Abstract
Due to the long durations spent inside by many humans, indoor air quality has become a growing concern. Biofiltration has emerged as a potential mechanism to clean indoor air of harmful volatile organic compounds (VOCs), which are typically found at concentrations higher indoors than outdoors. Root-associated microbes are thought to drive the functioning of plant-based biofilters, or biowalls, converting VOCs into biomass, energy, and carbon dioxide, but little is known about the root microbial communities of such artificially grown plants, how or whether they differ from those of plants grown in soil, and whether any changes in composition are driven by VOCs. In this study, we investigated how bacterial communities on biofilter plant roots change over time and in response to VOC exposure. Through 16S rRNA amplicon sequencing, we compared root bacterial communities from soil-grown plants with those from two biowalls, while also comparing communities from roots exposed to clean versus VOC-laden air in a laboratory biofiltration system. The results showed differences in bacterial communities between soil-grown and biowall-grown plants and between bacterial communities from plant roots exposed to clean air and those from VOC-exposed plant roots. Both biowall-grown and VOC-exposed roots harbored enriched levels of bacteria from the genus Hyphomicrobium. Given their known capacities to break down aromatic and halogenated compounds, we hypothesize that these bacteria are important VOC degraders. While different strains of Hyphomicrobium proliferated in the two studied biowalls and our lab experiment, strains were shared across plant species, suggesting that a wide range of ornamental houseplants harbor similar microbes of potential use in living biofilters.
INTRODUCTION
People in the United States and other industrialized countries inhabit buildings close to 90% of their time (1), and within those building they are exposed to volatile organic compounds (VOCs), which are often found at higher concentrations indoors than outdoors (2, 3). The hundreds of VOCs in indoor air can have a range of health effects, causing chronic health problems and reduced life expectancy (4, 5). VOCs can also cause acute sick building syndrome (SBS), which is a general term for building-specific illnesses involving headaches, dizziness, nausea, sore eyes and throat, or loss of concentration (6). Most indoor VOCs originate indoors from sources such as building materials, furnishings, cleaning agents, and consumer products (7–9). Since worker productivity decreases are correlated with low ventilation rates (10, 11), it is plausible that VOCs have impacts of economic as well as medical importance.
Different methods are used to reduce indoor VOCs. Most frequently, indoor-generated VOC concentrations are diluted with outdoor air by using mechanical ventilation systems (12, 13) in commercial buildings or relying on infiltration air for residences (14). However, this exchanged air must be conditioned to maintain the thermal comfort of the occupants, which increases the use of energy. Since buildings consume approximately 40% of the total U.S. energy supply (15), alternative methods are desirable. Source control is also used to reduce indoor VOCs, and methods of source control include reducing their presence in materials or products (9) or changing materials in existing buildings. However, the removal of pollutant sources can be quite costly and complex, and it is often impractical. Moreover, source control does not account for VOC or particle generation indoors by reactive chemistry, instigated, for instance, by ozone reactions with gases, such as terpenoids (16–18); building material surfaces (19); surface-sorbed compounds (20); or even human skin (21).
Biofiltration is an alternative method of removing VOCs generated indoors and purifying the indoor air (22). Biofilters clean the air by utilizing microbes to consume VOCs and remove them from contaminated airstreams (23). This is potentially advantageous, in that biofiltration may remove indoor VOCs while not requiring energy for conditioning ventilation air. Several laboratory-scale studies have shown that biofilters are capable of removing indoor VOCs present at typical concentrations, including total volatile organic compounds (TVOCs), formaldehyde, toluene, ethylbenzene, and xylene (24, 25).
The work in this study has been largely motivated by the installation of an indoor biofilter at Drexel University, called a biowall, which is shown in Fig. S1 in the supplemental material. Shown in other contexts to remove VOCs (26), the installation of such botanical biofilters has become more common in indoor spaces. The Drexel biowall is a five-story vertical wall with plants rooted into an inorganic, porous textile substrate. Behind this textile, recirculated water constantly trickles, keeping the plants' roots moist. The indoor air is drawn through the substrate by a fan behind the biowall so that VOCs can partition from the air to the water phase, wherein the VOCs should be consumed by the root-colonizing microbes, on the basis of prior findings of rhizosphere degradation activity (27, 28). Microbial consumption of these chemicals should create a steady driving force for the VOCs to move from the air to the water phase. This biofiltered air is then delivered to other zones in the building via the mechanical system. Our ongoing research aims to investigate the efficiency of air purification by such a biofiltration device at the building scale, though smaller, room-scale biofiltration devices have shown good removal (e.g., efficiencies of ∼0.25 to 0.9) of select VOCs (24, 25).
To date, no culture-independent study has investigated the microbial communities inhabiting indoor botanical biofilters. Therefore, we aimed to use next-generation sequencing to characterize the bacteria on plant roots in the Drexel biowall, in another biowall from Morristown, NJ, and in a laboratory-scale biofilter that we constructed. Specifically, we have studied how the root-associated bacteria in biofilters change both over time and in response to VOC challenge. In doing so, we addressed the questions of whether biowall growth consistently favors the proliferation of particular bacteria (objective 1), whether VOC exposure alters root bacterial communities in consistent ways (objective 2), and whether the changes are similar after VOC exposure and wall growth (objective 3). Affirmative findings for objective 3 would suggest that community-level responses to wall growth could, in fact, be direct responses to VOC exposure. Furthermore, consistency across different host plant species would suggest that bacteria likely to impact indoor VOC concentrations can be cultivated on any number of plant species.
MATERIALS AND METHODS
Biowall sampling design.
To investigate the effects of wall growth on microbial communities (objective 1), we took two approaches, referred to as the Dodge experiments and the Drexel experiments. For the Dodge experiments, we collected root samples from plants grown in a greenhouse facility at Parker Plants Inc. (Scotch Plains, NJ) on 17 February 2011. On the same day, roots from identical plant species were collected from plants in the Geraldine R. Dodge Foundation biowall (Morristown, NJ). Plants growing in this biowall had come from the aforementioned greenhouse facility, allowing an ideal comparison between soil-grown and biowall-grown plant root communities. To control for possible host specificity, only two host plants were targeted: Ficus elastica (rubber tree plant) and Schefflera arboricola “Gold Capella” (umbrella tree). Three 2- to 9-cm fine-root-tissue samples were obtained from each of three individual plants of each species from both the soil and biowall settings. While we have no definite way of knowing exactly how long the biowall-grown plants were actually in situ, this time was less than 25 months, as the Dodge biowall has existed since January 2009.
For the Drexel experiments, we sampled the roots of individual biowall-grown plants at two points in time. The first sampling for each plant, taken in July 2011, involved the collection of three 2- to 9-cm lengths of fine root tissues from plants that had been placed in the Drexel biowall for 0 to 24 h. The roots of these plants had been freed of soil after extensive rinsing in Philadelphia, PA, city water. The second round of fine root sampling from the same plants (tagged and individually labeled in July 2011) occurred in March 2012, with the same number of root samples being taken per plant. The design for this study involved nine individual plants from six different plant varieties that were each sampled twice: Croton “Mammy,” rubber tree, Schefflera arboricola “Gold Capella,” Schefflera arboricola, an unidentified Ficus species, and Algerian ivy.
Laboratory experiment design.
To identify whether and how exposure to VOCs alters bacterial root communities (objective 2), we conducted a set of manipulative laboratory experiments using a specially constructed bench-scale laboratory system. To achieve this, we devised a chamber system enabling plant roots to be exposed and challenged with a VOC-laden airstream through aeroponic growth systems. Aeroponics is the process of growing plants with the roots contained in a misted environment in the absence of any sort of medium, soil, water, or aggregate (29) and is akin to hydroponics, in which the plant roots are submerged in water. Biowalls use a trickling water flow to keep the plant roots moist, so our method does not exactly mimic that mechanism, but it is a practical method that may be used to challenge the plant roots with controlled VOCs over time and that allows root samples to be periodically taken for microbial study.
The experimental system, shown in Fig. S2 in the supplemental material, consisted of four identical aeroponic chambers (MicroGarden Aeroponic), capable of growing eight plants each, which we plumbed with 1/4-in. (outer diameter) stainless steel tubing to meet air delivery and VOC introduction/sampling needs. A zero air generator system supplied clean air, and this main airstream was split, so that VOCs could be introduced into one of the airstreams that supplied two of the chambers via a syringe pump and a heated injection system. Both airstreams were split again, so that two airstreams supplied VOC-laden air to the VOC chambers and two supplied clean air to the control chambers. The flow into each chamber was regulated at 1.5 liters/min (determined with an Aalborg FMA5518ST flow meter), and air was drawn out with a vacuum pump at flow rates regulated with rotameters. These flow rates were less than the inflow, so that the chambers were positively pressurized and no laboratory air infiltrated into the system. VOC concentrations up- and downstream of the chambers were measured with a gas chromatograph/flame ionization detector (GC/FID) equipped with an automatic sampling valve and thermal desorption system (catalog no. 8610C; SRI Instruments).
A solution of six VOCs, including propanol, hexanal, perchloroethene, d-limonene, benzene, and toluene, was used in the syringe pump, and those VOCs were delivered to the VOC chambers at mean concentrations of 210 to 14,740 μg/m3, which are about 10 to 100 times the typical concentrations of those VOCs in buildings. The VOCs were chosen because they are specifically detected in many indoor settings (30) and because they represent classes commonly found indoors, namely, alcohols, aldehydes, halogenates, terpenes, and aromatics.
The main events of this study, focused on Ficus elastica (rubber tree plant) and Epipremnum pinnatum cv. Aureum (pothos ivy), are described in Table 1. In short, sampling was performed 22 days prior to VOC exposure and at both 40 and 96 days after exposure was begun. VOC lines were then switched so that clean air-exposed plants were then exposed to dirty air, and vice versa. Roots were sampled 39 days after this switch.
TABLE 1.
Main events of laboratory experiments (objective 2), their dates, and event descriptions
| Event | Date (mo/day/yr) | Event description |
|---|---|---|
| 1 | 12/1/2011 | Placement of plants into chambers A, B, C, and D and provision of clean air only to all plants |
| 2 | 1/10/2012 | Microbial sampling 1 for plants in chambers A, B, C, and D |
| 3 | 2/1/2012 | Beginning of VOC exposure for chambers A and B and clean air in chambers C and D |
| 4 | 3/12/2012 | Microbial sampling 2 for plants in chambers A, B, C, and D |
| 5 | 5/7/2012 | Microbial sampling 3 for plants in chambers A, B, C, and D |
| 6 | 5/7/2012 | Beginning of VOC exposure for plants in chambers C and D and clean air for plants in chambers A and B |
| 7 | 6/15/2012 | Microbial sampling 4 for plants in chambers A, B, C, and D |
| 8 | 6/15/2012 | Cessation of VOC exposure and end of experiments |
General sampling methodology.
In all cases, fine root tissues were taken by researchers wearing sterile blue nitrile gloves. The scissors used to take root tissues were cleaned with ethanol between samplings. Approximately 2- to 9-cm lengths of root tissue were frozen at −80°C until processing, and upon thawing, ∼1- to 2-cm portions were excised from these master samples, with gloves being worn at all times and ethanol being used to wash razor blades between excisions. The samples were subsequently rinsed with sterile deionized water to remove particulate matter. From there, root tissues were macerated in sterile 1.5-ml tubes using sterile plastic pestles after the tissues were frozen in liquid nitrogen. DNA was extracted from these ground samples using a Powersoil DNA isolation kit (MO BIO Laboratories, Carlsbad, CA). After estimaton of the DNA concentration using a NanoDrop spectrophotometer, samples were normalized and combined. For objective 1, we pooled DNA from 3 roots per plant, yielding 12 pooled samples (1 sample from each of 12 plants) for the Dodge biowall-versus-greenhouse comparisons and 18 samples (2 samples from each of 9 plants) for the Drexel prebiowall-versus-postbiowall comparisons. For the VOC exposure extractions (i.e., objective 2), we pooled DNA from two roots per individual plant per time point, yielding 80 samples (i.e., 5 plants × 4 time points × 2 species × 2 treatments). Further details on sample metadata can be found in Table S1 in the supplemental material.
DNA sequencing and bioinformatics.
Amplicon pyrosequencing (performed by the Research and Testing Laboratory, Lubbock, TX) targeted the V1-V3 variable regions of the 16S rRNA gene, which were amplified with primers Gray28F (5′-GAGTTTGATCNTGGCTCAG) and Gray519R (5′-GTNTTACNGCGGCKGCTG). A total of 558,955 raw sequences from 112 samples were analyzed using the QIIME pipeline (version 1.6.0) (31). All low-quality or ambiguous reads were removed using the default quality control parameters of a minimum sequence quality score of 25 and a minimum read length of 200 bp. Sequences with mismatches in primer regions or with homopolymer tracts longer than 6 bp were also discarded. The remaining 497,336 reads were denoised by the QIIME denoiser and grouped into operational taxonomic units (OTUs) at 97% similarity using the uclust program (32). The most abundant sequence from each OTU was chosen as a representative and used to assign a taxonomic identity to each OTU using the RDP classifier (33) with a 50% bootstrap support cutoff. OTUs classified as chloroplasts were removed, and representative sequences of the remaining OTUs were aligned by PyNAST (34) and checked for chimeras against the Greengenes reference database of 16S rRNA gene sequences (http://greengenes.lbl.gov/cgi-bin/nph-index.cgi) with ChimeraSlayer. Potentially chimeric OTUs were removed from all subsequent analyses. Using this data set, we then utilized QIIME to produce a table of nonchimeric OTUs containing information on the abundance and taxonomy of bacteria from all samples in our study (see Table S2 in the supplemental material).
Statistical analyses of 454 pyrosequencing data.
To estimate the alpha diversity within samples, rarefaction curves were constructed from the estimated number of OTUs with 97% similarity in each individual sample with iterations of 10 and increments of 100 using QIIME (see Fig. S3 in the supplemental material). OTU tables were rarefied to 466 (the lowest read number among 115 samples) to equalize sampling depth. After rarefaction and normalization, we also computed species richness (Chao 1 estimator) and diversity (Shannon index) for each sequence library (see Table S2 in the supplemental material). Beta diversity, or the similarity of communities from different samples, was estimated from these normalized libraries using both the UniFrac and Bray-Curtis metrics in QIIME (35–37). Weighted UniFrac distances, which consider both the presence/absence and the relative abundance of lineages, were estimated on the basis of the fraction of the phylogenetic branch length shared by plant root bacterial communities. The maximum likelihood phylogeny for this analysis was inferred from an alignment of all representative sequences using FastTree (38). Weighted UniFrac distance and Bray-Curtis distance values were then used for principal coordinates analyses (PCoAs), and the results were visualized using Origin software (Microcal Software Inc., Northampton, MA). Overall, UniFrac results were similar to those from Bray-Curtis analyses for all analyses, so for simplicity, we present below only those measures from the latter.
Community variation among experimental classes of plant roots was assessed using the ADONIS program in the vegan package (39), implemented in R (version 2.02), which performed permutational multivariate analysis of variance tests on the basis of weighted UniFrac or Bray-Curtis distance matrices (1,000 permutations). Analysis of variance (ANOVA) tests (performed in QIIME, version 1.6.0) were separately used to identify OTUs with differing relative abundances between VOC-exposed (VOC+) plants and non-VOC-exposed (VOC−) plants, as well as between biowall-grown and soil-grown plants.
In addition to our focus on OTUs with 97% similarity for beta diversity analyses, we also compared root communities after classification of sequence reads to the family level, using the ADONIS and ANOVA approaches described in the previous paragraph. By considering community composition across multiple taxonomic scales, our approach allowed us to draw broader conclusions on the trends of beta diversity.
Nearly all of our statistical analyses were designed to test objectives 1, 2, and 3. The first objective was assessed by comparing wall and soil communities from the Drexel and Dodge experiments in both separate and pooled analyses. Objective 2 was achieved through analyses of the laboratory experiments, using VOC exposure at the time of collection as the independent variable. Objective 3 was achieved by comparing the communities of VOC+ and wall-grown plants to those of clean air-exposed- and soil-grown plants. We were mindful of the variation in plant species and its likely impact on community composition; hence, we examined whether patterns from our analyses were consistent across different plants. In summary, through the aforementioned ANOVA and beta diversity statistics, we were able to tell whether soil-grown root bacterial communities shifted in response to wall growth (objective 1), whether VOCs altered the profile of root bacterial communities (objective 2), and whether VOC exposure and wall growth had similar impacts on root microbial communities (objective 3).
Follow-up analyses on biowall and VOC+ proliferators.
The proliferation of Hyphomicrobium bacteria from three OTUs on biowall-grown and VOC-exposed plant roots led us to further explore the diversity of strains and species from this genus. To do so, we retrieved all raw sequence reads classifying to this taxon. Chimeras were removed, as were sequences with errors in primers or bar codes. The remaining reads were trimmed to a homologous 220- to 224-bp region. Sequences were aligned using Muscle in the SeaView package (32, 40) (the alignment is available upon request). From there, the program Codon Code Aligner (version 4.02; CodonCode Corporation, Centerville, MA) was used to generate a table of polymorphic sites, i.e., all those where the frequency of the minor allele exceeded 1% across the entire pool of raw, quality Hyphomicrobium sequence reads. We excluded variable sites arising due to indels in homopolymer regions of 3 bp or longer. Genotypes were then computed for each Hyphomicrobium sequence read by concatenating all variable sites. From there, we determined the fractional contribution of common genotypes/strains (i.e., those making up at least 8% of all raw, quality reads in at least one sequence library) to the total number of raw, quality reads from each library, comparing these among roots from different plant species and treatments.
A maximum likelihood phylogenetic analysis was performed to place these and other strains (i.e., all strains with at least 25 representative reads across the entire data set of raw, quality sequences) into a broader evolutionary context. Sequences were aligned as described above in SeaView, where maximum likelihood analysis was performed using the PhyML program with default parameters and 500 bootstrap replicates (41). The alignment matrix used for this analysis also included sequences identified from the NCBI database via BLASTn searches against the nr/nt database using a subset of our sequences as queries. We further included representative sequences from a previous phylogenetic analysis of the genus Hyphomicrobium (42). An outgroup 16S rRNA sequence from Escherichia coli was used to root the tree.
Nucleotide sequence accession numbers.
Single representatives of each analyzed Hyphomicrobium genotype from our amplicon sequencing libraries were deposited in the NCBI database under GenBank accession numbers KJ820976 to KJ820991. Also, sequences were deposited in NCBI's Sequence Read Archive under accession number SRP042016.
RESULTS
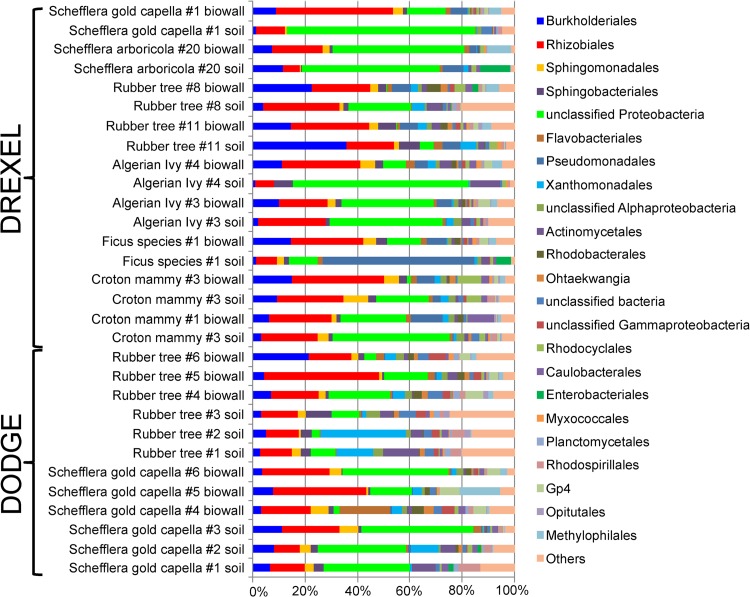
A total of 479,880 sequence reads from all samples were retained after quality control analyses. Chloroplast-free libraries ranged from 466 to 20,010 quality, denoised sequences, with a median of 2,301. At the 97% sequence identity level, a total of 4,633 OTUs were identified across all libraries, while a median of 225 OTUs was found in each sequence library. After normalizing all sequence libraries to 466 reads, this median value equaled 121 OTUs (see Table S2 in the supplemental material). Root communities from all experiments were dominated by Proteobacteria, with substantial fractions of Rhizobiales and Burkholderiales. Bacteroidetes, including Sphingobacteriales and Flavobacteriales, also made up sizeable fractions of these communities (Fig. 1; see also Fig. S4 in the supplemental material).
FIG 1.
Taxonomic makeup of root communities from the Drexel and Dodge (biowall) experiments. An ordinal-level classification is shown for nonnormalized sequence libraries of biowall- and soil-grown plant roots. Sequences not classifying to the level of order with 50% bootstrap support were placed into the next highest taxon meeting this threshold.
Community-level differences.
For objective, 1, separate statistical analyses on the Drexel and Dodge experiments revealed significantly different communities of root bacteria between biowall-grown and soil-grown plants. When pooled across all plant species, ADONIS analyses of Bray-Curtis values for OTUs with 97% similarity yielded P values of 0.001 and 0.004 for the Drexel and Dodge comparisons, respectively (Table 2; see also Table S3 in the supplemental material for the results of similar analyses focused on the family level for this comparison as well as those described below). Biowall-grown plant communities were also distinct from soil-grown communities in an analysis pooling samples from the Drexel and Dodge biowalls (P = 0.001; see Table S3 in the supplemental material). Strikingly, in a PCoA plot of this set of pooled data (see Fig. S5 in the supplemental material), nearly all biowall-grown root communities showed a rightward shift along the first axis compared to their counterparts from the same plant species (filled symbols) grown in soil. Biowall-grown plant root communities also clustered near the top of the second PCoA axis. Both trends were consistent, in spite of our inclusion of multiple host plant species and data from the two biowall environments, suggesting that biowall growth may have a consistent effect on bacterial communities that colonize plant roots.
TABLE 2.
Statistical analyses assessing root community differences at OTU 97% similarity level between plants under different rearing conditions
| Expt | Variable | Treatment 1 | Treatment 2 | ADONIS P value |
|---|---|---|---|---|
| Dodge biowall vs soil | Growth location | Soil growth | Wall growth | 0.004 |
| Drexel biowall vs soil | Growth location | Soil growth | Wall growth | 0.001 |
| Drexel and Dodge biowalls | Growth location | Soil growth | Wall growth | 0.001 |
| Laboratory pothos ivy | Air purity | VOC+ air | VOC− air | 0.001 |
| Laboratory rubber tree | Air purity | VOC+ air | VOC− air | 0.001 |
| All combined | Growth conditions/air purity | VOC+ air/wall growth | VOC− air/soil growth | 0.001 |
Like the shifts due to biowall growth, controlled exposure to indoor VOCs in the laboratory experiments had a similarly large effect on bacterial root communities in objective 2. When analyzed separately for the two plant species, which harbored notably different microbial communities, VOC-exposed communities from pothos ivy were found to cluster to the right of the first principal coordinate axis (see Fig. S6a in the supplemental material) and very tightly toward the middle of the second axis, suggesting that exposure to these compounds had a rather strong, homogenizing effect on root bacteria. Communities from VOC-exposed rubber tree roots clustered near the bottom of the third principal coordinate axis and toward the left end of the second axis (see Fig. S6b in the supplemental material). Statistical analyses accordingly showed that VOC treatment had effects on the root communities from both rubber tree and pothos ivy (see Table 2 for the results of analyses of OTUs with 97% similarity; see Table S3 in the supplemental material for analyses at the family level).
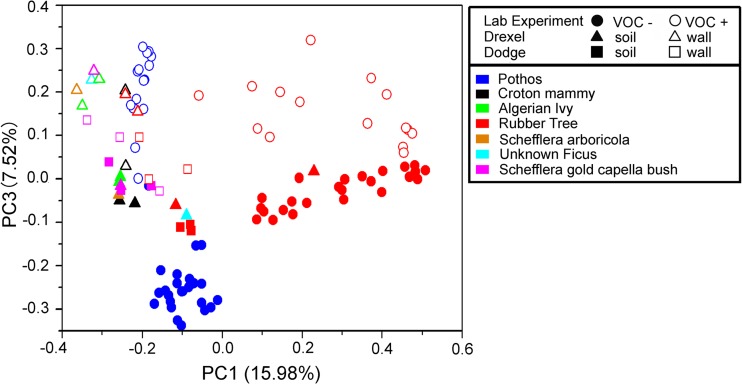
We hypothesized that biowall growth leads to alterations in bacterial root communities due to prolonged VOC exposure (objective 3). One prediction from this hypothesis is that controlled VOC exposure and biowall growth should lead to similar community shifts. To assess this, we performed statistical analyses of a combination of results for VOC-exposed plant root communities and biowall-grown plant root communities and, separately, results for communities from the roots of plants not exposed to VOCs and those from the roots of plants grown in soil. ADONIS statistics revealed a significant difference between these two experimental classes (P = 0.001). This difference matched the patterns in our PCoA plots, with biowall-grown and VOC+ plant root communities clustering at the top of the third axis (Fig. 2).
FIG 2.
Principal coordinates analysis reveals the similarities of bacterial root communities in VOC-exposed plants and those from biowalls. Open symbols, communities from VOC+ and biowall-grown roots, which generally clustered on the upper end of the third principal coordinate axis; filled symbols, VOC− and soil-grown roots. Different shapes reveal different experiments, whereas different colors help to distinguish the host plant species considered in this pooled analysis. While these results were based on community similarity measures obtained with the Bray-Curtis metric, weighted UniFrac distances yielded generally similar findings (data not shown).
Biowall and VOC+ proliferators.
A number of bacterial families proliferated in response to wall versus soil growth and in response to VOC exposure (see Table S4 in the supplemental material for statistics). The family showing the most notable parallel enrichment under VOC exposure and wall growth conditions (relative to the communities present under no VOC exposure and soil growth conditions) was the Hyphomicrobiaceae, which rose in frequency between 3.8 and 23.8% in a consistent fashion across plant species and biowalls. Bacteria from the Pseudomonadaceae, the Rhodobacteraceae, and Burkholderia incertae sedis all showed significant increases across three of four VOC+/wall growth -versus-VOC−/soil growth comparisons. In these cases, relative frequencies increased from 2.3 to 9.3%. Members of the Rhodospirillaceae were, in contrast, significantly less common in three of four VOC+/wall growth libraries (0.6 to 2.7% reduction).
ANOVA analyses of OTUs varying significantly across treatments revealed one OTU classified to the genus Hyphomicrobium (OTU4720) with a notably higher relative abundance in both VOC+ and biowall-grown plants than in VOC− and soil-grown conditions (2.1 to 11.6% enrichment). Two other OTUs classifying to the same genus (OTU12 and OTU4794) showed enrichment in roots from plants grown on one or both biowalls compared to their levels in the roots of plants grown in soil. In addition, four OTUs proliferating in response to VOC exposure came from the family Comamonadaceae. Additional OTUs with changing relative abundance across one or more treatments were more broadly scattered across genera and families, as shown in Table S5 in the supplemental material.
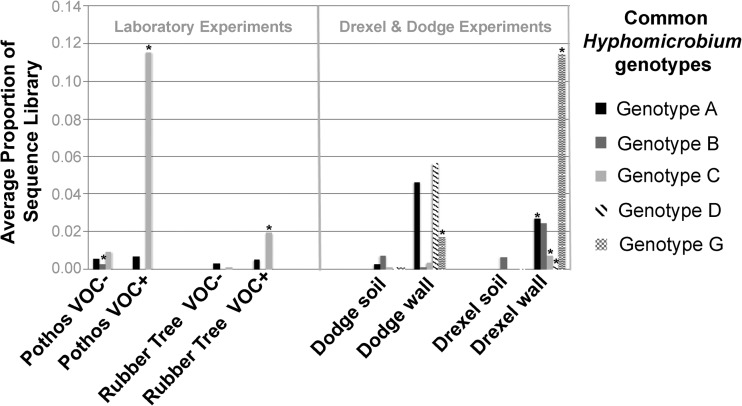
To identify the strains responsible for this proliferation, we calculated the relative abundance of 16S rRNA genotypes from all raw, quality reads classifying to the genus Hyphomicrobium (see Table S6 in the supplemental material). Several genotypes reached a high abundance within sequence libraries, adding up to ∼31% in the most dominated library. By focusing on those with at least 5% abundance in at least one library, we narrowed the range of dominant Hyphomicrobium genotypes to five, naming these genotypes A, B, C, D, and G, which variably dominated across different treatments (Table 3; Fig. 3). For instance, genotype G was significantly enriched in wall versus soil roots across plant species in the Drexel and Dodge experiments. Genotypes A, C, and D were also enriched after plants were replanted in the Drexel biowall, though the last two remained quite rare among the sampled microbes. Genotype C dominated in the laboratory experiment plants, for instance, making up an average of 11.5% of all raw, quality sequences from pothos ivy roots exposed to VOCs. In contrast to these trends, reads from the rare genotype B actually decreased after VOC exposure.
TABLE 3.
P values from t tests of Hyphomicrobium strain/genotype abundance across biowall versus soil growth or VOC presence versus absence
| Expt |
P value for the following genotype: |
||||
|---|---|---|---|---|---|
| A | B | C | D | G | |
| Laboratory expt (pothos ivy)a | 0.640 | 0.007 | 0.000 | NAb | NA |
| Laboratory expt (rubber tree)a | 0.300 | NA | 0.000 | NA | NA |
| Dodge exptc | 0.253 | 0.302 | 0.292 | 0.066 | 0.046 |
| Drexel exptc | 0.002 | 0.057 | 0.005 | 0.048 | 0.002 |
Comparisons are between VOC-exposed and clean air-exposed plants.
NA, not applicable.
Comparisons are between biowall-grown and soil-grown plants.
FIG 3.
Average relative abundance of Hyphomicrobium strains/genotypes across experiments. Values show the average proportion of the total number of quality reads made up by the named common genotypes (see Table S5 in the supplemental material for more details on each and for raw data on genotype abundance). *, means significantly different from those in the paired experimental treatment (i.e., wall- versus soil-grown plants or VOC+ versus VOC− plants). Note that in all but one case where the results were significantly different, Hyphomicrobium strains were more abundant under VOC exposure or biowall growth. Note also that different strains seemed to proliferate in different contexts.
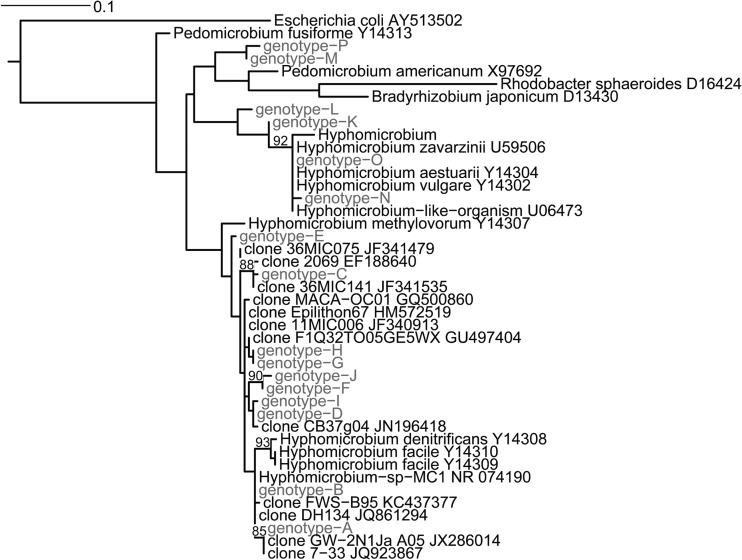
Our phylogenetic analysis (Fig. 4) revealed that the more common genotypes A to D and G were closely related to one another and to Hyphomicrobium denitrificans, H. facile, and H. methylovorum. Also related to our biowall and VOC+ proliferators (genotypes A, C, D, and G and genotype C, respectively) were various uncultured bacteria from different water sources, soil, and sewer biofilms. It should be noted that bootstrap support was low, overall, across the tree, and thus, many of these relationships remain tentative.
FIG 4.
Maximum likelihood phylogeny showing relationships among common and rare Hyphomicrobium species from plant roots. Those identified in this study are indicated in gray font. Bootstrap values of >80% (based on 500 maximum likelihood bootstrap replicates) are placed on their respective nodes.
DISCUSSION
Bacterial communities are known to shift in response to environmental variables (43–45). Such shifts may often correspond to competitive dynamics unfolding when certain bacteria are given access to ideal (or suboptimal) food, temperature, humidity, salinity, pH, etc. So, given the known capacities for bacteria to degrade and utilize carbon from VOCs, in the laboratory experiments described here we hypothesized that bacterial communities would be altered after VOC exposure (objective 2). Indeed, this hypothesis was supported on the basis of our observations of predictable community shifts of root communities. Consistency across replicate aeroponic chambers, across individual plants, and across plant species lent further support to this phenomenon.
It is quite possible that those microbes showing proliferation after VOC treatment are VOC degraders. Indeed, genome annotations from the Hyphomicrobium genus (www.genome.jp/kegg/), which includes representatives from plant rhizospheres (46), suggest that some species carry genes involved in toluene degradation. Similarly, some Hyphomicrobium strains can degrade formaldehyde, a capacity that has been harnessed by the biosensor industry (47). Other research on biofiltration has identified aromatic/halogenated compound breakdown by relatives of common plant root associates, including Hyphomicrobium, Alcaligenes, Acinetobacter, Burkholderia, Pseudomonas, and Xanthobacter (23). Thus, the capacity for root-associated VOC degradation activity by plants comes as little surprise (24, 25, 28).
The inclusion of benzene, toluene, and perchloroethene in our VOC mixture thus raises a candidate mechanism behind our observation of Hyphomicrobium species proliferation, namely, that the additional carbon sources were utilized by these bacteria, giving them a competitive edge. Further study of this possibility, of the fungal portion of root communities, and of the VOC-degrading capacities for other proliferating microbes (e.g., see Tables S4 and S5 in the supplemental material) should be undertaken to better understand the ways in which plants and their microbes might help to purify our air (or soils).
The proliferation of Hyphomicrobium on biowall- versus soil-grown plants provided an intriguing link between VOC treatment and wall growth. Similar patterns were gleaned for bacteria from the genus Rhizobacter (see Table S5 in the supplemental material), although those proliferating on biowalls versus those proliferating upon VOC exposure under aeroponic growth came from different OTUs. Similarly, when examined at the strain (i.e., 16S rRNA genotype) level, the dominant Hyphomicrobium variants differed between aeroponic and biowall conditions and to some extent between biowalls. This suggests that the growth environment could constrain the success of related microbes that are favored after VOC exposure. Nevertheless, the known degradation capacities of these bacterial groups and the expected exposure of wall-growing plant root communities to higher concentrations of VOCs than soil-grown plants hint that VOC exposure could be a main driver of community shifts in biowall-grown plants. The proliferation of VOC degraders in response to airborne chemicals should facilitate the abilities of biowalls to purify indoor air and is, in our opinion, quite worthy of future study.
Finally, it is worth noting how quickly the bacterial communities responded to changes in VOC exposure. In the laboratory experiments, microbial sample 2 (Table 1) was taken 40 days after VOC exposure commenced, yet bacterial communities had already shifted compared to those from the same plants in microbial sample 1. These communities did not change drastically for the 57 days (through the time of sampling of microbial sample 3) under constant VOC exposure, but after switching the VOC and control chambers, communities had essentially reversed in character within 40 days (i.e., by the time that microbial sample 4 was taken) (see Fig. S6 in the supplemental material). The speed of these community-level shifts is consistent with prior findings showing changes in VOC removal by bacterial communities after only days (48, 49). So, given the lack of a strong time lag in community shifts and the distribution of proliferating bacterial species and strains across plant varieties, it appears that VOC degraders may be poised for activity across a range of ornamental plants, enabling the use of many varieties in green biofilters.
Conclusions.
The risks posed by microbes and airborne chemicals necessitate the study of the indoor habitat as we attempt to create healthier work and living spaces. The recent drive to characterize the indoor microbiome (50, 51, 53) has often focused on pathogenic microbes (52), while the emphasis on air purification has often been placed on ventilation for VOC control (11, 13) and the use of nonliving filters for control of aerosols (54, 55). However, our work, combined with previous efforts (24, 25), suggests a hidden dimension of the indoor environment involving beneficial microbes. We advocate that VOC degraders thus be more broadly explored as residents of indoor habitats, as their natural colonization of building surfaces could serve to mitigate the harmful impacts of polluted indoor air.
Supplementary Material
ACKNOWLEDGMENTS
We thank Donna Murasko, Drexel's dean of the College of Arts and Sciences, for her advocacy and feedback on this work. Jay Satava and his team from Parker Plants were immensely helpful in enabling the sampling of plant roots. Cynthia Evans from the Geraldine R. Dodge Foundation was also a great help in this regard. Alan Darlington from Nedlaw Living Walls has provided useful feedback since the inception of this work. We owe further gratitude to Drexel students Young Kwang Lee, who helped to construct the laboratory apparatus, and Matthew Novin, who contributed helpful work and discussions in the early stages of our biowall research.
Funding for this work was provided by the Pennsylvania Department of Health Commonwealth Universal Research Enhancement Program (CURE).
Footnotes
Published ahead of print 30 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00595-14.
REFERENCES
- 1.Klepeis NE, Nelson WC, Ott WR, Robinson JP, Tsang AM, Switzer P, Behar JV, Hern SC, Engelmann WH. 2001. The national human activity pattern survey (NHAPS): a resource for assessing exposure to environmental pollutants. J. Expo. Anal. Environ. Epidemiol. 11:231–252. 10.1038/sj.jea.7500165 [DOI] [PubMed] [Google Scholar]
- 2.U.S. Environmental Protection Agency. 2006. Building assessment survey and evaluation (BASE). U.S. Environmental Protection Agency, Washington, DC: http://www.epa.gov/iaq/base Accessed 18 February 2013 [Google Scholar]
- 3.Weisel CP, Zhang J, Turpin BJ, Morandi MT, Colome S, Stock TH, Spektor DM. 2005. Relationships of indoor, outdoor, and personal air (RIOPA). Part I. Collection methods and descriptive analyses. HEI research report 130 and NUATRC research report 7. Heath Effects Institute, Boston MA, and Mickey Leland National Urban Air Toxics Research Center, Houston, TX: [PubMed] [Google Scholar]
- 4.Logue JM, McKone TE, Sherman MH, Singer BC. 2011. Hazard assessment of chemical air contaminants measured in residences. Indoor Air 21:92–109. 10.1111/j.1600-0668.2010.00683.x [DOI] [PubMed] [Google Scholar]
- 5.Logue JM, Price PN, Sherman MH, Singer BC. 2012. A method to estimate the chronic health impact of air pollutants in US residences. Environ. Health Perspect. 120:216–222. 10.1289/ehp.1104035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones AP. 1999. Indoor air quality and health. Atmos. Environ. 33:4535–4564. 10.1016/S1352-2310(99)00272-1 [DOI] [Google Scholar]
- 7.Nazaroff WW, Weschler CJ. 2004. Cleaning products and air fresheners: exposure to primary and secondary air pollutants. Atmos. Environ. 38:2841–2865. 10.1016/j.atmosenv.2004.02.040 [DOI] [Google Scholar]
- 8.Singer BC, Destaillats H, Hodgson AT, Nazaroff WW. 2006. Cleaning products and air fresheners: emissions and resulting concentrations of glycol ethers and terpenoids. Indoor Air 16:179–191. 10.1111/j.1600-0668.2005.00414.x [DOI] [PubMed] [Google Scholar]
- 9.Weschler CJ. 2009. Changes in indoor pollutants since the 1950s. Atmos. Environ. 43:153–169. 10.1016/j.atmosenv.2008.09.044 [DOI] [Google Scholar]
- 10.Fisk WJ, Black D, Brunner G. 2012. Changing ventilation rates in US offices: implications for health, work performance, energy, and associated economics. Build. Environ. 47:368–372. 10.1016/j.buildenv.2011.07.001 [DOI] [Google Scholar]
- 11.Sundell J, Levin H, Nazaroff WW, Cain WS, Fisk WJ, Grimsrud DT, Gyntelberg F, Li Y, Persily AK, Pickering AC, Samet JM, Spengler JD, Taylor ST, Weschler CJ. 2011. Ventilation rates and health: multidisciplinary review of the scientific literature. Indoor Air 21:191–204. 10.1111/j.1600-0668.2010.00703.x [DOI] [PubMed] [Google Scholar]
- 12.ASHRAE. 2013. ANSI/ASHRAE standard 62.1-2013—ventilation for acceptable indoor air quality. American Society for Heating, Refrigerating, and Air-Conditioning Engineers, Atlanta, GA [Google Scholar]
- 13.Rackes A, Waring MS. 2013. Modeling impacts of dynamic ventilation strategies on indoor air quality of offices in six US cities. Build. Environ. 60:243–253. 10.1016/j.buildenv.2012.10.013 [DOI] [Google Scholar]
- 14.Murray DM, Burmaster DE. 1995. Residential air exchange rates in the United States—empirical and estimated parametric distributions by season and climatic region. Risk Anal. 15:459–465. 10.1111/j.1539-6924.1995.tb00338.x [DOI] [Google Scholar]
- 15.Perez-Lombard L, Ortiz J, Pout C. 2008. A review on buildings energy consumption information. Energy Buildings 40:394–398. 10.1016/j.enbuild.2007.03.007 [DOI] [Google Scholar]
- 16.Waring MS. 6 January 2014. Secondary organic aerosol in residences: predicting its fraction of fine particle mass and determinants of formation strength. Indoor Air 10.1111/ina.12092 [DOI] [PubMed] [Google Scholar]
- 17.Waring MS, Siegel JA, Corsi RL. 2008. Ultrafine particle removal and generation by portable air cleaners. Atmos. Environ. 42:5003–5014. 10.1016/j.atmosenv.2008.02.011 [DOI] [Google Scholar]
- 18.Waring MS, Wells JR, Siegel JA. 2011. Secondary organic aerosol formation from ozone reactions with single terpenoids and terpenoid mixtures. Atmos. Environ. 45:4235–4242. 10.1016/j.atmosenv.2011.05.001 [DOI] [Google Scholar]
- 19.Wang H, Morrison GC. 2006. Ozone-initiated secondary emission rates of aldehydes from indoor surfaces in four homes. Environ. Sci. Technol. 40:5263–5268. 10.1021/es060080s [DOI] [PubMed] [Google Scholar]
- 20.Waring MS, Siegel JA. 2013. Indoor secondary organic aerosol formation initiated from reactions between ozone and surface-sorbed d-limonene. Environ. Sci. Technol. 47:6341–6348. 10.1021/es400846d [DOI] [PubMed] [Google Scholar]
- 21.Wisthaler A, Weschler CJ. 2010. Reactions of ozone with human skin lipids: sources of carbonyls, dicarbonyls, and hydroxycarbonyls in indoor air. Proc. Natl. Acad. Sci. U. S. A. 107:6568–6575. 10.1073/pnas.0904498106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guieysse B, Hort C, Platel V, Munoz R, Ondarts M, Revah S. 2008. Biological treatment of indoor air for VOC removal: potential and challenges. Biotechnol. Adv. 26:398–410. 10.1016/j.biotechadv.2008.03.005 [DOI] [PubMed] [Google Scholar]
- 23.Malhautier L, Khammar N, Bayle S, Fanlo JL. 2005. Biofiltration of volatile organic compounds. Appl. Microbiol. Biotechnol. 68:16–22. 10.1007/s00253-005-1960-z [DOI] [PubMed] [Google Scholar]
- 24.Darlington A, Chan M, Malloch D, Pilger C, Dixon MA. 2000. The biofiltration of indoor air: implications for air quality. Indoor Air 10:39–46. 10.1034/j.1600-0668.2000.010001039.x [DOI] [PubMed] [Google Scholar]
- 25.Darlington A, Dat JF, Dixon MA. 2001. The biofiltration of indoor air: air flux and temperature influences the removal of toluene, ethylbenzene and xylene. Environ. Sci. Technol. 35:240–246. 10.1021/es0010507 [DOI] [PubMed] [Google Scholar]
- 26.Chen W, Zhang JS, Zhang Z. 2005. Performance of air cleaners for removing multiple volatile organic compounds in indoor air. ASHRAE Trans. 111:1101–1114 [Google Scholar]
- 27.Wolverton BC, Johnson A, Bounds K. 1989. Interior landscape plants for indoor air pollution abatement, p 1–22, National Aeronautics and Space Administration, Washington, DC [Google Scholar]
- 28.Wolverton BC, McDonald RC, Watkins EA. 1984. Foliage plants for removing indoor air pollutants from energy-efficient homes. Econ. Bot. 38:224–228. 10.1007/BF02858837 [DOI] [Google Scholar]
- 29.Weathers PJ, Zobel RW. 1992. Aeroponics for the culture of organisms, tissues and cells. Biotechnol. Adv. 10:93–115. 10.1016/0734-9750(92)91353-G [DOI] [PubMed] [Google Scholar]
- 30.Brown SK, Sim MR, Abramson MJ, Gray CN. 1994. Concentrations of volatile organic compounds in indoor air: a review. Indoor Air 4:123–124. 10.1111/j.1600-0668.1994.t01-2-00007.x [DOI] [Google Scholar]
- 31.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 33.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145. 10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. 10.1093/bioinformatics/btp636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamady M, Lozupone C, Knight R. 2010. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 4:17–27. 10.1038/ismej.2009.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235. 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lozupone CA, Hamady M, Kelley ST, Knight R. 2007. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73:1576–1585. 10.1128/AEM.01996-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26:1641–1650. 10.1093/molbev/msp077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MRH, Wagner H. 2011. vegan: community ecology package. R package version 2.0-2 The R Project for Statistical Computing, Vienna, Austria [Google Scholar]
- 40.Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27:221–224. 10.1093/molbev/msp259 [DOI] [PubMed] [Google Scholar]
- 41.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- 42.Layton AC, Karanth PN, Lajoie CA, Meyers AJ, Gregory IR, Stapleton RD, Taylor DE, Sayler GS. 2000. Quantification of Hyphomicrobium populations in activated sludge from an industrial wastewater treatment system as determined by 16S rRNA analysis. Appl. Environ. Microbiol. 66:1167–1174. 10.1128/AEM.66.3.1167-1174.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gehring C, Flores-Rentería D, Sthultz CM, Leonard TM, Flores-Rentería L, Whipple AV, Whitham TG. 2014. Plant genetics and interspecific competitive interactions determine ectomycorrhizal fungal community responses to climate change. 23:1379–1391 Mol. Ecol. 10.1111/mec.12503 [DOI] [PubMed] [Google Scholar]
- 44.Hu Y, Łukasik P, Moreau CS, Russell JA. 2014. Correlates of gut community composition across an ant species (Cephalotes varians) elucidate causes and consequences of symbiotic variability. Mol. Ecol. 23:1284–1300. 10.1111/mec.12607 [DOI] [PubMed] [Google Scholar]
- 45.Oliveira V, Gomes NCM, Almeida A, Silva AMS, Simões MMQ, Smalla K, Cunha Â. 2014. Hydrocarbon contamination and plant species determine the phylogenetic and functional diversity of endophytic degrading bacteria. Mol. Ecol. 23:1392–1404. 10.1111/mec.12559 [DOI] [PubMed] [Google Scholar]
- 46.Crowley D. 2001. Function of siderophores in the plant rhizosphere. In Pinton R, Varanini Z, Nannipieri P. (ed), The rhizosphere: biochemistry and organic substances at the soil-plant interface. Marcel Dekker, New York, NY [Google Scholar]
- 47.Achmann S, Hermann M, Hilbrig F, Jerome V, Hammerle M, Freitag R, Moos R. 2008. Direct detection of formaldehyde in air by a novel NAD(+)- and glutathione-independent formaldehyde dehydrogenase-based biosensor. Talanta 75:786–791. 10.1016/j.talanta.2007.12.015 [DOI] [PubMed] [Google Scholar]
- 48.Mathur AK, Majumder CB, Dhananjay S, Shashi B. 2010. Biodegredation of mono-cholorobenzene by using a trickle bed air biofilter (TBAB). J. Environ. Biol. 31:445–451 [PubMed] [Google Scholar]
- 49.Shukla AK, Vishwakarma P, Singh RS, Upadhyay SN, Dubey SK. 2010. Bio-filtration of trichloroethylene using diazotrophic bacterial community. Bioresour. Technol. 101:2126–2133. 10.1016/j.biortech.2009.10.094 [DOI] [PubMed] [Google Scholar]
- 50.Adams RI, Miletto M, Taylor JW, Bruns TD. 2013. Dispersal in microbes: fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. ISME J. 7:1262–1273. 10.1038/ismej.2013.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jeon YS, Chun J, Kim BS. 2013. Identification of household bacterial community and analysis of species shared with human microbiome. Curr. Microbiol. 67:557–563. 10.1007/s00284-013-0401-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kembel SW, Jones E, Kline J, Northcutt D, Stenson J, Womack AM, Bohannan BJM, Brown GZ, Green JL. 2012. Architectural design influences the diversity and structure of the built environment microbiome. ISME J. 6:1469–1479. 10.1038/ismej.2011.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kettleson E, Kumar S, Reponen T, Vesper S, Meheust D, Grinshpun SA, Adhikari A. 2013. Stenotrophomonas, Mycobacterium, and Streptomyces in home dust and air: associations with moldiness and other home/family characteristics. Indoor Air 23:387–396. 10.1111/ina.12035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bekö G, Clausen G, Weschler CJ. 2008. Is the use of particle air filtration justified? Costs and benefits of filtration with regard to health effects, building cleaning and occupant productivity. Build. Environ. 43:1647–1657. 10.1016/j.buildenv.2007.10.006 [DOI] [Google Scholar]
- 55.Waring MS, Siegel JA. 2008. Particle loading rates for HVAC filters, heat exchangers, and ducts. Indoor Air 18:209–224. 10.1111/j.1600-0668.2008.00518.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.