Abstract
Lactose (1,4-O-β-d-galacto-pyranosyl-d-glucose) induces cellulolytic enzymes in Trichoderma reesei and is in fact one of the most important soluble carbon sources used to produce cellulases on an industrial level. The mechanism underlying the induction is, however, not fully understood. In this study, we investigated the cellular functions of the intracellular β-glucosidases CEL1a and CEL1b in the induction of cellulase genes by lactose in T. reesei. We demonstrated that while CEL1a and CEL1b were functionally equivalent in mediating the induction, the simultaneous absence of these intracellular β-glucosidases abolished cbh1 gene expression on lactose. d-Galactose restored the efficient cellulase gene induction in the Δcel1a strain independently of its reductive metabolism, but not in the Δcel1a Δcel1b strain. A further comparison of the transcriptional responses of the Δcel1a Δcel1b strain complemented with wild-type CEL1a or a catalytically inactive CEL1a version and the Δcel1a strain constitutively expressing CEL1a or the Kluyveromyces lactis β-galactosidase LAC4 showed that both the CEL1a protein and its glycoside hydrolytic activity were indispensable for cellulase induction by lactose. We also present evidence that intracellular β-glucosidase-mediated lactose induction is further conveyed to XYR1 to ensure the efficiently induced expression of cellulase genes.
INTRODUCTION
Cost-effective conversion of plant cell wall-derived polysaccharides holds the potential for production of an environmentally clean and renewable source of energy and platform chemicals (1). Trichoderma reesei (teleomorph Hypocrea jecorina) is well known for its high capacity to secrete large amounts of lignocellulosic enzymes that release fermentable sugars and has thus been developed into one of the most prolific industrial cellulase producers. High-yield production of the bulk of the plant cell wall-degrading machinery in T. reesei is, however, dependent on induction by insoluble substrates that include cellulose, hemicellulose, and mixtures of plant polymers. Considering the ease of manipulation and the complication of separating enzymes from insoluble plant cell wall materials, soluble inducing substrates are usually preferred or required (2). Among others, the disaccharide lactose (1,4-O-β-d-galacto-pyranosyl-d-glucose) is an important and economic soluble carbon source for cellulase production by T. reesei. However, the induced cellulase yields on lactose are usually lower than those on cellulose (3, 4). Understanding the differences in the inducing efficiency and the mode by which lactose triggers cellulase formation would be helpful for improving the performance of industrial strains.
In fungi, catabolism of lactose is thought to proceed either by extracellular hydrolysis and subsequent uptake of the resulting sugar monomers or by uptake of the disaccharide followed by intracellular hydrolysis (4). For T. reesei, it has been assumed that lactose metabolism relies on the first strategy, based on several findings, including the absence of apparent orthologs for lactose permease and intracellular β-galactosidase in the T. reesei genome (4, 5). The resulting d-galactose moiety from lactose can be converted either by the Leloir pathway before being channeled into the glycolytic pathway or by a second pathway, initiated by a xylose reductase-mediated reduction of galactose to galactitol (6). While deletion of the first gene of either pathway, gal1 or xyl1, drastically reduces cellulase gene expression during growth on lactose (7), it has been shown that d-galactose alone is not an inducer and that overexpression of bga1, encoding the major extracellular β-galactosidase, almost abolishes the cellulase induction on lactose (5, 8). On the other hand, it was recently reported that three major facilitator superfamily (MFS) transporters behave as lactose permeases and are involved in cellulase induction (9–11). Our previous results also demonstrated that the absence of the intracellular β-glucosidase CEL1a delayed the induction of cellulase gene expression on lactose (12). Eleven predicted β-glucosidase-encoding genes exist in the genome of Trichoderma reesei, including those for the well-characterized major extracellular β-glucosidase BglI as well as the intracellular β-glucosidases CEL1a and CEL1b (12, 13). However, it remains unclear how these intracellular β-glucosidases affect the induction of cellulases by lactose.
In the present study, we extrapolate from the previous observation that CEL1a affects the induction of cellulase gene expression on lactose. We demonstrate that simultaneous deletions of cel1a and cel1b almost abolish the induction. We further present evidence that CEL1a-associated hydrolytic activity participates in a process beyond lactose catabolism to initiate efficient cellulase induction. We discuss a possible role of intracellular β-glucosidases in contributing to cellulase biosynthesis induced by lactose.
MATERIALS AND METHODS
Strains and cultivation conditions.
Trichoderma reesei TU-6 (ATCC MYA-256) (14), a uridine-auxotrophic derivative of T. reesei QM9414 (ATCC 26921) with a mutant pyr4 gene, was used as the parental strain. Trichoderma reesei strains used are listed in Table 1. All the strains were maintained on malt extract agar supplemented with 10 mM uridine when necessary. Escherichia coli DH5α was used for conventional gene cloning and vector construction. Escherichia coli Origami B(DE3) was used as a host for production of recombinant proteins.
TABLE 1.
Trichoderma reesei strains used in this study
| Strain name or genotype | Reference |
|---|---|
| TU-6 | 14 |
| Δcel1a | 12 |
| Δcel1b | 12 |
| Δcel1a Δcel1b | 12 |
| ΔtriβG | 12 |
| Δcel1a Δcel1b CEL1b I174C | 12 |
| Δcel1a Δxyl1 | This study |
| Δcel1a Δbga1 | This study |
| Δcel1a Δcel1b CEcrt1 | This study |
| Δcel1a Δcel1b CExyr1 | This study |
| Δcel1a CElac4 | This study |
| Δcel1a CEcel1a | This study |
| Δcel1a CEcel1a (E367A) | This study |
| Δcel1a Δcel1b NEcel1a | This study |
| Δcel1a Δcel1b NEcel1a (E367A) | This study |
For transcription and cellulase production analysis, T. reesei strains were pregrown in 1-liter Erlenmeyer flasks on a rotary shaker (200 rpm) at 30°C in 250 ml Mandels-Andreotti medium with glycerol (1% [vol/vol]) as the carbon source for 48 h. Mycelia were harvested by filtration and washed twice with a medium without a carbon source. Equal amounts of mycelia were then transferred to a fresh medium containing 1% (wt/vol) lactose or other carbon sources, as indicated, without peptone, and incubation was continued for the indicated times.
Plasmids.
For expression of CEL1a and its mutant derivative in Escherichia coli, the coding sequence of cel1a or cel1a (E367A) was amplified from the cDNA of T. reesei or pNEcel1a (E367A) (see below) by use of primers harboring EcoRI and HindIII sites and ligated into pET32a(+) to obtain pET32a-cel1a or pET32a-cel1a (E367A).
For bga1 and xyl1 gene deletion in the Δcel1a strain, a hygromycin resistance cassette containing the gpd (glyceraldehyde-3-phosphate dehydrogenase) promoter from T. reesei and the hygromycin resistance gene from pRLMex30 (15) was amplified using primers hph-F and hph-R and then ligated into pMD19-T to obtain pMDhph. The 2-kb upstream and downstream flanking sequences of the bga1 gene were digested by SalI/SpeI and XbaI/NotI, respectively, and ligated into the corresponding sites of pMDhph sequentially to obtain pMDbga1hph. In the same way, the 2-kb upstream region of the xyl1 coding sequence was digested by BamHI/NotI, and the 2-kb downstream region was digested by SpeI/SalI, and the fragments were ligated into pMDhph to obtain pMDxyl1hph.
In order to complement the Δcel1a Δcel1b strain with CEL1a and CEL1a (E367A) under the control of the native cel1a promoter, the cel1a disruption vector pUCcel1apyr4 (12) was digested with XbaI and SalI to remove the pyr4 gene, followed by ligation with the hygromycin resistance cassette digested with the same enzymes to create pUCcel1ahph. This plasmid was digested with XbaI, dephosphorylated using shrimp alkaline phosphatase (TaKaRa), and further ligated with the coding region for CEL1a to obtain pNEcel1a. A mutant cel1a gene encoding a E367A substitution in wild-type (WT) CEL1a was created by oligonucleotide-mediated mutagenesis of the cel1a gene, using a two-step fusion PCR with pNEcel1a as the template. The resulting PCR product was ligated into pUCcel1ahph to obtain pNEcel1a (E367A).
To achieve the constitutive expression (CE) of xyr1, cel1a, cel1a (E367A), and cel1b, the coding sequences of these genes were amplified from the cDNA of T. reesei or the pNEcel1a (E367A) plasmid and were ligated into pFL3 (pIG1783 with the stop codon of egfp deleted), which contains a gpd promoter and a trpC terminator, to obtain pFL3xyr1, pFL3cel1a, pFL3cel1a (E367A), and pFL3cel1b (16). To achieve the constitutive expression of crt1 and lac4, the coding sequences of crt1 and lac4 were placed under the control of a constitutive pki promoter. DNA fragments corresponding to the pki promoter and the cbh2 terminator from T. reesei were amplified from pRLMex30, digested with HindIII/SphI and KpnI/EcoRI, respectively, and ligated into pUC19 to obtain pUCpki-cbh2. The coding sequences of crt1 and lac4 were further inserted into pUCpki-cbh2 after being digested with SphI/XbaI to generate pCEcrt1 and pCElac4, respectively. Oligonucleotides used in this study for plasmid construction, gene deletion, and probe preparation are listed in Table S1 in the supplemental material.
Production of recombinant CEL1a in E. coli.
For purification of CEL1a and its mutant derivative, E. coli Origami B(DE3) cells transformed with pET32a-cel1a or pET32a-cel1a (E367A) were grown at 37°C to an optical density at 600 nm (OD600) of 0.6. IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 100 μM, and the incubation was continued at 20°C for 16 h. The proteins were purified from the supernatant with Ni-nitrilotriacetic acid-agarose (Qiagen), essentially as previously described (12).
Construction of T. reesei strains.
For xyl1 and bga1 gene deletions in the Δcel1a strain, plasmids pMDxyl1hph and pMDbga1hph were linearized by SalI and XbaI, respectively, before transformation (see Fig. S1 in the supplemental material). For constitutive expression of xyr1 or crt1, the Δcel1a Δcel1b strain was transformed with plasmid pFL3xyr1 or pCEcrt1 (together with pRLMex30) to obtain the Δcel1a Δcel1b CExyr1 and Δcel1a Δcel1b CEcrt1 strains. For constitutive expression of cel1a, cel1a (E367A), and lac4 in the Δcel1a strain, the Δcel1a strain was transformed with plasmids pFL3cel1a, pFL3cel1a (E367A), and pCElac4 (together with pRLMex30) to obtain the Δcel1a CEcel1a, Δcel1a CEcel1a (E367A), and Δcel1a CElac4 strains, respectively. For constitutive expression of cel1a or cel1b in the Δcel1a Δcel1b strain, the Δcel1a Δcel1b strain was transformed with plasmid pFL3cel1a or pFL3cel1b to obtain the Δcel1a Δcel1b CEcel1a and Δcel1a Δcel1b CEcel1b strains. In order to express CEL1a and CEL1a (E367A) under the control of the native cel1a promoter, the Δcel1a Δcel1b strain was transformed with plasmid pNEcel1a or pNEcel1a (E367A) linearized by HindIII to obtain the Δcel1a Δcel1b NEcel1a and Δcel1a Δcel1b NEcel1a (E367A) strains (see Fig. S2). Transformation was carried out essentially as described by Penttila et al. (17). Transformants were selected on minimal medium for resistance to 100 μg/ml of hygromycin. For ectopic complementation via constitutive expression, all the recombinant strains were verified for the presence of the entire expression cassette by PCR, and at least two transformants were analyzed for the relevant phenotypes.
Nucleic acid isolation and hybridization.
Fungal mycelia were harvested by filtration, washed with tap water, and frozen in liquid nitrogen. Fungal genomic DNA was isolated according to the instructions of an EZNA fungal DNA miniprep kit (Omega Biotech). Total RNA was isolated by use of the TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Southern and Northern hybridization analyses were performed with a digoxigenin-based nonradioactive system from Roche Applied Science, as described previously (18). Relative transcription levels were normalized to that of the 18S rRNA control by densitometry, using the software program ImageJ (http://rsb.info.nih.gov/ij).
Quantitative RT-PCR (qRT-PCR).
Total RNA was further digested by use of a Turbo DNA-free kit (Ambion) to eliminate genomic DNA contamination. Reverse transcription (RT) was performed using a PrimeScript RT reagent kit (TaKaRa) according to the manufacturer's instructions. Quantitative PCRs were performed using SYBR green supermix (TaKaRa) on a Bio-Rad myIQ2 thermocycler (Bio-Rad). Reactions were performed in triplicate with a total volume of 20 μl, including 250 nM (each) forward and reverse primers and template cDNA. Data were analyzed using the relative quantitation/comparative threshold cycle (ΔΔCT) method and were normalized to an endogenous control (actin), with expression on glycerol as the reference sample.
Enzyme and protein assays.
Cellobiohydrolase and β-glucosidase activities were determined by measuring the amount of p-nitrophenol, using p-nitrophenyl-d-cellobioside (pNPC; Sigma) and p-nitrophenyl-β-d-glucopyranoside (pNPG; Sigma), respectively, as substrates. β-Galactosidase activity was measured with o-nitrophenyl-d-galactopyranoside (ONPG; Sigma) as the substrate. The assays were carried out in 200-μl reaction mixtures including 50 μl of culture supernatant or cell extract and 50 μl of the respective substrate (5 mM; pNPC, pNPG, or ONPG) in 50 mM sodium acetate buffer (pH 5.0). The reaction mixtures were incubated at 45°C for 30 min for pNPC and pNPG and at 30°C for 30 min for ONPG. Cellular extracts used for assay of intracellular β-glucosidase activity were prepared as follows. T. reesei strains were grown on Mandels-Andreotti medium with lactose or glycerol as the carbon source. The mycelia were harvested and washed twice with 0.9% NaCl. Lysates were prepared by grinding the mycelia into a fine powder under liquid nitrogen, and the powder was then suspended in 50 mM sodium phosphate buffer (pH 7.0) with protease inhibitors. Cell debris was removed by centrifugation at 14,000 × g for 10 min. One unit of activity corresponds to transformation of 1 μmol of substrate per min under the test conditions. The activity of CEL1a toward lactose was measured as follows. The reaction mixture (200 μl), containing 50 μl of a 4% lactose solution, 50 μl purified CEL1a protein (50 μg), and 100 μl 50 mM sodium phosphate buffer (pH 7.0), was incubated at 30°C for 30 min. The reaction was stopped by heating for 10 min in a boiling water bath. The products were analyzed by high-performance liquid chromatography (HPLC) as previously described (12).
Total secreted and intracellular proteins were determined using the method of the Bradford protein assay, with bovine serum albumin (BSA) as the standard. SDS-PAGE and Western blotting were performed according to standard protocols (19). CBH1 was immunoblotted using a polyclonal antibody raised against amino acids 426 to 446 of the protein, as previously described (12). Relative induced CBH1 production was analyzed semiquantitatively by densitometry, using the software program ImageJ (http://rsb.info.nih.gov/ij).
Biomass determination.
Mycelial dry weight was recorded by withdrawing 5-ml aliquots from the culture, centrifuging them at 10,000 × g for 10 min, and drying them to a constant weight at 80°C.
Statistical analyses.
Student's t tests were performed on all sets of data. All the differences mentioned fulfill the criterion of having a P value of <0.05.
RESULTS
The absence of intracellular β-glucosidases CEL1a and CEL1b compromises cellulase induction by lactose.
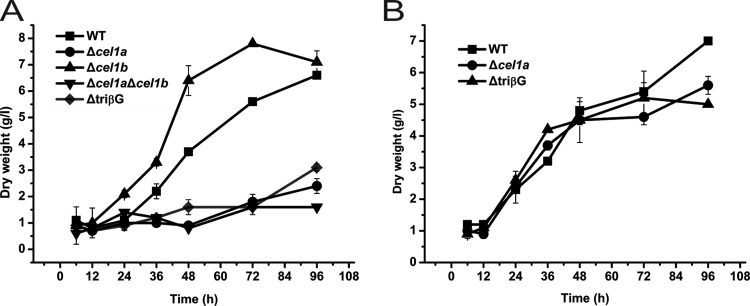
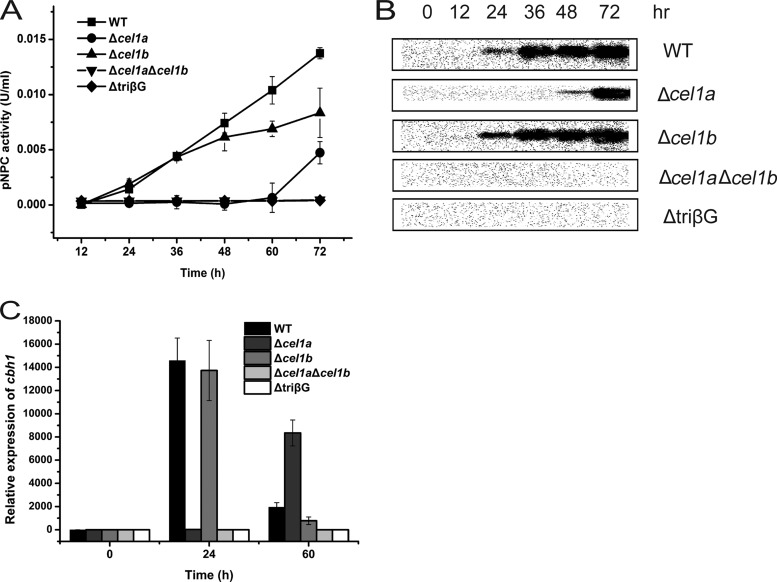
Our previous result showed that deletion of the major intracellular β-glucosidase-encoding gene, cel1a, delayed the induced expression of cellulase genes on lactose (12). To investigate the effects of the absence of other β-glucosidases on cellulase induction by lactose, a strain deleted for cel1b, encoding another highly expressed intracellular β-glucosidase upon lactose incubation, a double (Δcel1a Δcel1b) mutant strain, and a triple (ΔtriβG) mutant strain with simultaneous deletions in cel1a, cel1b, and bgl1 were used to analyze growth and cellulase gene expression. While the absence of CEL1b facilitated the growth on lactose compared with that of the WT, deletion of cel1a led to a severe growth defect (Fig. 1A). Additional deletions of cel1b and bgl1 did not build upon the compromised growth in the absence of CEL1a. In contrast, the Δcel1a and ΔtriβG strains grew as well as the WT on galactose (Fig. 1B). Analysis of the induced production of CBH1 on lactose demonstrated that compared with the case of the Δcel1a strain, the absence of CEL1b hardly affected the efficient induction and resulted only in slightly less production of CBH1 and a lower exoglucanase activity than those of the parental strain at the later stage of induction (Fig. 2A and B). Simultaneous absence of CEL1a and CEL1b, however, abolished the detection of secreted CBH1 and extracellular pNPC hydrolytic activity. A triple mutant carrying a deletion of bgl1 displayed the same noninduction phenotype as that of the Δcel1a Δcel1b strain. Further examination of the endogenous mRNA by qRT-PCR revealed that the defective cellulase gene induction observed in the deletion strains occurred at the transcriptional level, with the Δcel1a Δcel1b and ΔtriβG mutants showing no significantly induced transcription of cbh1 (Fig. 2C). Taken together, these results indicate that intracellular β-glucosidases CEL1a and CEL1b are critical for the efficient induction of cellulase gene expression on lactose.
FIG 1.
Disruption of cel1a, cel1b, and bgl1 affects growth of T. reesei on lactose but not on galactose. Growth of WT and β-glucosidase deletion strains in shake flasks with 1% (wt/vol) lactose (A) or 1% (wt/vol) galactose (B) as the carbon source was analyzed by measuring dry weight. Data shown are the means for results from three independent experiments. Error bars show the standard deviations (SD) for these replicates.
FIG 2.
Absence of intracellular β-glucosidases compromises cellulase induction by lactose. (A) Exoglucanase activity analysis of the culture supernatants of WT and mutant strains deleted for β-glucosidase genes grown on 1% (wt/vol) lactose. The differences between the WT and Δcel1b strains were found to be statistically relevant, by Student's t test, at the later stages of induction (P = 0.025 for 60 h and P = 0.029 for 72 h). (B) Western blot analysis of CBH1 secreted into the culture supernatants of WT and mutant strains grown on 1% (wt/vol) lactose. Equal amounts of culture supernatant relative to biomass were loaded for all strains. (C) Quantitative RT-PCR analysis of gene expression of cbh1 in WT and mutant strains after induction by 1% (wt/vol) lactose for different periods, as indicated. Values in this figure are the means for three biological replicates. Error bars show the SD for these replicates.
CEL1a and CEL1b are functionally equivalent in cellulase induction by lactose.
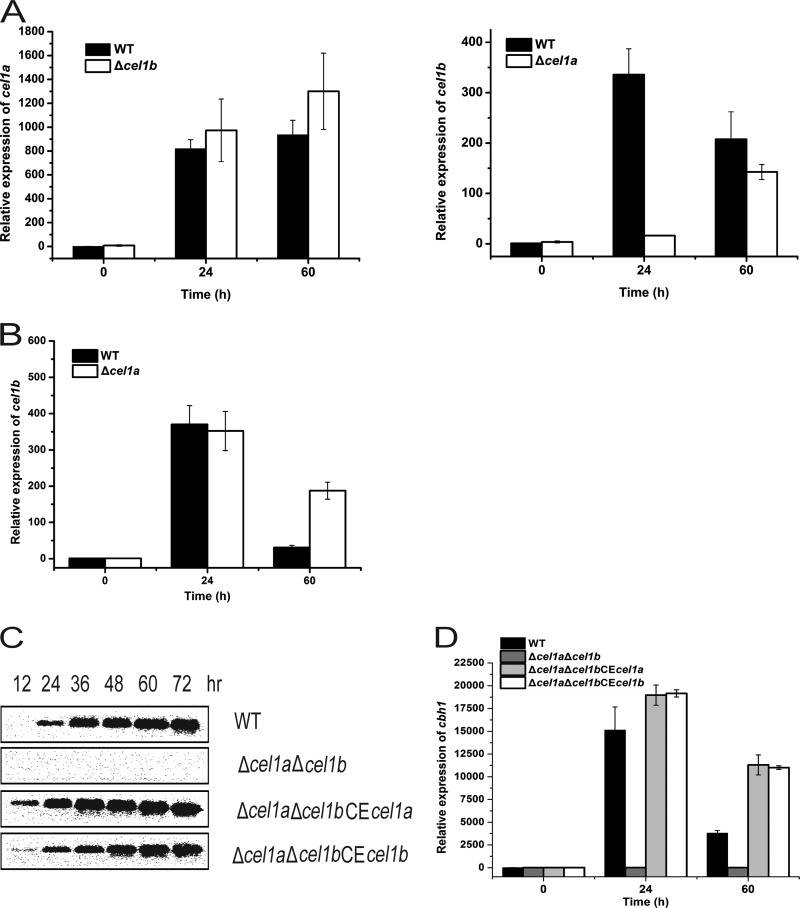
Given the very different phenotypes displayed by the single and double deletion mutants, we tried to probe the independent roles of CEL1a and CEL1b in the induction on lactose. Analysis of mRNA levels of cel1a and cel1b in the WT, Δcel1a, and Δcel1b strains revealed that while the absence of cel1b resulted in a slightly higher transcription level of cel1a, deletion of cel1a led to a significant delay in the transcription of cel1b on lactose (Fig. 3A). This delayed induction of cel1b was largely corrected by the addition of a small amount of galactose, which also restored the efficient cellulase induction in the Δcel1a strain (Fig. 3B) (see below). To further tease apart the differences in the effects of CEL1a and CEL1b, two recombinant strains were constructed to constitutively express either CEL1a or CEL1b in the Δcel1a Δcel1b strain. Analysis of the induced expression of cbh1 in the respective recombinant strains revealed that expression of either CLE1a or CEL1b alone was fully capable of mediating the induced expression of cbh1 on lactose (Fig. 3C and D). These results indicate that CEL1a and CEL1b are functionally equivalent in mediating the induction of cellulase and that CEL1b plays an important role in induced expression of cellulase genes in the absence of CEL1a.
FIG 3.
Either CEL1a or CEL1b is capable of inducing cellulase gene expression. (A) Transcription of cel1a (left) or cel1b (right) was analyzed by quantitative RT-PCR analysis of the WT, Δcel1a, and Δcel1b strains cultivated on 1% (wt/vol) lactose. Statistical analyses indicated significant differences in the transcription of cel1a between the WT and Δcel1b strains (P = 0.043 for 24 h and P = 0.026 for 60 h) as well as in the transcription of cel1b between the WT and Δcel1a strains (P = 0.0041 for 24 h). (B) Transcription of cel1b was analyzed by quantitative RT-PCR analysis of the WT and Δcel1a strains cultivated on 1% (wt/vol) lactose plus 0.1% (wt/vol) galactose. (C) Western blot analysis of CBH1 secreted into the culture supernatants of the WT, Δcel1a Δcel1b, Δcel1a Δcel1b CEcel1a, and Δcel1a Δcel1b CEcel1b strains grown on 1% (wt/vol) lactose. Equal amounts of culture supernatant relative to biomass were loaded for all strains. (D) Transcription of cbh1 was analyzed by quantitative RT-PCR analysis of the WT, Δcel1a Δcel1b, Δcel1a Δcel1b CEcel1a, and Δcel1a Δcel1b CEcel1b strains during growth on 1% (wt/vol) lactose. Values in this figure are the means of results from at least three biological replicates. Error bars show the SD for these replicates. For the Δcel1a Δcel1b CEcel1a and Δcel1a Δcel1b CEcel1b strains, at least three independent transformants were determined to possess similar phenotypes, and results for a representative transformant of each are shown.
Addition of galactose restores cellulase induction in the Δcel1a strain but not in the Δcel1a Δcel1b strain.
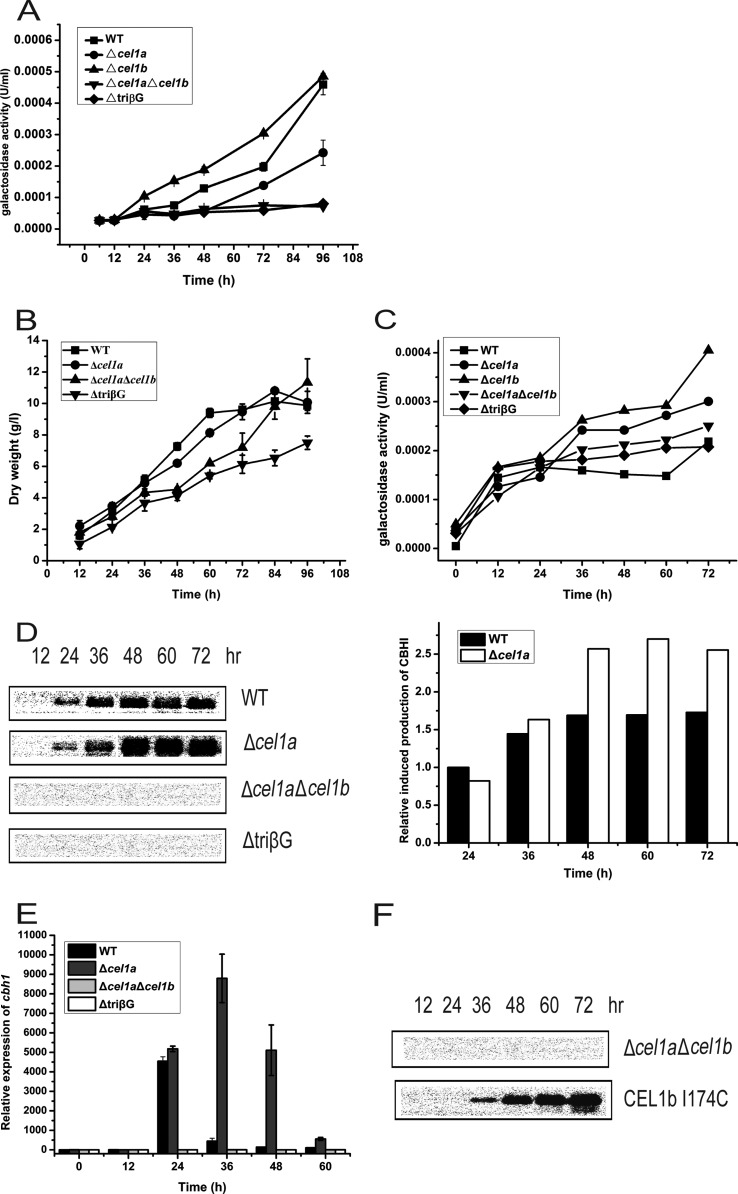
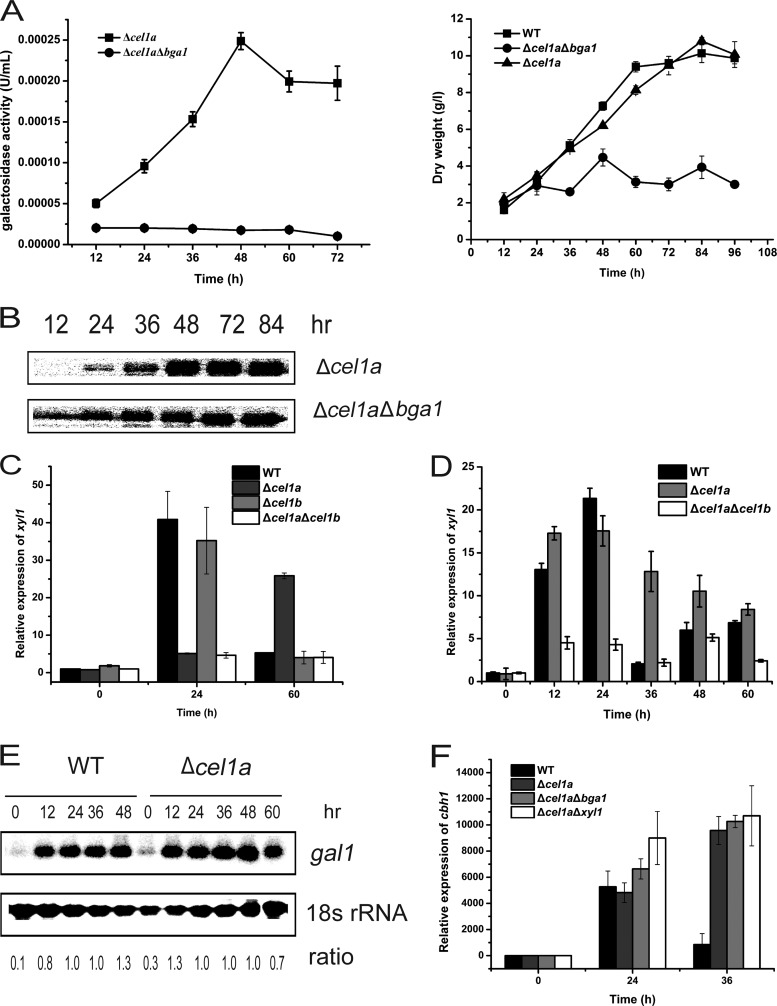
To ask whether the growth and cellulase induction defects on lactose displayed in the absence of CEL1a and CEL1b are due to the inefficient hydrolysis of lactose, we determined the extracellular β-galactosidase activity of the mutant strains. Analysis of the ONPG hydrolytic activity of the supernatant from the culture grown on lactose revealed that the extracellular β-galactosidase activity was drastically reduced in the Δcel1a, Δcel1a Δcel1b, and ΔtriβG strains compared with the WT and Δcel1b strains (Fig. 4A). These results correlated well with the observed growth phenotype of the mutant strains on lactose. We wondered whether the inefficient hydrolysis of lactose, and thus release of the d-galactose moiety, accounts for the cellulase induction defect in the β-glucosidase mutants. We therefore tested the effect of addition of different amounts of galactose to the lactose medium on the induced production of CBH1. Addition of galactose at a molar ratio of 1 to 5 relative to lactose restored the growth of the Δcel1a strain as well as the Δcel1a Δcel1b and ΔtriβG mutants on lactose (Fig. 4B). A significant increase in the extracellular β-galactosidase activity was also detected for the Δcel1a, Δcel1a Δcel1b, and ΔtriβG strains under such conditions compared with those induced only by lactose (Fig. 4C). However, an efficient induction of cbh1 expression, to a level even higher than that of the WT, was observed only for the Δcel1a strain, not for the double- and triple-deletion mutant strains (Fig. 4D and E). Galactose alone at such a concentration had no induction effect (see Fig. S3 in the supplemental material). Moreover, reintroduction of a CEL1b mutant (CEL1b I174C) with a higher pNPG hydrolytic activity than that of wild-type CEL1b into the Δcel1a Δcel1b strain (14) recovered the ability of the double mutant to induce cbh1 expression upon incubation with lactose and galactose (Fig. 4F). The restored induction was not the result of accelerated growth in the presence of galactose. Indeed, addition of glycerol had no effect on the induction defect in the Δcel1a and Δcel1a Δcel1b strains, despite their improved growth compared with that on lactose (see Fig. S4). These data suggest that inefficient extracellular hydrolysis of lactose may not account for the induction defect in the absence of CEL1a and CEL1b.
FIG 4.
Galactose restores cellulase induction in the Δcel1a strain but not in the Δcel1a Δcel1b strain. (A) Analysis of extracellular β-galactosidase activity in the culture supernatants of WT and mutant strains grown on 1% (wt/vol) lactose. (B) Growth of WT and β-glucosidase deletion strains in shake flasks with 1% (wt/vol) lactose plus 0.1% (wt/vol) galactose as the carbon source. (C) Analysis of extracellular β-galactosidase activity in the culture supernatants of WT and mutant strains grown on 1% (wt/vol) lactose plus 0.1% (wt/vol) galactose. Data points are the means of results from triplicates. SD are <10%. Statistical analyses indicated significant differences in the galactosidase activities of the Δcel1a, Δcel1a Δcel1b, and ΔtriβG strains between conditions with lactose and lactose plus galactose at all the relevant time points (P < 0.001). (D) (Left) Western blot analysis of CBH1 in the culture supernatants of the WT, Δcel1a, Δcel1a Δcel1b, and ΔtriβG strains after induction with 1% (wt/vol) lactose plus 0.1% (wt/vol) galactose. (Right) CBH1 expression was quantitated by scanning densitometry of the developed membranes. Equal amounts of culture supernatant relative to biomass were loaded for all strains. (E) Gene expression of cbh1 as analyzed by quantitative RT-PCR after induction with 1% (wt/vol) lactose plus 0.1% (wt/vol) galactose for different periods. The differences in transcription of cbh1 between the WT and Δcel1a strains were found to be statistically relevant by Student's t test (P = 0.015 at 24 h and P < 0.001 at 36 h, 48 h, and 60 h). (F) Western blot analysis of CBH1 in the culture supernatants of the Δcel1a Δcel1b strain and the Δcel1a Δcel1b strain complemented with CEL1b I174C after induction with 1% (wt/vol) lactose plus 0.1% (wt/vol) galactose. Equal amounts of culture supernatant relative to biomass were loaded for all strains. All values are the means of results from three biological replicates. Error bars show the SD for these replicates.
Neither extracellular hydrolysis of lactose nor metabolism of galactose is involved in the restored induction in the Δcel1a strain.
To further exclude the possibility that the restored cellulase induction upon addition of galactose in the absence of CEL1a is a result of enhanced extracellular lactose hydrolysis, we deleted bga1 from the Δcel1a strain and analyzed its response to lactose induction. While the absence of BGA1 drastically reduced the extracellular β-galactosidase activity and cell growth in the presence of galactose and lactose, its deletion did not interfere with the restored efficient induction observed in the Δcel1a strain (Fig. 5A, B, and F). Metabolism of galactose by either the reduction pathway or the Leloir pathway has been shown to be involved in the induced expression of cellulase genes on lactose (6, 7). In the absence of CEL1a, induction of xyl1, encoding a d-xylose reductase necessary for β-galactosidase and cellulase induction, was found to be delayed on lactose but was restored in the presence of lactose and galactose (Fig. 5C and D). In contrast, transcription of gal1, encoding galactose kinase, was not affected in the absence of CEL1a (Fig. 5E). We further tested the effect of deletion of xyl1 on the restored induction. As shown in Fig. 5F, in contrast with the results reported for the WT (7), the absence of XYL1 did not compromise the galactose-restored transcription of cbh1 in the Δcel1a strain. These data implicate that unlike the case in the WT, galactose metabolism may not be involved in the restored cellulase induction upon addition of galactose in the absence of CEL1a.
FIG 5.
Roles of bga1 and xyl1 in the restored cbh1 induction in the Δcel1a strain. (A) Extracellular β-galactosidase activities (left) and growth curves (right) for the Δcel1a and Δcel1a Δbga1 strains grown on 1% (wt/vol) lactose plus 0.1% (wt/vol) galactose. (B) Western blot analysis of extracellularly secreted CBH1 of the Δcel1a and Δcel1a Δbga1 strains grown on 1% (wt/vol) lactose plus 0.1% (wt/vol) galactose. Equal amounts of culture supernatant relative to biomass were loaded for all strains. (C and D) Quantitative RT-PCR analyses of xyl1 gene expression of wild-type and mutant strains induced by solely 1% (wt/vol) lactose (C) or 1% (wt/vol) lactose plus 0.1% (wt/vol) galactose (D). (E) Northern blot analysis of gal1 mRNA and 18S rRNA of the WT and Δcel1a strains cultured on 1% (wt/vol) lactose. The values below the panels indicate the ratios of the intensities of the gal1 signals measured by densitometry to that of the 18S rRNA control. (F) Quantitative RT-PCR analysis of cbh1 gene expression in the WT, Δcel1a, Δcel1a Δbga1, and Δcel1a Δxyl1 strains induced by 1% (wt/vol) lactose plus 0.1% (wt/vol) galactose for different periods, as indicated. All values are the means of results from three biological replicates. Error bars show the SD for these replicates.
Both CEL1a and its associated hydrolytic activity are responsible for the efficient induction of cellulase genes.
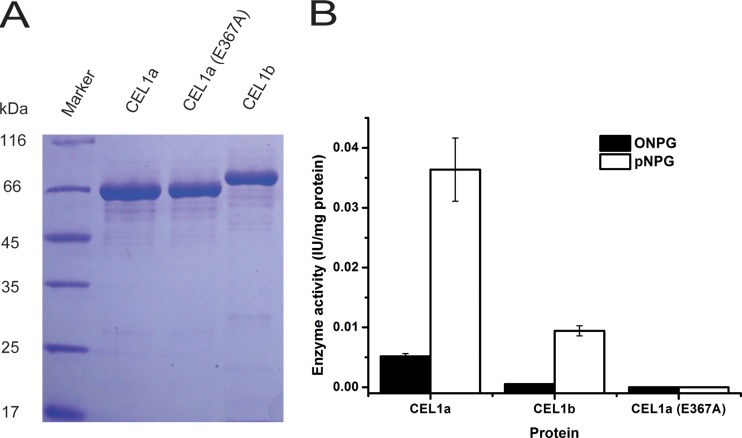
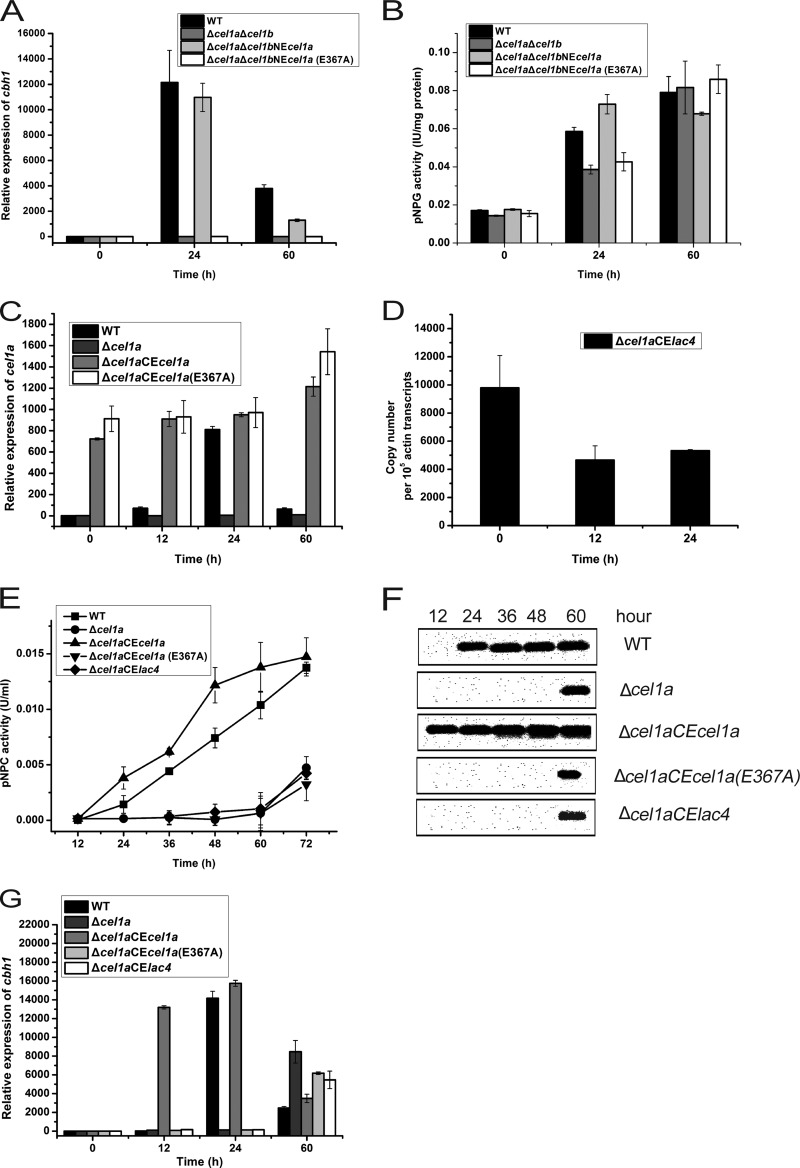
Although no apparent intracellular β-galactosidase-encoding gene exists in the T. reesei genome and negligible intracellular compared with extracellular β-galactosidase activity was detected during culture on lactose (5), we still wondered whether CEL1a and CEL1b are capable of hydrolyzing lactose. We heterologously expressed and purified CEL1a and CEL1b from Escherichia coli and tested their lactose hydrolytic activity. The purified CEL1a protein exhibited apparent hydrolytic activity toward ONPG and lactose, though the hydrolysis was much less efficient than that toward pNPG (Fig. 6A and B; see Fig. S5 and Table S2 in the supplemental material). As expected, targeted change of the catalytic residue Glu367 to Ala (E367A) in CEL1a resulted in the complete loss of both pNPG- and ONPG-targeted hydrolytic activities (Fig. 6A and B). To gain insight into the possibility that CEL1a-associated glycoside hydrolytic activity is involved in the induction process, we reintroduced the WT or catalytic mutant cel1a gene back into the endogenous cel1a locus so that the expression of the gene was under the control of its own promoter in the Δcel1a Δcel1b strain. Compared with the reintroduced WT CEL1a protein, which fully recapitulated the cellulase induction by lactose, CEL1a (E367A) was incapable of induction (Fig. 7A). Further analysis of the intracellular β-glucosidase activity demonstrated that a corresponding increase in the activity at the early stage of lactose induction (24 h) was observed only in cells expressing WT CEL1a, not for the CEL1a (E367A)-complemented strain (Fig. 7B). To further test whether CEL1a-associated hydrolytic activity toward lactose is fully responsible for cellulase induction, we constitutively expressed WT CEL1a, CEL1a (E367A), or the β-galactosidase LAC4 from Kluyveromyces lactis in the Δcel1a strain. Transcription of the reintroduced cel1a, cel1a (E367A), and lac4 genes occurred efficiently even before induction on lactose (Fig. 7C and D). Analysis of the induced expression of cbh1 revealed that while constitutive expression of WT CEL1a resulted in relatively more induced expression of cbh1 than that with the WT strain, cells expressing CEL1a (E367A) or LAC4 displayed the same kinetics of cbh1 induction as that in the Δcel1a strain on lactose (Fig. 7E to G). Altogether, these data suggest that either a specific kinetics of lactose hydrolysis achieved only by CEL1a and CEL1b or intracellular processing of lactose beyond sole hydrolysis mediated by these β-glucosidases may account for their important roles in cellulase induction by lactose.
FIG 6.
Enzymatic characterization of recombinant CEL1a, CEL1b, and CEL1a (E367A). (A) SDS-PAGE analysis of purified CEL1a, CEL1b, and CEL1a (E367A) produced in E. coli. (B) Recombinant CEL1a and CEL1b exhibit β-glucosidase activity and low levels of β-galactosidase activity, while CEL1a (E367A) completely lacks both activities. ONPG and pNPG were used as the substrates. Values are the means of results from three replicates. Error bars show the SD for these replicates.
FIG 7.
Expression of catalytically inactive CEL1a (E367A) or K. lactis LAC4 is unable to restore cellulase induction. Transcription levels of cbh1 (A) and intracellular β-glucosidase activities (B) in the WT, Δcel1a Δcel1b, Δcel1a Δcel1b NEcel1a, and Δcel1a Δcel1b NEcel1a (E367A) strains after induction with lactose (1% [wt/vol]) are shown. (C) Transcript levels of cel1a in T. reesei WT, Δcel1a, Δcel1a CEcel1a, and Δcel1a CEcel1a (E367A) strains after transfer to a lactose (1% [wt/vol])-containing medium. (D) Transcript levels of lac4 in the Δcel1a CElac4 strain induced by 1% (wt/vol) lactose were analyzed by quantitative RT-PCR. Copy numbers of lac4 transcripts relative to those of the actin gene are shown. (E) Exoglucanase activity analysis of the culture supernatants of the WT, Δcel1a, Δcel1a CEcel1a, Δcel1a CEcel1a (E367A), and Δcel1a CElac4 strains grown on 1% (wt/vol) lactose. pNPC was used as the substrate. (F) Western blot analysis of extracellularly secreted CBH1 proteins of the WT, Δcel1a, Δcel1a CEcel1a, Δcel1a CEcel1a (E367A), and Δcel1a CElac4 strains grown on 1% (wt/vol) lactose. Equal amounts of culture supernatant relative to biomass were loaded for all strains. (G) Transcript levels of cbh1 in T. reesei WT, Δcel1a, Δcel1a CEcel1a, Δcel1a CEcel1a (E367A), and Δcel1a CElac4 strains after transfer to a lactose (1% [wt/vol])-containing medium. Data shown are the means of results from three independent experiments. Error bars show the SD for these replicates.
Regulation of expression of xyr1 and crt1 essential for lactose induction by CEL1a and CEL1b.
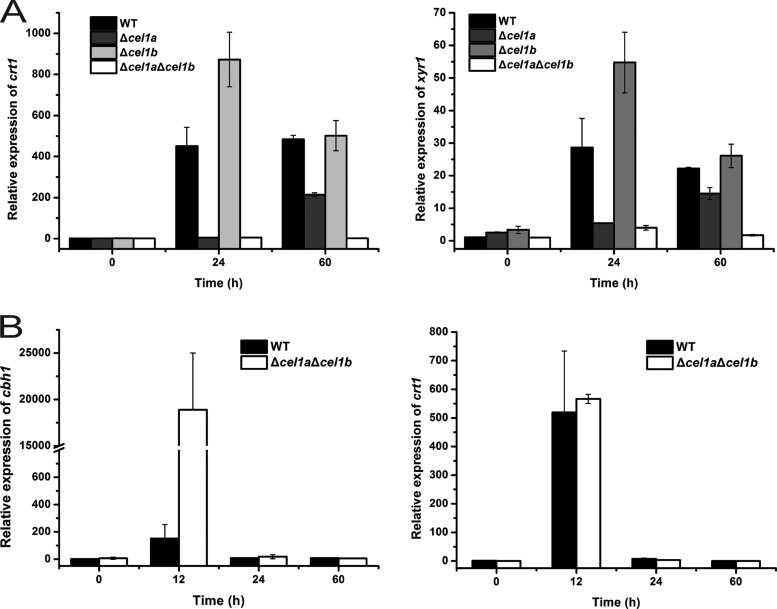
XYR1 has been established as the key regulator of the cellulolytic response to Avicel or lactose in T. reesei (20, 21). Our recent results, together with others, also identified an MFS sugar transporter, CRT1 (Tr_3405), that is essential for lactose- or Avicel-induced cellulase gene expression (9). We therefore tested whether the absence of CEL1a and CEL1b affected the expression of the respective genes in response to lactose. Compared with that of the WT, transcription of both xyr1 and crt1 was drastically reduced in the Δcel1a and Δcel1a Δcel1b strains during culture on lactose (Fig. 8A). In contrast to the case on lactose, induced transcription of crt1 was not affected, and the cbh1 transcript level was even significantly increased, in the Δcel1a Δcel1b strain on cellobiose (Fig. 8B), suggesting that the regulatory effect on the cellulolytic response by CEL1a and CEL1b is specific for lactose. To test whether the induction defect observed in the absence of CEL1a and CEL1b was solely due to the compromised expression of crt1 or xyr1, we expressed crt1 or xyr1 under the control of the pki or gpd promoter in the Δcel1a Δcel1b strain and analyzed its response to lactose. No induced expression of cbh1 was detected in the recombinant Δcel1a Δcel1b strain even under lactose induction, though the expression of crt1 and xyr1 in the recombinant strain occurred early on lactose and reached a level comparable to that of the WT (see Fig. S6 in the supplemental material), suggesting that, unlike the case for the WT strain with constitutive expression of Xyr1 (22), expression of crt1 or xyr1 is not sufficient to activate cellulolytic transcription, even upon induction, in the absence of CEL1a and CEL1b.
FIG 8.
Transcriptional analysis of xyr1, crt1, and cbh1 in response to lactose or cellobiose. (A) Transcription of xyr1 and crt1 in WT and β-glucosidase deletion strains cultured on 1% (wt/vol) lactose. Statistical analyses indicated significant differences in the transcription of xyr1 or crt1 between the WT and Δcel1a strains (P = 0.01 at 24 h and P = 0.002 at 60 h for xyr1; P = 0.001 at 24 h and P < 0.001 at 60 h for crt1) as well as between the WT and Δcel1a Δcel1b strains (P = 0.008 at 24 h and P < 0.001 at 60 h for xyr1; P = 0.001 at 24 h and P < 0.001 at 60 h for crt1). (B) Transcription of cbh1 and crt1 in WT and β-glucosidase deletion strains cultured on 0.25% (wt/vol) cellobiose. Values are the means of results from three biological replicates. Error bars show the SD for these replicates.
DISCUSSION
Although rarely present in its natural habitat, lactose is able to support the growth of cellulolytic Trichoderma reesei and to simultaneously induce its secretion of cellulases. Lactose has thus been one of the most important carbon sources for (hemi)cellulolytic enzyme production from T. reesei on a technical scale, despite the less-well-understood mechanism. In this study, we demonstrated that the absence of the intracellular β-glucosidases CEL1a and CEL1b abolishes cbh1 gene expression on lactose. We further presented evidence that both the CEL1a protein and its hydrolytic activity are responsible for the efficient cellulase induction by lactose. The intracellular β-glucosidase-mediated induction pathway may act downstream of intracellular delivery of lactose to activate XYR1, ensuring the successfully induced expression of cellulase genes.
While several lines of evidence support the notion that catabolism of lactose in T. reesei is realized only by extracellular hydrolysis and subsequent uptake of the resultant monomers (4), the most recent research identifies several MFS transporters that behave as lactose permeases and are involved in cellulase induction (9–11). In these scenarios, it is plausible to speculate that the intracellular presence of lactose may represent a key point in generating an inducing signal for cellulase gene expression. In the present report, we provide evidence supporting such an assumption. First, our data revealed that in addition to β-glucosidase activity, recombinant CEL1a also possesses weak hydrolytic activity toward lactose and ONPG. It is known that quite a few β-glucosidases belonging to glycosyl hydrolase (GH) family 1 also display β-galactosidase activity (23). Second, the absence of CEL1a and CEL1b almost abolishes cellulase gene induction by lactose, and catalytically inactive CEL1a is incapable of restoring efficiently induced cellulase gene expression. On the other hand, intracellular hydrolysis of lactose does not seem to fully account for the CEL1a- and CEL1b-mediated induction, since expression of a different GH2 galactosidase is incapable of restoring cellulase gene induction by lactose. Indeed, it has been shown that intracellular β-galactosidase activity is negligible, and slow metabolism of d-galactose alone is not sufficient to account for the efficient cellulase induction on lactose (8). Our result showing that galactose is not able to restore efficient induction in the absence of CEL1a and CEL1b reinforces such a point. However, the possibility that the subtle difference in the rates at which these two glycosidases catalyze the hydrolysis of lactose represents a very critical parameter for the efficient induction by lactose cannot be excluded. The possibility also exists that CEL1a and CEL1b could otherwise possess hydrolysis-associated activities, such as transglycosylation, to process lactose beyond hydrolysis. In this respect, it has been demonstrated that intracellular accumulation of galacto-oligosaccharides correlates with the level of cellulase formation during growth of T. reesei on lactose (24). Although the exact mechanism for how intracellular β-glucosidases contribute to the formation of inducer for cellulase gene expression warrants further study, the relatively earlier induction and higher level of cellulase gene expression conveyed by the constitutively expressed CEL1a protein are industrially relevant in view of cellulase production on lactose.
The lactose metabolism pathway has been shown to affect cellulase induction, though the involved mechanism is still elusive. An interesting point that somewhat contrasts with what has been reported for wild-type T. reesei can be made from our data. While the efficient transcription of cellulase genes is compromised in the Δcel1a strain, we found that full restoration or even a higher level of cellulase gene expression in the Δcel1a strain by adding galactose to the lactose medium was not influenced by further deletion of xyl1, whose absence results in a severe reduction in cellulase gene expression in WT T. reesei (7). This finding indicates that the restored cellulase gene expression can proceed independently of the second d-galactose-catabolizing pathway in the absence of CEL1a. On the other hand, transcription of gal1, the first gene of the Leloir pathway, is not affected during the growth of the Δcel1a strain on lactose. Although the possibility that GAL1 contributes to the restored induction upon galactose addition in the absence of CEL1a cannot be excluded at present, it is clear that intracellular β-glucosidases act independently of galactose metabolism in mediating the inducing cascade, considering that their absence has no effect on the growth on galactose. Finally, the induced cellulase gene expression in the Δcel1a strain upon galactose addition is not due to the restored extracellular hydrolysis of lactose, since the simultaneous absence of BGA1 has no effect on the recovered induction. The finding that addition of galactose is not able to restore the induction in the Δcel1a Δcel1b strain, although it restores its growth on lactose, further supports such a conclusion.
Our previous work and that of others have shown that CRT1, an MFS transporter protein, is critical for cellulase induction by lactose and cellulose (9). While transcription of crt1 is downregulated in the absence of intracellular glucosidases, constitutively expressing crt1 in the Δcel1a Δcel1b strain is not sufficient to recover its ability to respond to lactose induction. The failure to achieve efficient induction in these strains does not seem to result from the decreased expression of XYR1, since overexpression of XYR1 in the Δcel1a Δcel1b strain failed to initiate induction. We therefore hypothesize that intracellular processing of lactose by CEL1a and CEL1b acts downstream of intracellular delivery of lactose mediated by lactose permeases to achieve the efficient induction of cellulase genes (Fig. 9). The possibility cannot be excluded, however, that a potential signaling cascade initiated by CRT1 may exist and run relatively independent of β-glucosidase-mediated processing of lactose. Inducing signals transmitted by intracellular glucosidases and CRT1 both are thus necessary and may converge on XYR1 to ensure the successful cellulase gene expression on lactose.
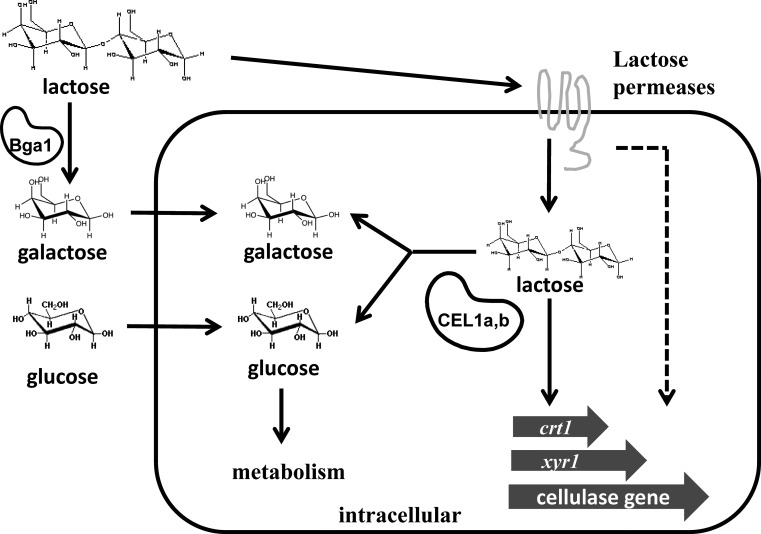
FIG 9.
Model of the role of intracellular β-glucosidases in cellulase induction by lactose in T. reesei. Upon growth of cells on lactose, lactose is either hydrolyzed extracellularly by BGA1 or transported intracellularly by lactose permeases. Further processing of lactose by the intracellular β-glucosidases CEL1a and CEL1b is necessary to initiate the transcriptional induction of genes, including xyr1, crt1, and cellulase genes. A possible signaling cascade may exist that initiates from CRT1 and converges on the β-glucosidase-mediated pathway to ensure efficient induction (dashed line).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Basic Research Program of China (grant 2011CB707402), the National Natural Science Foundation (grant 31270116 to W. Liu, grant 31300059 to W. Zhang, and grant 31240010 to Q. Zhou), New Century Excellent Talents in University (grant NCET-10-0546), and Shandong Provincial Funds for Distinguished Young Scientists (grant JQ201108). W. Zhang and Q. Zhou were also supported by grants for Young Excellent Scientists of Shandong Province (grants BS2013NJ021 and BS2013SW038). Q. Zhou also thanks the Key Laboratory of Industrial Fermentation Microbiology of the Ministry of Education and the Tianjin Key Lab of Industrial Microbiology, Tianjin University of Science & Technology (grant 2013IM001).
Footnotes
Published ahead of print 30 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00100-14.
REFERENCES
- 1.Lynd LR, Laser MS, Bransby D, Dale BE, Davison B, Hamilton R, Himmel M, Keller M, McMillan JD, Sheehan J. 2008. How biotech can transform biofuels. Nat. Biotechnol. 26:169–172. 10.1038/nbt0208-169 [DOI] [PubMed] [Google Scholar]
- 2.England G, Kelley A, Mitchinson C. June 2004. Induction of gene expression using a high concentration sugar mixture. US patent 20040121446
- 3.Andreotti RE, Medeiros JE, Roche C, Mandels M. 1980. Effects of strain and substrate on production of cellulases by Trichoderma reesei mutants, p 353–371 In Proceedings of the Bioconversion and Bioengineering Symposium. BERC, IIT, New Delhi, India [Google Scholar]
- 4.Seiboth B, Pakdaman BS, Hartl L, Kubicek CP. 2007. Lactose metabolism in filamentous fungi: how to deal with an unknown substrate. Fungal Biol. Rev. 21:42–48. 10.1016/j.fbr.2007.02.006 [DOI] [Google Scholar]
- 5.Seiboth B, Hartl L, Salovuori N, Lanthaler K, Robson GD, Vehmaanpera J, Penttila ME, Kubicek CP. 2005. Role of the bga1-encoded extracellular β-galactosidase of Hypocrea jecorina in cellulase induction by lactose. Appl. Environ. Microbiol. 71:851–857. 10.1128/AEM.71.2.851-857.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seiboth B, Hartl L, Pail M, Fekete E, Karaffa L, Kubicek CP. 2004. The galactokinase of Hypocrea jecorina is essential for cellulase induction by lactose but dispensable for growth on d-galactose. Mol. Microbiol. 51:1015–1025. 10.1046/j.1365-2958.2003.03901.x [DOI] [PubMed] [Google Scholar]
- 7.Seiboth B, Gamauf C, Pail M, Hartl L, Kubicek CP. 2007. The d-xylose reductase of Hypocrea jecorina is the major aldose reductase in pentose and d-galactose catabolism and necessary for β-galactosidase and cellulase induction by lactose. Mol. Microbiol. 66:890–900. 10.1111/j.1365-2958.2007.05953.x [DOI] [PubMed] [Google Scholar]
- 8.Karaffa L, Fekete E, Gamauf C, Szentirmai A, Kubicek CP, Seiboth B. 2006. d-Galactose induces cellulase gene expression in Hypocrea jecorina at low growth rates. Microbiology 152:1507–1514. 10.1099/mic.0.28719-0 [DOI] [PubMed] [Google Scholar]
- 9.Zhang W, Kou Y, Xu J, Cao Y, Zhao G, Shao J, Wang H, Wang Z, Bao X, Chen G. 2013. Two major facilitator superfamily sugar transporters from Trichoderma reesei and their roles in induction of cellulase biosynthesis. J. Biol. Chem. 288:32861–32872. 10.1074/jbc.M113.505826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porciuncula J, Furukawa T, Shida Y, Mori K, Kuhara S, Morikawa Y, Ogasawara W. 2013. Identification of major facilitator transporters involved in cellulase production during lactose culture of Trichoderma reesei PC-3-7. Biosci. Biotechnol. Biochem. 77:1014–1022. 10.1271/bbb.120992 [DOI] [PubMed] [Google Scholar]
- 11.Ivanova C, Bååth JA, Seiboth B, Kubicek CP. 2013. Systems analysis of lactose metabolism in Trichoderma reesei identifies a lactose permease that is essential for cellulase induction. PLoS One 8:e62631. 10.1371/journal.pone.0062631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Q, Xu J, Kou Y, Lv X, Zhang X, Zhao G, Zhang W, Chen G, Liu W. 2012. Differential involvement of β-glucosidases from Hypocrea jecorina in rapid induction of cellulase genes by cellulose and cellobiose. Eukaryot. Cell 11:1371–1381. 10.1128/EC.00170-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Häkkinen M, Arvas M, Oja M, Aro N, Penttilä M, Saloheimo M, Pakula TM. 2012. Re-annotation of the CAZy genes of Trichoderma reesei and transcription in the presence of lignocellulosic substrates. Microb. Cell Fact. 11:134. 10.1186/1475-2859-11-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruber F, Visser J, Kubicek CP, de Graaff LH. 1990. The development of a heterologous transformation system for the cellulolytic fungus Trichoderma reesei based on a pyrG-negative mutant strain. Curr. Genet. 18:71–76. 10.1007/BF00321118 [DOI] [PubMed] [Google Scholar]
- 15.Mach RL, Schindler M, Kubicek CP. 1994. Transformation of Trichoderma reesei based on hygromycin B resistance using homologous expression signals. Curr. Genet. 25:567–570. 10.1007/BF00351679 [DOI] [PubMed] [Google Scholar]
- 16.Pöggeler S, Masloff S, Hoff B, Mayrhofer S, Kück U. 2003. Versatile EGFP reporter plasmids for cellular localization of recombinant gene products in filamentous fungi. Curr. Genet. 43:54–61. 10.1007/s00294-003-0370-y [DOI] [PubMed] [Google Scholar]
- 17.Penttila M, Nevalainen H, Ratto M, Salminen E, Knowles J. 1987. A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene 61:155–164. 10.1016/0378-1119(87)90110-7 [DOI] [PubMed] [Google Scholar]
- 18.Hartl L, Kubicek CP, Seiboth B. 2007. Induction of the gal pathway and cellulase genes involves no transcriptional inducer function of the galactokinase in Hypocrea jecorina. J. Biol. Chem. 282:18654–18659. 10.1074/jbc.M700955200 [DOI] [PubMed] [Google Scholar]
- 19.Green MR, Sambrook J. 2012. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 20.Stricker AR, Steiger MG, Mach RL. 2007. Xyr1 receives the lactose induction signal and regulates lactose metabolism in Hypocrea jecorina. FEBS Lett. 581:3915–3920. 10.1016/j.febslet.2007.07.025 [DOI] [PubMed] [Google Scholar]
- 21.Stricker AR, Grosstessner-Hain K, Wurleitner E, Mach RL. 2006. Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and d-xylose metabolism in Hypocrea jecorina. Eukaryot. Cell 5:2128–2137. 10.1128/EC.00211-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mach-Aigner AR, Pucher ME, Steiger MG, Bauer GE, Preis SJ, Mach RL. 2008. Transcriptional regulation of xyr1, encoding the main regulator of the xylanolytic and cellulolytic enzyme system in Hypocrea jecorina. Appl. Environ. Microbiol. 74:6554–6562. 10.1128/AEM.01143-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhatia Y, Mishra S, Bisaria V. 2002. Microbial β-glucosidases: cloning, properties, and applications. Crit. Rev. Biotechnol. 22:375–407. 10.1080/07388550290789568 [DOI] [PubMed] [Google Scholar]
- 24.Karaffa L, Coulier L, Fekete E, Overkamp KM, Druzhinina IS, Mikus M, Seiboth B, Novák L, Punt PJ, Kubicek CP. 2013. The intracellular galactoglycome in Trichoderma reesei during growth on lactose. Appl. Microbiol. Biotechnol. 97:5447–5456. 10.1007/s00253-012-4667-y [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.