Abstract
A comparison of the daclatasvir (DCV [BMS-790052]) resistance barrier on authentic or hybrid replicons containing NS5A from hepatitis C virus (HCV) genotypes 1 to 6 (GT-1 to -6) was completed using a replicon elimination assay. The data indicated that genotype 1b (GT-1b) has the highest relative resistance barrier and genotype 2a (GT-2a M31) has the lowest. The rank order of resistance barriers to DCV was 1b > 4a ≥ 5a > 6a ≅ 1a > 2a JFH > 3a > 2a M31. Importantly, DCV in combination with a protease inhibitor (PI) eliminated GT-2a M31 replicon RNA at a clinically relevant concentration. Previously, we reported the antiviral activity and resistance profiles of DCV on HCV genotypes 1 to 4 evaluated in the replicon system. Here, we report the antiviral activity and resistance profiles of DCV against hybrid replicons with NS5A sequences derived from HCV GT-5a and GT-6a clinical isolates. DCV was effective against both GT-5a and -6a hybrid replicon cell lines (50% effective concentrations [EC50s] ranging from 3 to 7 pM for GT-5a, and 74 pM for GT-6a). Resistance selection identified amino acid substitutions in the N-terminal domain of NS5A. For GT-5a, L31F and L31V, alone or in combination with K56R, were the major resistance variants (EC50s ranging from 2 to 40 nM). In GT-6a, Q24H, L31M, P32L/S, and T58A/S were identified as resistance variants (EC50s ranging from 2 to 250 nM). The in vitro data suggest that DCV has the potential to be an effective agent for HCV genotypes 1 to 6 when used in combination therapy.
INTRODUCTION
Daclatasvir (DCV [BMS-790052]) is a cross-genotypic NS5A inhibitor with picomolar to low nanomolar potency in the replicon system (1, 2). The antiviral activity of DCV in vitro translated into clinical efficacy, with hepatitis C virus (HCV) RNA declines of ∼3 to 4 log10 observed in genotype 1a (GT-1a)-infected subjects treated once daily (QD) with 60 mg of DCV in a 14-day multiple ascending dose (MAD) monotherapy study (3, 4). Moreover, DCV was effective against GT-1b and -1a in combinations that include either pegylated interferon and ribavirin (PEG-IFN-RBV) or other direct-acting anti-HCV agents (DAAs) (5–8). There are large populations of viral quasispecies preexisting in infected individuals, and variants that confer resistance to antiviral agents can be rapidly enriched and/or selected during antiviral treatment (9–11). Since DCV resistance variants show no cross-resistance to other DAAs, DCV should rapidly suppress wild-type virus and variants resistant to other DAAs, thereby enhancing the effectiveness of other DAAs in combination therapies (2, 3). This effect is predicted to lead to higher rates of sustained viral response (SVR) and/or shorten the duration of treatment necessary to achieve SVR. Recent clinical results with DCV plus asunaprevir (ASV [BMS-650032]) in patients infected with GT-1b and with DCV plus sofosbuvir (SOF [GS-7977]) in patients infected with GT-1, -2, and 3 demonstrate the effectiveness of DCV in interferon-free DAA combination therapies (6, 12).
Prior studies using the in vitro replicon system indicated that DCV should be an effective antiviral agent against HCV genotypes 1 to 4 (2, 13–16). Here, we report the antiviral activity and resistance profiles of DCV against hybrid replicons with NS5A sequences derived from GT-5a and GT-6a clinical isolates. GT-5a, the only GT-5 subtype, accounts for ∼40% of HCV infections in South Africa and is also found at lower prevalence in other parts of the world (17, 18). GT-6 is prevalent in Asian countries, including China (especially Hong Kong), Vietnam, Myanmar, Thailand, Indonesia, and South Korea. GT-6 has 21 subtypes (GT-6a to GT-6u), with GT-6a being the most prevalent, while the others are rarely observed (18). DCV is an inhibitor of GT-5a and GT-6a hybrid replicon cell lines, with 50% effective concentrations (EC50s) of less than 75 pM. Resistance-associated amino acid substitutions in NS5A were observed at positions similar to those reported for other genotypes within the first 100 amino acids (aa) of NS5A. The availability of GT-5a and GT-6a NS5A hybrid replicons, together with authentic or hybrid replicons from genotypes 1 to 4 (2, 14–16), enabled direct comparison of the resistance barriers of DCV in these diverse genotypes by using replicon elimination assays (13). These studies revealed that GT-1b has the highest barrier of resistance to DCV, while GT-2a has the lowest resistance barrier, with a relative rank order of 1b > 4a ≥ 5a > 6a ≅ 1a > 2a JFH > 3a > 2a M31. Importantly, a combination of DCV with a protease inhibitor (PI) (MK-5172) (19) was effective in clearing replicon RNA from the most resistant genotype, GT-2a M31, at clinically relevant doses, while either agent alone was not. Overall, these in vitro results suggest that DCV has the potential to be effective in combination therapies targeting HCV genotypes 1 to 6.
MATERIALS AND METHODS
Molecular clones and HCV DAAs.
Viral RNA was isolated from HCV-positive sera from three GT-5a-infected subjects (patient ID no. 1209A016, 1209A017, and 1209A019; Boca Biolistics, Coconut Creek, FL) and one GT-6a-infected subject (lot no. 9256752; SeraCare Life Sciences, Milford, MA) by using a QIAamp minElute virus vacuum kit following the manufacturer's protocol (Qiagen, Germantown, MD). NS5A cDNA was obtained by reverse transcription-PCR (RT-PCR) by using a first-strand cDNA synthesis kit (Invitrogen, Carlsbad, CA) followed by amplification with Platinum Taq high-fidelity DNA polymerase (Invitrogen, Carlsbad, CA). The PCR primers were 5′-CAGTCTCCGGAACATGGCTTAGGGCCATTTG-3′ and 5′-GAAAGATCCGGATCCCCAGGCTCGCCCTCCAATG-3′ for GT-5a and 5′-CATGCTCCGGATCATGGTTACGCGACGTGTGGG-3′ and 5′-GACTGTCCGGATCACCAGGCTCCCCCTCAAG-3′ for GT-6a. Following digestion with BspEI restriction enzyme, the amplicons were ligated into BspEI sites in a JFH1 subgenomic, bicistronic replicon that expresses a Renilla luciferase reporter gene and neomycin (G418) resistance. A GT-2a M31 replicon was similarly constructed with sequences encoding amino acids 1 to 425 amplified by PCR from a pJ6CF infectious clone (20). The resulting replicons encoded NS5A proteins with amino acids 1 to 430, 1 to 431, and 1 to 425 derived from GT-5a, GT-6a, and GT-2a J6CF, respectively, and an additional 41 C-terminal amino acids from the parental JFH1 strain (15). Replicon sequences were confirmed by sequencing analysis. Replicon cell lines (GT-5a, GT-6a, and GT-2a M31 and NS5A variants) were selected as previously described (2). Replicon RNA was recovered from the cell lines, and NS5A sequences were verified by sequence analysis as described previously (2). Maintenance of the variant replicon cell lines did not require the presence of DCV. DCV and an NS3 protease inhibitor (PI), MK-5172 (19), were synthesized by Bristol-Myers Squibb.
Resistance selection and genotypic analysis.
For resistance selection, GT-5a or GT-6a replicon cells seeded in T-75 tissue culture flasks were maintained in complete growth medium (Dulbecco's modified Eagle's medium [DMEM] plus 10% fetal bovine serum) supplemented with 0.5 mg/ml G418 and the desired concentration of DCV (BMS-790052) diluted in dimethyl sulfoxide (DMSO) (Sigma-Aldrich Corp., St. Louis, MO). Control cells were maintained with an equivalent percent volume of DMSO. Cells were split or fed with fresh medium containing inhibitor twice weekly to maintain a subconfluent monolayer. After about 2 to 3 weeks, pooled cells were expanded for phenotypic and genotypic analyses.
For genotypic analysis of DCV-treated cells, total cellular RNA was isolated from replicon cells with TRIzol reagent (Invitrogen, Carlsbad, CA) and treated with DNase (Promega, Madison, WI) prior to being used as a template for reverse transcription-PCR (RT-PCR). First-strand NS5A cDNA was synthesized from random hexamers with a Superscript first-strand cDNA synthesis kit following the provided protocol (Invitrogen, Carlsbad, CA). Platinum Taq high-fidelity DNA polymerase was used for PCR amplification of NS5A cDNA (Invitrogen, Carlsbad, CA). The primer set was 5′-CTTACTATAACCAGCCTACTCAGAAGACTCCAC-3′ (forward) and 5′-CTCAAAGGGTTGATTGGCAACTTTTCCTCTTC-3′ (reverse). The resulting amplicons were used for direct sequencing analysis (population sequencing) and to generate NS5A cDNA clones via ligation into a TOPO TA cloning vector (Invitrogen, Carlsbad, CA).
Phenotypic analysis.
Mutations were introduced into the NS5A coding region of GT-5a and GT-6a replicons by recombinant PCR as previously described (2). All nucleotide changes were verified by sequence analysis. Stable cell lines bearing G418-resistant replicons were selected as described previously (2). Renilla luciferase assays were performed with a Renilla luciferase assay system (Promega, Madison, WI) as previously described (2). The 50% effective concentration (EC50) was calculated as previously described (2) and represents the concentration of inhibitor required for a 50% reduction in viral replication as measured by luciferase activity (2). For each experiment, values from DMSO-treated wells were treated as 0% inhibition, while values from wells treated with 2 μM protease inhibitor (the control inhibitor) were designated as having 100% inhibition.
Replicon elimination assay.
For replicon elimination, replicon cells (∼1.5 × 106) were plated in 10-cm-diameter dishes in complete DMEM containing 0.5 mg/ml G418 and maintained at 37°C in 5% CO2. After 24 h, medium was replaced with complete DMEM without G418 and containing the desired concentration of DCV or DMSO (control). After 3 days, cells were split, and 1/8 of the culture was maintained without G418 and with the same concentration of DCV. These cells were processed in the same way on days 7, 10, and 14. On day 14, the cells were split into two 10-cm dishes, and each dish was plated with 1/4 of the cells. One dish was maintained without inhibitor for 2 weeks in complete medium supplemented with 0.5 mg/ml G418 and maintained without further splitting to allow replicon-retaining cells to form colonies, which were stained with Coomassie brilliant blue R-250 staining solution (Bio-Rad, Hercules, CA). In the second dish, G418-resistant cells were amplified for phenotypic and genotypic analyses (13). For the study in which DCV was combined with a PI, cells were split on days 3 and 7 for colony formation, but they were not amplified for genotypic and phenotypic analyses.
Nucleotide sequence accession numbers.
Nucleotide sequences of the genes encoding aa 1 to 430 of GT-5a-6, GT-5a-7, and GT-5a-9 and aa 1 to 431 of GT-6a-16 are available in GenBank under accession no. KJ719452 to KJ719454 and KJ719455, respectively.
RESULTS
GT-5a and GT-6a NS5A hybrid replicons.
Hybrid replicons constructed from NS5A sequences derived from GT-5a and -6a clinical isolates were used to study the antiviral activities and resistance profiles of DCV. Three GT-5a replicon clones derived from three different subjects were constructed, while, a single GT-6a replicon clone was made (Fig. 1A). Previous studies have shown that the N-terminal region of NS5A is largely responsible for mediating the antiviral activity of DCV and other DCV-like NS5A inhibitors (1, 2, 13, 21). Alignments of the first 100 amino acids (aa) of NS5A from GT-5a hybrid replicons with SA13 and EUH1480, two GT-5a sequences deposited in GenBank (SA13, GenBank accession no. AF064490; and EUH1480, GenBank accession no. Y13184), revealed that the GT-5a strains were 91 to 96% identical to the reference sequences over this region (Fig. 1B), while the GT-6a NS5A sequence (aa 1 to 100) was 96% identical to two GT-6a reference sequences HK6a and EUHK2 (HK6a, GenBank accession no. HQ852465; and EUHK2, GenBank accession no. Y12083) (Fig. 1C). This comparison suggests that these patient isolates are representative GT-5a and GT-6a strains, respectively.
FIG 1.
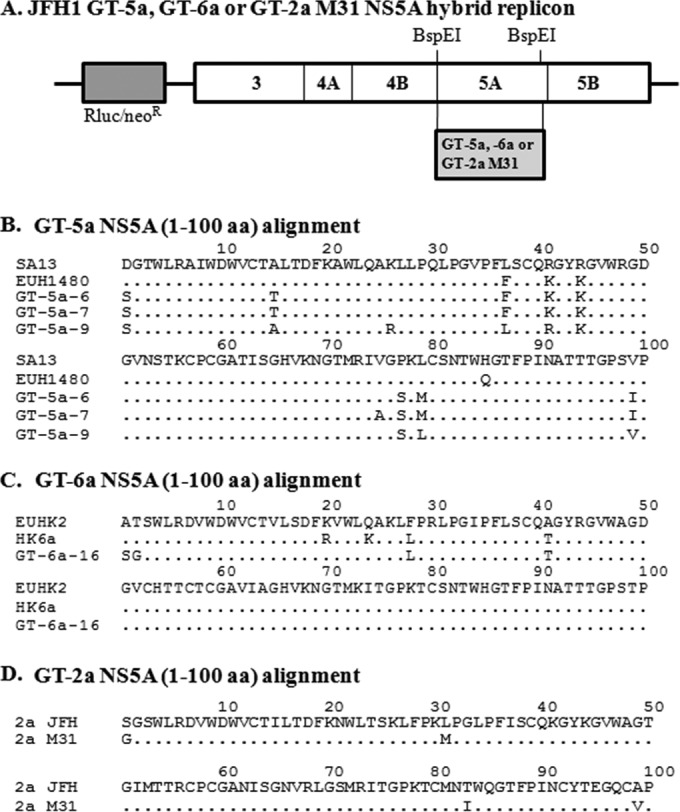
(A) Schematic diagram of a bicistronic JFH1 replicon with Renilla luciferase (Rluc) and neomycin resistance (neoR) genes. The 5′ untranslated region (5′-UTR) is derived from the GT-1b Con1 strain, and the nonstructural genes and 3′-UTR are derived from the GT-2a JFH1 strain. Hybrid replicons were constructed by replacing the indicated BspEI restriction fragment with sequences from GT-5a-6, GT-5a-7, GT-5a-9, and GT-6a-16 clinical isolates or the pJ6CF infectious clone (GT-2a M31). (B) Alignment of the N-terminal 100 amino acids of the NS5A from GT-5a-6, GT-5a-7, and GT-5a-9 isolates and two GT-5a NS5A reference sequences (SA13, GenBank accession no. AF064490; EUH1480, GenBank accession no. Y13184). (C) Alignment of the N-terminal 100 amino acids of the NS5A from the GT-6a-16 isolate and two GT-6a NS5A reference sequences (EUHK2, GenBank accession no. Y12083; HK6a, GenBank accession no. HQ852465). (D) Alignment of the N-terminal 100 amino acids of the NS5A from the JFH1 strain and the pJ6CF (GT-2a M31) infectious clone. Amino acid identities are indicated with dots.
In vitro activity of DCV on GT-5a NS5A.
Stable replicon cell lines harboring each of the GT-5a hybrid replicons (GT-5a-6, GT-5a-7, and GT-5a-9) were used to evaluate the antiviral activity of DCV. DCV displayed effective intrinsic antiviral activity toward each stable cell line, with EC50s ranging from 3 to 7 pM (Table 1). Since the GT-5a-7 and GT-5a-6 strains were 99% identical to each other over the NS5A first 100 amino acids, GT-5a-7 and GT-5a-9 replicon cell lines were chosen for resistance selection studies. Replicon cell lines were maintained for 2 to 3 weeks in the presence of DCV at concentrations of 1, 10, and 100 nM (∼200-, 2,000-, and 20,000-fold over the DCV EC50 in the parental cell line, respectively), with G418 included in the medium. No cells survived selection at 100 nM DCV. At 10 nM DCV, a few colonies were obtained from the GT-5a-7 replicon cell line, but no GT-5a-9 replicon cells survived. At 1 nM DCV, some colonies were obtained from both replicon cell lines. Colonies from each selection condition were pooled, expanded, and used to determine susceptibility to DCV. The DCV EC50s were ∼14 and ∼37 nM, respectively, for the 1 and 10 nM DCV-selected GT-5a-7 cells and 4.5 nM for the 1 nM DCV-selected GT-5a-9 cells (Table 1). Selected cells retained the same sensitivity as the parental cell lines to DAA inhibitors targeting NS3 and NS5B (data not shown). NS5A cDNA was amplified by RT-PCR and analyzed by bulk population sequencing analysis. In cDNA obtained from the 1 nM DCV-selected cells, a mixture of L31F and L31V amino acid substitutions was predicted, while in the GT-5a-7 cells selected with 10 nM DCV, L31F and K56R changes were predicted (Table 1). Analysis of individual NS5A cDNA clones confirmed the presence of these amino acid substitutions. Of 21 clones from the GT-5a-7 replicon selection with 1 nM DCV, 15 had nucleotide changes resulting in L31F and 3 had changes resulting in L31V. The remaining three clones encoded NS5A proteins with linked amino acid substitutions (L31F-K56R or L31V-K56R) (Table 2). Similarly, in the 1 nM DCV-selected GT-5a-9 cells, all of the clones encoded NS5A proteins with L31F or L31V changes. As predicted by the population sequencing results, NS5A cDNA clones derived from the 10 nM DCV-selected GT-5a-7 cells encoded proteins containing either L31F by itself (17 clones) or L31F linked with K56R (11 clones). L31 variants were previously identified in GT-1a, -1b, -2a, and -3a. For example, L31F was observed in GT-1b and -3a, L31M was observed in GT-1a, -1b, and -2a, and L31V was observed in GT-1a and -1b (1, 2, 13–15). K56R is a novel variant observed in the current study.
TABLE 1.
DCV activity on parental and selected GT-5a NS5A hybrid replicon cell lines
| Replicon | EC50 (nM) for parental cellsa | DCV selection concn (nM)b | EC50 (nM) for selected cellsa | NS5A amino acid substitution(s) (%)c |
|---|---|---|---|---|
| GT-5a-6 | 0.0072 ± 0.0007 | Not done | NAd | NA |
| GT-5a-7 | 0.0036 ± 0.0007 | DMSO | 0.0054 ± 0.0002 | None |
| 1 | 14 ± 0.22 | L31F/V (80/20) | ||
| 10 | 37 ± 0.51 | L31F (100), K56R (50) | ||
| 100 | No cells survived | NA | ||
| GT-5a-9 | 0.003 ± 0.0003 | DMSO | 0.0042 ± 0.0006 | None |
| 1 | 4.5 ± 0.35 | L31F/V (45/55) | ||
| 10 | No cells survived | NA | ||
| 100 | No cells survived | NA |
Values are means ± standard deviations (n ≥ 3).
An equivalent percent volume of DMSO alone was used as a control.
Substitutions were deduced from bulk NS5A sequence traces. Numbers in parentheses represent the estimated percentages of the indicated amino acids.
NA, not applicable.
TABLE 2.
NS5A amino acid substitutions in cDNA clones isolated from DCV-treated GT-5a replicon cells
| Replicon | Amino acid substitution(s) | No. of clones with indicated substitution at DCV selection concn of: |
|
|---|---|---|---|
| 1 nM | 10 nM | ||
| GT-5a-7 | L31F | 15 | 17 |
| L31V | 3 | 0 | |
| L31F-K56R | 2 | 11 | |
| L31V-K56R | 1 | 0 | |
| Total no. of clones | 21 | 28 | |
| GT-5a-9 | |||
| L31F | 9 | NAa | |
| L31V | 12 | NA | |
| Total no. of clones | 21 | NA | |
NA, not applicable.
To confirm and evaluate the resistance of GT-5a variants selected by DCV, nucleotide changes resulting in amino acid substitutions L31F, L31V, K56R, L31F-K56R, and L31V-K56R were introduced into the GT-5a-7 backbone, and an L31V-resulting change was introduced into the GT-5a-9 hybrid replicon. Because these variants replicated poorly in transient replication assays in the absence or presence of DCV, we were unable to calculate DCV EC50s (data not shown). Cell lines harboring GT-5a replicons with these substitutions were therefore selected and used to determine the sensitivity of these variants to DCV. As shown in Table 3, individual L31F and L31V amino acid changes conferred significant resistance to DCV with EC50s ranging from 2 to 7 nM. K56R by itself did not confer substantial resistance to DCV (∼3-fold), but, when combined with L31F or L31V, it enhanced resistance to DCV by ∼6- to 13-fold relative to the L31 changes alone (Table 3). The EC50s of L31V in GT-5a-7 and GT-5a-9 were very similar, indicating that L31V resistance was not affected by other polymorphic differences in these strains.
TABLE 3.
Susceptibility of GT-5a replicon variants to DCV
| Parent or variant | EC50 (nM)a | Fold resistance |
|---|---|---|
| GT-5a-7 (parent) | 0.0048 ± 0.0005 | 1 |
| GT-5a-7 L31F | 6.9 ± 1.0 | 1,438 |
| GT-5a-7 L31V | 2.2 ± 0.81 | 458 |
| GT-5a-7 K56R | 0.012 ± 0.0013 | 3 |
| GT-5a-7 L31F-K56R | 39.3 ± 13.1 | 8,188 |
| GT-5a-7 L31V-K56R | 29.4 ± 2.2 | 6,125 |
| GT-5a-9 (parent) | 0.0031 ± 0.0005 | 1 |
| GT-5a-9 L31V | 2.2 ± 0.05 | 710 |
Values are means ± standard deviations obtained from Huh-7 cells harboring the indicated GT-5a replicon variants (n ≥ 3).
In vitro activity of DCV on GT-6a NS5A.
A GT-6a NS5A hybrid replicon cell line, GT-6a-16 (EC50 for DCV of 74 pM) was used for resistance selection with DCV at concentrations of 1, 10, and 100 nM (∼14-, 140-, and 1,400-fold over the parental cell line EC50, respectively). Colonies obtained from each of the selection conditions were pooled and expanded for phenotypic and genotypic analyses. The selected cells showed reduced sensitivity to DCV, with EC50s of 6.4, 59.2, and 323 nM, respectively, for 1, 10, and 100 nM DCV (Table 4). Bulk population sequencing analysis of NS5A cDNA obtained from the DCV-selected replicon cells revealed a mixture of P32S and T58A/S NS5A amino acid substitutions in the replicon selected with 1 nM DCV and a mixture of L31M and P32L/S in the replicon selected with 10 nM DCV. The relative proportions of each change, as predicted from the bulk population chromatograms, are shown in Table 4. In contrast, P32L was the only change observed in NS5A cDNA recovered from the 100 nM DCV-selected cells (Table 4). To determine which of the substitutions observed in cDNAs from the 1 and 10 nM DCV selections are dominant and if any of them are linked, sequencing analysis of individual cDNA clones was performed (Table 5). From the 1 nM selection, 22 of 43 cDNA clones contained changes resulting in a T58A amino acid substitution, and 8 additional clones had changes resulting in P32S. Nucleotide changes resulting in a variety of amino acid substitutions were observed in the remaining clones, including Q24H, L31M, P32Q, T58S, Q24H-T58A, L31P-P32S, and T58A-T93N. Clones from the 10 nM-selected cells were more homogenous, with the majority encoding NS5A proteins with P32S (∼60%). Mutations resulting in individual P32L, L31M, P32A, or T58N amino acid substitutions were present in the remaining clones.
TABLE 4.
DCV activity on the parental and selected GT-6a NS5A hybrid replicon (GT-6a-16) cell lines
| Replicon | EC50 (nM) for parental cellsa | DCV selection concn (nM)b | EC50 (nM) for selected cellsa | NS5A amino acid substitution(s) (%)c |
|---|---|---|---|---|
| GT-6a-16 parent | 0.074 ± 0.005 | DMSO | 0.041 ± 0.004 | None |
| GT-6a-16 DCV selected | 1 | 6.4 ± 1.3 | P32S (25), T58A/S (45/10) | |
| 10 | 59.2 ± 8.2 | L31 M (20), P32L/S (10/70) | ||
| 100 | 323 ± 6.5 | P32L (100) |
Values are means ± standard deviations (n ≥ 3).
An equivalent percent volume of DMSO alone was used as a control.
Substitutions were deduced from bulk NS5A sequence traces. Numbers in parentheses represent the estimated percentages of the indicated amino acids.
TABLE 5.
NS5A amino acid substitutions in cDNA clones isolated from DCV-treated GT-6a-16 replicon cells
| Amino acid substitution | No. of clones with indicated amino acid substitution(s) at DCV selection concn of: |
|
|---|---|---|
| 1 nM | 10 nM | |
| Q24H | 3 | 0 |
| L31M | 2 | 6 |
| P32A | 0 | 1 |
| P32L | 0 | 9 |
| P32Q | 1 | 0 |
| P32S | 8 | 28 |
| T58A | 22 | 0 |
| T58N | 0 | 1 |
| T58S | 4 | 0 |
| Q24H-T58A | 1 | 0 |
| L31P-P32S | 1 | 0 |
| T58A-T93N | 1 | 0 |
| Total no. of clones | 43 | 45 |
To assess the antiviral activity of DCV on GT-6a resistance variants, stable replicon cell lines harboring GT-6a-16 NS5A with individual amino acid substitutions (Q24H, L31M, P32L, P32S, T58A, T58N, or T58S) were generated (Table 6). DCV inhibited replicons in these cell lines, with EC50s ranging from 2 nM (Q24H and T58A/S) to 250 nM (P32L), confirming that each of these substitutions confers resistance to DCV in this genetic background. The levels of resistance conferred by the different amino acid substitutions generally accord with the different selective pressures (Tables 4 and 6). For example, the relatively high level of resistance conferred by the P32L amino acid substitution is sufficient to explain the existence of this variant in cells that survived treatment with 100 nM DCV.
TABLE 6.
Susceptibility of GT-6a replicon variants to DCV
| Parent or variant | EC50 (nM)a | Fold resistance |
|---|---|---|
| GT-6a-16 (parent) | 0.050 ± 0.0048 | 1 |
| Q24H | 2.04 ± 0.5 | 41 |
| L31M | 43.9 ± 12.4 | 878 |
| P32L | 250 ± 69.4 | 5,000 |
| P32S | 19.1 ± 1.43 | 382 |
| T58A | 2.36 ± 0.69 | 47 |
| T58N | 26.6 ± 3.76 | 532 |
| T58S | 2.4 ± 0.4 | 48 |
Values are means ± standard deviations derived from Huh-7 cells harboring the indicated GT-6a-16 replicon variants (n ≥ 3).
DCV resistance barriers on NS5A from HCV genotypes 1 to 6.
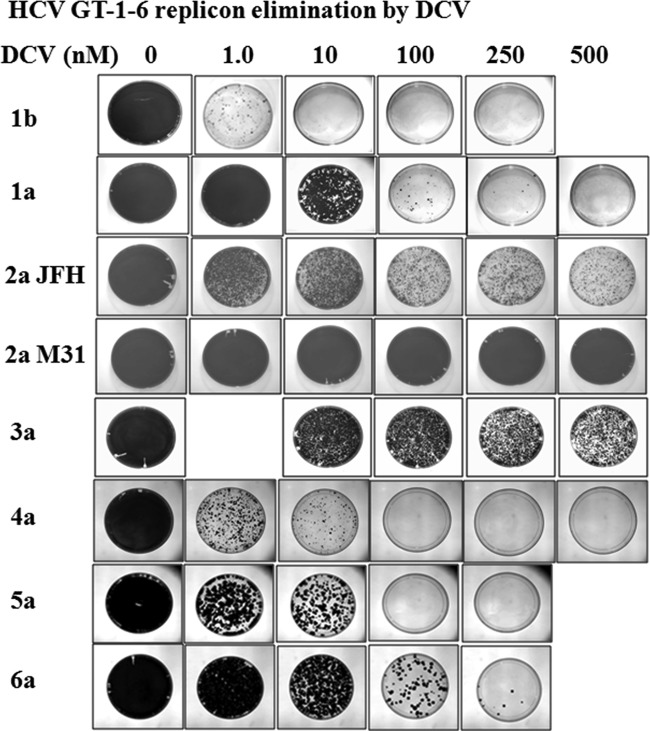
We previously reported the use of authentic or hybrid NS5A replicons to study the in vitro antiviral activity of DCV against HCV genotypes 1a, 1b, 2a JFH, 3a, and 4a (1, 2, 13–16). In addition, we have used replicon elimination assays (also referred to as replicon curing or colony formation assays) to examine the in vitro resistance barriers of GT-1a, -1b, and -2a JFH to DCV (13, 22). In replicon elimination assays, variants that are not sensitive to a specific concentration of DCV emerge as replicon-retaining colonies once selective pressure (G418 [500 μg/ml]) is applied following treatment with DCV for a given period of time (Fig. 2). These assays therefore provide a semiquantitative method to compare the relative sensitivity of each genotype to DCV based on the relative numbers of resistant colonies that remained. In the present study, resistance barriers were defined by the concentration of DCV required to eliminate or greatly reduce the number of emerging resistant replicon colonies following treatment with DCV for 14 days. For example, a genotype was considered to have a higher resistance barrier if few or no colonies remained following DCV treatment, and a genotype was considered to have a lower resistance barrier if more colonies remained after the same treatment. The availability of GT-5a and GT-6a hybrid replicons reported in this study allowed us to extend our study of the DCV resistance barrier to include replicons with NS5A derived from HCV genotypes 1 to 6. In addition to the GT-2a JFH strain, we also included a hybrid JFH-J6 GT-2a subgenomic replicon in this analysis (Fig. 1D). The JFH-J6 hybrid replicon, herein termed GT-2a M31, has a methionine at NS5A amino acid position 31 (20), while the parental JFH1 strain contains a leucine at this position (Fig. 1D). Since the majority of GT-2 NS5A sequences (∼80%) deposited in the database contain a methionine at NS5A amino acid position 31 (23)—a residue associated with resistance to DCV in several genotypes, including GT-2a (14)—the GT-2a M31 replicon is expected to be a better representative of GT-2a sequences in the clinical population. As mentioned above, we have previously reported results from replicon elimination studies for GT-1b and GT-1a replicon cell lines. In order to obtain a more direct comparison of the results, replicon elimination assays with these replicon cell lines were repeated in the present study, and results similar to those previously reported (13) were observed (Fig. 2).
FIG 2.
HCV replicon elimination with DCV. Huh-7 cells harboring GT-1 to -6 replicons were treated with the indicated concentrations of DCV for 14 days in the absence of G418. Cell cultures were maintained at subconfluency by splitting as described in Materials and Methods. After 14 days, DCV was removed, and a portion of the cell cultures were treated with medium containing G418 (500 μg/ml) for 2 weeks to allow cells that harbored the HCV replicon to form colonies. The colonies that formed were photographed. For GT-2a JFH, GT-2a M31, and GT-3a replicon cells, parallel cultures of cells were amplified for genotypic analysis. The replicon cell lines used in this study are as follows: 1b, Con1, and 1a, H77c (2, 13); 2a JFH, JFH-1 (AB047639); 2a M31 NS5A (aa 1 to 425), 2a infectious clone-pJ6CF (NC_009823.1); 3a NS5A (aa 1 to 429), HCV3a1 (JX944789); 4a NS5A, HCV4a-23 (JQ347515); 5a NS5A (aa 1 to 430), GT-5a-7 (KJ719453); and 6a NS5A (aa 1 to 431), GT-6a-16 (KJ719455).
Results from replicon elimination studies for GT-1a, -1b, -2a JFH, -2a M31, -3a, -4a, -5a, and -6a are shown in Fig. 2. The replicon cell lines included in the analysis, as well as DCV EC50s for parental and major DCV resistance variants, are shown in Table 7. For GT-3a and GT-4a, similar results were obtained with cells harboring hybrid replicons with NS5A sequences derived from an additional patient isolate (data not shown). The in vitro resistance barrier to DCV is lower for GT-1a than GT-1b, a result that correlates well with the observed in vivo efficacy of DCV (Fig. 2) (3, 4, 13). The current analysis indicates that the genetic barriers to resistance for GT-4a and GT-5a fall between those for GT-1b and GT-1a. For both of these genotypes, the number of replicon-retaining colonies decreased in a dose-dependent manner following 14 days of treatment with DCV, and each of these replicons was completely eliminated by 100 nM DCV (Fig. 2). In comparison, the GT-1b replicon was eliminated with 10 nM DCV, while GT-1a required 500 nM DCV. The results with the GT-6a replicon were similar to those obtained with GT-1a, with a few colonies observed when cells were treated with 250 nM DCV (Fig. 2).
TABLE 7.
Comparison of DCV activities on HCV genotype 1 to 6 replicon cells of parental and major resistance variants
| Genotype | Parental EC50 (nM)a | Major resistance variantb (EC50 nM)a | Reference(s) |
|---|---|---|---|
| 1b (Con1) | 0.0031 ± 0.0016 | L31V-Y93H (250 ± 100) | 2, 3, 13 |
| 1a (H77c) | 0.022 ± 0.002 | Q30E (259.1 ± 38), Y93N (354.2 ± 119.1), or linkage | 2, 3, 13, 24 |
| 2a JFH | 0.011 ± 0.001 | F28S (1,852 ± 25) | 14; this study |
| 2a M31 | 13 ± 3.8 | F28S-L31M (>2,000)c | This study |
| 3a | 0.53 ± 0.15 | Y93H (1,451 ± 344) or E92K (NDd) | 15; this study |
| 4a | 0.013 ± 0.0001 | L30H (15.8 ± 7.2) or R30G (12.1 ± 2.0) | 16 |
| 5a | 0.0048 ± 0.0005 | L31F (6.9 ± 1.0) or L31F-K56R (39.3 ± 13.1) | This study |
| 6a | 0.074 ± 0.005 | P32L (250 ± 69.4) | This study |
Values are means ± standard deviations (n ≥ 3).
Major resistance variant identified in clinically relevant concentration or highest concentration in replicon elimination assay, in vitro resistance selection or clinical study.
Data derived from F28S-L31M in the GT-2a JFH backbone as F28S in the GT-2a M31 backbone had poor replication and was unable to construct a replicon cell line.
ND, not determined.
In contrast, the GT-2a and GT-3a replicons displayed lower genetic barriers for resistance to DCV, and in each of these cases, multiple colonies were obtained after treatment for 14 days with 500 nM DCV. Since these replicons were the most resistant to DCV, genotypic analysis was performed on colonies that were collected and expanded from parallel plates at the higher DCV concentrations (250 or 500 nM). For GT-2a JFH and GT-2a M31 replicons, sequencing analysis of NS5A cDNA recovered from pooled colonies revealed the consensus F28S amino acid substitution. A 2a JFH F28S replicon variant was shown previously to confer a high level of resistance to DCV (Table 7) (14). An F28S variant in the 2a M31 hybrid replicon backbone was not analyzed successfully because it replicated poorly. Instead, a replicon variant with a combination of F28S and L31M in the 2a JFH1 backbone was evaluated and found to be very resistant to DCV (DCV EC50, >2 μM) (Table 7), thus explaining the ability of the replicon cells to survive treatment at 500 nM DCV. Genotypic analysis of the GT-3a replicons from surviving colonies revealed that a majority contained an NS5A Y93H amino acid substitution (∼90%) but also revealed the presence of an E92K substitution (∼10%). The Y93H variant of GT-3a was previously reported and was found to confer high-level resistance to DCV (EC50, ∼1.5 μM) (Table 7) (15), but efforts to determine the level of resistance to DCV conferred by E92K were unsuccessful due to poor replicative ability (data not shown). An A92K substitution in the GT-1a background has been shown to confer high-level resistance to DCV (DCV EC50, ∼1 μM) (C. Wang and R. A. Fridell, unpublished data).
Based on the replicon elimination results, the GT-2a M31 variant was the least sensitive to DCV. Even though little replicon clearing is apparent from the plates photographed in Fig. 2, substantial cell death was observed when G418 was added to the cells following 14 days of treatment with DCV at ≥100 nM, suggesting that DCV partially eliminated replicon RNA from these cells. Consistent with this observation, genotypic analysis revealed that all of the surviving replicon cells under treatment with 500 nM DCV had a resistant amino acid substitution (F28S), indicating that DCV effectively suppressed the parental replicon.
To determine if DCV could effectively eliminate GT-2a M31 replicon in combination therapy with a second DAA, we evaluated DCV plus a PI in the replicon elimination assay as described in Materials and Methods (Fig. 3). Treatment of GT-2a M31 replicon cells for 3 or 7 days with 50 or 100 nM PI was not sufficient to eradicate the replicon RNA. When DCV (250 nM) was combined with a PI (100 nM), the GT-2a M31 replicon was effectively eliminated in 7 days. Furthermore, the number of surviving colonies was substantially reduced when 250 nM DCV was used in combination with either 50 or 100 nM PI following 3 days of treatment, compared to cells treated for the same period with PI alone (Fig. 3). These results demonstrate that despite a relatively low resistance barrier for the GT-2a 31M replicon, DCV still has the potential to be effective in combination therapy against this least susceptible HCV strain.
FIG 3.
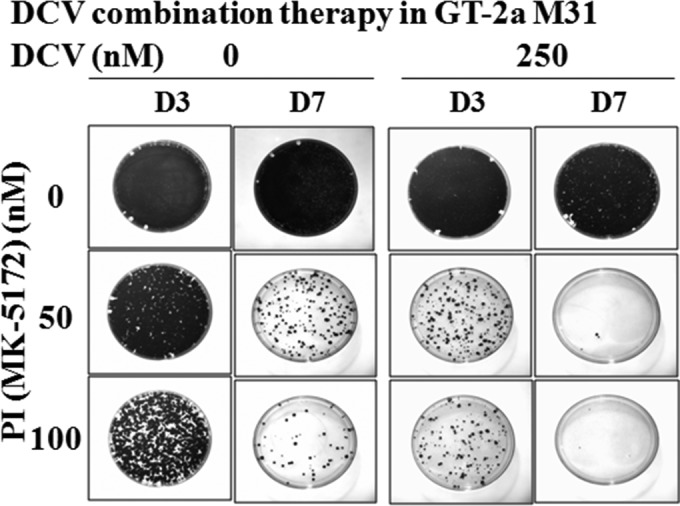
GT-2a M31 replicon cells were treated for 3 or 7 days with the indicated concentration of a PI, MK-5172, in the absence or presence of 250 nM DCV and without G418. After either 3 or 7 days of treatment (D3 and D7, respectively), inhibitors were removed and cells were split and maintained in medium containing G418 (500 μg/ml) for 2 weeks to allow replicon-retaining cells to form colonies.
DISCUSSION
DCV is an HCV NS5A inhibitor with picomolar to low-nanomolar potency in replicon assays and has demonstrated effective antiviral activity in the clinic (1–4). An initial viral RNA decline of greater than 3.0 log10 was achieved by DCV in a 14-day multiple ascending dose (MAD) study with doses of 30 to 100 mg QD in genotype 1 HCV-infected subjects (3, 4). In phase I monotherapy studies, DCV was able to completely eradicate wild-type and low-level-resistance variants with ∼3- to 4-log10 viral RNA declines while selecting or enriching more highly resistant variants that persisted for at least 24 to 48 weeks during posttreatment follow-up (3, 8, 24). With its ability to achieve rapid and substantial viral load declines and its promising clinical safety profile, DCV has the potential to be a valuable component of HCV combination therapies. Recent clinical results with combinations of DCV and the NS3 protease inhibitor asunaprevir (ASV [BMS-650032]) in subjects infected with GT-1b or DCV plus sofosbuvir (SOF [GS-7977]) in subjects infected with GT-1, -2, and -3 have confirmed the effectiveness of DCV in all-oral DAA combination regimens (6, 12).
In the present study, we have shown that DCV inhibits hybrid replicons with NS5A sequences derived from GT-5a clinical isolates with single-digit pM EC50s. Resistance selection and phenotypic analysis in the GT-5a hybrid replicons identified L31F, L31V, and L31F/V-K56R as DCV resistance-associated NS5A amino acid substitutions. L31F or L31V variants have also been found to be associated with resistance to DCV in GT-1b, -1a, or -3a replicons (2, 13, 15). Prior studies also identified amino acid substitutions at NS5A residues 54, 58, or 62 in association with DCV resistance in genotype 1 replicons (2, 3, 13, 25), but the K56R change observed here is the first indication that substitutions at residue 56 can contribute to resistance. By itself, K56R did not confer resistance to DCV, but it did enhance the level of resistance conferred by L31F (∼6-fold) and L31V (∼13-fold). The impact of K56R in GT-5a is similar to the impact of the E62D variant in GT-1a, which when linked with Q30R increased the level of resistance conferred by Q30R (∼15-fold) (25). In resistance selection studies, GT-5a replicon variants did not emerge when cells were maintained in the presence of 100 nM DCV and G418 selective pressure (Table 1). The most resistant GT-5a replicon variant identified in the present study was L31F-K56R, which was selected by treatment with 10 nM DCV and was inhibited by DCV with an EC50 of ∼40 nM. This finding was consistent with the replicon elimination results where treatment with 100 nM DCV for 14 days completely cleared the GT-5a replicon RNA from Huh-7 cells (Fig. 2).
DCV displays effective antiviral activity toward a GT-6a hybrid replicon with an EC50 of 74 pM. Resistance selection and phenotypic analysis identified Q24H, L31M, P32L/S, and T58A/N/S as resistance variants. With the exception of position 24, all of these NS5A residue positions have been found in association with DCV resistance in other genotypes. Most of the GT-6a variants identified in this study conferred modest levels of resistance to DCV, with EC50s ranging from 2 to 44 nM (Table 6). In contrast, the P32L amino acid substitution conferred a higher level of resistance to DCV (EC50, ∼250 nM). A proline at NS5A position 32 is highly conserved among all HCV sequences deposited in the European HCV database and is present in all 16 GT-6a sequences in the database (23), suggesting that preexisting variants at this position could be relatively rare. Moreover, DAAs directed against other HCV targets (for example NS3 protease inhibitors) retain wild-type antiviral activity against the GT-6a P32L variant (data not shown), so the P32L variant would be expected to be suppressed in combination therapies. In this sense, the P32L variant may be similar to a Q30E variant in GT-1a. The Q30E variant in GT-1a is similarly resistant to DCV (EC50, 259 nM) (Table 7) and is readily enriched or selected in DCV monotherapy or in vitro replicon elimination studies (3, 13, 24). However, DCV is still effective against GT-1a in combination therapies. Furthermore, the pattern of GT-6a replicon elimination was very similar to that of GT-1a, indicating that GT-6a and GT-1a have similar resistance barriers to DCV.
The overall results from replicon elimination assays (Fig. 2) suggest the following relative rank order for the resistance barrier to DCV: 1b > 4a ≥ 5a > 6a ≅ 1a > 2a JFH > 3a > 2a M31. As the resistance barriers for GT-1b, -4a, -5a, and -6a are similar to or higher than that of GT-1a, and since DCV effectiveness against GT-1a has been established in the clinic in combination therapy studies (5, 7, 12), our studies suggest that DCV will display antiviral activity on GT-4a, -5a, and -6a in combination therapies.
The potential clinical effectiveness of DCV in subjects infected with GT-2a and GT-3a is less clear, since there are no clinical data available for DCV monotherapy, and GT-2a and -3a replicons have lower resistance barriers to DCV than GT-1a. Consistent with the antiviral activity of DCV toward the parental replicons (DCV EC50, 0.011, 13.0, and 0.53 nM for GT-2a JFH, -2a M31, and -3a, respectively), 100% of GT-2a JFH, -2a M31, and -3a replicon RNA recovered from cells that survived treatment at higher concentrations of DCV (GT-2a JFH and -3a at 250 and 500 nM; GT-2a M31 at 500 nM) contained a resistance-associated amino acid substitution predicted to confer a high level of resistance to DCV. This observation indicates that the replicons containing wild-type sequences and variants with low-level resistance were eradicated, while highly resistant variants were enriched or selected (F28S for GT-2a JFH and -2a M31 and Y93H or E92K for GT-3a). In a recent report, Scheel et al. examined the activity of DCV against recombinant HCV strains with NS5A sequences derived from genotypes 1 to 7 (26). Consistent with the results reported here, they observed that GT-2a M31 (J6) and GT-3a strains were the least sensitive to DCV. However, they did not directly examine the relative resistance barriers of the hybrid viruses to DCV.
The effectiveness of DCV in a combination therapy against the replicon with the lowest resistance barrier, GT-2a M31 (Fig. 3), was examined by pairing DCV with a PI (19). While monotherapy with DCV was poorly effective against the GT-2a M31 replicon (Fig. 2), DCV clearly enhanced the effectiveness of the PI, and the combination of DCV and the PI completely eradicated the replicon RNA at clinically relevant concentrations. Gottwein et al. also demonstrated that DCV in combination with asunaprevir (ASV [BMS-650032]) was effective against GT-2a and GT-3a recombinant viruses (27). In clinical studies, DCV has been found to be effective in combination with sofosbuvir for GT-2- and GT-3-infected subjects (12). These findings provide both in vitro and in vivo evidence that DCV can be an effective agent in combination therapies directed against GT-2a and GT-3a.
Previous genetic data confirming the NS5A inhibitor's genotypic specificity implicated that DCV or DCV-like inhibitors target NS5A domain I (2, 21). Other studies found that DCV-like inhibitors interact with NS5A in replicon cells (1, 28, 29) and further demonstrated that a DCV-like inhibitor could be cross-linked to a region of NS5A comprising amino acids 21 to 30 (28). Recently, Ascher et al. reported a direct interaction between DCV and purified NS5A domain 1 protein from GT-1b (Con1) and showed that DCV was able to disrupt an NS5A-RNA interaction (30). Furthermore, the authors observed that the common resistance substitutions L31V and Y93H reduced the binding affinity of DCV to NS5A (30). These studies and the location of NS5A resistance variants to the N terminus of NS5A (1–3, 13, 21, 24, 25) are consistent with a hypothesis that DCV and DCV-like NS5A inhibitors directly target NS5A. The results support the use of hybrid replicons with NS5A sequences derived from different strains to evaluate the antiviral profile of DCV or DCV-like NS5A inhibitors.
Overall, DCV has been shown to inhibit replicons with NS5A sequences derived from HCV genotypes 1 to 6 with picomolar to low-nanomolar potencies (Table 7). The antiviral activities, resistance profiles, and resistance barriers observed in this and other studies (1–4, 13–16, 31) support further exploration of DCV as an agent for HCV cross-genotypic combination therapy.
ACKNOWLEDGMENTS
We thank Mark Cockett, Nicholas Meanwell, and Makonen Belema for valuable discussions and critical reading of the manuscript.
This study was supported by Bristol-Myers Squibb.
Footnotes
Published ahead of print 16 June 2014
REFERENCES
- 1.Gao M, Nettles RE, Belema M, Snyder LB, Nguyen VN, Fridell RA, Serrano-Wu MH, Langley DR, Sun J-H, O'Boyle DR, II, Lemm JA, Wang C, Knipe JO, Chien C, Colonno RJ, Grasela DM, Meanwell NA, Hamann LG. 2010. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature 465:96–100. 10.1038/nature08960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fridell RA, Qiu D, Wang C, Valera L, Gao M. 2010. Resistance analysis of the hepatitis C virus NS5A inhibitor BMS-790052 in an in vitro replicon system. Antimicrob. Agents Chemother. 54:3641–3650. 10.1128/AAC.00556-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fridell RA, Wang C, Sun J-H, O'Boyle DR, Nower P, Valera L, Qiu D, Roberts S, Huang X, Kienzle B, Bifano M, Nettles RE, Gao M. 2011. Genotypic and phenotypic analysis of variants resistant to hepatitis C virus nonstructural protein 5A replication complex inhibitor BMS-790052 in humans: in vitro and in vivo correlations. Hepatology 54:1924–1935. 10.1002/hep.24594 [DOI] [PubMed] [Google Scholar]
- 4.Nettles RE, Gao M, Bifano M, Chung E, Persson A, Marbury TC, Goldwater R, DeMicco MP, Rodriguez-Torres M, Vutikullird A, Fuentes E, Lawitz E, Lopez-Talavera JC, Grasela DM. 2011. Multiple ascending dose study of BMS-790052, a nonstructural protein 5A replication complex inhibitor, in patients infected with hepatitis C virus genotype 1. Hepatology 54:1956–1965. 10.1002/hep.24609 [DOI] [PubMed] [Google Scholar]
- 5.Pol S, Ghalib RH, Rustgi VK, Martorell C, Everson GT, Tatum HA, Hézode C, Lim JK, Bronowicki J-P, Abrams GA, Bräu N, Morris DW, Thuluvath PJ, Reindollar RW, Yin PD, Diva U, Hindes R, McPhee F, Hernandez D, Wind-Rotolo M, Hughes EA, Schnittman S. 2012. Daclatasvir for previously untreated chronic hepatitis C genotype-1 infection: a randomised, parallel-group, double-blind, placebo-controlled, dose-finding, phase 2a trial. Lancet Infect. Dis. 12:671–677. 10.1016/S1473-3099(12)70138-X [DOI] [PubMed] [Google Scholar]
- 6.Chayama K, Takahashi S, Toyota J, Karino Y, Ikeda K, Ishikawa H, Watanabe H, McPhee F, Hughes E, Kumada H. 2012. Dual therapy with the nonstructural protein 5A inhibitor, daclatasvir, and the nonstructural protein 3 protease inhibitor, asunaprevir, in hepatitis C virus genotype 1b-infected null responders. Hepatology 55:742–748. 10.1002/hep.24724 [DOI] [PubMed] [Google Scholar]
- 7.Lok AS, Gardiner DF, Lawitz E, Martorell C, Everson GT, Ghalib R, Reindollar R, Rustgi V, McPhee F, Wind-Rotolo M, Persson A, Zhu K, Dimitrova DI, Eley T, Guo T, Grasela DM, Pasquinelli C. 2012. Preliminary study of two antiviral agents for hepatitis C genotype 1. N. Engl. J. Med. 366:216–224. 10.1056/NEJMoa1104430 [DOI] [PubMed] [Google Scholar]
- 8.McPhee F, Hernandez D, Yu F, Ueland J, Monikowski A, Carifa A, Falk P, Wang C, Fridell R, Eley T, Zhou N, Gardiner D. 2013. Resistance analysis of hepatitis C virus genotype 1 prior treatment null responders receiving daclatasvir and asunaprevir. Hepatology 58:902–911. 10.1002/hep.26388 [DOI] [PubMed] [Google Scholar]
- 9.Cuevas JM, Gonzalez-Candelas F, Moya A, Sanjuan R. 2009. Effect of ribavirin on the mutation rate and spectrum of hepatitis C virus in vivo. J. Virol. 83:5760–5764. 10.1128/JVI.00201-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guedj J, Rong L, Dahari H, Perelson AS. 2010. A perspective on modelling hepatitis C virus infection. J. Viral Hepat. 17:825–833. 10.1111/j.1365-2893.2010.01348.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simmonds P. 2004. Genetic diversity and evolution of hepatitis C virus—15 years on. J. Gen. Virol. 85:3173–3188. 10.1099/vir.0.80401-0 [DOI] [PubMed] [Google Scholar]
- 12.Sulkowski MS, Gardiner DF, Rodriguez-Torres M, Reddy KR, Hassanein T, Jacobson I, Lawitz E, Lok AS, Hinestrosa F, Thuluvath PJ, Schwartz H, Nelson DR, Everson GT, Eley T, Wind-Rotolo M, Huang S-P, Gao M, Hernandez D, McPhee F, Sherman D, Hindes R, Symonds W, Pasquinelli C, Grasela DM. 2014. Daclatasvir plus sofosbuvir for previously treated or untreated chronic HCV infection. N. Engl. J. Med. 370:211–221. 10.1056/NEJMoa1306218 [DOI] [PubMed] [Google Scholar]
- 13.Wang C, Huang H, Valera L, Sun J-H, O'Boyle DR, Nower PT, Jia L, Qiu D, Huang X, Altaf A, Gao M, Fridell RA. 2012. Hepatitis C virus RNA elimination and development of resistance in replicon cells treated with BMS-790052. Antimicrob. Agents Chemother. 56:1350–1358. 10.1128/AAC.05977-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fridell RA, Qiu D, Valera L, Wang C, Rose RE, Gao M. 2011. Distinct functions of NS5A in hepatitis C virus RNA replication uncovered by studies with the NS5A inhibitor BMS-790052. J. Virol. 85:7312–7320. 10.1128/JVI.00253-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang C, Valera L, Jia L, Kirk MJ, Gao M, Fridell RA. 2013. In vitro activity of daclatasvir on hepatitis C virus genotype 3 NS5A. Antimicrob. Agents Chemother. 57:611–613. 10.1128/AAC.01874-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang C, Jia L, Huang H, Qiu D, Valera L, Huang X, Sun J-H, Nower PT, O'Boyle DR, Gao M, Fridell RA. 2012. In vitro activity of BMS-790052 on hepatitis C virus genotype 4 NS5A. Antimicrob. Agents Chemother. 56:1588–1590. 10.1128/AAC.06169-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smuts HE, Kannemeyer J. 1995. Genotyping of hepatitis C virus in South Africa. J. Clin. Microbiol. 33:1679–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naamani KA, Sinani SA, Deschênes M. 2013. Epidemiology and treatment of hepatitis C genotypes 5 and 6. Can. J. Gastroenterol. Hepatol. 27:e8–e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Summa V, Ludmerer SW, McCauley JA, Fandozzi C, Burlein C, Claudio G, Coleman PJ, DiMuzio JM, Ferrara M, Di Filippo M, Gates AT, Graham DJ, Harper S, Hazuda DJ, McHale C, Monteagudo E, Pucci V, Rowley M, Rudd MT, Soriano A, Stahlhut MW, Vacca JP, Olsen DB, Liverton NJ, Carroll SS. 2012. MK-5172, a selective inhibitor of hepatitis C virus NS3/4a protease with broad activity across genotypes and resistant variants. Antimicrob. Agents Chemother. 56:4161–4167. 10.1128/AAC.00324-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yanagi M, Purcell RH, Emerson SU, Bukh J. 1999. Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology 262:250–263. 10.1006/viro.1999.9889 [DOI] [PubMed] [Google Scholar]
- 21.Lemm JA, O'Boyle D, II, Liu M, Nower PT, Colonno R, Deshpande MS, Snyder LB, Martin SW, St Laurent DR, Serrano-Wu MH, Romine JL, Meanwell NA, Gao M. 2010. Identification of hepatitis C virus NS5A inhibitors. J. Virol. 84:482–491. 10.1128/JVI.01360-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Boyle DR, II, Nower PT, Sun J-H, Fridell R, Wang C, Valera L, Gao M. 2013. A 96-well based analysis of replicon elimination with the HCV NS5A replication complex inhibitor daclatasvir. J. Virol. Methods 193:68–76. 10.1016/j.jviromet.2013.04.024 [DOI] [PubMed] [Google Scholar]
- 23.Combet C, Garnier N, Charavay C, Grando D, Crisan D, Lopez J, Dehne-Garcia A, Geourjon C, Bettler E, Hulo C, Mercier PL, Bartenschlager R, Diepolder H, Moradpour D, Pawlotsky J-M, Rice CM, Trepo C, Penin F, Deleage G. 2007. euHCVdb: the European Hepatitis C Virus database. Nucleic Acids Res. 35:D363–D366. 10.1093/nar/gkl970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang C, Sun J-H, O'Boyle DR, Nower P, Valera L, Roberts S, Fridell RA, Gao M. 2013. Persistence of resistant variants in hepatitis C virus-infected patients treated with the NS5A replication complex inhibitor daclatasvir. Antimicrob. Agents Chemother. 57:2054–2065. 10.1128/AAC.02494-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun J-H, O'Boyle DR, II, Zhang Y, Wang C, Nower P, Valera L, Roberts S, Nettles RE, Fridell RA, Gao M. 2012. Impact of a baseline polymorphism on the emergence of resistance to the hepatitis C virus nonstructural protein 5a replication complex inhibitor, BMS-790052. Hepatology 55:1692–1699. 10.1002/hep.25581 [DOI] [PubMed] [Google Scholar]
- 26.Scheel TKH, Gottwein JM, Mikkelsen LS, Jensen TB, Bukh J. 2011. Recombinant HCV variants with NS5A from genotypes 1–7 have different sensitivities to an NS5A inhibitor but not interferon-α. Gastroenterology 140:1032–1042e1036. 10.1053/j.gastro.2010.11.036 [DOI] [PubMed] [Google Scholar]
- 27.Gottwein JM, Jensen SB, Li Y-P, Ghanem L, Scheel TKH, Serre SBN, Mikkelsen L, Bukh J. 2013. Combination treatment with hepatitis C virus protease and NS5A inhibitors is effective against recombinant genotype 1a, 2a, and 3a viruses. Antimicrob. Agents Chemother. 57:1291–1303. 10.1128/AAC.02164-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Boyle DR, II, Sun J-H, Nower PT, Lemm JA, Fridell RA, Wang C, Romine JL, Belema M, Nguyen VN, Laurent DRS, Serrano-Wu M, Snyder LB, Meanwell NA, Langley DR, Gao M. 2013. Characterizations of HCV NS5A replication complex inhibitors. Virology 444:343–354. 10.1016/j.virol.2013.06.032 [DOI] [PubMed] [Google Scholar]
- 29.Targett-Adams P, Graham EJS, Middleton J, Palmer A, Shaw SM, Lavender H, Brain P, Tran TD, Jones LH, Wakenhut F, Stammen B, Pryde D, Pickford C, Westby M. 2011. Small molecules targeting hepatitis C virus-encoded NS5A cause subcellular redistribution of their target: insights into compound modes of action. J. Virol. 85:6353–6368. 10.1128/JVI.00215-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ascher DB, Wielens J, Nero TL, Doughty L, Morton CJ, Parker MW. 2014. Potent hepatitis C inhibitors bind directly to NS5A and reduce its affinity for RNA. Sci. Rep. 4:4765. 10.1038/srep04765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y-P, Ramirez S, Humes D, Jensen SB, Gottwein JM, Bukh J. 2014. Differential sensitivity of 5′UTR-NS5A recombinants of hepatitis C virus genotypes 1–6 to protease and NS5A inhibitors. Gastroenterology 146:812–821.e4. 10.1053/j.gastro.2013.11.009 [DOI] [PubMed] [Google Scholar]