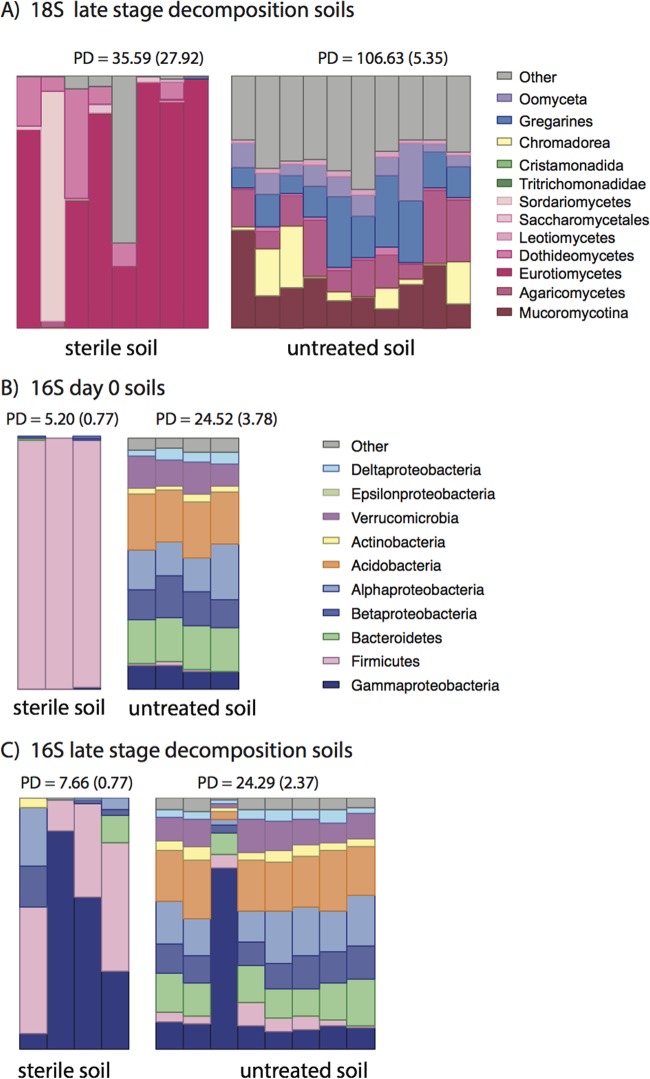
First, we assessed the effect that our soil sterilization treatment had on the endogenous soil microbial community. Our sterilization treatment resulted in a significant decrease in the α diversity of the treated soil relative to the untreated soil (Fig. 2) (see Table S2 in the supplemental material). In fact, for eukaryotic communities, soil samples did not result in robust data (e.g., no site had >2 samples working for a time point) (see Table S3 in the supplemental material) until day 20, after rupture. Thus, only active and advanced decay decomposition eukaryotic samples were analyzed (for the sterile group, days 20 and 34). The effect of sterilization was nonetheless evident even at the active and advanced decay stages of decomposition, as α diversity (Fig. 2; see Table S2) in the sterilized soil was significantly lower than that in the untreated soil and was dominated by two fungal taxa comprising 40% of the eukaryotic community (Eurotiomycetes, 27%; Leotiomycetes, 13%,) across all late decomposition samples in this study. Although still dominated by fungal taxa, untreated soils hosted abundant taxa (>10% of total community in most samples) from Metamonada, Metazoa, Apicomplexa, and Stramenopiles. These groups generally represented less than 1% of the eukaryotic community in sterilized soils.
FIG 2.
Effects of sterilization treatment on soils. Shown is the relative abundance of microbial eukaryotic (18S) late-stage decomposition soils (A), bacterial (16S) taxa in day 0 soils (B), and bacterial taxa in late-stage decomposition soils (C). Each stacked bar shows the relative abundance of eukaryotes or bacteria in a single sample. Microbial eukaryotic DNA was not detected in day 0 soils after sterilization treatment. For 18S, taxa are shaded by phylum: Fungi, red/pink; Excavata, green; Nematoda, yellow; Alveolata, blue; Stramenopiles, purple. Other low-abundance taxa are represented as gray.
The soil bacterial community was also significantly affected by the sterilization treatment. Marked decreases in α diversity and shifts in taxonomic composition were observed in sterilized compared to untreated soils. α diversity was reduced 5-fold (from 24.5 to 5.2, Faith's PD) in the sterilized soil compared to the untreated soil at time zero (Fig. 2; see Table S2 in the supplemental material). Taxonomically the Firmicutes (mostly of the endospore-forming genus Bacillus) became dominant members of the soil bacterial community after sterilization (0.7 to 99%), while the unsterilized soils contained similar abundances of major taxa (Fig. 2). Archaea were in very low abundance in all samples regardless of soil treatment and stage of decomposition.
The effect of the sterilization treatment on mouse decomposition rates.
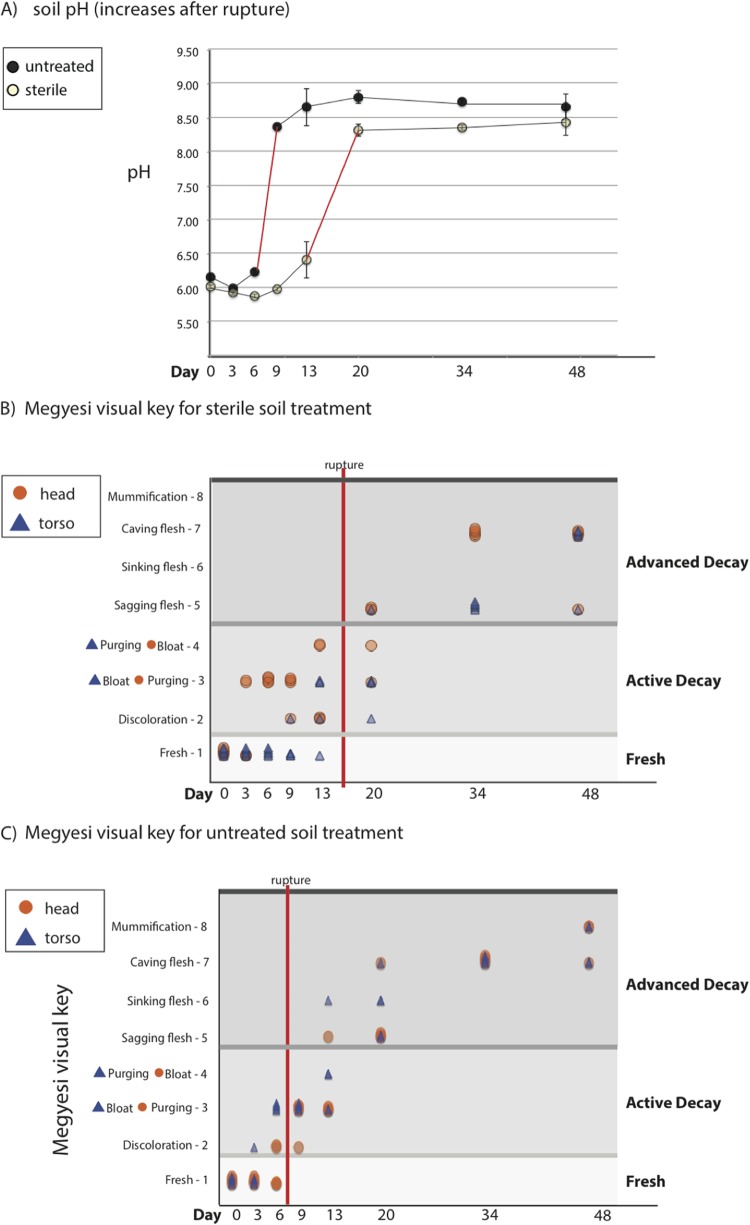
Mice decomposing on the sterilized soil decomposed notably more slowly than mice decomposing on soils with an intact microbial community. For the untreated soil, rupture occurred between days 6 and 9, while for the sterile soil treatment samples, rupture occurred between days 13 and 20 (Fig. 3). The timing of rupture coincides with a predictable increase in soil pH indicative of rupture as ammonia-rich fluids are released into the soil (24). Furthermore, the stage of decomposition was assessed with visual score data following Megyesi et al. (9). We discovered that mice decomposing on untreated soil were in active decay, which includes caving in of flesh and mummification (see Table S1 in the supplemental material), for a much shorter window of time than mice on sterilized soil (Fig. 3). Mice on sterilized soil did not reach advanced decay (see Table S1) until day 20, whereas mice on untreated soil reached advanced decay by day 13.
FIG 3.
(A) Average pH values are shown for untreated (dark circles) and sterilization-treated (open circles) soil for each sampling event (in days since time zero) with standard error bars, for which n = 5. (B and C) Megysei visual keys and points for the skin of the head (orange circles) and the skin of the torso (blue triangles) on sterile (B) and untreated (C) soil. Fresh, 1 point; discoloration, 2 points; bloat-purging, 3 points; purging-bloat, 4 points; sagging flesh, 5 points; sinking flesh, 6 points; caving flesh, 7 points; mummification, 8 points. Purging of the torso (3 points) preceded that of the head (4 points). The general state of carcass decomposition is indicated by the horizontal lines to separate fresh, active, and advanced decay. These data suggest that rupture occurred sometime before 9 days in the untreated experimental group and before 20 days in the sterilization-treated soil experimental group.
The effect of sterilized soils on host microbial communities during decomposition.
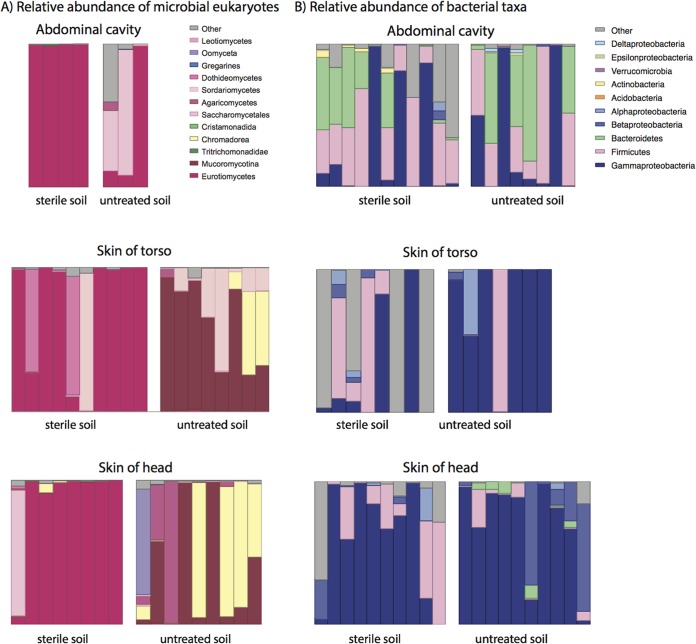
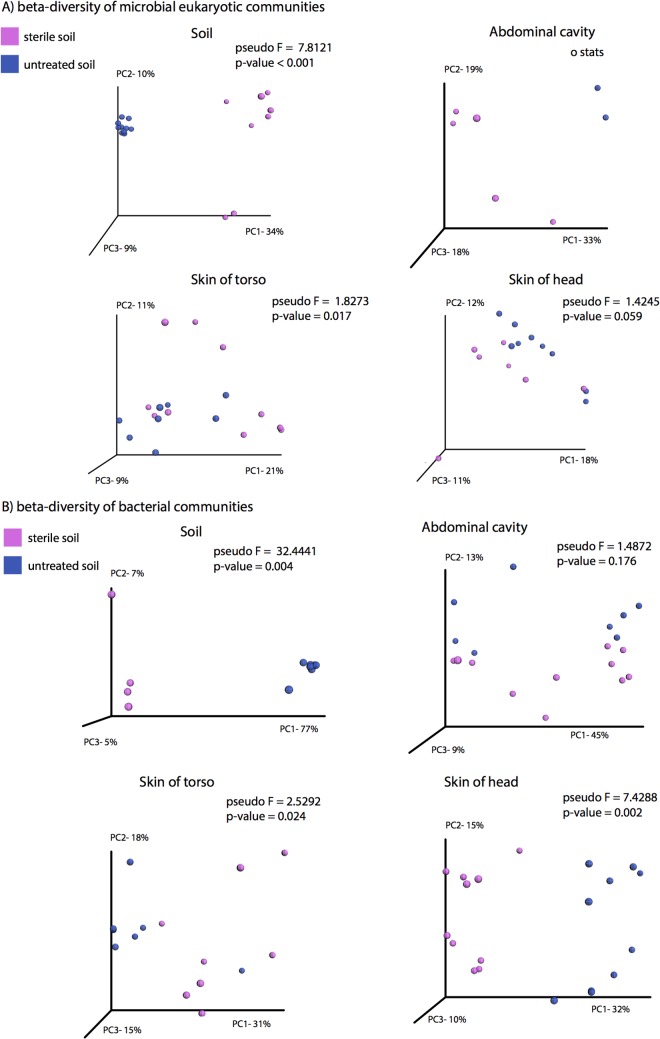
We compared the microbial communities of each treatment group at the two time points following rupture (sterile group, days 20 and 34; untreated group, days 13 and 20) to assess the effect of sterilization on late-stage decomposition during active and advanced decay (Fig. 4 and 5). PERMANOVA comparison of the microbial eukaryotic community unweighted UniFrac (Fig. 5A) distance showed that the skin of the torso (P = 0.017) and soil communities (P = 0.001) were significantly different between the treatments during active/advanced decay, while the skin on the head community was marginally but not significantly affected by sterilization (P = 0.059). These observations are reflected in the shifts in taxonomic composition, in which taxa in the Eurotiomycetes and Mucoromycotina dominated samples from the sterilized and untreated soil treatments, respectively. Eurotinomyetes dominated in all samples from the sterile treatment (Fig. 2; see Table S4 in the supplemental material), comprising a minimum 66% of the eukaryotic community within each sample type. Further, the Eurotinomyetes were by far the most abundant eukaryote in the abdominal cavity and skin of the head, representing greater than 98% of the community during active and advanced decay (Fig. 2; see Table S4). The Mucoromycotina, on the other hand, were most prevalent in samples from the untreated soil, with abundances ranging from 17% in the soil to 62% on the skin of the torso (Fig. 2; see Table S4), being, on average, the most dominant eukaryote from this treatment. Additionally, nematodes of the class Chromadorea, which were identified by Metcalf et al. (7) as the species Oscheius tipulae by sequencing long reads of 16S rRNA genes on the PacBio platform, were more abundant in host-associated and gravesoil samples in the untreated soil group, although they were only significantly more abundant in gravesoil samples when both days 13 and 20 were considered. In the study by Metcalf et al. (7), this nematode was demonstrated to bloom during advanced decomposition (days 20, 34, and 48), likely in response to an increase in bacterial biomass during active decay. Nematodes were not detected in samples from the sterile soil treatment, with the exception of a single sample of skin of head at an abundance of ∼6%, which likely represented contamination. The abundances of both the Eurotiomycetes and Mucoromycotina were significantly affected by the sterilization treatment at late-stage decomposition, as were the abundances of most soil eukaryotes, with the exception of the Dothideomycetes and Sordariomycetes (see Table S4).
FIG 4.
(A) Relative abundance of microbial eukaryotic taxa is shown at the highest level of taxonomic resolution for each sample site and each treatment (sterile versus untreated soil substrate). Each stacked bar shows the relative abundance of eukaryotes or bacteria in a single sample. Taxa are shaded by phylum: Fungi, red/pink; Excavata, green; Nematoda, yellow; Alveolata, blue; Stramenopiles, purple. Other low-abundance taxa are represented as gray. (B) Relative abundance of host-associated, late-stage decomposition bacterial communities with taxa abundance shown at the phylum level. Each bar in the figure represent the relative abundance of a given taxa for a single sample.
FIG 5.
(A) PCoA plot based on unweighted UniFrac distances of microbial eukaryotic communities sampled during late-stage decomposition showing sterile treated soil (pink) versus untreated soil (blue). (B) PCoA plot based on unweighted UniFrac distances of microbial bacterial communities sampled during late-stage decomposition showing sterile treated soil (pink) versus untreated soil (blue).
Similar to the microbial eukaryotic communities, the active and advanced decay bacterial communities of skin and soils were different between the treatments (P ≤ 0.024, PERMANOVA), while those in the abdomen and the skin of the belly were not significantly affected by substrate type (Fig. 4B). There were several consistent trends within treatments in the abundance of bacterial taxa when comparing early and late-stage decomposition (day 0 compared to active/advanced decay). Burkholderia, Novosphingobium, Staphylococcus, and Stenotrophomonas were more abundant on the skin of mice and gravesoils associated with the sterilized soil treatment during active and advanced decay (see Table S5 in the supplemental material). These taxa generally comprised greater than 2% of the bacterial communities but were, for the most part, undetectable in samples from mice placed on untreated soil. In the untreated soil group, Morganella increased in abundance during decomposition in all host-associated communities, representing at least 30% of the bacterial community in each active/advanced decay skin or abdominal sample. Furthermore, Morganella was more abundant in untreated (6.5%) compared to sterilized (0%) gravesoil during active and advanced decay. However, the higher proportion of Morganella in the untreated group was only significant at the skin of the torso (see Table S5). The abundance of Proteus spp. was higher on mice placed on untreated soil, and abundance was significantly different between treatments in both skin sites (head and torso). We also discovered that Bacillus spp. were significantly higher in active and advanced decay sterile soil communities, while “Candidatus Nitrososphaera DA101” (belonging to the Verrucomicrobia), and “Candidatus Solibacter” were more abundant in the untreated soil (see Table S5).
Sources of microbial decomposer communities.
We estimated the potential sources of the active and advanced decay decomposer communities using the Bayesian source tracking software SourceTracker (23) to estimate the proportion of each day 0 community (i.e., soil, abdominal, or skin) that is detectible in each late-stage community. A test of distinctiveness of our sources revealed that each bacterial community was highly distinct and discernible from other source communities (i.e., untreated soil, sterile soil, abdominal cavity, cecum, fecal, skin of torso, or skin of head) (Fig. 6). Late-stage decomposition communities were similar to day 0 skin communities and an unknown community (a community not well represented by any source) in both skin sites as well as the abdominal site. The active and advanced decay untreated gravesoil communities were very similar to the starting soil community. In contrast, the active and advanced decay sterilized soil communities were highly influenced by skin. Interestingly, overall the bacterial communities associated with active and advanced decay (all sites) in the sterile treatment appeared more influenced by bacteria from the skin of the head, while those associated with the untreated soil sites tended to be influenced by bacteria from skin of the torso.
FIG 6.
Bayesian source tracking analysis. Each rectangle is composed of 100 columns representing the mixture of sources estimated in one of 100 Gibbs sampling draws (23). (A) “Leave-one-out” source predictions for day 0 source communities. The distinctness of source communities was tested by leaving each source sample out and estimating its source community. Sources were highly distinct, with a small proportion of misassignments between fecal and cecum communities, which is not surprising given that fecal material passes through the cecum. (B) Bayesian source tracking results of late-stage decomposition bacterial communities using source communities shown in panel A. In cases in which proportions of the late-stage decomposition bacterial community were not accurately represented by a day 0 source, it was assigned to an “unknown community” (shown in gray).
DISCUSSION
Lower rate of decomposition in the absence of a soil microbial community.
Carcass decomposition presents a scenario in which nutrients appear in a pulse and are ephemeral in nature, favoring the growth of any organism that can quickly utilize the resources derived from the carcass. When a carcass decomposes on a soil substrate, the diverse decomposer microbial community includes taxa from across the tree of life, including nematodes, fungi, and bacteria from multiple phyla (Fig. 2). In the absence of a soil microbial community, mice did not reach active and advanced decay until 7 to 10 days after those placed on soil with an endogenous microbial community. The lower rate of carrion decomposition in the sterile group was associated with a less diverse microbial decomposer community (Fig. 2). This observation is not surprising given that many soil microbes can be saprotrophic, especially the fungi, which are well known for their decomposing activities of other types of detritus (25). Furthermore, soil invertebrates play a critical role in breaking down much of the plant detritus deposited on the soil surface (26, 27), and this study suggests that they could also have an impact on the rate at which mammalian carcasses decompose as they were, for the most part, nonexistent in the sterilized soils in this study. Additionally, many bacteria have the capability of producing extracellular enzymes (proteases, cellulases, and nucleases) capable of mineralizing complex macromolecular structures that would be commonly found in a mammalian carcass (28).
Microbial communities differ between the treatment groups during active and advanced decay.
To better understand the potential role of soil microbes in decomposition, we compared community compositions during active and advanced decay across treatments to identify taxa that may be associated with increased rates of decomposition in the untreated group. Several microbial eukaryotes were present in active and advanced decay decomposer communities in the untreated group that were largely absent in the sterile group, such as the fungal group Mucoromycotina. Fungi in this group are known to thrive in nutrient-rich environments and to become abundant during the latter stages of plant litter decomposition (29). These life history strategies appear to be evident in our data as the Mucoromycotina are the dominant members of the eukaryotic community on the skin and in the soil during active and advanced decay, and this suggests that their role as decomposers is not limited to plant material. Additionally, the Eurotiomycetes, which were most abundant during active and advanced decay in samples from the sterilized group, are also commonly found on decomposing plant material. These results suggest that taxa involved in plant decomposition may serve as generalist consumers of labile organic material in the environment.
Assessing the bacterial community, we discovered that only a few genera were different in abundance between treatments during active and advanced decay. Morganella and Proteus were in greater abundance in the untreated group during late decomposition compared to both sterile soil group samples as well as day 0 untreated soil group samples. These proteobacterial genera include as taxa many potential decomposers that are ureolytic and saprophytic (30). Decomposition studies by Pechal et al. and Hyde et al. (6, 8), however, showed that Proteobacteria in general declined and Firmicutes increased in abundance after rupture on swine carcasses and human cadavers, respectively. These samples were derived from different body sites than those used in this study, which may explain the contrasting results. Alternatively, Morganella and Proteus may be opportunistic members of the decomposer community specific to our experiment. Additional studies of bacterial community change associated with decomposing mammalian taxa under different environmental conditions will provide insight to the specificity or universality of bacterial decomposer communities.
Another important factor that may explain the difference between decomposition rates between our treatment groups is α diversity of microbial communities, or the number of microbial taxa participating in the decomposition process. The speed of carrion decomposition may increase in the presence of a more diverse microbial community with a more diverse set of metabolic capabilities. Therefore, soils and other microbially diverse substrates will likely increase decomposition. Moreover, in outdoor scenarios, insects and scavengers may increase the diversity of microbial decomposer communities by introducing microbes through body fluids, defecation, and skin contact.
Source of late-stage microbial decomposer communities.
The dramatic influence of the endogenous soil community on carcass decomposition may indicate that soil microbes are the source of the main decomposer community. Surprisingly, we did not detect a major influence of soil bacteria on carrion skin. Even though the presence of an endogenous soil bacterial community significantly changes the composition of the late-stage skin decomposer community, the soil is not clearly the source of the difference—a more complicated interaction may be responsible. Additionally, we did not detect an influence of abdominal cavity bacteria on the late-stage soil bacterial community, even though abdominal cavity fluids had purged into the soil. It is possible that the abdominal cavity bacteria of larger mammalian taxa, such as deer (31) or bison (32), may have a more significant influence on the soil communities.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Office of Justice Programs award 2011-DN-BX-K533, the Earth Microbiome Project (http://www.earthmicrobiome.org/), and in part by the Howard Hughes Medical Institute. We thank the Colorado University—Boulder MCDB Transgenic Facility and J. Huntley and the Colorado University Next-Generation Sequencing facility. Research capacity and infrastructure at Chaminade University of Honolulu are supported by NIH-BRIC P20MD006084.
Footnotes
Published ahead of print 6 June 2014
Supplemental material for this article may be found at htpp://dx.doi.org/10.1128/AEM.00957-14.
REFERENCES
- 1.Carter DO, Yellowlees D, Tibbett M. 2007. Cadaver decomposition in terrestrial ecosystems. Naturwissenschaften 94:12–24. 10.1007/s00114-006-0159-1 [DOI] [PubMed] [Google Scholar]
- 2.Payne JA. 1965. A summer carrion study of the baby pig Sus scrofa Linneaeus. Ecology 46:592–602. 10.2307/1934999 [DOI] [Google Scholar]
- 3.Dilly O, Bartsch S, Rosenbrock P, Buscot F, Munch JC. 2001. Shifts in physiological capabilities of the microbiota during the decomposition of leaf litter in a black alder (Alnus glutinosa (Gaertn.) L.) forest. Soil Biol. Biochem. 33:921–930. 10.1016/S0038-0717(00)00239-X [DOI] [Google Scholar]
- 4.Sinsabaugh RL, Lauber CL, Weintraub MN, Ahmed B, Allison SD, Crenshaw C, Contosta AR, Cusack D, Frey S, Gallo ME, Gartner TB, Hobbie SE, Holland K, Keeler BL, Powers JS, Stursova M, Takacs-Vesbach C, Waldrop MP, Wallenstein MD, Zak DR, Zeglin LH. 2008. Stoichiometry of soil enzyme activity at global scale. Ecol. Lett. 11:1252–1264. 10.1111/j.1461-0248.2008.01245.x [DOI] [PubMed] [Google Scholar]
- 5.de Boer W, Folman LB, Summerbell RC, Boddy L. 2005. Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 29:795–811. 10.1016/j.femsre.2004.11.005 [DOI] [PubMed] [Google Scholar]
- 6.Hyde ER, Haarmann DP, Lynne AM, Bucheli SR, Petrosino JF. 2013. The living dead: bacterial community structure of a cadaver at the onset and end of the bloat stage of decomposition. PLoS One 8:e77733. 10.1371/journal.pone.0077733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Metcalf JL, Parfrey LW, Gonzalez A, Lauber CL, Knights D, Ackermann G, Humphrey GC, Gebert MJ, Van Treuren W, Berg-Lyons D, Keepers K, Guo Y, Bullard J, Fierer N, Carter DO, Knight R. 2013. A microbial clock provides an accurate estimate of the postmortem interval in a mouse model system. eLife 2:e01104. 10.7554/eLife.01104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pechal JL, Crippen TL, Benbow ML, Tarone AM, Dowd S, Tomberlin JK. 2013. The potential use of bacterial community succession in forensics as described by high throughput metagenomic sequencing. Int. J. Legal Med. 128:193–205. 10.1007/s00414-013-0872-1 [DOI] [PubMed] [Google Scholar]
- 9.Megyesi MS, Nawrocki SP, Haskell NH. 2005. Using accumulated degree-days to estimate the postmortem interval from decomposed human remains. J. Forensic Sci. 50:618–626 [PubMed] [Google Scholar]
- 10.Amaral-Zettler LA, McCliment EA, Ducklow HW, Huse SM. 2009. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS One 4:e6372. 10.1371/journal.pone.0006372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6:1621–1624. 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vestheim H, Jarman SN. 2008. Blocking primers to enhance PCR amplification of rare sequences in mixed samples—a case study on prey DNA in Antarctic krill stomachs. Front. Zool. 5:12. 10.1186/1742-9994-5-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fierer N, Hamady M, Lauber CL, Knight R. 2008. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc. Natl. Acad. Sci. U. S. A. 105:17994–17999. 10.1073/pnas.0807920105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig WG, Peplies J, Glockner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196. 10.1093/nar/gkm864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. 10.1093/bioinformatics/btp636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berger SA, Krompass D, Stamatakis A. 2011. Performance, accuracy, and web server for evolutionary placement of short sequence reads under maximum likelihood. Syst. Biol. 60:291–302. 10.1093/sysbio/syr010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145. 10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26:1641–1650. 10.1093/molbev/msp077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. 2013. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10:57–59. 10.1038/nmeth.2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235. 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faith DP. 1992. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61:1–10. 10.1016/0006-3207(92)91201-3 [DOI] [Google Scholar]
- 23.Knights D, Parfrey LW, Zaneveld J, Lozupone C, Knight R. 2011. Human-associated microbial signatures: examining their predictive value. Cell Host Microbe 10:292–296. 10.1016/j.chom.2011.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer J, Anderson B, Carter DO. 2013. Seasonal variation of carcass decomposition and gravesoil chemistry in a cold (Dfa) climate. J. Forensic Sci. 58:1175–1182. 10.1111/1556-4029.12169 [DOI] [PubMed] [Google Scholar]
- 25.Sagara N. 1995. Association of ectomycorrhizal fungi with decomposed animal wastes in forest habitats—a cleaning symbiosis. Can. J. Bot. 73:S1423–S1433 [Google Scholar]
- 26.Dadour IR, Harvey ML. 2008. The role of invertebrates in terrestrial decomposition: forensic applications. CRC Press, Boca Raton, FL [Google Scholar]
- 27.Smith KGV. 1986. A manual of forensic entomology. British Museum (Natural History) and Cornell University Press, London, United Kingdom [Google Scholar]
- 28.Carter DO, Yellowlees D, Tibbett M. 2010. Moisture can be the dominant environmental parameter governing cadaver decomposition in soil. Forensic Sci. Int. 200:60–66. 10.1016/j.forsciint.2010.03.031 [DOI] [PubMed] [Google Scholar]
- 29.Voriskova J, Baldrian P. 2013. Fungal community on decomposing leaf litter undergoes rapid successional changes. ISME J. 7:477–486. 10.1038/ismej.2012.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manos J, Belas R. 2006. The genera Proteus, Providencia, and Morganella. Springer, New York, NY [Google Scholar]
- 31.Bump JK, Webster CR, Vucetich JA, Peterson RO, Shields JM, Powers MD. 2009. Ungulate carcasses perforate ecological filters and create biogeochemical hotspots in forest herbaceous layers allowing trees a competitive advantage. Ecosystems 12:996–1007. 10.1007/s10021-009-9274-0 [DOI] [Google Scholar]
- 32.Towne EG. 2000. Prairie vegetation and soil nutrient responses to ungulate carcasses. Oecologia 122:232–239. 10.1007/PL00008851 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.