LETTER
Specific resistance to lincosamides (L phenotype) is the result of modification and inactivation by lincosamide nucleotidyltransferase enzymes encoded by members of the lnu (previously lin) gene family (1, 2), of which six different types, lnu(A), lnu(B), lnu(C), lnu(D), lnu(E), and lnu(F), are currently recognized (1, 3–7). In Streptococcus agalactiae, the genetic environment in which the gene lnu(B) is located has not been explored; in contrast, a genetic element carrying lnu(C) has been reported in this species (1), as well as different elements harboring lnu(A) and lnu(B) in Staphylococcus aureus (8, 9), lnu(B) in Enterococcus faecium (10), lnu(C) in Streptococcus anginosus (11), and a truncated copy of lnu(E) in Streptococcus suis (7).
In 2008, an S. agalactiae strain (SGB76) was obtained from a pregnant female outpatient at Mater Dei Hospital in Buenos Aires. By Etest, the isolate showed susceptibility to erythromycin (MIC of 0.06 μg/ml) and resistance to clindamycin (MIC of 6 μg/ml). Total DNA was used as the template for PCR screening of erm(A), erm(B), mef(A), and lnu(B) genes. Of these, only lnu(B) was detected and confirmed by sequencing. To determine the genetic environment of the lnu(B) gene, two strategies were used, thermal asymmetric interlaced PCR (TAIL-PCR) in combination with PCR mapping using primers designed based on the previously reported structures from Enterococcus faecalis and S. aureus (GenBank accession numbers AF408195.1, JQ861958.1, and JX560992) (see Table S1 in the supplemental material).
The 12,076-bp lnu(B)-carrying fragment from strain SGB76 contained 11 open reading frames (ORFs) of at least 100 amino acids (accession number KF772204). Other resistance genes detected in this structure include aadE (streptomycin resistance), spw (spectinoycin resistance), and lsa(E) (pleuromutilin, lincosamide, and streptogramin A resistance) (Fig. 1). Basic Local Alignment Search Tool (BLAST) analysis revealed that this sequence exhibited similarity to the lnu(B)-containing structures previously identified in S. aureus (JQ861959 and JX560992) and E. faecalis (AF408195) (Fig. 1) (8, 9). An insertion sequence (IS1216E) is located at the left-hand end, similar to IS1216 in the structure described in S. aureus (JX560992) and in a variant of the latter, IS1216v, in E. faecalis. However, two copies of IS257 are flanking the lnu(B)-carrying element in the structure described in S. aureus by Lozano et al. (accession number JQ861959) (9). The region of 5,982 bp located upstream from the lnu(B) gene is 99% identical to the structures described in S. aureus and E. faecalis (Fig. 1). The lsa(E) gene located immediately upstream from the lnu(B) gene encodes an ABC transporter involved in active efflux of lincosamides, streptogramins A, and pleuromutilins (10, 12). However, the region of 2,331 bp situated downstream from the lnu(B) gene showed 99% nucleotide identity to the S. aureus (but not to the E. faecalis) structure, which includes one copy of the gene encoding a putative transposase of the ISL3 family (tnp) (Fig. 1). Besides the occurrence of this cluster in E. faecalis and S. aureus, a sequence from an Enterococcus faecium isolate of swine origin that was recently released (KF421157.1) (10) shows 99% nucleotide identity.
FIG 1.
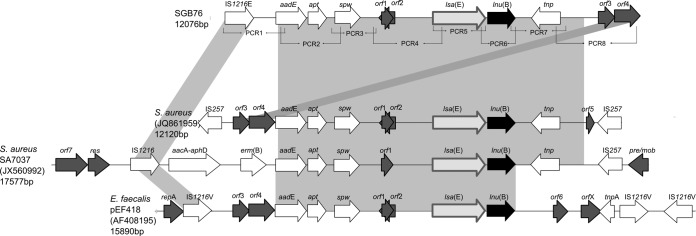
Genetic environment of the lnu(B) gene in S. agalactiae SGB76 (accession number KF772204) and structural comparison with the corresponding regions identified in S. aureus (accession numbers JQ861959 and JX560992) and E. faecalis pEF418 (accession number AF408195). Gray areas indicate regions with at least 99% nucleotide sequence identity. The arrows represent the position and orientation of the depicted genes as they were proposed in the original references. The amplicons of the 8 PCRs used to investigate the genetic environment of the lnu(B) gene in SGB76 are indicated below the top structure (see Table S1 in the supplemental material). orf1 to orf7 represent open reading frames with either unknown or unconfirmed functions. aadE, aminoglycoside adenyltransferase E; aacA-aphD, aminoglycoside acetyltransferase A and aminoglycoside phosphotransferase D; apt, adenine phosphoribosyltransferase; erm(B), methyltransferase; IS, insertion sequence; lnu(B), lincosamide nucleotidyltransferase; lsa(E), ATP binding protein; res and mob, mobilization elements; spw, spectinomycin resistance gene; tnp, transposase.
At the right-hand end, a region containing two genes (orf3 and orf4) is 98% identical to the orf3 and orf4 region described in S. aureus (JQ861959); however, in the latter it is located at the left-hand end, associated with IS257. The description of this multiresistance cluster in S. agalactiae represents another example of resistance genes shared by enterococci, staphylococci, and streptococci (10).
Even if S. agalactiae SGB76 was susceptible to quinupristin-dalfopristin, as susceptibility to pleuromutilins and streptogramin A could not be assayed, a possible contribution of the lsa(E) gene to clindamycin resistance, as was described in S. aureus, could not be disregarded (8, 12). Further experiments will assess the contribution of this resistance marker in S. agalactiae.
Nucleotide sequence accession number.
Sequence data were deposited in the GenBank/EMBL nucleotide databases under accession number KF772204.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by grants from the University of Buenos Aires, Argentina (number 20020100100510), and CONICET Argentina (PIP, 11220110100707CO). M.M., G.G., and L.B. are members of CONICET.
Footnotes
Published ahead of print 23 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02630-14.
REFERENCES
- 1.Achard A, Villers C, Pichereau V, Leclercq R. 2005. New lnu(C) gene conferring resistance to lincomycin by nucleotidylation in Streptococcus agalactiae UCN36. Antimicrob. Agents Chemother. 49:2716–2719. 10.1128/AAC.49.7.2716-2719.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberts MC. 2008. Update on macrolide-lincosamide-streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol. Lett. 282:147–159. 10.1111/j.1574-6968.2008.01145.x [DOI] [PubMed] [Google Scholar]
- 3.Bozdogan B, Berrezouga L, Kuo MS, Yurek DA, Farley KA, Stockman BJ, Leclercq R. 1999. A new resistance gene, linB, conferring resistance to lincosamides by nucleotidylation in Enterococcus faecium HM1025. Antimicrob. Agents Chemother. 43:925–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brisson-Noël A, Delrieu P, Samain D, Courvalin P. 1988. Inactivation of lincosaminide antibiotics in Staphylococcus. Identification of lincosaminide O-nucleotidyltransferases and comparison of the corresponding resistance genes. J. Biol. Chem. 263:15880–15887 [PubMed] [Google Scholar]
- 5.de Azavedo JC, McGavin M, Duncan C, Low DE, McGeer A. 2001. Prevalence and mechanisms of macrolide resistance in invasive and noninvasive group B streptococcus isolates from Ontario, Canada. Antimicrob. Agents Chemother. 45:3504–3508. 10.1128/AAC.45.12.3504-3508.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petinaki E, Guerin-Faublee V, Pichereau V, Villers C, Achard A, Malbruny B, Leclercq R. 2008. Lincomycin resistance gene lnu(D) in Streptococcus uberis. Antimicrob. Agents Chemother. 52:626–630. 10.1128/AAC.01126-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Q, Wendlandt S, Li H, Li J, Wu C, Shen J, Schwarz S, Wang Y. 2014. Identification of the novel lincosamide resistance gene lnu(E) truncated by ISEnfa5-cfr-ISEnfa5 insertion in Streptococcus suis: de novo synthesis and confirmation of functional activity in Staphylococcus aureus. Antimicrob. Agents Chemother. 58:1785–1788. 10.1128/AAC.02007-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li B, Wendlandt S, Yao J, Liu Y, Zhang Q, Shi Z, Wei J, Shao D, Schwarz S, Wang S, Ma Z. 2013. Detection and new genetic environment of the pleuromutilin-lincosamide-streptogramin A resistance gene lsa(E) in methicillin-resistant Staphylococcus aureus of swine origin. J. Antimicrob. Chemother. 68:1251–1255. 10.1093/jac/dkt015 [DOI] [PubMed] [Google Scholar]
- 9.Lozano C, Aspiroz C, Saenz Y, Ruiz-Garcia M, Royo-Garcia G, Gomez-Sanz E, Ruiz-Larrea F, Zarazaga M, Torres C. 2012. Genetic environment and location of the lnu(A) and lnu(B) genes in methicillin-resistant Staphylococcus aureus and other staphylococci of animal and human origin. J. Antimicrob. Chemother. 67:2804–2808. 10.1093/jac/dks320 [DOI] [PubMed] [Google Scholar]
- 10.Li XS, Dong WC, Wang XM, Hu GZ, Wang YB, Cai BY, Wu CM, Wang Y, Du XD. 2014. Presence and genetic environment of pleuromutilin-lincosamide-streptogramin A resistance gene lsa(E) in enterococci of human and swine origin. J. Antimicrob. Chemother. 69:1424–1426. 10.1093/jac/dkt502 [DOI] [PubMed] [Google Scholar]
- 11.Gravey F, Galopin S, Grall N, Auzou M, Andremont A, Leclercq R, Cattoir V. 2013. Lincosamide resistance mediated by lnu(C) (L phenotype) in a Streptococcus anginosus clinical isolate. J. Antimicrob. Chemother. 68:2464–2467. 10.1093/jac/dkt255 [DOI] [PubMed] [Google Scholar]
- 12.Wendlandt S, Lozano C, Kadlec K, Gomez-Sanz E, Zarazaga M, Torres C, Schwarz S. 2013. The enterococcal ABC transporter gene lsa(E) confers combined resistance to lincosamides, pleuromutilins and streptogramin A antibiotics in methicillin-susceptible and methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 68:473–475. 10.1093/jac/dks398 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


