Abstract
The presence of methicillin-susceptible Staphylococcus aureus (MSSA) was analyzed in different free-living wild animals to assess the genetic diversity and predominant genotypes on each animal species. Samples were taken from the skin and/or nares, and isolates were characterized by spa typing, multilocus sequence typing (MLST) and antimicrobial susceptibility testing. The proportion of MSSA carriers were 5.00, 22.93, 19.78, and 17.67% in Eurasian griffon vulture, Iberian ibex, red deer, and wild boar, respectively (P = 0.057). A higher proportion of isolates (P = 0.000) were recovered from nasal samples (78.51%) than skin samples (21.49%), but the 9.26% of red deer and 18.25% of wild boar would have been undetected if only nasal samples had been tested. Sixty-three different spa types were identified, including 25 new spa types. The most common were t528 (43.59%) in Iberian ibex, t548 and t11212 (15.79% and 14.04%) in red deer, and t3750 (36.11%) in wild boar. By MLST, 27 STs were detected, of which 12 had not been described previously. The most frequent were ST581 for Iberian ibex (48.72%), ST425 for red deer (29.82%), and ST2328 for wild boar (42.36%). Isolates from Eurasian griffon vulture belong to ST133. Host specificity has been observed for the most frequent spa types and STs (P = 0.000). The highest resistance percentage was found against benzylpenicillin (average, 22.2%), although most of the S. aureus isolates were susceptible to all antimicrobial tested. Basically, MSSA isolates were different from those MRSA isolates previously detected in the same animal species.
INTRODUCTION
Staphylococcus aureus is a commensal microorganism in animals and humans (1) that colonizes mainly the nares but also the throat and skin (2–4). S. aureus is also the causative agent of several diseases, such as pneumonia and wound and bloodstream infections (3), and colonization has been associated with clinical infection (5, 6).
Methicillin-resistant S. aureus (MRSA) emerged by the integration of resistance mechanisms in methicillin-susceptible S. aureus (MSSA) (7, 8). The acquisition of mecA (9) or mecC (10) is a public health concern due to limited options for treatment. Moreover, MRSA infections are related to longer hospitalization stays and higher mortality (11, 12).
MRSA have been detected in domestic animals (13, 14). The genetic background and antimicrobial resistance of S. aureus have been associated with host specificity in livestock (8, 15, 16). Companion animals are normally colonized by human-related genotypes, although some studies have described colonization factors that determine host specificity (8, 13). MRSA detection in free-living wild animals in Spain has revealed a very low prevalence but genotypes related to livestock and humans (17, 18). The genetic diversity of MSSA has been studied in domestic animals, also revealing predominant genotypes in different hosts (19, 20). However, little is known about the molecular epidemiology of the susceptible S. aureus population in free-living wild animals. In the present study, we investigated the presence of MSSA in free-living wild animals (Eurasian griffon vulture, Iberian ibex, red deer, and wild boar) to assess the genetic diversity and predominant genotypes of MSSA on each animal species. This study would generate new knowledge concerning niches of S. aureus that have not been previously investigated.
MATERIALS AND METHODS
Sampling.
Apparently healthy animals were captured (box trapping) or hunted between March 2009 and November 2011 in 10 different Spanish provinces. Animals were sampled with sterile swabs through the nares and/or swabbing on ∼1 cm2 of skin (ears or inguinal-mammal area). In total, 2,230 samples from 1,183 animals were tested, including Eurasian griffon vulture ( Gyps fulvus), Iberian ibex (Capra pyrenaica), red deer ( Cervus elaphus), and wild boar (Sus scrofa). The number of individuals per animal species, and the numbers of samples tested are shown in Table 1. Sampling details were previously described (17).
TABLE 1.
S. aureus detection and characterization by spa typing and MLST
| Animal species | No. of animals |
No. of samples/no. of isolates |
MLSTa | spa type(s)b (no. of isolates) | ||
|---|---|---|---|---|---|---|
| Tested | Positive | Nasal samples | Skin samples | |||
| Eurasian griffon vulture | 40 | 2 | 40/2 | 0/0 | ST133 | t7304 (2) |
| Iberian ibex | 157 | 36 | 157/36 | 103/3 | ST5 | t002 (4) |
| ST130 | t1736 (3) | |||||
| ST425 | t3369 (1) | |||||
| ST581 | t528 (16), t843 (2), t1535 (1) | |||||
| ST2328 | t3750 (1), t11501 (1) | |||||
| ST2637 | t11221 (7) | |||||
| ST2639 | t7229 (1), t11216 (1) | |||||
| ST2673 | t528 (1) | |||||
| Red deer | 273 | 54 | 269/49 | 273/8 | ST1 | t098 (1), t127 (2), t11223 (1) |
| ST5 | t548 (9), t11210 (1) | |||||
| ST30 | t342 (1) | |||||
| ST133 | t2678 (2) | |||||
| ST350 | t11215 (5) | |||||
| ST398 | t571 (1) | |||||
| ST425 | t1077 (1), t6386 (1), t6909 (1), t11208 (3), t11212 (8), t11228 (1), t11231 (2) | |||||
| ST522 | t528 (1), t1534 (1), t3576 (3) | |||||
| ST2640 | t742 (2) | |||||
| ST2671 | t11211 (2), t11226 (3), t11233 (3) | |||||
| ST2681 | t015 (1), t11217 (1) | |||||
| Wild boar | 713 | 126 | 694/103 | 694/41 | ST1 | t098 (2), t127 (5), t607 (2), t1407 (2), t2601 (1), t11223 (1) |
| ST5 | t548 (18), t2516 (2), t7174 (2), t11210 (5), t11214 (1), t11219 (3) | |||||
| ST15 | t084 (1) | |||||
| ST96 | t11218 (1) | |||||
| ST130 | t6220 (1) | |||||
| ST133 | t3583 (7), t10476 (2), t11220 (1) | |||||
| ST188 | t189 (2) | |||||
| ST398 | t034 (2) | |||||
| ST425 | t742 (3), t6909 (1), t11222 (1), t11225 (1), t11232 (1) | |||||
| ST1643 | t10712 (7) | |||||
| ST2328 | t3750 (52), t11227 (1), t11230 (8) | |||||
| ST2641 | t11229 (1) | |||||
| ST2672 | t359 (1) | |||||
| ST2675 | t11209 (2) | |||||
| ST2678 | t11502 (1) | |||||
| ST2681 | t015 (1) | |||||
| ST2682 | t6384 (1) | |||||
| ST2729 | t011 (1) | |||||
ST, sequence type. At least one isolate per spa type per host reservoir was selected to perform MLST (n = 88), the rest being inferred.
Partially published previously (37). New spa types and STs identified in the present study are indicated in boldface.
Isolation and identification.
Recovery of S. aureus (MSSA) was achieved by direct plating on Baird Parker agar with rabbit plasma fibrinogen (bioMérieux) to obtain black colonies with an opaque halo around them as presumptive S. aureus. One colony per sample was confirmed as MSSA (S. aureus mecA and mecC negative) by PCR as previously described (21).
Molecular characterization.
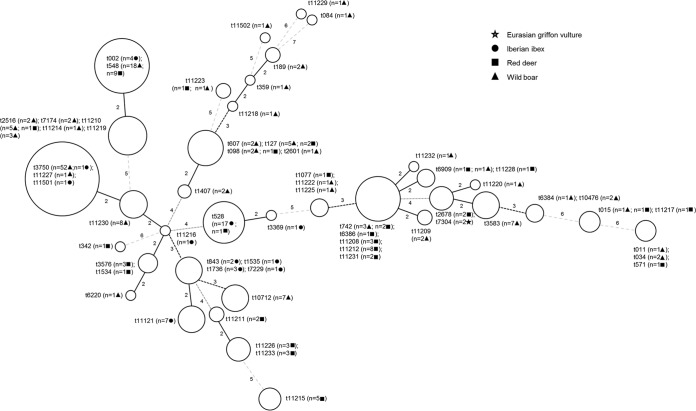
All confirmed S. aureus isolates were characterized by spa typing sequencing the variable fragment of protein A (22). In order to examine further the variation in spa types identified, the spa types were analyzed by the minimal spanning tree algorithm (Bionumerics 6.0) (Fig. 1). Multilocus sequence typing (MLST) was performed according to the protocol of Enright et al. (23), with the exception of self-designed primers for arc, tpi, and yqiL housekeeping genes (18). MLST was performed on representative isolates from all spa types found per host. At least one isolate per spa type and host, together with S. aureus with spa types containing less than three repeats (24), were characterized by MLST (n = 88). Based on these results, sequence types (STs) were assigned.
FIG 1.
Clustering of spa types by minimal spanning tree algorithm. Circles grouped related spa types, and the sizes are proportionate to the number of isolates identified for each group. Lines between circles represent the distance coding between the different spa types (maximum neighbor distance, 1.00).
Phenotypic antimicrobial resistance.
Isolates were also tested for antimicrobial susceptibility by broth microdilution (20). Briefly, the MIC was determined by microdilution using Sensititre Staphylococcus plate EUST (Trek Diagnostic Systems) and interpreted according to the epidemiological cutoffs established by the European Committee on Antibiotic Susceptibility Testing (http://www.eucast.org/). The antimicrobials tested are shown in Table 2. Only one isolate per animal with the same spa type was tested for antimicrobial susceptibility.
TABLE 2.
Antimicrobial susceptibility and resistance of S. aureus isolates using the indicated cutoff value
| Antimicrobial | Value (μg/ml)a |
No. of resistant isolates/no. of tested isolatesb |
||||
|---|---|---|---|---|---|---|
| Cutoff | Tested range | Eurasian griffon vulture (n = 2) | Iberian ibex (n = 36) | Red deer (n = 57) | Wild boar (n = 135) | |
| Benzylpenicillin | 0.125 | 0.12–2 | 0/2 | 4/36 | 11/57 | 36/135 |
| Cefoxitin | 4 | 0.5–16 | 0/2 | 0/36 | 0/57 | 0/135 |
| Chloramphenicol | 16 | 4–64 | 0/2 | 0/36 | 0/57 | 1/135 |
| Ciprofloxacin | 1 | 0.25–8 | 0/2 | 0/36 | 0/57 | 0/135 |
| Clindamycin | 0.25 | 0.12–4 | 0/2 | 0/36 | 0/57 | 0/135 |
| Erythromycin | 1 | 0.25–8 | 0/2 | 0/36 | 0/57 | 0/135 |
| Fusidic acid | 0.5 | 0.5–4 | 0/2 | 0/36 | 0/57 | 0/135 |
| Gentamicin | 2 | 1–16 | 0/2 | 0/36 | 0/57 | 0/135 |
| Kanamycin | 8 | 4–64 | 0/2 | 0/36 | 0/57 | 0/135 |
| Linezolid | 4 | 1–8 | 0/2 | 0/36 | 0/57 | 0/135 |
| Mupirocin | 0.5 | 0.5–2,256 | 0/2 | 0/36 | 0/57 | 0/134 |
| Quinupristin-dalfopristin | 1 | 0.5–4 | 0/2 | 0/36 | 0/57 | 0/135 |
| Rifampin | 0.032 | 0.016–0.5 | 0/2 | 0/36 | 0/57 | 0/135 |
| Streptomycin | 16 | 4–32 | 0/2 | 0/36 | 5/56 | 15/133 |
| Sulfamethoxazole | 128 | 64–512 | 0/2 | 1/33 | 1/53 | 0/135 |
| Tetracycline | 1 | 0.5–16 | 0/2 | 0/36 | 0/57 | 4/135 |
| Tiamulin | 2 | 0.5–4 | 0/2 | 0/36 | 0/57 | 0/135 |
| Trimethoprim | 2 | 2–32 | 0/2 | 0/36 | 2/57 | 1/135 |
| Vancomycin | 2 | 1–16 | 0/2 | 0/36 | 0/57 | 0/135 |
The range of studied concentrations is indicated. Cutoff values were as reported online (http://www.eucast.org/mic_distributions/ [last access, 19 May 2014]).
Boldfacing indicates that phenotypic resistance was detected. (The corresponding antimicrobial is also boldfaced in column 1.)
Statistical analysis.
Comparison of the proportions of positive animals among Eurasian Griffon vultures, Iberian ibex, red deer, and wild boars was performed. A Pearson chi-squared test and the confidence intervals (CIs) at 95% were calculated by using SPSS 20 software and an online tool developed by WinEpi (http://www.winepi.net/sp/disease/cprev1.asp), respectively. Detection of S. aureus in nasal samples and skin samples was compared by using the McNemar test (SPSS 20).
Simpson's index of diversity (SID) and Jackknife 95% CI pseudovalues were used to estimate the genetic diversity of MSSA isolates based on spa types and MLST (http://darwin.phyloviz.net/ComparingPartitions/index.php?link=Tool) (25), except for Eurasian griffon vultures due to the limited number of S. aureus isolates.
A Fisher exact test (SPSS 20) was calculated to analyze the relationship between hosts and spa types and STs. Comparison was performed for the most frequent ones in the collection (>5% of the isolates) except for Eurasian griffon vultures because of the limited number of S. aureus isolates. The proportion of phenotypic resistance to antimicrobials was compared in Iberian ibex, red deer, and wild boars using the Fisher exact test (SPSS 20).
RESULTS
In total, 242 MSSA isolates were obtained (Table 1). Animals were considered positive regardless of whether S. aureus was isolated from skin or nasal samples. The proportion of positive animals was 5.00% (95% CI = 0.00 to 11.75) in Eurasian griffon vultures (2 positive animals out of 40 tested), 22.93% (95% CI = 16.35 to 29.51) in Iberian ibex (36/157), 19.78% (95% CI = 15.05 to 24.51) in red deer (54/273), and 17.67% (95% CI = 14.87 to 20.47) in wild boars (126/473). No significant differences between species (P = 0.057) were detected.
A higher proportion of isolates (P = 0.000) were recovered from nasal samples (78.51%; 190/242) than from skin samples (21.49%; 52/242) in all animal species where comparison was possible (all except vultures). In red deer and especially in wild boars, double sampling increased the proportion of positive animals detected. If only nasal samples had been tested, five positive red deer (5/54; 9.26%) and 23 positive wild boars (23/126; 18.25%) would have been overlooked. In Iberian ibex, the three positive animals in skin samples were also positive in nasal samples. Half of the animals (n = 12) with both nasal and skin samples that were positive (3 Iberian ibex, 3 red deer, and 18 wild boars) presented the same spa type in both samples. The other 12 (3 red deer and 9 wild boars) presented different spa types.
Sixty-three different spa types were identified, including 25 spa types not yet reported (Table 1). Distribution of spa types is shown in the Fig. 1, where 41 spa types clustered in 14 complexes (n = 169 isolates) and 22 spa types (n = 73) not grouped. The most common spa types detected were t528 (17/39; 43.59%) in Iberian ibex, t548 (9/57; 15.79%) and t11212 (8/57; 14.04%) in red deer, and t3750 (53/144; 36.11%) in wild boars. The two isolates from Eurasian griffon vulture belonged to spa type t7304 (Table 1). In general, spa types were mostly detected only in a single host (Table 1). One exception would be t548, which was the most frequent spa type detected in red deer and the second most frequent spa type in wild boars (Table 1). However, the differences between the most frequent spa types and hosts were statistically significant (P = 0.000).
MLST analysis yielded 27 different STs, 12 of which had not been described previously (Table 1). The most common STs were ST581 for Iberian ibex (19/39; 48.72%), ST425 for red deer (17/57; 29.82%), and ST2328 for wild boar (61/144; 42.36%). Isolates from Eurasian griffon vultures belonged to ST133 (Table 1). As described above for spa types, STs were mostly found in a single host, although some of them (ST1, ST5, and ST133) were sporadically detected in more than one host (Table 1). One exception would be ST425 (Table 1), which was detected mostly in red deer (n = 17) but also in wild boar (n = 7) and Iberian ibex (n = 1). Nevertheless, the differences between the most common STs and hosts were statistically significant (P = 0.000).
Based on the spa types, the SID was 0.928 (95% CI = 0.907 to 0.949). SID revealed that the genetic diversity was significantly higher (P < 0.05) for isolates from red deer (0.913; 95% CI = 0.943 to 0.972) than for isolates from Iberian ibex (0.656; 95% CI = 0.775 to 0.894) and from wild boars (0.794; 95% CI = 0.846 to 0.899) (Table 1). Accordingly, the SID based on MLST was higher for red deer (0.851; 95% CI = 0.797 to 0.905) than for Iberian ibex (0.726; 95% CI = 0.593 to 0.859) and wild boars (0.761; 95% CI = 0.705 to 0.817). Differences were also statistically significant between red deer and wild boars (P = 0.0207) but not significant for the comparisons performed for red deer and Iberian ibex or for wild boars and Iberian ibex (P > 0.05).
Antimicrobial susceptibly testing revealed that the most of the isolates were susceptible to all antimicrobial tested (Table 2). Nonsignificant differences were detected between the proportion of resistance in isolates from Iberian ibex, red deer, and wild boar (P > 0.050) for any of the antimicrobials tested. The most noteworthy resistance percentage was found against benzylpenicillin, with 11.11% (95% CI = 0.84 to 21.38) of isolates from Iberian ibex, 19.30% (95% CI = 9.05 to 29.54) of isolates from red deer, and 26.67% (95% CI = 19.21 to 34.13) of isolates from wild boars being resistant to this antimicrobial (Table 2). Isolates with any resistance to antimicrobials in our panel (n = 62) presented 11 phenotypic resistance patterns. The most common ones were resistant only to benzylpenicillin (35 isolates out of 230 isolates tested; 15.22%), resistant to benzylpenicillin-streptomycin (10/230; 4.35%), and resistant only to streptomycin (7/230; 3.04%). The remaining resistance patterns were represented only by one or two isolates with a maximum of resistance to three antimicrobials (benzylpenicillin-streptomycin-tetracycline). Most of the isolates with benzylpenicillin resistance (n = 51) belonged to ST5 (30/51; 58.82%). A comparison between ST5 and the proportion of benzylpenicillin resistance showed statistically significant differences (P = 0.000).
DISCUSSION
In this study, the genetic background of MSSA isolates from wild animals was determined in order to identify predominant genetic lineages on different free-living wild animal species. The carriage of S. aureus has been evaluated revealing that S. aureus colonization is common in free-living artiodactyls (Table 1). However, the carriage rates detected in the present study are lower than those reported in different domestic animals such as pigs (36%), small ruminants (from 29 to 64%), donkeys (50%), and rabbits (56%) (26–31). In contrast to the results obtained for MRSA (17), the detection rate of MSSA in the Eurasian griffon vulture was lower than in the Iberian ibex, red deer, and wild boar, although the differences were not significant (P = 0.057). The lower number of griffon vultures tested might explain the lack of statistical significance.
Most of the animals simultaneously sampled in nares and skin for isolating S. aureus were positive only in one sample (174/198; 87.88%), with nasal swabs being the better option for sampling (172/198; 86.87%). Despite the higher rate of detection in nasal samples, some red deer (5/54; 9.26%) and even more wild boars (23/126; 18.25%) would have tested negative if only nasal samples had been tested. Therefore, double sampling (both nares and skin) is recommended in studies dealing with the detection of MSSA carriers. Similar results have been observed for MRSA (17, 32). The spa types detected in nasal and skin samples were different in 50% of the positive animals in both samples, indicating that double sampling increases the diversity of spa types found. This should be considered an additional benefit when studying the genetic diversity of MSSA.
The most common spa types and STs identified in MSSA isolates in this study (Table 1) were host associated (P = 0.000), suggesting host specificity as previously observed by other authors (33, 34). However, some of these spa types and STs (and related spa types or STs) have been previously isolated in other animal species, although usually at low frequencies. Thus, ST2328-t9857 (a spa type related to t3750) was found in sheep in Denmark (28); t3750 was previously described in Spain in 2006, although the host was not recorded (http://spa.ridom.de/ [accessed 19 May 2014]); ST5-t548 was found in humans and in pigs in the United States and in the United Kingdom (35, 36); ST425-t6386 and t742 were found in humans in the United Kingdom (10); ST581 was found in bulk milk of caprine origin in Portugal in 2003 (www.mlst.net [accessed 19 May 2014]); and ST1740 (a single-locus variant of ST581)-t528 was described in small ruminants in Spain (20). Based on the data available on the MLST webpage (www.mlst.net [accessed 19 May 2014]) ST2328, ST425, ST5, and ST133 are distributed worldwide. However, ST581 and related STs (ST1740, ST1758, ST2490, and ST2673) has been detected in countries in the south of Europe, such as Spain, Portugal, and France.
The dispersion of spa types shown in Fig. 1 reflects the high genetic diversity found among MSSA isolates. We compared the genetic diversity of MSSA isolates examined in the present study to the MRSA isolates recovered from the same free-living animal species, where MRSA belonged to ST398 (t011 and t1451) and ST1 (t127) (17). The genetic diversity calculated with the SID based on spa types was much higher for MSSA (0.928; 95% CI = 0.907 to 0.949) than for MRSA isolates (0.410; 95% CI = 0.041 to 0.780; P = 0.0052). This higher genetic heterogeneity observed in MSSA isolates in free-living wild animals agrees with the higher genetic diversity exhibited by MSSA in previous studies performed in Europe with human isolates (7, 37). When comparing SID (based on spa types and STs) between the animal species included in the study (all except vultures), the genetic diversity of MSSA detected in red deer was higher than in wild boars and Iberian ibex. However, the justification for such differences remains unclear. The spa types and STs detected among MRSA isolates in free-living wild animals (17) were identified only sporadically in MSSA isolates (Table 1). Thus, only seven isolates belonging to genotype ST1-t127 (Table 1) represented the 2.89% of the MSSA isolates. Similarly, only three MSSA isolates (1.24%) belonged to ST398 (Table 1), and none of them belonged to the spa types t011 or 1451 detected in MRSA isolates (17). The single t011 isolate (0.41%) was ST2729, a single-locus variant of ST398.
Most of the MSSA isolates from free-living wild animals presented very low proportions of phenotypic resistance, which is probably linked to the absence of selective pressure due to no antimicrobial use in these animals (38). The highest resistance percentage found in our study was against benzylpenicillin (22.17%; 95% CI = 16.81 to 27.54%). Although the origin of this resistance is unknown, previous studies showed that it is broadly disseminated in food animals (20, 26, 27) and humans (39–41). None of the ST1 and ST398 MSSA isolates exhibited the antimicrobial susceptibility patterns observed in MRSA (tetracycline resistance in ST398 and ciprofloxacin susceptibility plus clindamycin-erythromycin-tetracycline resistance in ST1 [17]).
Overall, the results presented here indicate that the MSSA population in the Eurasian griffon vulture, Iberian ibex, red deer, and wild boar differed in genetic diversity (i.e., SID), genotypes, and antimicrobial resistance patterns from MRSA in the same animal species.
ACKNOWLEDGMENTS
This study was partially supported by the Spanish Ministry of Science and Innovation (FAU200600011; FAU2008-00021) and by the Autonomous Community of Madrid (S0505/AGR-0265; S2009/AGR-1489). Eurasian griffon vultures were captured within the Territorial Cooperation Programme Spain-France-Andorra (NECROPIR-EFA 130/09).
We thank J. A. Carriço (Molecular Microbiology and Infection Unit, Instituto de Medicina Molecular Instituto de Microbiologia, Faculdade de Medicina de Lisboa, Universidade de Lisboa) for help in using the Web tool designed to calculate the SID and confidence intervals, and we thank the technicians M. C. Comerón, N. Maasoumi, and L. del Moral (VISAVET) for their invaluable assistance in the isolation, identification, and characterization of the isolates.
Footnotes
Published ahead of print 6 June 2014
REFERENCES
- 1.Quinn PJ, Carter ME, Markey B, Carter GR. 1999. Staphylococcus species, p 118–126 In Quinn PJ, Carter ME, Markey BK, Carter GR. (ed), Clinical veterinary microbiology. Mosby, Edinburgh, Scotland [Google Scholar]
- 2.Williams RE. 1963. Healthy carriage of Staphylococcus aureus: its prevalence and importance. Bacteriol. Rev. 27:56–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lowy FD. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520–532. 10.1056/NEJM199808203390806 [DOI] [PubMed] [Google Scholar]
- 4.Fluit AC. 2012. Livestock-associated Staphylococcus aureus. Clin. Microbiol. Infect. 18:735–744. 10.1111/j.1469-0691.2012.03846.x [DOI] [PubMed] [Google Scholar]
- 5.Frank DN, Feazel LM, Bessesen MT, Price CS, Janoff EN, Pace NR. 2010. The human nasal microbiota and Staphylococcus aureus carriage. PLoS One 5:e10598. 10.1371/journal.pone.0010598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, Nouwen JL. 2005. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 5:751–762. 10.1016/S1473-3099(05)70295-4 [DOI] [PubMed] [Google Scholar]
- 7.Grundmann H, Aanensen DM, van den Wijngaard CC, Spratt BG, Harmsen D, Friedrich AW. 2010. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Med. 7:e1000215. 10.1371/journal.pmed.1000215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pantosti A. 2012. Methicillin-resistant Staphylococcus aureus associated with animals and its relevance to human health. Front. Microbiol. 3:127. 10.3389/fmicb.2012.00127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers HF. 1997. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin. Microbiol. Rev. 10:781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.García-Alvárez L, Holden MT, Lindsay H, Webb CR, Brown DF, Curran MD, Walpole E, Brooks K, Pickard DJ, Teale C, Parkhill J, Bentley SD, Edwards GF, Girvan EK, Kearns AM, Pichon B, Hill RL, Larsen AR, Skov RL, Peacock SJ, Maskell DJ, Holmes MA. 2011. Methicillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect. Dis. 11:595–603. 10.1016/S1473-3099(11)70126-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kock R, Schaumburg F, Mellmann A, Koksal M, Jurke A, Becker K, Friedrich AW. 2010. Methicillin-resistant Staphylococcus aureus (MRSA): burden of disease and control challenges in Europe. Euro Surveill. 15:19688 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19688 [DOI] [PubMed] [Google Scholar]
- 12.Otto M. 2012. MRSA virulence and spread. Cell Microbiol. 14:1513–1521. 10.1111/j.1462-5822.2012.01832.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fitzgerald JR. 2012. Livestock-associated Staphylococcus aureus: origin, evolution, and public health threat. Trends Microbiol. 20:192–198. 10.1016/j.tim.2012.01.006 [DOI] [PubMed] [Google Scholar]
- 14.Cuny C, Friedrich A, Kozytska S, Layer F, Nubel U, Ohlsen K, Strommenger B, Walther B, Wieler L, Witte W. 2010. Emergence of methicillin-resistant Staphylococcus aureus (MRSA) in different animal species. Int. J. Med. Microbiol. 300:109–117. 10.1016/j.ijmm.2009.11.002 [DOI] [PubMed] [Google Scholar]
- 15.Franco A, Hasman H, Lurescia M, Lorenzetti R, Stegger M, Pantosti A, Feltrin F, Ianzano A, Porrero MC, Liapi M, Battisti A. 2011. Molecular characterization of spa type t127, sequence type 1 methicillin-resistant Staphylococcus aureus from pigs. J. Antimicrob. Chemother. 66:1231–1235. 10.1093/jac/dkr115 [DOI] [PubMed] [Google Scholar]
- 16.Price LB, Stegger M, Hasman H, Aziz M, Larsen J, Andersen PS, Pearson T, Waters AE, Foster JT, Schupp J, Gillece J, Driebe E, Liu CM, Springer B, Zdovc I, Battisti A, Franco A, Zmudzki J, Schwarz S, Butaye P, Jouy E, Pomba C, Porrero MC, Ruimy R, Smith TC, Robinson DA, Weese JS, Arriola CS, Yu F, Laurent F, Keim P, Skov R, Aarestrup FM. 2012. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. mBio 3:e00305-11. 10.1128/mBio.00305-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porrero MC, Mentaberre G, Sánchez S, Fernández-Llario P, Gómez-Barrero S, Navarro-González N, Serrano E, Casas-Díaz E, Marco I, Fernández-Garayzábal JF, Mateos A, Vidal D, Lavín S, Domínguez L. 2013. Methicillin-resistant Staphylococcus aureus (MRSA) carriage in different free-living wild animal species in Spain. Vet. J. 198:127–130. 10.1016/j.tvjl.2013.06.004 [DOI] [PubMed] [Google Scholar]
- 18.Porrero MC, Valverde A, Fernández-Llario P, Díez-Guerrier A, Mateos A, Lavín S, Cantón R, Fernández-Garayzábal JF, Domínguez L. 2014. Staphylococcus aureus carrying mecC gene in animals and urban wastewater, Spain. Emerg. Infect. Dis. 20:899–901. 10.3201/eid2005.130426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasman H, Moodley A, Guardabassi L, Stegger M, Skov RL, Aarestrup FM. 2010. spa type distribution in Staphylococcus aureus originating from pigs, cattle, and poultry. Vet. Microbiol. 141:326–331. 10.1016/j.vetmic.2009.09.025 [DOI] [PubMed] [Google Scholar]
- 20.Porrero MC, Hasman H, Vela AI, Fernández-Garayzábal JF, Domínguez L, Aarestrup FM. 2012. Clonal diversity of Staphylococcus aureus originating from the small ruminants goats and sheep. Vet. Microbiol. 156:157–161. 10.1016/j.vetmic.2011.10.015 [DOI] [PubMed] [Google Scholar]
- 21.Stegger M, Andersen PS, Kearns A, Pichon B, Holmes MA, Edwards G, Laurent F, Teale C, Skov R, Larsen AR. 2012. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecA (LGA251). Clin. Microbiol. Infect. 18:395–400. 10.1111/j.1469-0691.2011.03715.x [DOI] [PubMed] [Google Scholar]
- 22.Harmsen D, Claus H, Witte W, Rothganger J, Turnwald D, Vogel U. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442–2548. 10.1128/JCM.41.12.5442-5448.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakwinska O, Giddey M, Moreillon M, Morisset D, Waldvogel A, Moreillon P. 2011. Staphylococcus aureus host range and human-bovine host shift. Appl. Environ. Microbiol. 77:5908–5915. 10.1128/AEM.00238-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carrico JA, Silva-Costa C, Melo-Cristino J, Pinto FR, de Lencastre H, Almeida JS, Ramirez M. 2006. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J. Clin. Microbiol. 44:2524–2532. 10.1128/JCM.02536-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gharsa H, Ben Sallem R, Ben Slama K, Gomez-Sanz E, Lozano C, Jouini A, Klibi N, Zarazaga M, Boudabous A, Torres C. 2012. High diversity of genetic lineages and virulence genes in nasal Staphylococcus aureus isolates from donkeys destined to food consumption in Tunisia with predominance of the ruminant associated CC133 lineage. BMC Vet. Res. 8:203. 10.1186/1746-6148-8-203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gharsa H, Ben Slama K, Lozano C, Gomez-Sanz E, Klibi N, Ben Sallem R, Gomez P, Zarazaga M, Boudabous A, Torres C. 2012. Prevalence, antibiotic resistance, virulence traits and genetic lineages of Staphylococcus aureus in healthy sheep in Tunisia. Vet. Microbiol. 156:367–373. 10.1016/j.vetmic.2011.11.009 [DOI] [PubMed] [Google Scholar]
- 28.Eriksson J, Espinosa-Gongora C, Stamphoj I, Larsen AR, Guardabassi L. 2013. Carriage frequency, diversity, and methicillin resistance of Staphylococcus aureus in Danish small ruminants. Vet. Microbiol. 163:110–115. 10.1016/j.vetmic.2012.12.006 [DOI] [PubMed] [Google Scholar]
- 29.Vautor E, Abadie G, Guibert JM, Chevalier N, Pepin M. 2005. Nasal carriage of Staphylococcus aureus in dairy sheep. Vet. Microbiol. 106:235–239. 10.1016/j.vetmic.2004.11.019 [DOI] [PubMed] [Google Scholar]
- 30.Selva L, Viana D, Penadés JR, Corpa JM. 2008. Staphylococcus aureus nasal carriage in rabbits, p 1079–1083 9th World Rabbit Congress, Verona, Italy [Google Scholar]
- 31.Oppliger A, Moreillon P, Charriere N, Giddey M, Morisset D, Sakwinska O. 2012. Antimicrobial resistance of Staphylococcus aureus strains acquired by pig farmers from pigs. Appl. Environ. Microbiol. 78:8010–8014. 10.1128/AEM.01902-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pletinckx LJ, De Bleecker Y, Dewulf J, Rasschaert G, Goddeeris BM, De Man I. 2012. Evaluation of salt concentrations, chromogenic media and anatomical sampling sites for detection of methicillin-resistant Staphylococcus aureus in pigs. Vet. Microbiol. 154:363–368. 10.1016/j.vetmic.2011.07.027 [DOI] [PubMed] [Google Scholar]
- 33.Guinane CM, Ben Zakour NL, Tormo-Mas MA, Weinert LA, Lowder BN, Cartwright RA, Smyth DS, Smyth CJ, Lindsay JA, Gould KA, Witney A, Hinds J, Bollback JP, Rambaut A, Penades JR, Fitzgerald JR. 2010. Evolutionary genomics of Staphylococcus aureus reveals insights into the origin and molecular basis of ruminant host adaptation. Genome Biol. Evol. 2:454–466. 10.1093/gbe/evq031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sung JM, Lloyd DH, Lindsay JA. 2008. Staphylococcus aureus host specificity: comparative genomics of human versus animal isolates by multi-strain microarray. Microbiology 154:1949–1959. 10.1099/mic.0.2007/015289-0 [DOI] [PubMed] [Google Scholar]
- 35.Jackson CR, Davis JA, Barrett JB. 2013. Prevalence and characterization of methicillin-resistant Staphylococcus aureus isolates from retail meat and humans in Georgia. J. Clin. Microbiol. 51:1199–1207. 10.1128/JCM.03166-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frana TS, Beahm AR, Hanson BM, Kinyon JM, Layman LL, Karriker LA, Ramirez A, Smith TC. 2013. Isolation and characterization of methicillin-resistant Staphylococcus aureus from pork farms and visiting veterinary students. PLoS One 8:e53738. 10.1371/journal.pone.0053738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Porrero MC, Mentaberre G, Sánchez S, Fernández-Llario P, Gómez-Barrero S, Navarro-González N, Serrano E, Casas-Díaz E, Marco I, Fernández-Garayzábal JF, Mateos A, Vidal D, Lavín S, Domínguez L. 2013. Staphylococcus aureus in wild animals: healthy carriers and genetic diversity, p 44. Med-Vet-Net Association International Science Conference, Lyngby, Denmark [Google Scholar]
- 38.Navarro-Gonzalez N, Porrero MC, Mentaberre G, Serrano E, Mateos A, Dominguez L, Lavin S. 2013. Antimicrobial resistance in indicator Escherichia coli isolates from free-ranging livestock and sympatric wild ungulates in a natural environment (northeastern Spain). Appl. Environ. Microbiol. 79:6184–6186. 10.1128/AEM.01745-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lowy FD. 2003. Antimicrobial resistance: the example of Staphylococcus aureus. J. Clin. Invest. 111:1265–1273. 10.1172/JCI18535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chatterjee SS, Otto M. 2013. Improved understanding of factors driving methicillin-resistant Staphylococcus aureus epidemic waves. Clin. Epidemiol. 5:205–217. 10.2147/CLEP.S37071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tong SY, Chen LF, Fowler VG., Jr 2012. Colonization, pathogenicity, host susceptibility, and therapeutics for Staphylococcus aureus: what is the clinical relevance? Semin. Immunopathol. 34:185–200. 10.1007/s00281-011-0300-x [DOI] [PMC free article] [PubMed] [Google Scholar]