Abstract
Biological Mn oxidation is responsible for producing highly reactive and abundant Mn oxide phases in the environment that can mitigate metal contamination. However, little is known about Mn oxidation in low-pH environments, where metal contamination often is a problem as the result of mining activities. We isolated two Mn(II)-oxidizing bacteria (MOB) at pH 5.5 (Duganella isolate AB_14 and Albidiferax isolate TB-2) and nine strains at pH 7 from a former uranium mining site. Isolate TB-2 may contribute to Mn oxidation in the acidic Mn-rich subsoil, as a closely related clone represented 16% of the total community. All isolates oxidized Mn over a small pH range, and isolates from low-pH samples only oxidized Mn below pH 6. Two strains with different pH optima differed in their Fe requirements for Mn oxidation, suggesting that Mn oxidation by the strain found at neutral pH was linked to Fe oxidation. Isolates tolerated Ni, Cu, and Cd and produced Mn oxides with similarities to todorokite and birnessite, with the latter being present in subsurface layers where metal enrichment was associated with Mn oxides. This demonstrates that MOB can be involved in the formation of biogenic Mn oxides in both moderately acidic and neutral pH environments.
INTRODUCTION
Oxidation of soluble Mn(II) to insoluble Mn(III, IV) oxide minerals, although thermodynamically favored, tends to be slow in natural environments (1). Microorganisms, bacteria and fungi, catalyze Mn oxidation and increase the reaction rate by several orders of magnitude relative to abiotic reactions (1–3). Therefore, environmental Mn oxides, such as birnessite, are presumed to be of biological origin, e.g., biogenic (3). Mn(II)-oxidizing bacteria (MOB) are phylogenetically diverse and are detected in many different environments, such as soils, water (fresh to marine), and sediments (4). While microorganisms readily oxidize Mn(II) and precipitate Mn oxides at pH ∼7 under oxic and hypoxic conditions (5), little is known about biological oxidation at acidic pH. Under these conditions, chemical Mn oxidation with oxygen is predicted to be thermodynamically unfavorable and Mn(II) is stable in the environment (2, 5–7). All MOB isolated to date are neutrophiles, including the model organisms Roseobacter sp. AzwK-3b (8), Bacillus sp. SG-1 (9), Leptothrix sp (10), Pseudomonas putida GB-1 (11), and Aurantimonas manganoxydans (12), and have been studied for mechanisms of Mn oxidation and biomineral formation. The only reports of microbial Mn oxidization at low pH have come from the work of Bromfield, who analyzed cell extracts of Streptomyces spp. (13) and soil microorganisms (14) that oxidized Mn(II) at pH levels of <6, and from Ivarson, who reported that the fungus Cephalosportium sp. oxidized Mn(II) at pH 4.5 (15). Although to the best of our knowledge no MOB has been isolated from low-pH environments, there is evidence for biologically driven Mn oxidation at pH levels of <7 in the environment, such as that observed by Sparrow (16) for acidic soils.
Mn oxidation is catalyzed by heterotrophic organisms, and both direct and indirect mechanisms are identified for Mn oxidation (as summarized in reference 2). Indirect mechanisms are when the microbe modifies the pH or redox of the local environment or releases a chemical oxidant for Mn, such as superoxide (17). Direct oxidation, on the other hand, can occur via the production of polysaccharides or enzymatic activity (as summarized in reference 2). Although Mn(IV) formation could generate energy for organisms, this has not been definitively shown for any organism (4, 5). However, Mn oxidation was shown to increase the survival of Pseudomonas putida GB-1 in the presence of reactive oxygen species (ROS) (18). In addition, Mn oxidation has been speculated to provide other advantages to the cell, such as abiotic oxidation of refractory organic matter by Mn oxides (3) or controlling the availability of toxic metal ions (2, 3, 19). Biogenic Mn oxides are effective metal sorbents (2, 20–23), with two to five times more Pb-adsorbing capacities than abiotic Mn oxides and crystalline MnO2 minerals (20). However, this previous work was performed with model microorganisms cultivated at neutral pH, and little is known about the advantages of MOB in low-pH metal-contaminated environments.
In the former uranium mining area near Ronneburg, Germany, there is evidence for the natural attenuation of metal contaminants in Mn-rich subsurface layers with pH levels of 4 to 5 (24–26). In these layers, metal contaminants (e.g., Cd, Ni, and Co) are primarily associated with Mn oxides (24), suggesting a role for the oxides as geochemical barriers, with selected elements retained in local epigenetic zones of soils or sediments inhibiting further transport of such elements (27). Thus, it is important to determine the mechanisms behind the formation of these geochemical barriers in order to maintain the status quo or exploit the attenuation process for more widespread remediation of contaminants. We hypothesized that MOB contribute to the formation of Mn oxides and are thereby affecting the fate of metals in the Ronneburg mining area. Therefore, we aimed in this study to (i) isolate MOB from the contaminated mining site at a variety of pH levels, (ii) evaluate the biological formation of Mn oxides, and (iii) characterize microbial communities associated with Mn oxide-rich geochemical barriers.
MATERIALS AND METHODS
Site and sampling description.
The study sites are located within the former Ronneburg uranium mining district in Thuringia, Germany. Uranium was leached from low-grade black shale with acid mine drainage (AMD) and sulfuric acid from the 1970s to 1989, and this resulted in widespread environmental contamination with acid mine drainage and high concentrations of metals. Physical remediation was completed in 2004, although contaminated groundwater still threatens nearby ecosystems; for a more detailed description of the site history, see references 25, 26, 28, and 29. Contaminated Mn-rich samples were collected in 2010 from three locations: the Gessenwiese test field (GTF; location E 4510469, N 5635476 [Gauss-Krueger Potsdam coordinate system]), the bed of the Gessen Creek (R3; location E 4510271, N 5635859), and soil material from the Gessen Creek bank (B; location, E 4510121, N 5635807), as shown in Fig. S1 in the supplemental material. A soil depth profile was excavated at the GTF a few meters southeast from the formerly described profile of Burkhardt et al. (24), and soil materials were collected from three layers: an Mn-rich layer (gray-black soil layer located at a depth of 56 to 70 cm), top (above the Mn layer), and bottom (reddish soil layer below the Mn-rich layer) (see Fig. S1). The Gessen Creek location was chosen for study as it is located downhill from the former leaching heap (site GTF) and is currently receiving contaminated groundwater inputs due to flooding of the former mines. Samples from site GTF were used for geochemical characterization, microbial community characterization, and isolation; the soil solution was collected from water-saturated subsurface layers by using Rhizon dialysis samplers. Site R3 and B samples were used for isolation and geochemical characterization. Samples for microbial analyses were collected aseptically and stored at 4°C until analysis or stored at −80°C for microbial community characterization.
Geochemical characterization.
The redox potential (Eh) was measured in situ by using a Pt4800-M5-S7/80 redox probe (Mettler-Toledo). At site R3, the pH was measured in situ, whereas for sites B and GTF the pH of the soil and sediment was measured by the CaCl2 method as previously described (30). Total element content was determined for air-dried, sieved (<2 mm), and mortared sediment and soil samples after digestion using ICP-OES (inductively coupled plasma-optical emission spectrometry; 725 ES; Varian) and ICP-MS (inductively coupled plasma-mass spectrometry; XSeries II; Thermo Fisher Scientific), as previously described (24). Total organic carbon (TOC) was measured with a CN analyzer (varioMAXCN; Elementar, Germany) as previously described (31). Soil solution samples were collected by using miniature plastic suction cups (ecoTech). Samples were filtered directly (<0.2 μm) via the polymeric membrane of the suction tubes. Samples for cation analysis were additionally acidified with HNO3 (68%; Suprapur; Merck) to a pH of <2. The solutions were then analyzed using ICP-OES and ICP-MS.
Isolation of Mn(II)-oxidizing bacteria.
Soil and sediment samples were serially diluted (10−1 to 10−5) in 0.7% NaCl, and 100 μl of each dilution was plated onto three types of MnCO3-containing agar at pH 5.5, 6.5, or 7.0. Basic MnCO3 agar (BM; modified Leptothrix medium [32]) contained (per liter): 2.0 g MnCO3, 0.5 g meat extract, 0.5 g yeast extract, 0.28 g Fe(NH4)2(SO4)2·6H2O, 0.17 g sodium citrate, 5.0 μg vitamin B12, 2 ml trace element solution SL9 (33), and 50 mg cycloheximide (to inhibit fungal growth). BM7 refers to plates at pH 7.0 buffered with 10 mM HEPES (pH 7.0), whereas BM5.5 refers to plates at pH 5.5 buffered with 10 mM morpholineethanesulfonic acid (MES). All plates were incubated at 15°C in the dark.
MOB were identified by using leucoberbelin blue (LBB) and benzidine, which react with Mn(III/IV) oxides, in colorimetric assays (34). In brief, a drop of the LBB or benzidine reagent was applied directly to a brownish-black colony, and the mixture was incubated for 20 min at room temperature in the dark prior to a visual inspection for color change. An additional spot of reagent was placed on a spare area of the same agar plate as a control for abiotic oxidation. No quantification was possible with the visual method.
LBB-positive colonies were transferred at least 5 times, and purity was controlled based on colony morphology. pH was monitored before and after incubation in the medium by homogenizing a subsample of the agar medium from plates and then using a pH electrode. No change in the bulk pH of the medium was observed during the time of incubation (1 week to 3 months).
Isolate 16S rRNA gene analysis.
Genomic DNA was isolated from pure cultures by boiling a loopful of bacterial cells in 100 μl 5% Chelex 100 solution (Bio-Rad) for 10 min. After 2 min of centrifugation at full speed, the supernatant was used as the template for PCR amplification. 16S rRNA genes were amplified with the universal primers 27F and 1492R, as previously described (35). Isolates were screened by amplified rRNA gene restriction analysis (ARDRA), and representative operational taxonomic units (OTUs) were chosen for Sanger sequencing (performed at Macrogen, Inc., South Korea and The Netherlands). Sequences were assembled using Geneious Pro version 4.6.4 (36), and isolates were grouped together if colony morphologies were identical and 16S rRNA gene sequences had >97% sequence similarity. Sequence identity was determined using the BLAST algorithm against the GenBank database available from NCBI (37). The closest type strains to the representative strains were determined by using the EzTaxon server (38). Sequences were aligned using the Fast Aligner algorithm within the ARB software package (39). Phylogenetic trees incorporating a Jukes-Cantor distance correction with the neighbor-joining algorithm and 1,000 bootstrap replicates were constructed using Geneious v5.6.5 (36).
pH and metal tolerance.
The pH ranges for growth and Mn(II) oxidation were determined in BM agar plates with the pH adjusted to 4.0, 5.0, 5.5, 6.0, 7.0, or 8.0 with NaOH or HCl and without the addition of a buffer. Isolates were plated in triplicate and incubated at 15°C in the dark. pH was monitored before and after incubation (growth) in the bulk medium, as described above, for control and inoculated plates, and no change in the pH was observed. Growth was monitored visually based on the appearance of colonies. Mn(II) oxidation was confirmed via the LBB and benzidine spot tests, as described above.
Isolates AB_14, AB_18, and TB-2 were evaluated for their tolerance to 3 concentrations of cadmium, copper, and nickel in liquid medium. Liquid BM medium was prepared as described above without agar and with 100 μM MnCl2 instead of MnCO3 as the Mn(II) source. AB_14 and TB-2 were cultivated at pH 5.5 with MES as a buffer, whereas AB_18 was cultivated at pH 7.0 with HEPES as a buffer. Cadmium was added to growth media at final concentrations of 0.01, 0.05, and 0.5 mM; copper was added at final concentrations of 0.01, 0.05, and 1.0 mM; nickel was added to final concentrations of 0.1, 1.0, and 2.0 mM. Cultures were grown in triplicate with metals added to the liquid BM medium, and growth was monitored based on the optical density at 600 nm (OD600) measurements on a DR6000 UV-Vis spectrophotometer (Hach Lange GmbH, Germany). Mn(II) oxidation was evaluated by measuring the formation of Mn oxides in a quantitative LBB colorimetric assay after 742 h of growth (40). In brief, a 0.1-ml culture sample was added to 0.5 ml of 0.04% leucoberbelin blue in 45 mM acetic acid and the mixture was incubated for 10 min in the dark at room temperature. The reaction was filtered (0.22 μm), and then the absorbance was measured at 620 nm on a Hach DR6000 UV-Vis spectrophotometer. Standard curves were constructed with KMnO4. Mn oxides in control cultures (medium alone with metals added) were also measured, in order to account for background interactions between the medium and the metals. The data presented represent the amount of Mn oxide produced in the presence of bacteria minus the measured Mn oxide in control cultures.
LA-ICP-MS.
Elemental distributions within colonies of MOB strains were determined qualitatively by using laser ablation-inductively coupled plasma-mass spectrometry (LA-ICP-MS). Six Gessen Creek strains (AB_7, AB_8, AB_9, AB_11, AB_13, and AB_14) were inoculated onto MnCO3 agar with the same conditions used for isolation by streaking inoculum over the whole plate in a zigzag pattern and incubated for 2 weeks at 15°C. Additional plates were prepared for AB_9 and AB_14 without Fe in the medium to evaluate the influence of Fe on Mn oxidation. Agar plates were dried at 40°C in a drying oven for 7 to 21 days, until an even, flat surface with a thickness of ∼100 μm was obtained. Sections of dried agar (4 by 2 cm) were excised from the plate and fixed on a microscope slide with double-sided adhesive tape. Profiles (length, 2.5 to 3.1 cm) covering biomass and agar alone were ablated with an Nd:YAG laser (Microprobe II; Merchantek, Fremont, CA) coupled to an X-Series II ICP-MS apparatus (Thermo Fisher Scientific, Bremen, Germany). All scans were normalized to carbon (13C) as an internal standard.
FTIR spectroscopy.
Samples of MOB biomass collected from agar plates and uninoculated agar were freeze-dried at −60°C and then ground into a fine powder for Fourier-transform infrared (FTIR) spectroscopy. FTIR spectra were recorded using a Nicolet iS10 spectrometer (Thermo Fisher Scientific, Dreieich, Germany). Mortared samples were mixed with KBr at a ratio of 1:100 and pressed into pellets. The pellets were studied in transmission mode in the mid-infrared range, between 4,000 and 400 cm−1, for a total of 16 scans at a resolution of 4 cm−1. Spectra were baseline corrected by subtracting a straight line running between the two minima of each spectrum and normalized by dividing each data point by the spectrum's maximum.
Transmission electron microscopy.
For transmission electron microscropy (TEM), the MOB biomass was collected from agar plates and fixed in a freshly prepared, 0.2-μm-pore filter-sterilized solution of 2.5% glutaraldehyde (electron microscopy grade; Roth) in phosphate-buffered saline (PBS) buffer. Cell suspensions were centrifuged to a pellet at 16,000 × g for 5 min and brought to the Electron Microscopy Center at the University Clinic in Jena, Germany, for resin embedding and ultramicrotomy. In brief, the samples were washed three times with PBS buffer, stained with 1% OsO4 in triply distilled water, and washed again three times with PBS buffer. Two dehydration steps of 30 and 50% ethanol were followed by 2% uranyl acetate in 50% alcohol (for staining) and four dehydration steps of 50%, 70%, 96%, and 100% ethanol. Absolute dehydration was guaranteed with propylene oxide before embedding with araldite resin (1:2, 1:1, 2:1, and 100% araldite:propylene oxide steps). Resin-embedded samples were cured at 60°C before slicing to a 50- to 80-nm thickness with an Ultratome III (LKB, Stockholm, Sweden). Ultramicrotomed resin slices were placed on Cu TEM grid films, poststained with lead citrate, and analyzed on a variety of TEMs, including a Zeiss EM 900 operated at 80 kV (with digitized photo film), an FEI Tecnai G2 operated at 200 kV, and an FEI Titan operated at 300 kV in STEM mode. In addition to images, elemental information was obtained by energy-dispersive X-ray spectroscopy (EDS), and structural information was obtained by selected area electron diffraction (SAED). Analysis times, especially on the 300-kV TEM, were kept to a minimum due to the occurrence of beam damage (loss of mineral crystallinity and unstable resin) within a few minutes.
Microbial community 16S rRNA gene analysis and quantitative PCR.
DNA was extracted from 1 g of subsurface soil from the top (above the Mn-rich layer), Mn (Mn-rich layer), and bottom (below the Mn-rich layer) layers according to the method of Zhou et al. (41). Genomic DNA was PCR amplified with universal bacteria primers 27F and 1492R, which target the 16S rRNA gene, as described above. PCR amplicons were purified and then cloned into the pGEM T-Easy vector according to the manufacturer's instructions (Promega). Clone PCR was performed with M13F/R primers, and positive clones were Sanger sequenced with the universal 907R primer at Macrogen, Inc. (South Korea and The Netherlands). Sequences were trimmed by using Geneious Pro version 4.6.4 (36). OTUs (based on a 97% sequence similarity cutoff), taxonomic affiliation, and statistical analyses were performed within mothur (42).
Total bacteria and Albidiferax ferrireducens were quantified using quantitative PCR (qPCR) targeting universal bacterial 16S rRNA genes with the primers Uni-338F-RC and Uni-907R (43) and Albidiferax ferrireducens-specific primers Rdo R-RC (modified from reference 44) and Uni-907R, respectively. Purified genomic DNA (10 ng) was used as the template in the Maxima SYBR green qPCR master mix (Fermentas, Germany). Plasmids CB54 (accession number HE604015) and NR27 (accession number HE604080) were used as standards (45). Thermocycling was performed with the following temperature program: 10 min at 95°C and 50 cycles of 30 s at 95°C, 45 s at 57°C, 60 s at 72°C, and 10 s at 78°C. Fluorescence measurements were made at 61, 72, and 78°C for each cycle, followed by a dissociation curve analysis with 1 min at 95°C, 30 s at 55°C, and heating to 95°C at a rate of 0.01°C s−1.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of isolates obtained in this study were deposited in the GenBank or EMBL database under the accession numbers JQ033385 (AB_11), JQ033386 (AB_13), JQ033387 (AB_7), JQ033388 (AB_8), JQ033389 (AB_9), JQ033390 (AB_17), JQ033391 (AB_18), JQ033393 (AB_14), HG423345 (TB-1) and HG003356 (TB-2). Sequences obtained from total microbial community analysis were deposited in the EMBL database under accession numbers HG003357 to HG003566.
RESULTS AND DISCUSSION
Isolation of Mn-oxidizing bacteria.
A total of 24 MOB isolates were obtained from three sites within the former Ronneburg uranium mining area (see Fig. S1 in the supplemental material): the Mn-rich geochemical barrier from the Gessenwiese test field (GTF), the sediment of the Gessen Creek (R3), and topsoil material from the Gessen Creek bank (B). These sites were predicted to vary in their favorability for the growth of Mn oxidizers, based on their pH and organic carbon contents. The Mn-rich soil layer obtained from a depth of 56 to 70 cm at site GTF contained 33,000 μg Mn per g of soil (see Fig. S2 in the supplemental material) had a pH of 4.8, and hydrous Mn oxides were detected by FTIR spectroscopy (26). Sites R3 and B at the Gessen Creek had neutral pH (pH 6.3) and higher TOC contents (R3, 3.9% TOC [31]; B, 5.4% TOC [46]) than site GTF (0.26% TOC) and therefore were predicted to favor growth of neutrophilic heterotrophic MOB. Site R3 sediment contained 667 μg of Mn/g of sediment (31), whereas site B contained 884 μg of Mn per g of soil. Analysis of 16S rRNA genes grouped these isolates into 10 representative strains (Table 1).
TABLE 1.
Mn(II)-oxidizing bacterial strains isolated from the former uranium mining district Ronneburg
| Sampling locationa | Representative strain | Accession no. | No. of isolates | Soil dilution | pH of isolation | Closest type strainb (accession no., % identity) |
|---|---|---|---|---|---|---|
| Gessen Creek site R3 | AB_9 | JQ033389 | 3 | 10−4 | 7.0 | Duganella zoogloeoides IAM 12670T (D14256, 97.8) |
| AB_14 | JQ033393 | 1 | 10−1 | 5.5 | Duganella zoogloeoides IAM 12670T (D14256, 97.8) | |
| Gessen Creek site B | AB_7 | JQ033387 | 1 | 10−1 | 7.0 | Janthinobacterium lividum DSM1522T (Y08846, 99.6) |
| AB_8 | JQ033388 | 2 | 10−1 | 7.0 | Flavobacterium aquidurense WB-1.1.56T (AM177392, 98.2) | |
| AB_11 | JQ033385 | 1 | 10−1 | 7.0 | Janthinobacterium lividum DSM1522T (Y08846, 100) | |
| AB_13 | JQ033386 | 1 | 10−1 | 7.0 | Pseudomonas putida PhyCEm-187 (AM921634, 99.7) | |
| Former leaching heap site GTF, Mn-rich layer | AB_17 | JQ033390 | 7 | 10−1 | 7.0 | Burkholderia sordidicola S5-BT (AF512826, 98.4) |
| AB_18 | JQ033391 | 6 | 10−1 | 7.0 | Arthrobacter stackebrandtii CCM 2783T (AJ640198, 97.5) | |
| TB-1 | HG423345 | 1 | 10−2 | 7.0 | Frigoribacterium sp. LPPA 548 (HE613773, 100) | |
| TB-2 | HG003356 | 1 | 10−2 | 5.5 | Albidiferax ferrireducens T118 (CP000267, 99) |
Samples were collected from three locations within the former uranium mining district Ronneburg: the Gessenwiese test field (GTF), the bed of the Gessen Creek (R3), and soil material from the Gessen Creek bank (B) (see Fig. S1 in the supplemental material).
The type strains closest to the representative strains were determined by using the EzTaxon server (38).
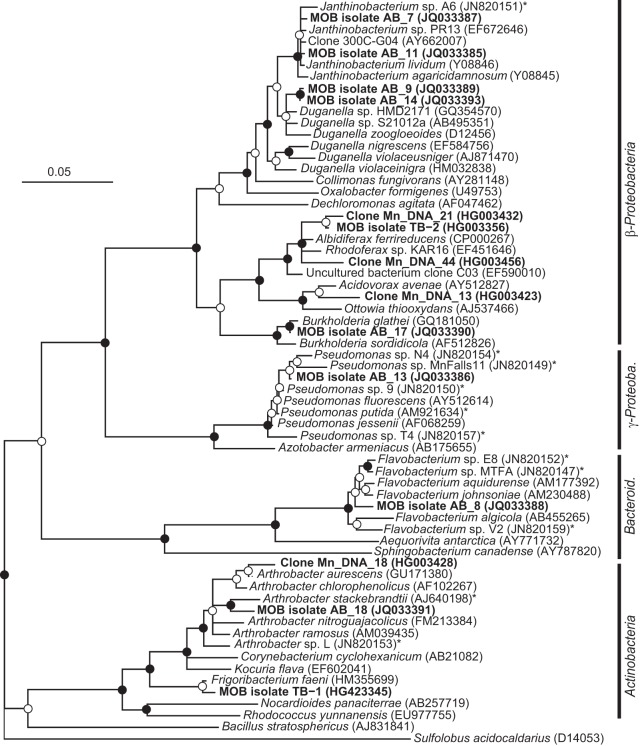
Two of the representative MOB strains were isolated at pH 5.5 (strains AB_14 and TB-2 within the Betaproteobacteria), whereas the other eight strains were isolated at pH 7 (members of the Beta- and Gammaproteobacteria, Actinobacteria, Bacteroidetes, and Firmicutes phyla) (Table 1 and Fig. 1). All isolated strains oxidized Mn, as shown by a positive LBB spot test, and Mn accumulation around colony biomass, as observed using LA-ICP-MS (Fig. 3). The pH did not change in the bulk medium during incubation.
FIG 1.
Neighbor-joining phylogenetic tree of 16S rRNA gene sequences of Mn(II)-oxidizing bacterial isolates and related clone sequences obtained from Mn-rich soil (indicated by boldface type). Isolates are designated by MOB followed by the strain number; clones are designated by Mn_DNA followed by the clone number. Known MOB are designated with an asterisk. Sulfolobus acidocaldarius was used as the outgroup. Branch points supported by bootstrap resampling (1,000 replicates) are indicated by filled circles (bootstrap values of >90%) and open circles (bootstrap values of >50%). Bar, 0.05 change per nucleotide position. Bacteroid., Bacteroidetes; γ-Proteob., Gammaproteobacteria.
FIG 3.
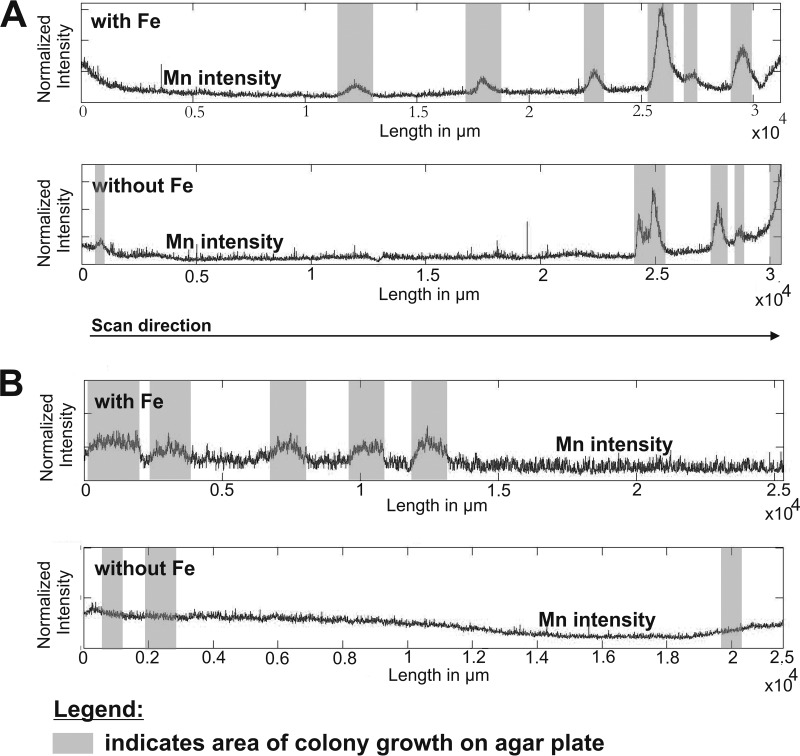
Elemental distributions of strains AB_14 (pH 5.5) (A) and AB_9 (pH 7) (B) grown in BM medium with or without iron, determined using LA-ICP-MS. The graphs show the signal intensities of Mn depending on the position of bacterial biomass on the agar plate (gray bars) and the availability of Fe. The intensities were normalized to an internal standard (carbon).
The low-pH isolate TB-2 was a member of the Comamonadaceae family, whereas strain AB_14 fell into the Oxalobacteraceae family. TB-2 was most closely related to Albidiferax ferrireducens T118 (99% sequence identity) (Table 1), which is a known Fe(III) reducer (47, 48) that has not been tested for its Mn-oxidizing capacities. However, our strain was not found to reduce Fe(III) or Mn(IV) under laboratory conditions (data not shown). As TB-2 is within the Comamonadaceae family, it joins Caldimonas manganoxidans (49) as the second organism within this family that is known to oxidize Mn. Strain AB_14 was most closely related to Duganella zoogloeoides (97.8% sequence identity) (Table 1) within the Oxalobacteraceae family and was also closely related to strain AB_9, which was isolated at pH 7. However, Duganella sp. AB_14 and AB_9 are considered separate strains, as they differ in their pH range for growth and Mn oxidation (Table 2 and below). Members of the genus Duganella were not previously shown to oxidize Mn. However, within the Oxalobacteraceae family there is a high degree of phylogenetic similarity between members of the genera Janthinobacterium, Oxalobacter, Duganella, and Herbaspirillum, as shown by DNA-RNA hybridization studies and small-subunit rRNA gene sequence analysis (50). Recently, Mn oxidation was described for isolate Janthinobacterium sp. A6 (51), which was obtained at neutral pH from ferromanganese deposits in caves. However, little is known to date of the mechanisms used by members of this family for Mn oxidation.
TABLE 2.
pH ranges for growth and Mn oxidation by MOB isolatesa
| Strain | pH of isolation | pH range toleratedb |
||||
|---|---|---|---|---|---|---|
| 5.0 | 5.5 | 6.0 | 7.0 | 8.0 | ||
| AB_7 | 7.0 | −/LBB− | −/LBB− | −/LBB− | +/LBB+ | −/LBB− |
| AB_11 | 7.0 | −/LBB− | −/LBB− | −/LBB− | +/LBB+ | +/LBB− |
| AB_9 | 7.0 | −/LBB− | −/LBB− | −/LBB− | +/LBB+ | +/LBB− |
| AB_17 | 7.0 | −/LBB− | −/LBB− | −/LBB− | +/LBB+ | −/LBB− |
| AB_13 | 7.0 | −/LBB− | −/LBB− | −/LBB− | +/LBB+ | −/LBB− |
| AB_18 | 7.0 | −/LBB− | −/LBB− | +/LBB+ | +/LBB+ | +/LBB+ |
| TB-1 | 7.0 | −/LBB− | −/LBB− | −/LBB− | +/LBB+ | −/LBB− |
| AB_8 | 7.0 | −/LBB− | −/LBB− | −/LBB− | +/LBB+ | −/LBB− |
| AB_14 | 5.5 | +/LBB+ | +/LBB+ | +/LBB− | +/LBB− | −/LBB− |
| TB-2 | 5.5 | −/LBB− | +/LBB+ | +/LBB+ | +/LBB− | −/LBB− |
Isolates were incubated on BM medium adjusted to different pHs; the pH levels remained stable during incubation.
Growth (+, colonies present after 1 month of incubation) or no growth (−, no colonies present after 1 month)/presence (LBB+) or absence (LBB−) of Mn oxides in LBB spot test.
Of the 8 isolates obtained at pH 7, only two were closely related to well-characterized MOB strains: AB_13 (99% sequence identity to Pseudomonas putida) and AB_18 (97.5% sequence identity to Arthrobacter stackebrantii) (Table 1 and Fig. 1). However, strains AB_7 and AB_8 were related to the newly discovered MOB strains Janthinobacterium sp. A6 (99.4% sequence identity) and Flavobacterium sp. E (98% sequence identity), respectively, suggesting that Mn oxidation may be common in these genera and not limited only to cave environments, where they were first observed to oxidize Mn (51).
Our study further broadens the known phylogenetic diversity of Mn-oxidizing bacteria, as our two low-pH and three neutral-pH MOB isolates were members of genera that were not previously shown to harbor Mn oxidizers. As research on MOB expands, additional phylogenetic groups might be identified. The presence of new phylogenetic groups may indicate the presence of unknown mechanisms for Mn oxidation, especially at low pH.
Physiology and in situ relevance of MOB isolates.
Tests for the pH growth range and Mn oxidation by the 10 representative strains revealed that only isolates Duganella sp. AB_14, Albidiferax sp. TB-2, and Arthrobacter sp. AB_18 were capable of growing and oxidizing Mn below pH 7.0 (Table 2). AB_14 could grow over 2 pH units (pH 5.0 to 7.0) but only oxidized Mn at pH 5.0 and 5.5. Similarly, TB-2 could grow at pH 5.5, 6.0, and 7.0 but only oxidized Mn at pH 5.5 and 6.0. Strain AB_18 grew and oxidized Mn at pH 6.0 to 8.0. Thus, we isolated two MOB strains that preferentially oxidize Mn at low pH: Albidiferax sp. TB-2 and Duganella sp. AB_14.
Bacterial Mn oxidation at low pH is unique, as Mn(II) oxidation under acidic conditions is predicted to be thermodynamically unfavorable (3, 5–7). At neutral to slightly alkaline pH (pH 7 to 9), Mn oxidation is thermodynamically favorable but occurs very slowly in the absence of bacteria (5, 6). However, to carry out Mn oxidation at low pH it is expected that a large activation energy is needed, which may not be favorable for a microorganism, especially as Mn oxidation is not thought to provide energy for the cell (2, 5).
Metal tolerance of MOB isolates.
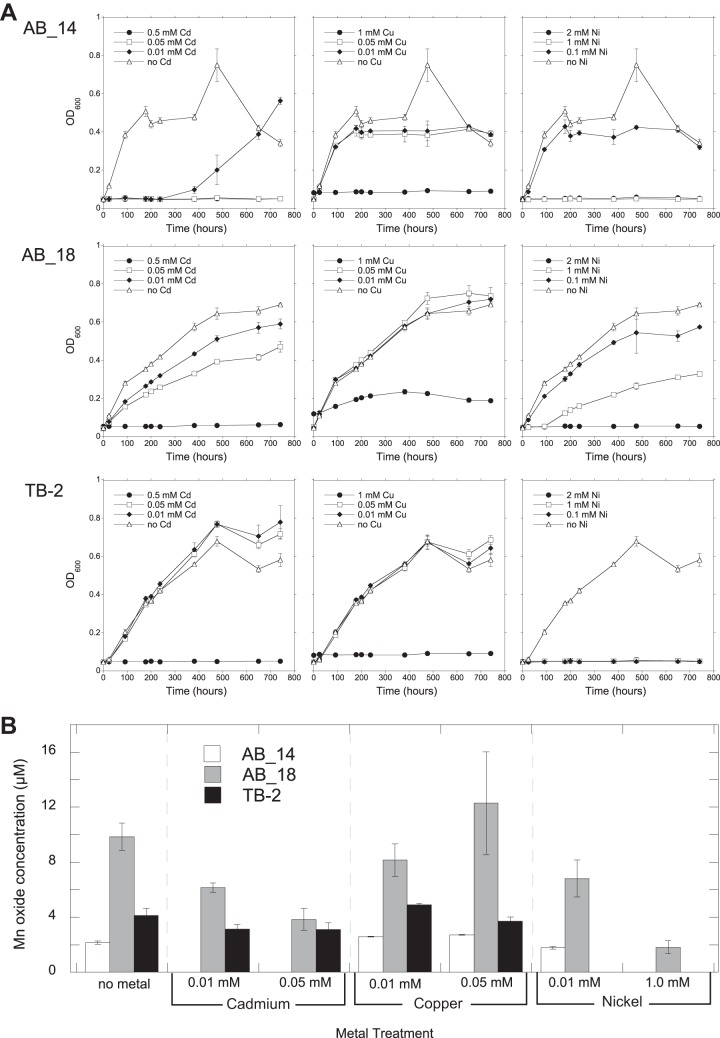
Metal ions affect the growth and Mn-oxidizing ability of Mn-oxidizing organisms (52). We evaluated the metal tolerance of Duganella sp. AB_14, Albidiferax sp. TB-2, and Arthrobacter sp. AB_18, which can oxidize Mn at pH levels that are relevant in situ (Mn-rich soil had a pH of 4.8; site R3 had a pH of 6.3). All three isolates grew and oxidized Mn in the presence of metals (Ni, Cd, and Cu) (Table 3 and Fig. 2). With Ni and Cu, AB_14 grew faster than the other isolates, reaching stationary phase at around 200 h (Fig. 2A). While AB_14 was sensitive to Cd in the early stages of growth, it grew with Cd after a 10-day lag phase (Fig. 2A). AB_18 tolerated all 3 metals but grew the slowest and did not reach stationary phase until ∼500 h (Fig. 2A). Interestingly, although Arthrobacter sp. AB_18 grew in the presence of 1.0 mM Cu, it only oxidized Mn in less than 0.05 mM Cu (Table 3 and Fig. 2B). Additionally, AB_18 oxidized more Mn than the other strains (Fig. 2B).
TABLE 3.
Metal tolerance of MOB isolates AB_14, AB_18, and TB-2 in the presence of various concentrations of Cd, Cu, or Nia
| Strain | Metal toleranceb |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cd |
Cu |
Ni |
|||||||
| 0.01 mM | 0.05 mM | 0.5 mM | 0.01 mM | 0.05 mM | 1.0 mM | 0.1 mM | 1.0 mM | 2.0 mM | |
| AB_14 | +/LBB− | −/LBB− | −/LBB− | +/LBB+ | +/LBB+ | −/LBB− | +/LBB+ | −/LBB− | −/LBB− |
| AB_18 | +/LBB+ | +/LBB+ | −/LBB− | +/LBB+ | +/LBB+ | +/LBB− | +/LBB+ | +/LBB+ | −/LBB− |
| TB-2 | +/LBB+ | +/LBB+ | −/LBB− | +/LBB+ | +/LBB+ | −/LBB− | −/LBB− | −/LBB− | −/LBB− |
Growth over time was monitored based on OD600 measurements for isolates cultured in liquid medium (AB_14 and TB-2 at pH 5.5 and AB_18 at pH 7).
Growth (+) or no growth (−)/presence of Mn oxides (LBB+) or no evidence of Mn oxidation (LBB−).
FIG 2.
Growth of (A) and Mn oxide production by (B) strains AB_14, AB_18, and TB-2 in the presence of various concentrations of Cd, Cu, and Ni. Growth over time was monitored by OD600 measurements of isolates cultured in liquid medium (AB_14 and TB-2 at pH 5.5, AB_18 at pH 7). Mn oxides were measured using the LBB colorimetric assay after 742 h of growth. All values are means of triplicate cultures minus the concentrations of control cultures (uninoculated media) ± standard deviations. Omitted values indicate no growth under the experimental conditions.
Soil pore water concentrations of Ni (170 μM), Cu (6 μM), and Cd (0.5 μM) at GTF (see Fig. S2 in the supplemental material) and water (8.5 μM Ni and 0.013 μM Cd; Cu was not determined) from Gessen Creek (31) both had lower metal concentrations than those tolerated by our strains. However, our isolates tolerated higher metal concentrations than what typically inhibits Mn oxidation by the model MOB, Leptothrix discophora SS-1 [Zn(II) and Co(II) at 10 μM and Cu(II) and Ni(II) at 100 μM] (52). This suggests that our strains are well adapted to the metal-contaminated conditions at the Ronneburg sites.
Although members of the Duganella genus, which contains our AB_14 isolate, have been isolated from metal-contaminated environments, little is known about the specific metal tolerance of these species. For example, Duganella strain EK-I49, which was isolated from metal-contaminated soils in Slovakia, contains the metal resistance (nccA) gene (53). Similarly, HgCl2-contaminated soils yielded another isolate with high sequence similarity to D. violaceusnigra (54). The metal tolerance we observed in Arthrobacter isolate AB_18 is consistent with other Arthrobacter strains that are well known for their extremely high metal tolerance (55–57). Albidiferax ferrireducens, the closest relative of isolate TB-2, is yet to be evaluated for metal tolerance; therefore, TB-2 tolerance to up to 0.05 mM Cd and Cu expands the known physiology of this organism.
The enzymes responsible for microbial Mn oxidation include calcium-binding heme peroxidases (MopA) and multicopper oxidases (MCO) (summarized in reference 4). MCO catalyze the transfer of electrons not only to Mn but also to Fe(II) and other substrates (58). Copper ions are necessary for MCO enzyme function, and our isolates AB_14, AB_18, and TB-2 grew and oxidized Mn in the presence of 0.01 and 0.05 mM Cu in the medium. These strains were also less sensitive to copper than nickel at the same concentrations. Further work is needed to determine whether the addition of copper enhances Mn oxidation in these strains, which could indicate the presence of an MCO or other oxidant.
Elemental distribution in MOB biomass.
LA-ICP-MS was used to obtain spatially resolved information on the elemental distribution of Mn and Fe within MOB biomass and surrounding agar medium. Generally, high Mn intensities were associated with microbial biomass for all investigated strains (as displayed for two examples in Fig. 3). In contrast, the LA-ICP-MS transect showed that areas of agar without biomass had constant element intensities except for the beginning and end regions for the Mn measurement. The cause of these unusual Mn intensities is likely the sample preparation, which resulted in higher dehydration of the agar pieces on the end, causing them to show higher Mn background levels.
As the medium initially used to isolate our organisms also contained Fe(II), the ability of the organisms to also oxidize Fe(II) may have led to enzymatic Fe oxidation that then may have triggered Mn(II) oxidation (59). Thus, LA-ICP-MS was used to evaluate if the coenrichment of Mn and Fe was due to oxidation of the Fe in the medium by MOB strains AB_9 (pH 7) and AB_14 (pH 5.5). The LA-ICP-MS profile for strain AB_14 (pH 5.5) (Fig. 3A) showed an enrichment of Mn in the bacterial biomass both with and without Fe in the medium, suggesting that secondary abiotic oxidation of Mn was not occurring. However, the model Mn(II) oxidizer P. putida GB-1 uses different pathways for Mn oxidation, depending on iron availability (60, 61). Under iron-replete conditions, enzymatic Mn oxidation is hypothesized to predominate, whereas under iron-limited conditions siderophore production is linked to abiotic oxidation (61). Further work is needed to determine whether AB_14 uses a single pathway for Mn oxidation or whether this strain produces siderophores that contribute to abiotic Mn oxidation. In contrast to AB_14, the other Duganella sp. strain, AB_9, isolated at pH 7 (Fig. 3B) did not accumulate Mn in its biomass in the absence of Fe. This suggests a linkage between Fe and Mn oxidation in strain AB_9, either through biological Fe(II) oxidation followed by secondary abiotic oxidation of Mn or via the iron-dependent enzymatic Mn(II) oxidation that is observed in P. putida GB-1 (61). In addition, this observation highlighted the physiological differences between strains AB_9 and AB_14, such as the presence of different Mn oxidation pathways and siderophore production in AB_14. Determining the mechanisms for Mn oxidation at low pH by isolate Duganella sp. AB_14 will be aided by genome queries, as the full-genome sequencing is under way in collaboration with the Joint Genome Institute.
Identification of biogenic Mn oxides.
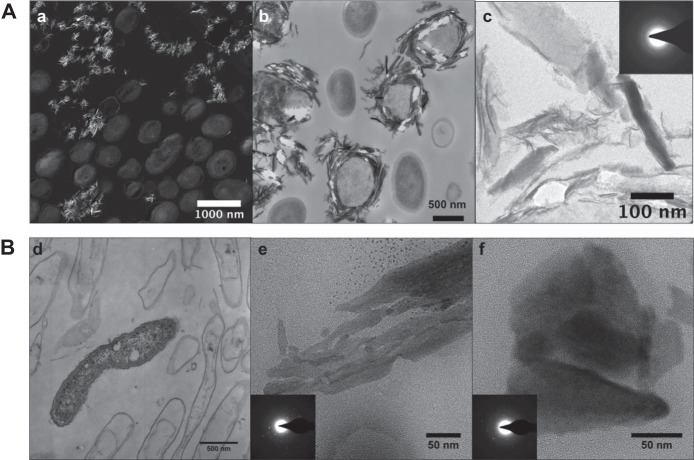
Mn oxides produced by Ronneburg isolates were identified using FTIR spectroscopy and TEM. FTIR spectra of all representative strains that produced precipitates showed a slightly higher absorbance between 750 and 400 cm−1, which is characteristic for the Mn-O vibrations of the MnO6 octahedral framework in Mn oxides (see Fig. S3 in the supplemental material). TEM revealed mineral-cell associations for the Mn oxides produced by isolates Arthrobacter sp. AB_18 and Duganella sp. AB_14 (Fig. 4). Many AB_18 cells were encrusted with Mn oxides (Fig. 4B); in contrast, AB_14 cells were not encrusted and Mn oxide precipitates were only observed away from the cells (Fig. 4A). EDS showed that the precipitates were composed of Mn and O along with other elements from the embedding resin (see Fig. S4 in the supplemental material). Analysis of SAED patterns showed crystallographic d-spacings similar to those of common Mn oxides (birnessite and todorokite), although they could not be conclusively identified (see Table S1 in the supplemental material).
FIG 4.
TEM images of AB_18 and AB_14 cells and Mn oxides. (A) AB_18. (a) STEM high-angle annular dark-field (HAADF) image (the brighter areas indicate higher Z elements); (b) TEM image of Mn oxide minerals surrounding some cells; (c) TEM image of a cluster of minerals and their selected area diffraction patterns, showing some crystallinity (inset). (B) AB_14. TEM images with diffraction patterns (insets) of live and mostly dead bacteria with few obvious minerals around them (d) or aggregates of Mn oxide particles (e and f). Diffraction areas were larger than the images shown.
The observed locations of the minerals on the bacteria may also indicate a potential role and/or mechanism for Mn oxidation. Our Arthrobacter sp. AB_18 was encrusted with a difficult-to-identify Mn oxide mineral (see Table S1), suggesting a direct mechanism of oxidation such as enzymatic oxidation on the outer membrane (2). The encrusted Mn oxides on the cell may be acting as a protective barrier from toxic metal ions, which would impart a selective advantage for such organisms in the metal-contaminated environments at the Ronneburg area. Mn oxides from Duganella sp. AB_14 differed in morphology from those from strain AB_18 and were not observed in close proximity to the cell. This may indicate that low-pH oxidation proceeds via a different enzymatic pathway than that used by Mn-oxidizing organisms found in neutral-pH samples, or that Mn oxidation occurs via indirect mechanisms, such as the production of superoxides (17). Recently, extracellular superoxide production was found to be widespread among heterotrophic bacteria, although the two Betaproteobacteria species tested were not shown to produce superoxide (62). However, additional Betaproteobacteria strains may prove to produce superoxide or other reactive oxygen species, as their production is ubiquitous among both bacteria and fungi (62).
Microbial community analysis.
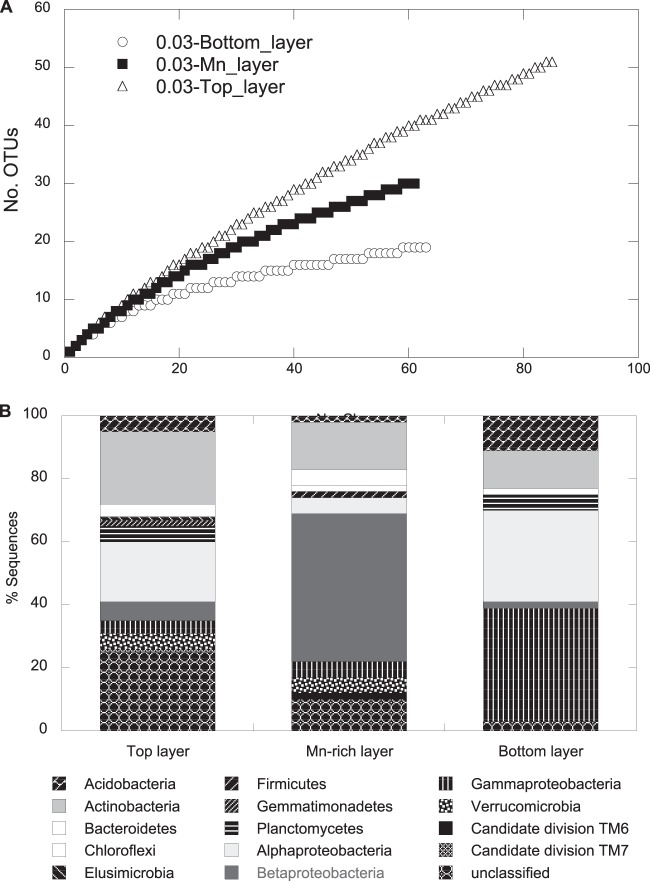
Total relative bacterial abundance was low in subsurface soils at site GTF with the soil layer above (top), the Mn-rich geochemical barrier, and the layer below (bottom), as we found 6.6 × 104, 8.5 × 105, and 6.1 × 105 16S rRNA gene copies per gram (wet weight) of soil, respectively (determined using qPCR). For comparative microbial community analysis, a 16S rRNA gene cloning and sequencing approach revealed a total of 209 sequences that grouped into 91 OTUs, with 4 OTUs present in all 3 libraries. Rarefaction analysis indicated saturation of sampling for the bottom sample but not for the Mn-rich or top samples (Fig. 5A), and coverage had a similar pattern (see Table S2 in the supplemental material). The community composition in the Mn-rich geochemical barrier differed distinctly from those of the top and bottom communities (Fig. 5B), with a dominance of Betaproteobacteria (47% of total clones). The Betaproteobacteria-related clone Mn_DNA_21 was most closely related to isolate TB-2 from the Mn-rich layer and to A. ferrireducens and represented ∼16% of total clones, suggesting a high in situ prevalence of this isolate at the former leaching heap site GTF (Fig. 1). The abundance of A. ferrireducens in the Mn-rich geochemical barrier was ∼105 16S rRNA gene copies per g (wet weight) of sediment as revealed by qPCR, representing 13.5% of the total bacterial community. As shown in a parallel study by Fabisch et al. (31), this bacterial group represents 27% of the total community in the sediment of the Gessen Creek site (R3), where bacterial 16S rRNA gene copy numbers per g (wet weight) of sediment are 5 orders of magnitude higher than at site GTF. Members of the Alphaproteobacteria dominated the bottom layer (29% of total clones). Clones related to members of the Actinobacteria represented 23%, 15%, and 12% of total clones in the top, Mn-rich, and bottom layers, respectively (Fig. 5B).
FIG 5.
Rarefaction curves (A) and frequencies of bacterial phylogenetic lineages (B) for the 16S rRNA gene clone libraries obtained for the three subsurface layers, Mn-rich, top, and bottom at the GTF site. A total of 85, 61, and 63 clones were analyzed for the top, Mn-rich, and bottom layers, respectively. Phylogenetic affiliations of OTUs (based on a >97% sequence similarity cutoff) were determined by using mothur against the SILVA database. Frequencies were based on the total number of clones associated with OTUs of sequenced representatives at the phylum level or class level for Proteobacteria.
Some RNA- and DNA-derived 16S rRNA clones from Gessen Creek sediment R3 obtained in the study of Fabisch et al. (31) are related to known MOB and to two of the MOB isolates obtained in this study. The RNA-derived clones included members from Actinobacteria (Arthrobacter related), Bacteroidetes (Flavobacterium sp.), Gammaproteobacteria (Acinetobacter sp.), and Betaproteobacteria (Albidiferax sp.) (31). The detection of these organisms on the RNA level indicates their potential activities within the environment. The presence of a DNA-based clone related to the Oxalobacteraceae family (31) was interesting, as this is the same family as our low-pH isolate AB_14 and suggests that this organism has the potential to be active at the Gessen Creek site.
We have provided evidence that MOB isolates obtained from the former Ronneburg uranium mining area could impact the fate of metal contaminants via adsorption to biogenic Mn oxide minerals formed at both acidic and neutral pHs. In many metal-contaminated environments, biological Mn oxidation is associated with a decrease in aqueous metal load. The biogenic minerals produced by our isolates at pH 5.5 (Duganella AB_14) and 7.0 (Arthrobacter AB_18) were difficult to conclusively identify. However, birnessite and todorokite comprised the Mn-rich layer at site GTF (M. Händel, personal communication, and (26)) where accumulation of metals was associated with Mn oxides. Although we do not show direct evidence that MOB formed the Mn oxides in the acidic Mn-rich geochemical barrier, the stability of Mn(II) under the environmental conditions is such that Mn oxides are not predicted to form under solely abiotic conditions (2, 5, 6). In addition, our Albidiferax strain TB-2 (pH 5.5) isolate was closely related to a clone from the Mn-rich layer that had a high relative contribution (16% total clones) to the in situ community. As strain TB-2 oxidizes Mn in the lab under pH levels that are relevant to in situ conditions, we can infer that this organism may be involved in biological Mn oxidation at the site, and further work is needed to assess the potential for biogenic Mn oxides to contribute to metal attenuation in the Mn-rich layer.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Fabisch, F. Beulig, P. Bouwma, and S. Lu (Aquatic Geomicrobiology, FSU Jena) for help with sampling. S. Nietzsche and C. Kämnitz of the Electron Microscopy Center at the University Clinic, FSU Jena, performed resin embedding and some TEM. The Ernst-Ruska Centre for Microscopy and Spectroscopy with Electrons user facility in Jülich, Germany, provided free access to and assistance with TEM. We thank Darren Dunlap for helpful discussions.
We also thank the German Science Foundation (DFG GRK 1257) for funding.
Any use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Footnotes
Published ahead of print 13 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01296-14.
REFERENCES
- 1.Morgan JJ. 2005. Kinetics of reaction between O2 and Mn(II) species in aqueous solutions. Geochim. Cosmochim. Acta 69:35–48. 10.1016/j.gca.2004.06.013 [DOI] [Google Scholar]
- 2.Tebo BM, Bargar JR, Clement BG, Dick GJ, Murray KJ, Parker D, Verity R, Webb SM. 2004. Biogenic manganese oxides: properties and mechanisms of formation. Annu. Rev. Earth Planet. Sci. 32:287–328. 10.1146/annurev.earth.32.101802.120213 [DOI] [Google Scholar]
- 3.Tebo B, Clement B, Dick G. 2007. Biotransformations of manganese, p 1223–1238 In Hurst CJ. (ed), Manual of environmental microbiology, 3rd ed. ASM Press, Washington, DC [Google Scholar]
- 4.Geszvain K, Butterfield C, Davis R, Madison A, Lee S, Parker D, Soldatova A, Spiro T, Luther G, Tebo B. 2012. The molecular biogeochemistry of manganese(II) oxidation. Biochem. Soc. Trans. 40:1244–1248. 10.1042/BST20120229 [DOI] [PubMed] [Google Scholar]
- 5.Nealson KH. 2006. The manganese-oxidizing bacteria, p 222–231 In Balows A, Trüper HG, Dworkin M, Harder W, Schleifer K-H. (ed), The prokaryotes. Springer, New York, NY [Google Scholar]
- 6.Luther GW. 2010. The role of one-and two-electron transfer reactions in forming thermodynamically unstable intermediates as barriers in multi-electron redox reactions. Aquat. Geochem. 16:395–420. 10.1007/s10498-009-9082-3 [DOI] [Google Scholar]
- 7.von Langen PJ, Johnson KS, Coale KH, Elrod VA. 1997. Oxidation kinetics of manganese (II) in seawater at nanomolar concentrations. Geochim. Cosmochim. Acta 61:4945–4954. 10.1016/S0016-7037(97)00355-4 [DOI] [Google Scholar]
- 8.Hansel C, Francis C. 2006. Coupled photochemical and enzymatic Mn(II) oxidation pathways of a planktonic Roseobacter-like bacterium. Appl. Environ. Microbiol. 72:3543–3549. 10.1128/AEM.72.5.3543-3549.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Francis CA, Tebo BM. 1999. Marine Bacillus spores as catalysts for oxidative precipitation and sorption of metals. J. Mol. Microbiol. Biotechnol. 1:71–78 [PubMed] [Google Scholar]
- 10.Boogerd F, de Vrind J. 1987. Manganese oxidation by Leptothrix discophora. J. Bacteriol. 169:489–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francis CA, Tebo BM. 2001. cumA multicopper oxidase genes from diverse Mn(II)-oxidizing and non-Mn(II)-oxidizing Pseudomonas strains. Appl. Environ. Microbiol. 67:4272–4278. 10.1128/AEM.67.9.4272-4278.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson C, Dick G, Chu M, Cho J, Davis R, Bräuer S, Tebo B. 2009. Aurantimonas manganoxydans, sp. nov. and Aurantimonas litoralis, sp. nov.: Mn(II) oxidizing representatives of a globally distributed clade of alpha-Proteobacteria from the order Rhizobiales. Geomicrobiol. J. 26:189–198. 10.1080/01490450902724840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bromfield SM. 1979. Manganous ion oxidation at pH values below 5.0 by cell-free substances from Streptomyces sp. cultures. Soil Biol. Biochem. 11:115–118. 10.1016/0038-0717(79)90086-5 [DOI] [Google Scholar]
- 14.Bromfield S. 1956. Oxidation of manganese by soil microorganisms. Aust. J. Biol. Sci. 9:238–252 [Google Scholar]
- 15.Ivarson K, Heringa P. 1972. Oxidation of manganese by microorganisms in manganese deposits of Newfoundland soil. Can. J. Soil Sci. 52:401–416. 10.4141/cjss72-052 [DOI] [Google Scholar]
- 16.Sparrow LA, Uren NC. 1987. Oxidation and reduction of Mn in acidic soils: effect of temperature and soil pH. Soil Biol. Biochem. 19:143–148. 10.1016/0038-0717(87)90073-3 [DOI] [Google Scholar]
- 17.Learman DR, Voelker BM, Vazquez-Rodriguez AI, Hansel CM. 2011. Formation of manganese oxides by bacterially generated superoxide. Nat. Geosci. 4:95–98. 10.1038/ngeo1055 [DOI] [Google Scholar]
- 18.Banh A, Chavez V, Doi J, Nguyen A, Hernandez S, Ha V, Jimenez P, Espinoza F, Johnson HA. 2013. Manganese (Mn) oxidation increases intracellular Mn in Pseudomonas putida GB-1. PLoS One 8:e77835. 10.1371/journal.pone.0077835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tebo BM, Johnson HA, McCarthy JK, Templeton AS. 2005. Geomicrobiology of manganese(II) oxidation. Trends Microbiol. 13:421–428. 10.1016/j.tim.2005.07.009 [DOI] [PubMed] [Google Scholar]
- 20.Nelson Y, Lion L, Ghiorse W, Shuler M. 1999. Production of biogenic Mn oxides by Leptothrix discophora SS-1 in a chemically defined growth medium and evaluation of their Pb adsorption characteristics. Appl. Environ. Microbiol. 65:175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peña J, Kwon KD, Refson K, Bargar JR, Sposito G. 2010. Mechanisms of nickel sorption by a bacteriogenic birnessite. Geochim. Cosmochim. Acta 74:3076–3089. 10.1016/j.gca.2010.02.035 [DOI] [Google Scholar]
- 22.Wang W, Shao Z, Liu Y, Wang G. 2009. Removal of multi-heavy metals using biogenic manganese oxides generated by a deep-sea sedimentary bacterium Brachybacterium sp. strain Mn32. Microbiology 155:1989–1996. 10.1099/mic.0.024141-0 [DOI] [PubMed] [Google Scholar]
- 23.Toner B, Manceau A, Webb S, Sposito G. 2006. Zinc sorption to biogenic hexagonal-birnessite particles within a hydrated bacterial biofilm. Geochim. Cosmochim. Acta 70:27–43. 10.1016/j.gca.2005.08.029 [DOI] [Google Scholar]
- 24.Burkhardt E, Meißner S, Merten D, Büchel G, Küsel K. 2009. Heavy metal retention and microbial activities in geochemical barriers formed in glacial sediments subjacent to a former uranium mining leaching heap. Chem. Erde Geochem. 69:21–34. 10.1016/j.chemer.2008.12.003 [DOI] [Google Scholar]
- 25.Carlsson E, Büchel G. 2005. Screening of residual contamination at a former uranium heap leaching site, Thuringia, Germany. Chem. Erde Geochem. 65:75–95. 10.1016/j.chemer.2005.06.007 [DOI] [Google Scholar]
- 26.Schäffner F. 2013. Natural attenuation of heavy metals in near-surface supergene Mn deposits occurring at the basement of a former leaching heap, Ronneburg, Germany. Ph.D. dissertation Friedrich-Schiller University Jena, Jena, Germany [Google Scholar]
- 27.Perel'man AI. 1967. Geochemistry of epigenesis, p 213–234 Plenum Press, New York, NY [Google Scholar]
- 28.Grawunder A, Lonschinski M, Merten D. 2009. Distribution and bonding of residual contamination in glacial sediments at the former uranium mining leaching heap of Gessen/Thuringia, Germany. Chem. Erde Geochem. 69:5–19. 10.1016/j.chemer.2008.06.001 [DOI] [Google Scholar]
- 29.Küsel K, Ewald E-M, Sitte J. 2008. Effect of metal-reducing microorganisms on element fluxes in a former uranium-mining district, p 128–137 In Liu S-J, Drake HL. (ed), Microbes in the environment: perspectives and challenges. Science Press, Beijing, China [Google Scholar]
- 30.Deutsches Institut fur Normung. 2000. 19684-1:1977-02. Methods of soil analysis for water management for agricultural purposes: chemical laboratory tests; determination of pH-value of the soil and lime requirement. Handbuch der Bodenuntersuchung. Beuth Verlag, Berlin, Germany [Google Scholar]
- 31.Fabisch M, Beulig F, Akob DM, Küsel K. 2013. Surprising abundance of Gallionella-related iron oxidizers in creek sediments at pH 4.4 or at high heavy metal concentrations. Front. Microbiol. 4:390. 10.3389/fmicb.2013.00390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mulder EG, van Veen WL. 1963. Investigations on the Sphaerotilus-Leptothrix group. Antonie Van Leeuwenhoek 29:121–153 [DOI] [PubMed] [Google Scholar]
- 33.Benz M, Brune A, Schink B. 1998. Anaerobic and aerobic oxidation of ferrous iron at neutral pH by chemoheterotrophic nitrate-reducing bacteria. Arch. Microbiol. 169:159–165. 10.1007/s002030050555 [DOI] [PubMed] [Google Scholar]
- 34.Krumbein W, Altmann H. 1973. A new method for the detection and enumeration of manganese oxidizing and reducing microorganisms. Helgol. Mar. Res. 25:347–356 [Google Scholar]
- 35.Lu S, Gischkat S, Reiche M, Akob DM, Hallberg KB, Küsel K. 2010. Ecophysiology of Fe-cycling bacteria in acidic sediments. Appl. Environ. Microbiol. 76:8174–8183. 10.1128/AEM.01931-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drummond A, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J, Kearse M, Markowitz S, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A. 2009. Geneious v4.6.0. Biomatters, Auckland, New Zealand [Google Scholar]
- 37.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 38.Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, Lim YW. 2007. EzTaxon: a Web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 57:2259–2261. 10.1099/ijs.0.64915-0 [DOI] [PubMed] [Google Scholar]
- 39.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar Buchner A, Lai T, Steppi S, Jobb G, Forster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, Konig A, Liss T, Lussmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer K-H. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371. 10.1093/nar/gkh293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Vrind JP, Boogerd FC, de Vrind-de Jong EW. 1986. Manganese reduction by a marine Bacillus species. J. Bacteriol. 167:30–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou JZ, Bruns MA, Tiedje JM. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schloss PD, Westcott S, Ryabin T, Hall J, Hartmann M, Hollister E, Lesniewski R, Oakley B, Parks D, Robinson C, Sahl J, Stres B, Thallinger G, Van Horn D, Weber C. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lane DJ. 1991. 16S/23S rRNA sequencing in E. coli, p 115–175 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, NY [Google Scholar]
- 44.Zhou J. 2008. The development of molecular tools for the evaluation of the bioremediation of chlorinated solvents. Ph.D. dissertation Utah State University, Logen, UT [Google Scholar]
- 45.Lu S, Chourey K, Reiche M, Nietzsche S, Shah MB, Neu TR, Hettich RL, Küsel K. 2013. Insights into the structure and metabolic function of microbes that shape pelagic iron-rich aggregates (“iron snow”). Appl. Environ. Microbiol. 79:4272–4281. 10.1128/AEM.00467-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Löffler S. 2007. Charakterisierung der geochemischen Barrieren im Gessenbach. Ph.D. dissertation Friedrich Schiller University Jena, Jena, Germany [Google Scholar]
- 47.Finneran KT, Johnsen CV, Lovley DR. 2003. Rhodoferax ferrireducens sp. nov., a psychrotolerant, facultatively anaerobic bacterium that oxidizes acetate with the reduction of Fe(III). Int. J. Syst. Evol. Microbiol. 53:669–673. 10.1099/ijs.0.02298-0 [DOI] [PubMed] [Google Scholar]
- 48.Ramana CV, Sasikala C. 2009. Albidoferax, a new genus of Comamonadaceae and reclassification of Rhodoferax ferrireducens (Finneran et al., 2003) as Albidoferax ferrireducens comb. nov. J. Gen. Appl. Microbiol. 55:301–304. 10.2323/jgam.55.301 [DOI] [PubMed] [Google Scholar]
- 49.Takeda M, Kamagata Y, Ghiorse WC, Hanada S, Koizumi J-I. 2002. Caldimonas manganoxidans gen. nov., sp. nov., a poly(3-hydroxybutyrate)-degrading, manganese-oxidizing thermophile. Int. J. Syst. Evol. Microbiol. 52:895–900. 10.1099/ijs.0.02027-0 [DOI] [PubMed] [Google Scholar]
- 50.Lincoln SP, Fermor TR, Tindall BJ. 1999. Janthinobacterium agaricidamnosum sp. nov., a soft rot pathogen of Agaricus bisporus. Int. J. Syst. Bacteriol. 49:1577–1589. 10.1099/00207713-49-4-1577 [DOI] [PubMed] [Google Scholar]
- 51.Carmichael MJ, Carmichael SK, Santelli CM, Strom A, Bräuer SL. 2013. Mn(II)-oxidizing bacteria are abundant and environmentally relevant members of ferromanganese deposits in caves of the Upper Tennessee River Basin. Geomicrobiol. J. 30:779–800. 10.1080/01490451.2013.769651 [DOI] [Google Scholar]
- 52.Miyata N, Tani Y, Sakata M, Iwahori K. 2007. Microbial manganese oxide formation and interaction with toxic metal ions. J. Biosci. Bioeng. 104:1–8. 10.1263/jbb.104.1 [DOI] [PubMed] [Google Scholar]
- 53.Karelová E, Harichová J, Stojnev T, Pangallo D, Ferianc P. 2011. The isolation of heavy-metal resistant culturable bacteria and resistance determinants from a heavy-metal-contaminated site. Biologia 66:18–26. 10.2478/s11756-010-0145-0 [DOI] [Google Scholar]
- 54.Mera N, Iwasaki K. 2007. Use of plate-wash samples to monitor the fates of culturable bacteria in mercury- and trichloroethylene-contaminated soils. Appl. Microbiol. Biotechnol. 77:437–445. 10.1007/s00253-007-1152-0 [DOI] [PubMed] [Google Scholar]
- 55.Margesin R, Schinner F. 1996. Bacterial heavy metal-tolerance—extreme resistance to nickel in Arthrobacter spp. strains. J. Basic Microbiol. 36:269–282. 10.1002/jobm.3620360410 [DOI] [Google Scholar]
- 56.Benyehuda G, Coombs J, Ward PL, Balkwill D, Barkay T. 2003. Metal resistance among aerobic chemoheterotrophic bacteria from the deep terrestrial subsurface. Can. J. Microbiol. 49:151–156. 10.1139/w03-012 [DOI] [PubMed] [Google Scholar]
- 57.Abou-Shanab RA, van Berkum P, Angle JS. 2007. Heavy metal resistance and genotypic analysis of metal resistance genes in gram-positive and gram-negative bacteria present in Ni-rich serpentine soil and in the rhizosphere of Alyssum murale. Chemosphere 68:360–367. 10.1016/j.chemosphere.2006.12.051 [DOI] [PubMed] [Google Scholar]
- 58.Kosman D. 2010. Multicopper oxidases: a workshop on copper coordination chemistry, electron transfer, and metallophysiology. J. Biol. Inorg. Chem. 15:15–28. 10.1007/s00775-009-0590-9 [DOI] [PubMed] [Google Scholar]
- 59.Junta JL, Hochella MF., Jr 1994. Manganese(II) oxidation at mineral surfaces: a microscopic and spectroscopic study. Geochim. Cosmochim. Acta 58:4985–4999. 10.1016/0016-7037(94)90226-7 [DOI] [Google Scholar]
- 60.Parker DL, Sposito G, Tebo BM. 2004. Manganese(III) binding to a pyoverdine siderophore produced by a manganese(II)-oxidizing bacterium. Geochim. Cosmochim. Acta 68:4809–4820. 10.1016/j.gca.2004.05.038 [DOI] [Google Scholar]
- 61.Parker DL, Morita T, Mozafarzadeh ML, Verity R, McCarthy JK, Tebo BM. 2007. Inter-relationships of MnO2 precipitation, siderophore-Mn(III) complex formation, siderophore degradation, and iron limitation in Mn(II)-oxidizing bacterial cultures. Geochim. Cosmochim. Acta 71:5672–5683. 10.1016/j.gca.2007.03.042 [DOI] [Google Scholar]
- 62.Diaz JM, Hansel CM, Voelker BM, Mendes CM, Andeer PF, Zhang T. 2013. Widespread production of extracellular superoxide by heterotrophic bacteria. Science 340:1223–1226. 10.1126/science.1237331 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.