Abstract
To design and discover new antimicrobial peptides (AMPs) with high levels of antimicrobial activity, a number of machine-learning methods and prediction methods have been developed. Here, we present a new prediction method that can identify novel AMPs that are highly similar in sequence to known peptides but offer improved antimicrobial activity along with lower host cytotoxicity. Using previously generated AMP amino acid substitution data, we developed an amino acid activity contribution matrix that contained an activity contribution value for each amino acid in each position of the model peptide. A series of AMPs were designed with this method. After evaluating the antimicrobial activities of these novel AMPs against both Gram-positive and Gram-negative bacterial strains, DP7 was chosen for further analysis. Compared to the parent peptide HH2, this novel AMP showed broad-spectrum, improved antimicrobial activity, and in a cytotoxicity assay it showed lower toxicity against human cells. The in vivo antimicrobial activity of DP7 was tested in a Staphylococcus aureus infection murine model. When inoculated and treated via intraperitoneal injection, DP7 reduced the bacterial load in the peritoneal lavage solution. Electron microscope imaging and the results indicated disruption of the S. aureus outer membrane by DP7. Our new prediction method can therefore be employed to identify AMPs possessing minor amino acid differences with improved antimicrobial activities, potentially increasing the therapeutic agents available to combat multidrug-resistant infections.
INTRODUCTION
Antimicrobial peptides (AMPs) are produced by multicellular organisms to defend against microbial infections. Along with potent antimicrobial activity, many AMPs also have the ability to enhance immunity by functioning as immunomodulators (1–3). AMPs therefore have excellent therapeutic potential, especially in light of increased drug resistance to many conventional antibiotic therapies. A number of naturally occurring peptides and synthetic derivatives have been developed or are currently in development (2, 4–6). Although AMPs vary in length, amino acid composition, and structure, they share some similarities, such as electrical charge and amphipathicity (3, 7). To determine the characteristics that are important in antimicrobial activity, bioinformatic tools and prediction methods have been developed (8), all based to some extent on the sequence similarities between peptides (9–11).
To optimize the antimicrobial activity of identified AMPs and to predict novel peptide sequences, we present a machine-learning method based on the concept of an antimicrobial activity contribution score for each amino acid. Here, we consider that each amino acid in a peptide sequence possesses a different level of importance for the biological activity of that peptide, and this is represented by an assigned score. By calculation of each amino acid's contribution score, we can predict the antimicrobial activity of an AMP.
To verify our results, we tested some of our designed AMPs in an antimicrobial assay with a range of bacterial strains. One of these peptides, DP7, was selected for further testing. The cytotoxicity of DP7 against HEK 293 and human epithelial fibroblasts and cells was evaluated, and experiments using a Staphylococcus aureus-infected mouse model were also performed to evaluate a suitable method of AMP administration. To investigate the antimicrobial mechanism employed by this peptide, we used transmission electron microscopy (TEM) and scanning electron microscopy (SEM) to elucidate the effect of DP7 on S. aureus in vitro.
MATERIALS AND METHODS
Data sources and preparation.
Twelve-mer peptide substitution data were obtained from the published literature (12, 13). Nine-mer peptide sequences and activities were obtained from the patent WO 2008/022444 application. The relative 50% effective concentration values for peptide Bac034 substitution data were calibrated to the proxy 50% inhibitory concentration value. Before contribution score calculations were determined, the logarithms of the values were calculated to standardize activity.
Calculation of the antimicrobial activity contribution score of each amino acid.
To determine the contribution of each amino acid to the antimicrobial activity of the peptide, we developed the following equation: , where C is the standardized activity of an antimicrobial peptide, n is the length of the antimicrobial peptide, AA is the antimicrobial activity contribution score of an amino acid, i represents the type of amino acid, j represents the position of the amino acid within the peptide sequence, and a is a constant that is assigned a value of 1 when this position is occupied with an amino acid and a value of 0 when this position is unoccupied.
Amino acid antimicrobial activity contribution score matrix.
The data for one peptide can be applied to the above-mentioned equation. We regard all these equations as one equation of the matrix. To the right of the equation would be the matrix containing the amino acid matrix which comprises the AAij of each antimicrobial peptide. To the left of the equation would be the matrix containing the standardized antimicrobial activity of the antimicrobial peptide in the left matrix. To solve equations, MATLAB (version 7.12.0, R2011a) was used. After calculation, the results were further transformed into a matrix, designated the amino acid antimicrobial activity contribution score matrix, in which the row indicated the position in the whole peptide and the column indicated the amino acid type.
Prediction method.
From the sequence data, 10 to 20% of the sequences were selected by random sampling as the testing set, and the rest of the sequences were used as the training set to build the amino acid contribution score matrix. The score matrix equation presented above was used to predict the activities of novel AMPs, with a higher score corresponding to higher activity. Short peptides were considered sub-sequences of the longer peptides, and their scores were calculated in every possible sub-sequence position. The sub-sequence with the highest score was then selected as the most probable sub-sequence.
Experimental materials.
To determine the MICs of the newly generated antimicrobial peptides, bacterial strains of Pseudomonas aeruginosa (PAO1), S. aureus (ATCC 25923 and ATCC 33591), and Escherichia coli (ATCC 25922) were purchased from the American Type Culture Collection (Rockville, MD). Salmonella enterica serovar Typhi strains were obtained from Xiang and colleagues (14). Clinical isolates were obtained from the Burn Unit, Southwest Hospital of China (Sichuan, China). The antibiotic linezolid (research compound; Airsea Pharmaceutical, Ltd., Taizhou, China), methanesulfonic acid levofloxacin injection (Resources Double-crane Pharmaceutical Co., Ltd., Beijing, China), and gentamicin sulfate injection (Hubei Pharmaceutical Co., Ltd., Hubei, China) were used as positive controls. Mueller-Hinton broth (MHB) and Mueller-Hinton agar (MHA), used to culture bacteria, were purchased from Qingdao Hope Bio-Technology Co., Ltd. (Shandong, China).
Peptide synthesis.
Peptides were synthesized by using fluorenylmethyloxycarbonyl (Fmoc) chemistry at Shanghai Science Peptide Biological Technology Co., Ltd. (Shanghai, China). Synthesized peptides were purified by high-performance liquid chromatography to 95% purity, and their molecular weights were confirmed by mass spectrometry. The peptide concentration was estimated from the weighed sample, and the peptide was resuspended in water or phosphate-buffered saline (PBS) to a concentration of 10 mg/ml and stored at −20°C.
Antimicrobial activity assay.
The antimicrobial activity assay was performed according to the National Committee for Clinical Laboratory Standards M7-A7 method. In general, the peptide was serially diluted in MHB and 50 μl of each concentration was added to replicate wells of a 96-well flat-bottom tissue culture plate. Bacteria, grown on MHA for 20 h, were diluted to ∼1 × 107 CFU ml−1 in MHB. Then, 50-μl aliquots of these bacterial suspensions were added in triplicate to wells containing antimicrobial peptide. The final peptide concentrations were in the range of 128 to 0.125 mg/liter. The solutions were then incubated at 37°C for 20 h, and their absorbances at 630 nm were measured using a microplate reader (Multiskan MK3; Thermo). The lowest concentration that showed 80% growth inhibition relative to the growth control determined the MIC.
Hemolysis assay.
Human erythrocytes collected from blood samples of healthy humans were harvested by centrifugation for 10 min at 2,000 rpm (400 × g) and washed three times in PBS (8.5 g NaCl, 2.2 g Na2HPO4, 0.4 g NaH2PO4, in 1 liter of solution; pH 7.4). A 20% (vol/vol) suspension of human erythrocytes in PBS was then stored at 4°C. When needed, the suspension was diluted 1:20 in PBS and 100 μl was added in triplicate to 100 μl of a 2-fold serial dilution series of peptide in a 96-well plate. Total hemolysis was achieved with 1% Tween 20. The plates were incubated at 37°C for 1 h and centrifuged for 10 min at 3,000 rpm (900 × g). Then, 160 μl of the supernatant was transferred to a new 96-well plate to measure the absorbance at 405 nm by using a microplate reader (Multiskan MK3; Thermo), and the percent hemolysis was calculated.
In vitro cytotoxicity assay.
HEK 293 cells (human embryo kidney cells; ATCC CRL-1573) and human epithelial fibroblasts (primary culture from human skin) were cultured in a complete medium (Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum) in a humidified atmosphere of 5% CO2 at 37°C. Trypsin-EDTA (0.05%) was used to detach cells in subcultures. Cells were seeded onto 96-well plates at a density of 5,000 cells/well in 0.1 ml of complete medium. After 20 h of incubation, cells were treated with serially diluted peptide (final concentrations in the range of 20 to 640 mg/liter) and incubated for another 24 h. Various shades of orange, which represented the amount of living cells, were measured with the CCK-8 cell counting kit (Yeasen, Shanghai, China), and the absorbance at 450 nm was measured with a microplate reader (Multiskan MK3; Thermo) to calculate the cell viability.
Electron microscopy.
The morphology of S. aureus cells incubated with DP7 (1× MIC) for 0, 10, 30, and 60 min was visualized by SEM (JSM-7500F; FEI). The morphology of S. aureus (ATCC 25923) cells incubated with DP7 (1/4× MIC, 1/2× MIC, and 1× MIC) for 1 h was analyzed by TEM (Tecnai G2 F20 S-Twin; FEI). In general, 1 × 108 mid-logarithmic-phase S. aureus cells were treated with peptide, and a no-peptide control was included. After incubation, bacteria were pelleted by centrifugation at 3,000 rpm (900 × g) for 10 min, followed by washing twice with 1 M PBS. Cells were then fixed with 2.5% glutaraldehyde–PBS solution at −4°C for 4 h. For SEM, the bacteria were further treated with 0, 20, 50, 80, and 100% ethyl alcohol for 20 min and centrifuged at 13,000 rpm (17,000 × g) for 10 min for dehydration. The bacterial pellet was resuspended in 100% ethyl alcohol and air dried. The prepared bacterial samples were sent to the Center of Forecasting and Analysis of Sichuan University (Sichuan, China) for imaging.
Animal models.
Male BALB/c mice, 6 to 8 weeks old, purchased from Chengdu Dossy Experimental Animals Co., Ltd. (Sichuan, China), were maintained and experiments were performed according to protocols approved by the Ethics Review Committee for Animal Experimentation of Sichuan University. Before infection, S. aureus (methicillin-resistant S. aureus; ATCC 33591) was incubated in MHB overnight and washed with normal saline three times (3,600 rpm for 5 min) before being resuspended to 1 × 107 CFU/ml in normal saline. Mice were then infected with 1 ml bacterial suspension by intraperitoneal (i.p.) injection. After 1 h, drugs diluted to the indicated concentrations with normal saline were inoculated through the indicated delivery route. After 20 h, 20-μl aliquots of serial dilutions of the peritoneal lavage fluid from individual mice were plated onto MHA. The plates were incubated overnight at 37°C, and bacterial colonies were enumerated by using an automatic color colony counter (Scan 500; Interscience).
Statistical analysis.
The significant difference method was used to determine whether statistically significant differences occurred among the mean values obtained. Data were analyzed with the Student t test using software SPSS 15.0. A P value of <0.05 was considered statistically significant.
RESULTS
Prediction method and construction of prediction model.
Because the peptide sequences of the training set did not always contain the same kinds of amino acids in the same positions, we determined that the electrical charge and amphipathicity of the amino acids were the most important factors influencing the activity of the AMPs. Amino acids exhibiting a similar charge or hydrophobicity were therefore merged as one parameter.
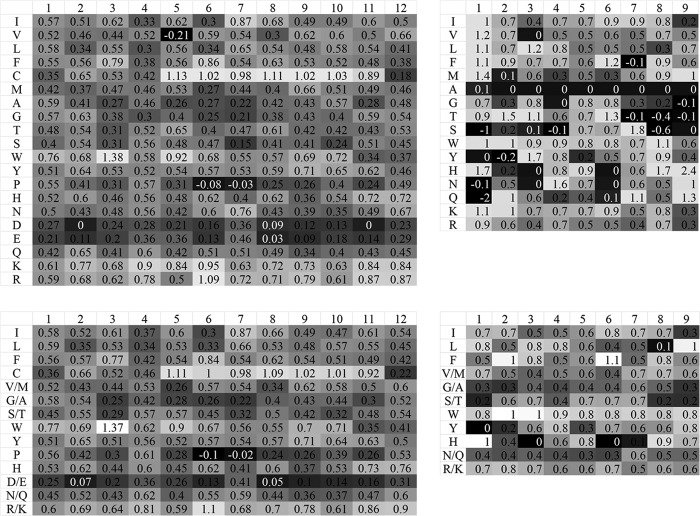
From the sequence data, we obtained 180 9-mer peptides and 424 12-mer peptides. With these peptides and their activity data, we trained and evaluated the machine-learning prediction model to build a relatively full score matrix (Fig. 1). When some of the amino acids were merged according to hydrophobicity, it was clear that the features of the contribution score matrix (Fig. 1, top right matrix) were mainly maintained, showing good prediction accuracy (P > 0.05) (Fig. 2, left graph). However, when a different group of peptides were predicted using the 9-mer AMPs for comparison, the prediction accuracy was lower (Fig. 2, middle graph).
FIG 1.
Amino acid antimicrobial activity contribution score matrix of the 12-mer peptides and 9-mer peptides. The first row of the matrix gives the positions in the peptide, and the first column gives the amino acid species. Shown are contribution score matrices generated by the same training data of 12-mer peptides and 9-mer peptides; for the matrices on the right we used the amino acids merging method (the merged amino acids are also listed in the first column).
FIG 2.
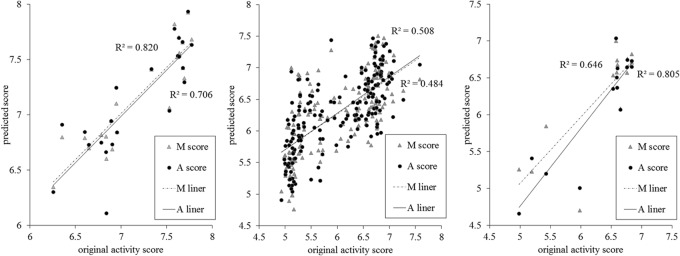
Prediction accuracy based on the antimicrobial activity contribution scores of the amino acids. The x coordinate is the predicted score for the peptides, and the y coordinate is the score for the activity of the original peptide. The liner regression trend lines of the scores generated from the contribution score matrix for all amino acids (A score) and the merged contribution score matrix for some of the amino acids (M score) are labeled A liner and M liner, respectively. The left and middle graphs show the predicted results for the 12-mer peptide testing set and the 9-mer peptides from the model built with 12-mer peptides, respectively; the graph on the right shows the predicted result of the 9-mer peptide testing set from the model built with 9-mer peptides.
To further assess the prediction ability of our method with more general sequence data, the 9-mer AMP sequences were used. These peptide sequences shared similar amino acid compositions, but unlike the 12-mer AMPs, they varied greatly in sequence. From these data, we obtained a contribution score matrix that exhibited good prediction performance, which differed from that obtained with the 12-mer AMPs (Fig. 1, bottom left matrix). No statistical difference was observed when some of the amino acids were merged (P > 0.05) (Fig. 2, right).
For the model sequence, AMP HH2 (3′-SQLRIRVAVIRA-NH2) was selected. This sequence shares the same ancestor as the other sequences used in this study, it has relatively improved activity, and it has been employed previously as an adjuvant (15, 16). We replaced two or three amino acids to produce new peptides which were then subjected to activity predictions in an established 12-mer peptide prediction model. Peptides that scored highly and exhibited an appropriate isoelectric point were selected for further testing.
Peptide antimicrobial activities in vitro.
Ten AMPs, selected from the class of novel peptides with the highest predicted scores, were used in an antimicrobial activity assay to test their activity against four bacterial strains (Table 1). Most of the peptides showed good antimicrobial activity, even with a MIC of 16 mg/liter (DP3, DP5, DP7, DP8, and DP11).
TABLE 1.
Antimicrobial assay of 10 12-mer novel peptides and control peptides
| Peptide | Sequence | MIC (mg/liter)a |
|||
|---|---|---|---|---|---|
| S. aureus (25923) | E. coli (25922) | P. aeruginosa (PAO1) | S. enterica serovar Typhi | ||
| HH2 | VQLRIRVAVIRA | 64 | 32 | 128 | 32 |
| Bac2A | RLARIVVIRVAR | 64 | 64 | 64 | 64 |
| Indolicidin | ILPWKWPWWPWRR | 16 | 32 | 64 | 16 |
| DP2 | VQWRIRVCVIRA | 32 | 32 | 32 | 32 |
| DP3 | VQWRIRIAVIRA | 8 | 8 | 32 | 4 |
| DP4 | VCWRIRVAVIRA | 32 | 64 | 64 | 64 |
| DP5 | VQLRIRVCVIRR | 16 | 16 | 32 | 16 |
| DP6 | KQWRIRVAVIRA | 32 | 32 | 128 | 64 |
| DP7 | VQWRIRVAVIRK | 16 | 16 | 16 | 8 |
| DP8 | VQLRIRVCVIRK | 16 | 16 | 16 | 8 |
| DP9 | KQWRIRVCVIRA | 32 | 32 | 64 | 32 |
| DP10 | VQLRCRVCVIRK | 64 | 32 | 64 | 32 |
| DP11 | VQWRIRIAVIRK | 16 | 16 | 32 | 16 |
The data are averages of three independent experiments performed in triplicate.
To further test the four peptides that exhibited the highest antimicrobial effect (DP3, DP5, DP7, and DP8), the antimicrobial assay was repeated using 39 clinically isolated, antibiotic-resistant strains. These strains included nine S. aureus isolates, nine Escherichia coli isolates, 10 Acinetobacter baumannii isolates, and 11 P. aeruginosa isolates. Relative to the control peptide HH2, all of the selected novel AMPs showed increased antimicrobial activity against P. aeruginosa, S. aureus, and A. baumannii while displaying activities against E. coli isolates comparable to that of HH2 (Table 2).
TABLE 2.
Distribution of the MICs of DP3, DP5, DP7, and DP8 for clinically isolated, antibiotic-resistant strains
| Peptide | MIC (mg/liter) for clinical isolatesa |
|||
|---|---|---|---|---|
| S. aureus | E. coli | A. baumannii | P. aeruginosa | |
| HH2 | 128 (64 to 128) | 64 (64 to 128) | 64 (32 to 128) | 128 (64 to >128) |
| DP3 | 32 (16 to 32) | 64 (32 to >128) | 32 (16 to 64) | 32 (32 to 64) |
| DP5 | 16 (16 to 32) | 128 (128 to >128) | 32 (32 to 64) | 32 (16 to 64) |
| DP7 | 16 (16 to 32) | 32 (32 to >128) | 16 (8 to 32) | 16 (16) |
| DP8 | 16 (8 to 16) | 64 (32 to 64) | 32 (16 to 64) | 16 (8 to 32) |
The MICs with the highest frequencies for each species. Values are averages from three independent experiments performed in triplicate, with ranges in parentheses.
Peptide cytotoxicity against human cells.
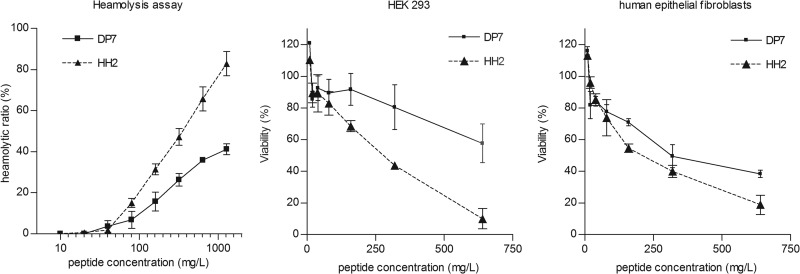
Among the novel AMPs, DP7 was determined to be the most effective antimicrobial peptide. Next, the cytotoxicities of DP7 and HH2 against human cells were tested. Although they differ by two amino acids, no significant difference was observed between DP7 and HH2 in their secondary structural composition, both being mostly random coils, as measured by circular dichroism (see Fig. S1 in the supplemental material). However, DP7 was less hemolytic to human blood than HH2 (50% hemolytic concentration, >1,280 mg/liter for DP7, versus 415 mg/liter for HH2) (Fig. 3, left graph), indicating that DP7 is less cytotoxic and therefore safer than HH2 with regard to human erythrocytes. Then, the cytotoxicities of DP7 and HH2 to HEK 293 cells and human epithelial fibroblast cells were assessed. Viability remained at the same level when cells were treated with DP7 or HH2 when the peptide concentration was lower than 80 mg/liter, but DP7 had lower cytotoxicity than HH2 at higher peptide concentrations (Fig. 3, middle and right graphs). The viability of both cell types after treatment with 20 mg/liter DP7 was about 90% (MIC, 16 mg/liter).
FIG 3.
Hemolytic activity and cytotoxicity of DP7 and HH2. The x coordinate is the concentration of peptides in the 2-fold serial dilution series that started from 1,280 mg/liter (hemolysis assay) or 640 mg/liter (cytotoxicity assay). The y coordinate is the degree of hemolysis relative to the total hemolysis control (hemolysis assay) or the viability of cells (cytotoxicity assay). (left) Release of hemoglobin was monitored to estimate the degree of erythrocyte lysis caused by the peptide. (Middle and right) Living HEK 293 and human epithelial fibroblasts after treatment with peptides were measured using a CCK-8 kit, and viability was calculated relative to the untreated control. All data represent means ± standard deviations from three independent experiments performed in triplicate.
DP7 antimicrobial activity in vivo.
To assess the antimicrobial effect of DP7 in vivo, a murine model for infection with S. aureus through i.p. injection was employed. DP7 was administered either locally by i.p. injection or at distant sites by intramuscular (i.m.), subcutaneous (s.c.), or intravenous (i.v.) injection (at a concentration of 20 mg/kg of body weight), 1 h after bacterial challenge. DP7 had almost no effect on bacterial counts via the i.p., i.m., or s.c. therapeutic routes (Fig. 4a). However, toxicity was severe when DP7 was administered through i.v. injection, with only two mice (out of six) surviving. The anatomical evidence showed that the liver was hyperemic. DP7 administered through s.c. injection also led to hemorrhaging at the injection site, while no health problems were observed with the other therapeutic routes (data not shown). Different concentrations of DP7 (from 20 to 60 mg/kg) were administered by i.p. to identify an effective dosage, equal doses of HH2 were used as a control, and 20 mg/kg of vancomycin was used as a positive control. DP7 showed a more positive therapeutic effect than HH2 in vivo, and a 60-mg/kg dose of DP7 showed a similar effect as 20 mg/kg of vancomycin (Fig. 4b). As a result, we evaluated the route of administration and found that DP7 injected via the i.m. and i.p. routes resulted in no detectable health problems and that DP7 injected via the i.p. route could effectively reduce the CFU count in the S. aureus-infected mouse model.
FIG 4.
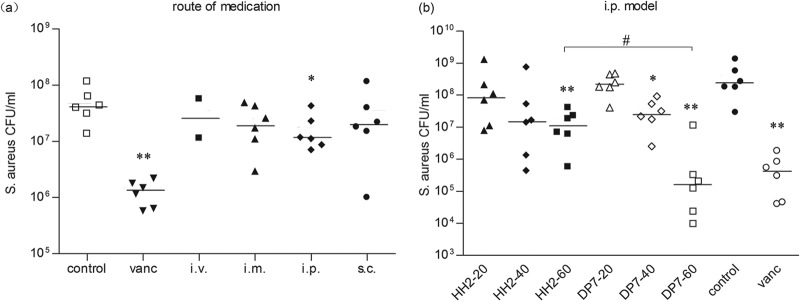
Efficacy of peptides in the S. aureus-infected mouse model. BALB/c mice were infected with 1 × 107 CFU S. aureus strain 33591 by i.p. injection, and antimicrobial drug was administered after 1 h. Bacterial loads in the peritoneal lavage fluid from individual mice after 20 h of infection are shown. (a) 20 mg/kg DP7 administered by i.p., i.m., s.c., or i.v, injection. For controls, 20 mg/kg vancomycin or normal saline was administered by i.p. injection. (b) Peptides (20, 40, and 60 mg/kg), vancomycin (20 mg/kg), and normal saline were administered by i.p. injection. Horizontal bars show standard deviations. *, P < 0.05; **, P < 0.01 (peptide versus normal saline control). #, P < 0.05 (60 mg/kg DP7 versus 60 mg/kg HH2).
Mechanism behind the DP7 antimicrobial effect on S. aureus.
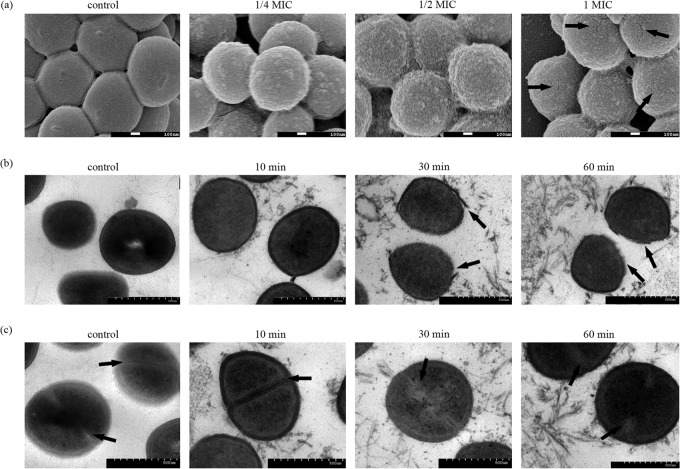
With their amphiphilic properties, AMPs can easily insert into the membranes of bacteria, and this is their main antimicrobial mechanism. To elucidate how the novel peptides tested in this study affect the outer layers of bacteria, we examined the morphological changes that occur in S. aureus as a result of treatment with DP7 under various conditions. S. aureus cells were treated with different concentrations of DP7 for 1 h and then visualized by SEM. Small surface protuberances occurred on the cells treated with 1/4× MIC of DP7, and these were caused by the attachment and insertion of peptides into the outer layer. When treated with 1/2× MIC, the cells displayed wrinkled surfaces, with many cracks appearing as the concentration increased to 1× MIC, indicating that the outer layer of the S. aureus cells was totally disrupted (Fig. 5a). The damaging effect of DP7 on S. aureus was also observed by TEM imaging. Different incubation times, with a range of 10 min to 1 h, were tested, and although no blebs were observed, the disruption of the outer membranes and the disturbance of the inner membranes were obvious (Fig. 5b). Furthermore, membrane formation of dividing cells was affected by peptides protruding from the cells (Fig. 5c). Taken together, these results indicate that the mechanism of DP7 killing is distinct from that of most cationic peptides, which usually influence membrane permeability or even form holes in the membrane and seriously disrupt cell morphology (17, 18). There has been a previous report of disruption of the cytomembrane by a defensin protein (19); in this case, the outer membrane was disrupted and the peptidoglycan layer and cytomembrane became wrinkled, but no cell dissociation occurred. In our study, the wrinkled surface observed with SEM was the exposed peptidoglycan layer, which maintained the cell shape but possessed a few cracks with DP7 at a concentration of 1× MIC. To determine whether DP7 can change membrane permeability, leading to the release of DNA or protein into the cytoplasm, the levels of extracellular DNA and protein were measured after incubation of S. aureus with DP7 or HH2 for up to 90 min. However, no changes in their concentrations were observed when the incubation time was increased (see Fig. S2 in the supplemental material), which indicated that no protein or DNA was released from the cytoplasm. Our study therefore provided evidence that DP7 can induce bacterial suppression by disrupting the outer membrane without affecting the permeability of the S. aureus cytomembrane.
FIG 5.
TEM and SEM image of S. aureus. (a) S. aureus cells treated with different concentrations of DP7 after 1 h were visualized by SEM. Magnification, ×50,000. Arrows (1× MIC panel) indicate where, as the concentration of DP7 increased, the smooth surface of S. aureus cells became plicated and some cracks appeared. (b and c) S. aureus cells treated with 1× MIC of DP7 for different time periods were visualized by TEM. Magnification, ×10,000. Over time, the outer membranes became disrupted as indicated and substances were present in the intercellular fluid (arrows in panel b). From the equator region of dividing cells in the control and the 10-min-treated samples, clear and stable membranes were evident, but over 30 min of treatment resulted in blurry and disturbed membranes. The membranes at the equatorial cortical region are indicated (arrows in panel c).
DISCUSSION
The design and discovery of effective AMPs play important roles in responding to the demand for novel efficient antimicrobial therapies. In this study, using the amino acid-based prediction method, we constructed a model to predict AMP activity with high accuracy. Our model offers various advantages over currently existing models for peptide predictions. First, in contrast to other peptide prediction methods (such as quantitative structure-activity relationships, artificial neural networks, and Hidden Markov models) that focus on the identification of common features between peptides, with the relevance of these features not always being completely apparent (10, 20–22), our method is based on the clear principle that each type of amino acid possesses a different level of importance with respect to peptide activity. Second, our method is easy to perform and quicker than other peptide prediction methods. Third, in other models peptide data from databases that vary both in length and sequence composition are invariably used to train these prediction models. Although the accuracy of predicting AMPs from diverse sequences can be improved, a strategy that considers peptides from different categories with distinct antimicrobial mechanisms would appear less likely to succeed. Our study focused on changing only a few of amino acids from the model peptide to improve antimicrobial activity, allowing for comparisons between peptides with just one amino acid difference.
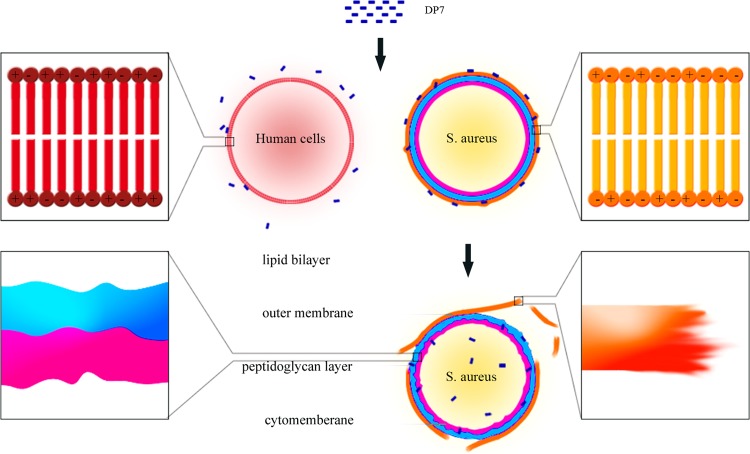
In this study, we developed AMP HH2 by using our method and enhanced both the antimicrobial effect and pathogen specificity by changing just two amino acids. In an antimicrobial activity assay, we found that most of our designed peptides possessed higher antimicrobial activities than the model peptide. In particular, DP7 showed the strongest antimicrobial activity to both standard microbial strains and isolated antibiotic-resistant strains, while displaying lower cytotoxicity to human erythrocytes, HEK 293 cells, and human epithelial fibroblast cells. Unlike most AMPs, which form pores in the cytoplasmic membrane that lead to cell lysis or leakage of the cytoplasm (19, 23, 24), Bac2A has been reported to kill S. aureus without cell lysis, but cross-wall formation was affected (25). As a derivative of Bac2A, DP7 is also a liner peptide (26–28), but it appears to operate via a distinct mechanism (Fig. 6). Outer membrane formation in the cytoplasm was found to be suppressed, and DP7 severely disrupted the outer membrane and disturbed the inner membranes of S. aureus. DP7 administered via i.p. injection reduced bacterial load in an S. aureus-infected mouse model, and host cytotoxicity levels were acceptable.
FIG 6.
Mechanism of DP7 killing of S. aureus. The charge on the cytomembrane of human normal cells is neutral, but the cell wall of a bacterium contains teichoic acids, which result in a strong negative charge (shown in the enlarged squares on the top left and top right). The strong positive charge of DP7 allows it to specifically attach to the outer membrane of bacteria because of the charge on the bacterial surface and its amphipathic properties, but DP7 cannot bind to mammalian cells. Upon binding to bacterial cells, DP7 inserts into the outer membrane, disrupting the membrane. The enlarged square on the right bottom shows that the outer layer has been broken and the fault surface is rather coarse. DP7 can also disturb the inner membrane but does not lyse the cell. The enlarged square on the left bottom indicates that the inner membrane and peptidoglycan layer were not as smooth as in untreated cells.
The synergistic action of AMPs with other AMPs or antibiotics can reduce the need for high dosages or minimize side effects, both of which are beneficial attributes for therapeutic strategies to fight multidrug-resistant bacteria (29–31). The novel AMPs developed in this study were derived from Bac2A and suppress bacterial growth via an outer membrane disruption mechanism. This finding could potentially be applied to enhance the antimicrobial activities of AMPs with different sequences or structures (31), and our novel AMPs may show some synergistic effects with antibiotics that display alternative antimicrobial mechanisms. Such issues will be investigated in future studies.
In summary, our study provides a new method for AMP design and prediction. Peptides generated by our method were found to possess effective antibacterial functions both in vitro and in vivo. In addition, our findings provide insight into the antimicrobial mechanism of the AMP DP7, which may have important implications for future studies into the development and application of novel antimicrobial therapies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Shengyong Yang for guidance in the early stages of the algorithm design and Zhijie Han for pointing our attention to the in vitro test cases.
The manuscript was written with contributions from all of the authors. All authors have given approval to the final version of the manuscript.
We declare no competing financial interest.
The study was supported mainly by the National Major Scientific and Technological Special Project for “Significant New Drugs Development” (grant 2013ZX09102030), Sichuan Outstanding Youth Science & Technology Funding (2012JQ0014), and partly by the National Major Scientific and Technological Special Project for “Significant New Drugs Development” (grant 2012ZX09103101-036).
Footnotes
Published ahead of print 30 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02823-14.
REFERENCES
- 1.Easton DM, Nijnik A, Mayer ML, Hancock RE. 2009. Potential of immunomodulatory host defense peptides as novel anti-infectives. Trends Biotechnol. 27:582–590. 10.1016/j.tibtech.2009.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hancock RE, Sahl HG. 2006. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24:1551–1557. 10.1038/nbt1267 [DOI] [PubMed] [Google Scholar]
- 3.Zasloff M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389–395. 10.1038/415389a [DOI] [PubMed] [Google Scholar]
- 4.Thaker HD, Som A, Ayaz F, Lui D, Pan W, Scott RW, Anguita J, Tew GN. 2012. Synthetic mimics of antimicrobial peptides with immunomodulatory responses. J. Am. Chem. Soc. 134:11088–11091. 10.1021/ja303304j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jenssen H, Fjell CD, Cherkasov A, Hancock REW. 2008. QSAR modeling and computer-aided design of antimicrobial peptides. J. Peptide Sci. 14:110–114. 10.1002/psc.908 [DOI] [PubMed] [Google Scholar]
- 6.Loose C, Jensen K, Rigoutsos I, Stephanopoulos G. 2006. A linguistic model for the rational design of antimicrobial peptides. Nature 443:867–869. 10.1038/nature05233 [DOI] [PubMed] [Google Scholar]
- 7.Epand RM, Vogel HJ. 1999. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta 1462:11–28. 10.1016/S0005-2736(99)00198-4 [DOI] [PubMed] [Google Scholar]
- 8.Fjell CD, Hiss JA, Hancock RE, Schneider G. 2012. Designing antimicrobial peptides: form follows function. Nat. Rev. Drug Discov. 11:37–51. 10.1038/nrd3591 [DOI] [PubMed] [Google Scholar]
- 9.Frank K, Sippl MJ. 2008. High-performance signal peptide prediction based on sequence alignment techniques. Bioinformatics 24:2172–2176. 10.1093/bioinformatics/btn422 [DOI] [PubMed] [Google Scholar]
- 10.Ebalunode JO, Zheng W, Tropsha A. 2011. Application of QSAR and shape pharmacophore modeling approaches for targeted chemical library design. Methods Mol. Biol. 685:111–133. 10.1007/978-1-60761-931-4_6 [DOI] [PubMed] [Google Scholar]
- 11.Foran DJ, Berg RA. 1994. A method for quantitative image assessment based on redundant feature measurements and statistical reasoning. Comput. Methods Programs Biomed. 45:291–305. 10.1016/0169-2607(94)01590-C [DOI] [PubMed] [Google Scholar]
- 12.Hilpert K, Elliott MR, Volkmer-Engert R, Henklein P, Donini O, Zhou Q, Winkler DF, Hancock RE. 2006. Sequence requirements and an optimization strategy for short antimicrobial peptides. Chem. Biol. 13:1101–1107. 10.1016/j.chembiol.2006.08.014 [DOI] [PubMed] [Google Scholar]
- 13.Hilpert K, Hancock RE. 2007. Use of luminescent bacteria for rapid screening and characterization of short cationic antimicrobial peptides synthesized on cellulose using peptide array technology. Nat. Protoc. 2:1652–1660. 10.1038/nprot.2007.203 [DOI] [PubMed] [Google Scholar]
- 14.Zhou H, Luo Y, Mizutani M, Mizutani N, Reisfeld RA, Xiang R. 2005. T cell-mediated suppression of angiogenesis results in tumor protective immunity. Blood 106:2026–2032. 10.1182/blood-2005-03-0969 [DOI] [PubMed] [Google Scholar]
- 15.Kindrachuk J, Jenssen H, Elliott M, Townsend R, Nijnik A, Lee SF, Gerdts V, Babiuk LA, Halperin SA, Hancock RE. 2009. A novel vaccine adjuvant comprised of a synthetic innate defence regulator peptide and CpG oligonucleotide links innate and adaptive immunity. Vaccine 27:4662–4671. 10.1016/j.vaccine.2009.05.094 [DOI] [PubMed] [Google Scholar]
- 16.Cao D, Li H, Jiang Z, Xu C, Cheng Q, Yang Z, Cao G, Zhang L. 2010. Synthetic innate defence regulator peptide enhances in vivo immunostimulatory effects of CpG-ODN in newborn piglets. Vaccine 28:6006–6013. 10.1016/j.vaccine.2010.06.103 [DOI] [PubMed] [Google Scholar]
- 17.Brogden KA. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3:238–250. 10.1038/nrmicro1098 [DOI] [PubMed] [Google Scholar]
- 18.Bechinger B, Lohner K. 2006. Detergent-like actions of linear amphipathic cationic antimicrobial peptides. Biochim. Biophys. Acta 1758:1529–1539. 10.1016/j.bbamem.2006.07.001 [DOI] [PubMed] [Google Scholar]
- 19.Nakajima Y, Ishibashi J, Yukuhiro F, Asaoka A, Taylor D, Yamakawa M. 2003. Antibacterial activity and mechanism of action of tick defensin against Gram-positive bacteria. Biochim. Biophys. Acta 1624:125–130. 10.1016/j.bbagen.2003.10.004 [DOI] [PubMed] [Google Scholar]
- 20.Jones DT. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195–202. 10.1006/jmbi.1999.3091 [DOI] [PubMed] [Google Scholar]
- 21.Zhang G, Hu YM, Eddy Patuwo B, Indro CD. 1999. Artificial neural networks in bankruptcy prediction: general framework and cross-validation analysis. Eur. J. Oper. Res. 116:16–32. 10.1016/S0377-2217(98)00051-4 [DOI] [Google Scholar]
- 22.Nielsen M, Lundegaard C, Worning P, Hvid CS, Lamberth K, Buus S, Brunak S, Lund O. 2004. Improved prediction of MHC class I and class II epitopes using a novel Gibbs sampling approach. Bioinformatics 20:1388–1397. 10.1093/bioinformatics/bth100 [DOI] [PubMed] [Google Scholar]
- 23.Kim H, Jang JH, Kim SC, Cho JH. 2014. De novo generation of short antimicrobial peptides with enhanced stability and cell specificity. J. Antimicrob. Chemother. 69:121–132. 10.1093/jac/dkt322 [DOI] [PubMed] [Google Scholar]
- 24.Rakowska PD, Jiang H, Ray S, Pyne A, Lamarre B, Carr M, Judge PJ, Ravi J, Gerling UI, Koksch B, Martyna GJ, Hoogenboom BW, Watts A, Crain J, Grovenor CR, Ryadnov MG. 2013. Nanoscale imaging reveals laterally expanding antimicrobial pores in lipid bilayers. Proc. Natl. Acad. Sci. U. S. A. 110:8918–8923. 10.1073/pnas.1222824110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedrich CL, Rozek A, Patrzykat A, Hancock RE. 2001. Structure and mechanism of action of an indolicidin peptide derivative with improved activity against gram-positive bacteria. J. Biol. Chem. 276:24015–24022. 10.1074/jbc.M009691200 [DOI] [PubMed] [Google Scholar]
- 26.Wu M, Hancock RE. 1999. Improved derivatives of bactenecin, a cyclic dodecameric antimicrobial cationic peptide. Antimicrob. Agents Chemother. 43:1274–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hilpert K, Volkmer-Engert R, Walter T, Hancock RE. 2005. High-throughput generation of small antibacterial peptides with improved activity. Nat. Biotechnol. 23:1008–1012. 10.1038/nbt1113 [DOI] [PubMed] [Google Scholar]
- 28.Scruten E, Kovacs-Nolan J, Griebel PJ, Latimer L, Kindrachuk J, Potter A, Babiuk LA, Littel-van den Hurk S, Napper S. 2010. Retro-inversion enhances the adjuvant and CpG co-adjuvant activity of host defence peptide Bac2A. Vaccine 28:2945–2956. 10.1016/j.vaccine.2010.02.015 [DOI] [PubMed] [Google Scholar]
- 29.Giacometti A, Cirioni O, Del Prete MS, Paggi AM, D'Errico MM, Scalise G. 2000. Combination studies between polycationic peptides and clinically used antibiotics against Gram-positive and Gram-negative bacteria. Peptides 21:1155–1160. 10.1016/S0196-9781(00)00254-0 [DOI] [PubMed] [Google Scholar]
- 30.Livermore DM, Warner M, Mushtaq S. 2013. Activity of MK-7655 combined with imipenem against Enterobacteriaceae and Pseudomonas aeruginosa. J. Antimicrob. Chemother. 68:2286–2290. 10.1093/jac/dkt178 [DOI] [PubMed] [Google Scholar]
- 31.Yan H, Hancock RE. 2001. Synergistic interactions between mammalian antimicrobial defense peptides. Antimicrob. Agents Chemother. 45:1558–1560. 10.1128/AAC.45.5.1558-1560.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.