Abstract
Carbapenems are one of the last lines of defense for Gram-negative pathogens, such as members of the Enterobacteriaceae. Despite the fact that most carbapenems are resistant to extended-spectrum β-lactamase (ESBL), emerging metallo-β-lactamases (MBLs), including New Delhi metallo-β-lactamase 1 (NDM-1), that can hydrolyze carbapenems have become prevalent and are frequently associated with the so-called “superbugs,” for which treatments are extremely limited. Crystallographic study sheds light on the modes of antibiotic binding to NDM-1, yet the mechanisms governing substrate recognition and specificity are largely unclear. This study provides a connection between crystallographic study and the functional significance of NDM-1, with an emphasis on the substrate specificity and catalysis of various β-lactams. L1 loop residues L59, V67, and W87 were important for the activity of NDM-1, most likely through maintaining the partial folding of the L1 loop or active site conformation through hydrophobic interaction with the R groups of β-lactams or the β-lactam ring. Substitution of alanine for L59 showed greater reduction of MICs to ampicillin and selected cephalosporins, whereas substitutions of alanine for V67 had more impact on the MICs of carbapenems. K224 and N233 on the L3 loop played important roles in the recognition of substrate and contributed to substrate hydrolysis. These data together with the structure comparison of the B1 and B2 subclasses of MBLs revealed that the broad substrate specificity of NDM-1 could be due to the ability of its wide active site cavity to accommodate a wide range of β-lactams. This study provides insights into the development of efficient inhibitors for NDM-1 and offers an efficient tactic with which to study the substrate specificities of other β-lactamases.
INTRODUCTION
Beta-lactams, the antimicrobial agents that kill bacteria by inhibiting the peptidoglycan layer synthesis of cell walls, represent the most widely used clinical antibiotics due to their efficacy and safety. The major mechanism of antimicrobial resistance is associated with the production of versatile β-lactamases, causing dissemination of multidrug resistance observed in various pathogenic microorganisms (1, 2). Based on their amino acid sequences and functional characteristics, β-lactamases have been classified into four classes, of which classes A, C, and D are serine β-lactamases in which a serine residue is involved in the catalysis in the active site and class B are metallo-β-lactamases (MBLs) that contain one to two divalent ions, such as Zn2+, in the active site (3, 4). The majority of the MBL genes in Enterobacteriaceae are found as gene cassettes on integrons, facilitating the transfer of these resistant genes among different microorganisms. Most of the MBLs have broad-spectrum activity, hydrolyzing penicillins, cephalosporins, and carbapenems, except subclass B2 MBLs, which conditionally hydrolyze carbapenems but not penicillins and cephalosporins (5, 6). Although MBLs are incapable of hydrolyzing monobactams, in most cases, bacteria harboring the mobile MBL genes also carry genes that encode enzymes that hydrolyze monobactams, making it difficult to kill those bacteria. Unlike the serine β-lactamases, MBLs are insensitive to β-lactamase inhibitors, such as clavulanic acid, sulbactam, and tazobactam, and no clinically proven inhibitor of MBLs is available to date (7). Recently, alarm has been raised over the high spread rate of blaNDM-1 and other plasmid-borne MBLs disseminated among Gram-negative microorganisms since the mortality of patients suffering from infections by these so-called “superbugs” is high and the choices of treatments are limited (1, 4, 8).
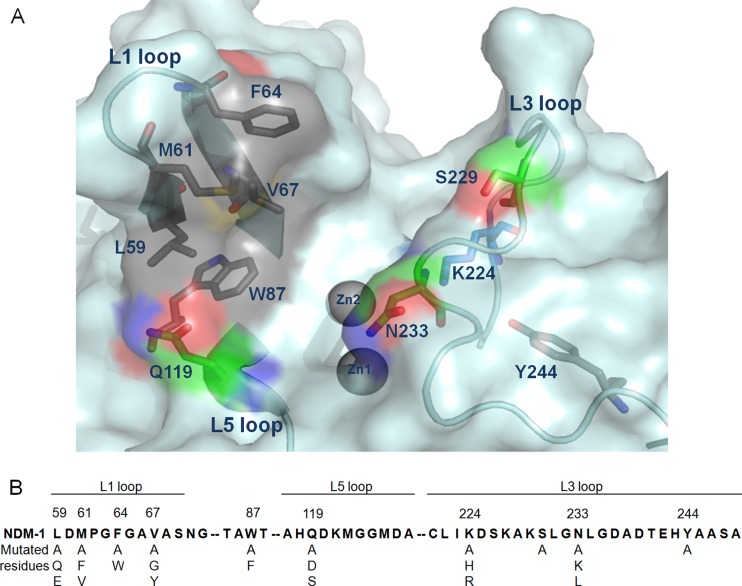
The MBL numbering has been used to designate the residues on different MBLs and was used throughout this study (6, 9). The crystal structures of several class B1 MBLs, including VIM-2, IMP-1, SPM-1, BcII, and NDM-1 (New Delhi metallo-β-lacamase 1), have been determined (10–14). Despite the divergence of the amino acid sequence, these MBLs share similar conservative active sites, containing two zinc ions, each coordinated by the 3H site (H116, H118, and H196) and DCH site (D120, C221, and H263) for Zn1 and Zn2, respectively (15). Two of the loops flank the active site, namely, the L1 loop, containing hydrophobic residues on the groove side, and the L3 loop, harboring residues K224 and N233, which are implicated in substrate binding, as well C221, which is involved in Zn2 binding (Fig. 1A) (14). A shared water or hydroxide ion existing between two zinc ions is proposed to be responsible for nucleophilic attack at the carbonyl carbon of β-lactams (16). The complex structures with antibiotics of MBLs suggest a possible mechanism of substrate recognition, where the substrate carbonyl oxygen atom coordinates with Zn1 and in some cases may form a hydrogen bond with the highly conserved N233, the substrate carboxylate coordinates with Zn2 through one oxygen atom and forms a hydrogen bond with the side chain of K224 through another oxygen atom, and the hydrophobic flapping loop (L1) interacts with the hydrophobic ring of some substrates (14, 17). Several residues in the L1 loop, in particular W64, S121 on the L5 loop, F218, and S262, have been shown to play important roles in substrate hydrolysis in IMP-1 (18, 19). In contrast, the residues in the L1 loop of VIM-2, F61, A64, and Y67, made less of a contribution to substrate hydrolysis; instead, the residues in the L3 loop Y224, R228, and N233 contributed to substrate hydrolysis, suggesting different mechanisms of substrate recognition among B1 MBLs (20, 21). Recently, the cocrystal structures of NDM-1 with several penicillins and meropenem suggested Q119, K224, and Y244 may contribute to substrate hydrolysis by stabilizing the intermediate products (14, 17). These studies shed some light on unraveling the mechanism of catalysis by NDM-1. Nevertheless, whether these residues play critical roles is not clear, and the information linking the crystal structures and the functional properties is still lacking. In this study, several residues in the L1, L3, and L5 loops of NDM-1 have been mutated to determine their contributions to the recognition and catalysis of different substrates.
FIG 1.
The important residues involved in substrate recognition and specificity of NDM-1. The important residues studied in this work are shown as gray, green, and blue sticks (A), and the point mutants generated are shown in panel B.
MATERIALS AND METHODS
Antibiotics and media.
Isopropyl β-d-1-thiogalactopyranoside (IPTG), ampicillin, and kanamycin were purchased from IBI, Inc. (Boca Raton, FL). Ceftriaxone, cephalothin, cefepime, meropenem, imipenem, ertapenem, aztreonam, and thrombin were purchased from Sigma Chemical Co. (St. Louis, MO). Luria broth (LB) was purchased from BD Co. (Franklin Lakes, NJ). Mueller-Hinton broth (MHB) was purchased from Oxoid Co. (Hampshire, United Kingdom).
Recombinant DNA technology and site-directed mutagenesis.
The blaNDM-1 gene was synthesized, PCR amplified, and constructed in pET28b in our lab (described elsewhere). Escherichia coli Tg1 was transformed with the constructed plasmid. Three versions of the blaNDM-1 constructs were made, namely, the full-length gene, pET28-blaNDM-1, the full-length gene carrying a C-terminal His6 tag, pET28-blaNDM-1-H6, and a truncated gene encoding G36 to R270 and carrying an N-terminal His6 tag, pET28-blaH6-mNDM-1. E. coli DH5α was transformed with the plasmids carrying mutations of blaNDM-1, which were generated using the GeneArt site-directed mutagenesis kit (Invitrogen Co., Grand Island, NY). The sequences were confirmed, and the transformants were selected from an LB plate supplemented with 50 μg/ml of kanamycin. E. coli BL21(DE3) was transformed with the clones carrying selected mutations for overexpression and purification.
Overexpression and purification of NDM-1 and mutants.
Overnight-incubated transformants of E. coli carrying pET28-blaNDM-1-H6 and pET28-blaH6-mNDM-1 were subcultured to 2 liters and 0.5 liter of LB, respectively, followed by induction with 1 mM IPTG, and incubated at 16°C overnight starting from an optical density at 600 nm (OD600) of 0.6. The cells were recovered, resuspended in lysis buffer (10 mM Tris-HCl [pH 7.6], 0.3 M sucrose, 1% NP-40, 0.5% Triton X-100, 0.5% Tween 20), and broken with French press (Thermo Scientific, Inc., Waltham, MA). The soluble fractions were passed through a Ni-nitrilotriacetic acid (NTA) column and eluted with a mixture of 20 mM Tris-HCl (pH 7.9), 500 mM NaCl, and 250 mM imidazole. The eluted proteins were further passed through a Sephacryl S-200 gel filtration column (GE Healthcare, Pittsburgh, PA) with a running buffer consisting of 10 mM Tris-HCl (pH 7.9)–50 mM NaCl. The His6 tag was removed by incubating the purified enzyme with His-tagged thrombin at a ratio of 100 μg of protein/unit of thrombin for overnight, followed by purification via passing through a Ni-NTA column. The purified proteins were concentrated using an Amicon Ultra-15 centrifugal filter device (nominal molecular weight limit [NMWL], 10,000), and the buffer was exchanged with 50 mM phosphate buffer (pH 7.0)–50 μM ZnSO4 during the process. The protein yields for the NDM-1-H6 and H6-mNDM-1 were approximately 1 and 8 mg/liter, respectively.
MIC.
The constructs containing blaNDM-1-H6, blaNDM-1, and mutants were grown on a Mueller-Hinton agar (MHA) plate, incubated at 37°C overnight and transferred to MHB medium supplemented with a serial concentration of selected β-lactams the next day. Antimicrobial susceptibility testing of transformants was interpreted according to the CLSI guidelines, and the results were determined as MICs (22).
Kinetic parameters.
The kinetic constants of MBLs were determined by incubation of the enzyme with different concentrations of β-lactams at 25°C in 500 μl of assay buffer (50 mM phosphate buffer [pH 7.0], 50 μM ZnSO4). The rate of hydrolysis of substrates was measured by observing the changes in absorption from the opening of the β-lactam ring at the specific wavelengths using a spectrometer (PerkinElmer Lambda Bio20) (19). The initial velocities versus substrate concentrations were measured and fitted to the Michaelis-Menten equation using the GraphPad Prism5 (GraphPad Software, Inc., San Diego, CA) (see Fig. S1 in the supplemental material). The initial velocities were measured at least in duplicate and averaged to determine Km and kcat.
Determination of zinc content.
The zinc content of the enzyme was measured by the inductively coupled plasma optical emission spectroscopy (ICP-OES) (Santa Clara, CA). The purified native enzymes were dialyzed with 50 mM HEPES (pH 7.6) overnight to remove the loosely bound zinc ions and concentrated using an Amicon Ultra-15 centrifugal filter device. The enzymes were further adjusted to ∼10 μM in the same buffer and used in the ICP-OES. Standard calibration curves were obtained using a series of BDH metal standards with correlation coefficients of >0.999. Emission wavelengths of 213.857 and 202.548 nm and 451.131 and 410.176 nm were used for the detection of zinc and indium (internal control), respectively.
Molecular docking.
Molecular docking was performed using the AutoDock Vina program from The Scripps Research Institute (23). The antibiotics were obtained from the ZINC database of the University of California, San Francisco website (24). The torsion trees of the ligands were determined using the AutoDock Tools 1.5.4. The nonhydrolyzed forms of β-lactams were docked in either the monomeric NDM-1 forms adopted from the dimer crystal structures 3Q6X and 4EYL. The docking results for each β-lactam with the lowest binding energies were used for comparison with the cocrystal structures of NDM-1, visualized and analyzed using the PyMol program.
RESULTS AND DISCUSSION
Earlier studies suggested that the L1, L3, and L5 loops and the consensus residue W87 were important for the substrate recognition and specificity of MBLs. Structural analysis of NDM-1 has identified several residues with potential functional importance in each of the three loops (Fig. 1A). To depict the roles of these residues of NDM-1 in the recognition of different substrates, a point mutation was introduced into pET28-blaNDM-1, pET28-blaNDM-1-H6, and pET28-blaH6-mNDM-1. The MICs of different β-lactams for E. coli that carried pET28-blaNDM-1 and pET28-blaNDM-1-H6 were determined to check if the His6 tag affects the β-lactam's activity. Compared with NDM-1, NDM-1-H6 showed at least 2-fold reduction in MICs of most of the β-lactams tested in this study, except for meropenem and aztreonam (see Table S1 in the supplemental material). These observations suggested the presence of the C-terminal His6 tag may interfere with the activity of this enzyme. Due to such concern, the MICs of different β-lactams were determined for E. coli strains that carried different point mutations of pET28-blaNDM-1 as the first-line screening of the effect of the mutation on NDM-1 substrate hydrolysis. Each of the residues was mutated to alanine or other corresponding residues in VIM-1 and IMP-1 (Fig. 1B). The expression levels of NDM-1-H6 and its derivatives in E. coli were determined by probing the total lysate with anti-His6 tag antibody. As shown in Fig.S1 in the supplemental material, NDM-1 and most of the mutants showed stable and similar levels of expression. V67A is a notable exception, and hence V67G was used for the MIC determination. The majority of the NDM-1 and mutants detected were present in the total lysate as the processed form and small amount of the full-length NDM-1 were observed from the immunoblot membrane with a longer exposure time (see Fig. S1A). The kinetic parameters were determined using purified H6-mNDM-1 and its derivatives since the presence of an N-terminal His6 tag had only a slight influence on activity (see Table S2 in the supplemental material). The structures of different β-lactam antibiotics are shown in Fig. S2 in the supplemental material. The zinc content of native wild-type (WT) NDM-1 showed an equivalent of one zinc, suggesting a monozinc form of NDM-1 was obtained in our purification (see Table S3 in the supplemental material). Furthermore, the kinetic assay indicated that NDM-1 showed approximately 2-fold increase of hydrolytic activity on ampicillin and meropenem in the presence of 50 μM ZnSO4 compared to the condition in the absence of zinc in the assay buffer (data not shown). Our kinetic constants determined under our assay condition supplemented with 50 μM ZnSO4 were similar to those of other reports (25), suggesting that under our assay condition, the NDM-1 protein was likely in its dizinc form.
Roles of the L1 loop in recognition of different substrates.
Mutations of NDM-1 L1 loop residues M61A, M61V, F64A, and F64W showed a slight or no effect on the ampicillin MICs (Table 1), suggesting the limited role of M61 and F64 in the recognition of ampicillin. Consistently, structural analysis of NDM-1 and hydrolyzed ampicillin (3Q6X) and benzylpenicillin (4EYF) showed that M61and F64 were too far away to have a direct interaction with the R group of penicillins. In contrast, L59 and V67 were close to and could have a potential hydrophobic interaction with the R group of ampicillin, which may explain the reduced ampicillin MICs of L59A and V67G (Fig. 2A and Table 1), in which the latter showed both increased Km and kcat compared to the wild type (Table 2). These observations were somewhat different from those from a previous study showing F61, A64, and Y67 of VIM-type enzyme were rather tolerant to substitution on hydrolyzing ampicillin (21). W87 was shown to be important for the ampicillin resistance and the proper folding of the structure but not for the catalytic activity of VIM-2 (21). In the case of NDM-1, however, W87A showed reduced ampicillin hydrolysis, and the purified W87A was too inactive to determine its kinetic constants for ampicillin (Tables 1 and 2). Interestingly, W87A showed lower zinc content than the wild type (see Table S3 in the supplemental material). W87F retained similar activity on ampicillin, implying the hydrophobicity of this residue is critical for the ampicillin hydrolysis by NDM-1 (Table 1). These data suggested that while L59 and V67 could be involved in the recognition of the R group of ampicillin, the influence of the W87 mutation may be due to its role in maintaining the correct coordination of the zinc ion through stabilizing the L1 loop.
TABLE 1.
MICs for NDM mutations against different β-lactams in this study
| MBL | MIC (μg/ml) ofa: |
|||||||
|---|---|---|---|---|---|---|---|---|
| Amp | Mer | Imi | Ert | Cep | Cro | Cpm | Azt | |
| Vector control | <4 | <1 | <1 | <0.25 | <4 | <1 | <0.06 | <0.5 |
| NDM-1 | ||||||||
| WT | 512 | 16 | 128 | 32 | 256 | 128 | 2 | <0.5 |
| Mutant | ||||||||
| L59A | 32 | 8 | 64 | 16 | 64 | 32 | 0.25 | <0.5 |
| M61A | 1,024 | 8 | 64 | 8 | 128 | 32 | 0.25 | <0.5 |
| M61V | 512 | 8 | 32 | 8 | 128 | 64 | 4 | <0.5 |
| F64A | 1,024 | 8 | 64 | 8 | 128 | 64 | 2 | <0.5 |
| F64W | 512 | 8 | 64 | 32 | 128 | 64 | 4 | <0.5 |
| V67G | 128 | <1 | 16 | 2 | 128 | 64 | <0.125 | <0.5 |
| V67Y | 512 | 4 | 32 | 8 | 64 | 32 | <0.125 | <0.5 |
| W87A | <4 | <1 | 4 | <1 | 8 | <4 | <0.125 | <0.5 |
| W87F | 256 | <1 | 64 | 16 | 128 | 64 | <0.125 | <0.5 |
| Q119A | 128 | 4 | 32 | 16 | 128 | 64 | <0.125 | <0.5 |
| Q119D | 16 | <1 | 4 | <1 | 16 | <2 | <0.125 | <0.5 |
| Q119S | 128 | 4 | 64 | 16 | 128 | 64 | <0.125 | <0.5 |
| K224A | <4 | <1 | 4 | <1 | 128 | 64 | <0.125 | <0.5 |
| K224R | 512 | 8 | 16 | 8 | 128 | 64 | <0.125 | <0.5 |
| K228A | 512 | 8 | 64 | 16 | 256 | 128 | 4 | <0.5 |
| S229A | 128 | 8 | 64 | 16 | 64 | 32 | <0.125 | <0.5 |
| N233A | 128 | 4 | 8 | 8 | 32 | 16 | <0.125 | <0.5 |
| Y244A | 512 | 8 | 128 | 16 | 128 | 64 | 2 | <0.5 |
Amp, ampicillin; Mer, meropenem; Imi, imipenem; Ert, ertapenem; Cep, cephalothin; Cro, ceftriaxone; Cpm, cefepime; Azt, aztreonam.
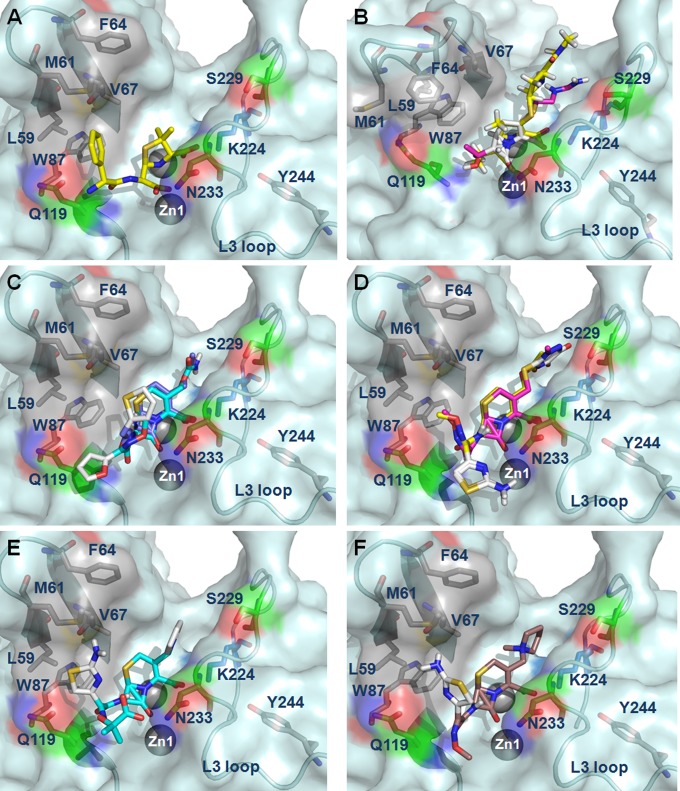
FIG 2.
Roles of the L1, L3, and L5 loops in recognition of different substrates from NDM-1 structures or docking output of β-lactams with the lowest binding energy in NDM-1. (A) Ampicillin (3Q6X); (B) meropenem (4EYL [yellow]) and imipenem (magenta); (C) ertapenem; (D) cephalothin; (E) ceftriaxone; (F) cefepime.
TABLE 2.
Kinetic constants of NDM mutants toward five β-lactams
| Drug | Constant | Result for: |
|||||
|---|---|---|---|---|---|---|---|
| WT | Mutant |
||||||
| V67G | W87A | Q119D | K224A | N233A | |||
| Ampicillin | Km (μM) | 270 ± 53 | 1,169 ± 322 | NDa | 170 ± 44 | ND | 266 ± 50 |
| kcat (s−1) | 834 ± 86 | 1,807 ± 399 | ND | 117 ± 13 | ND | 625 ± 62 | |
| kcat/Km (μM−1 s−1) | 3.09 | 1.55 | ND | 0.69 | ND | 2.35 | |
| Cefepime | Km (μM) | 319 ± 44 | ND | 538 ± 147 | 180 ± 39 | ND | 149 ± 39 |
| kcat (s−1) | 28 ± 2 | ND | 0.4 ± 0.1 | 3.8 ± 0.4 | ND | 1.2 ± 0.2 | |
| kcat/Km (μM−1 s−1) | 0.09 | ND | 0.00 | 0.02 | ND | 0.01 | |
| Meropenem | Km (μM) | 73 ± 17 | ND | ND | 32 ± 7 | ND | ND |
| kcat (s−1) | 398 ± 44 | ND | ND | 32 ± 2 | ND | ND | |
| kcat/Km (μM−1 s−1) | 5.45 | ND | ND | 1.00 | ND | ND | |
| Imipenem | Km (μM) | 313 ± 94 | 680 ± 279 | ND | 90 ± 15 | ND | 687 ± 327 |
| kcat (s−1) | 278 ± 54 | 576 ± 186 | ND | 42 ± 3 | ND | 45 ± 17 | |
| kcat/Km (μM−1 s−1) | 0.89 | 0.85 | ND | 0.47 | ND | 0.07 | |
| Ertapenem | Km (μM) | 121 ± 15 | 264 ± 56 | ND | 19 ± 2 | ND | 84 ± 27 |
| kcat (s−1) | 118 ± 8 | 224 ± 34 | ND | 4 ± 0 | ND | 16 ± 3 | |
| kcat/Km (μM−1 s−1) | 0.98 | 0.85 | ND | 0.21 | ND | 0.19 | |
ND, the kinetic constants could not be determined due to the low activity of the mutations.
In the case of carbapenem recognition, mutations of L59, M61, and F64 to alanine or other residues showed a limited effect on the MICs of meropenem and imipenem, while M61A, M61V, and F64A showed a 4-fold reduction of ertapenem MICs. Mutations of V67G and W87A caused dramatic attenuated MICs of these three carbapenems, whereas V67Y and W87F exhibited less effect on imipenem and ertapenem (Table 1). Kinetic data showed that V67G increased both Km and kcat for imipenem and ertapenem, while W87A was too inactive to achieve the saturation of fitting curves (Table 2; see Fig. S3E in the supplemental material). Structural analysis of the complex structure of NDM-1 and meropenem, 4EYL, suggested the potential hydrophobic interaction between V67 and the R3 group of meropenem (Fig. 2B; see Fig. S2B in the supplemental material) (17). Docking models of NDM-1 with imipenem and ertapenem placed the R3 group in close proximity to V67, between which the potential hydrophobic interaction may form (Fig. 2B and C). Taken together, these data suggested that the effect of the V67 mutation could be due to its hydrophobic interaction through the side chain with the R3 group of carbapenems, while the significance of W87 may rely on its role in maintaining the correct coordination of the zinc ion through stabilizing the L1 loop (see Table S3 in the supplemental material).
In addition to the hydrophobic R1 groups as penicillins, cephalosporins have an additional R2 group (see Fig. S2C in the supplemental material). It is widely accepted that a hydroxide/water between Zn1 and Zn2 is ready for the nucleophilic attack in a binding geometry where the carbonyl oxygen of the β-lactam ring associates with Zn1, while one of the carboxylate oxygens of the fused ring coordinates with Zn2 (14, 17, 25, 26). The approach of molecular docking following the suggested binding mode of β-lactam moieties to two Zn ions enabled us to model the promising structures of NDM-1 with different cephalosporins. Our docking models of NDM-1 structure and several cephalosporins showed that the β-lactam rings of cephalosporins fit in the active site in a quite rigid geometry in spite of the varied R groups (Fig. 2D to F). Furthermore, the R1 groups appear to be more mobile, whereas the R2 groups show a relatively fixed orientation (see Fig. S4 in the supplemental material). Mutations of the L1 loop residues L59A and M61A showed at least 4-fold-reduced MICs to ceftriaxone and cefepime, while the other mutations, including M61V, F64A, and F64W, showed limited effects (Table 1). These results were compatible with our docking structures using cephalosporins with the lowest binding energies, which showed the R1 group of cephalothin was located in close proximity to L59, whereas those of ceftriaxone and cefepime were located between L59 and M61 of the L1 loop, which could form a potential hydrophobic interaction with the R1 groups of these antibiotics (Fig. 2D to F). The effects of mutations of W87 on cephalosporin MICs were the same as those for penicillin and carbapenem antibiotics, suggesting a similar role of this residue in NDM-1 (Table 1). Kinetic characterization showed that V67G was much less active on cefepime, and thus the fitted data could not be obtained, implying that a potential interaction, mostly likely the hydrophobic interaction, may form between this residue and the R1 group of cefepime.
Different from other B1 MBLs, such as IMP-1 and CcrA, in which the L1 loop residue W64 has been shown to be important for both substrate binding and promotion of catalysis (19, 27), the L1 loop residue F64 of NDM-1 did not have a direct effect on the recognition of different β-lactams: instead, L59 and V67 were more important for optimal substrate recognition, most likely through the hydrophobic interactions with the R, R3, and R1 groups of ampicillin, carbapenems, and cephalosporins, respectively.
Roles of the L3 loop in recognition of different substrates.
The cocrystal structures of NDM-1 (3Q6X and 4EYL) suggested potential hydrogen bonds may form between the side chain of the highly conserved N233 and the β-carbonyl oxygen atoms of ampicillin and meropenem. There has been a controversy over the role of N233 among all MBLs. While this residue has been proposed to play a critical role in stabilizing the transitional intermediate formed during β-lactam hydrolysis, in some cases, such as IMP-1, it was shown to be less important (28–30). In this study, N233A showed attenuated MICs for the ampicillin, carbapenems, and cephalosporins tested, ranging from 4- to 16-fold reductions (Table 1). The kinetic data of N233A varied from those of the wild type, with different effects depending on the antibiotics being tested (Table 2). Different from the increased catalytic efficiency toward ampicillin and similar activity toward other β-lactams by the N233A of IMP-1, the same mutation showed similar activity toward ampicillin and lower catalytic efficiency toward cefepime, imipenem, and ertapenem due to much lower kcat values than those of the wild-type NDM-1 (28, 30). In addition to N233, previous studies suggested that K224 of other MBLs contributes to the substrate binding or catalysis, possibly by interacting with the carboxylate oxygen of a β-lactam-fused ring through a hydrogen bond (29, 30) (Fig. 2A). K224A attenuated MICs of ampicillin, cefepime, meropenem, imipenem, and ertapenem drastically, while it slightly reduced MICs of cephalothin and ceftriaxone. Mutation of K224A may abolish the hydrogen bond between K224 and the β-lactam carboxylate oxygen atom, while K224R could retain the hydrogen bond at this site and restore partial resistance to these drugs (Table 1). The complex structure of NDM-1 (4EYL) and docking structures revealed that K224 and N233 showed similar binding geometries for the carbapenems as that for ampicillin (Fig. 2B and C). Unfortunately, we were unable to obtain the kinetic constants of K224A due to the difficulty of obtaining the saturation of velocity (Table 2). Our data also suggested that N233 interacted with carbapenems most likely through its side chain since substitution of alanine for this residue showed lower kcat values (∼6- to 7-fold) for imipenem and ertapenem, implying its role in the contribution of the substrate catalysis of the carbapenems (Fig. 2B and Table 2). In addition to these two residues, the complex structures and docking structures of NDM-1 with several β-lactams suggested possible interactions between the hydroxyl oxygen of S229 and the R2 group of cephalosporins (Fig. 2; see Fig. S2 in the supplemental material). Substitution of Ala for this residue showed 4-fold reduction of cephalothin and ceftriaxone MICs, implying the presence of the predicted interaction. However, its contribution to both drugs was moderate.
Our data indicated the significance of L3 loop residues N233 and K224 in the recognition and hydrolysis of different β-lactams. This recognition is a common and important mechanism of substrate recognition for a variety of substrates by NDM-1.
Roles of the L5 loop in recognition of different substrates.
The L5 loop is composed of several important residues, including H116, H118, and D120, that are involved in zinc binding and are essential for the activities of MBLs. The structures of NDM-1 and hydrolyzed ampicillin revealed that a weak hydrogen bond may form between residue Q119 and the oxygen atom near the R group of ampicillin to stabilize the interaction between the L1 loop and the R group (see Fig. S5 in the supplemental material). Mutation of Q119A showed reduced MICs of ampicillin, meropenem, imipenem, and cefepime, while Q119D showed overall attenuated MICs of all drugs tested. Kinetic parameters of Q119D showed both lower Km and kcat for all substrates tested (Table 2). However, Q119D showed 70% reduced zinc content (see Table S3 in the supplemental material) compared to WT-NDM-1, suggesting that the importance of this residue may rely on its role in maintaining the proper orientation of its adjacent residues, H118 and D120, to coordinate with Zn1 rather than direct involvement in the catalysis.
Mechanisms of NDM-1 substrate recognition.
Although recognition of different categories of β-lactams was shown to be driven mostly by the zinc ions, different residues in the active site could contribute to the substrate binding and catalysis. The complex structures of NDM-1 (3Q6X) indicated the R group of ampicillin was in close proximity to L59, for which the mutation may destabilize the hydrophobic interaction between them and attenuated ampicillin MIC. The consensus W87 was involved in forming a hydrophobic patch that stabilized the L1 loop and zinc coordination for stable substrate recognition. In some MBLs, tyrosine was found in residue 67 (VIM-2 and BlaB) and phenylalanine in residue 87 (IMP-1) to stabilize the L1 loop and the active site (3). Ampicillin was proposed to form hydrogen bonds with both the main-chain N atom and Oε atom of Q119, our data showed an attenuation of the ampicillin MIC from the mutations of Q119, yet the effect was more likely due to the mutations caused by improper orientation of its adjacent residues, H118 and D120, and to affect the binding of Zn1 (14).
Distinct from ampicillin, both N233 and K224 were critical for the substrate binding and catalysis of carbapenems. The complex structure (4EYL) and docking models of NDM-1 demonstrated that the R3 groups of carbapenems were closed to V67, and substitution of alanine for this residue compromised the MICs of meropenem, imipenem, and ertapenem. Y244 of NDM-1 was proposed to play a role in stabilizing loop L10 and the conformation of the active site (14), yet our data showed that the substitution of alanine for tyrosine in NDM-1 had no effect on the MIC, suggesting the limited role this residue played in substrate hydrolysis.
Our docking models suggested that the β-lactam rings of cephalosporins dock in the active site in a rigid and fixed geometry with N233 and K244 as two anchoring points, despite their varied R groups. Furthermore, the R1 groups and R2 groups of cephalosporins were arranged in close proximity to the residues L59 and V67, respectively. The substrate carbonyl was associated with Zn1 and the side chain of N233, while the substrate carboxyl oxygen atoms were associated with Zn2 and the side chain of K224.
Conclusions.
This study showed that the importance of N233 depends on the antibiotics tested for NDM-1, consistent with earlier findings for IMP-1 (28). The dramatic loss of activity by K224A to most of the substrates tested in this study indicated the critical role of this residue in substrate catalysis, agreeing with the previous study showing that this mutation disrupts the hydrolytic activity of NDM-1 to meropenem (31). NDM-1 appeared to recognize its substrates through the coordination of the β-lactam carbonyl, Zn1, and N233, as well as the coordination of the carboxylate oxygen atom of the substrate by K224. Different from VIM-2, L1 residue L59 could form potential hydrophobic interactions with the R groups of penicillins or the R1 groups of cephalosporins. Similarly, V67 could form a potential hydrophobic interaction with the R2 groups of cephalosporins or R3 groups of carbapenems. The significance of W87 most likely relied on its importance in maintaining the stability and zinc coordination of this enzyme. NDM-1 broadened its spectrum of substrates through possession of a wide active site cavity, which may explain the broad spectrum of substrate specificity of B1 MBLs. The subclass B2 MBLs have a relatively narrow active site pocket marked by the presence of an extended α3 loop and a shorter L1 loop at the active site (32) (see Fig. S6 in the supplemental material). The restricted space adjacent to the L1 loop of B2 MBLs has insufficient room for the R groups of penicillins or R1 groups of cephalosporins, which may explain the narrow-spectrum activities of subclass B2 MBLs for only carbapenems but not penicillins and cephalosporins. The understanding of the molecular mechanisms of substrate specificity of NDM-1 provides insights into the development of efficient inhibitors, such as compounds carrying a sulfate to chelate the zinc ions and two other functional groups to target K224 and N233.
Supplementary Material
ACKNOWLEDGMENTS
We thank Edward Chan for critical reading of the manuscript.
This work was supported by the Chinese National Key Basic Research and Development (973) Program (2013CB127200) and the Research Fund for the Control of Infectious Diseases of the Food and Health Bureau, the Government of the Hong Kong SAR (12111612 to S.C.).
Footnotes
Published ahead of print 30 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01977-13.
REFERENCES
- 1.Bush K. 2010. Alarming beta-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr. Opin. Microbiol. 13:558–564. 10.1016/j.mib.2010.09.006 [DOI] [PubMed] [Google Scholar]
- 2.Bush K, Jacoby GA, Medeiros AA. 1995. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211–1233. 10.1128/AAC.39.6.1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush K, Jacoby GA. 2010. Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 54:969–976. 10.1128/AAC.01009-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bush K, Fisher JF. 2011. Epidemiological expansion, structural studies, and clinical challenges of new beta-lactamases from Gram-negative bacteria. Annu. Rev. Microbiol. 65:455–478. 10.1146/annurev-micro-090110-102911 [DOI] [PubMed] [Google Scholar]
- 5.Hernandez Valladares M, Felici A, Weber G, Adolph HW, Zeppezauer M, Rossolini GM, Amicosante G, Frere JM, Galleni M. 1997. Zn(II) dependence of the Aeromonas hydrophila AE036 metallo-beta-lactamase activity and stability. Biochemistry 36:11534–11541. 10.1021/bi971056h [DOI] [PubMed] [Google Scholar]
- 6.Garau G, Garcia-Saez I, Bebrone C, Anne C, Mercuri P, Galleni M, Frere JM, Dideberg O. 2004. Update of the standard numbering scheme for class B beta-lactamases. Antimicrob. Agents Chemother. 48:2347–2349. 10.1128/AAC.48.7.2347-2349.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drawz SM, Bonomo RA. 2010. Three decades of beta-lactamase inhibitors. Clin. Microbiol. Rev. 23:160–201. 10.1128/CMR.00037-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054. 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galleni M, Lamotte-Brasseur J, Rossolini GM, Spencer J, Dideberg O, Frere JM. 2001. Standard numbering scheme for class B beta-lactamases. Antimicrob. Agents Chemother. 45:660–663. 10.1128/AAC.45.3.660-663.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Saez I, Docquier JD, Rossolini GM, Dideberg O. 2008. The three-dimensional structure of VIM-2, a Zn-beta-lactamase from Pseudomonas aeruginosa in its reduced and oxidised form. J. Mol. Biol. 375:604–611. 10.1016/j.jmb.2007.11.012 [DOI] [PubMed] [Google Scholar]
- 11.Concha NO, Janson CA, Rowling P, Pearson S, Cheever CA, Clarke BP, Lewis C, Galleni M, Frere JM, Payne DJ, Bateson JH, Abdel-Mequid SS. 2000. Crystal structure of the IMP-1 metallo beta-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor. Biochemistry 39:4288–4298. 10.1021/bi992569m [DOI] [PubMed] [Google Scholar]
- 12.Murphy TA, Catto LE, Halford SE, Hadfield AT, Minor W, Walsh TR, Spencer J. 2006. Crystal structure of Pseudomonas aeruginosa SPM-1 provides insights into variable zinc affinity of metallo-beta-lactamases. J. Mol. Biol. 357:890–903. 10.1016/j.jmb.2006.01.003 [DOI] [PubMed] [Google Scholar]
- 13.Carfi A, Duee E, Galleni M, Frere JM, Dideberg O. 1998. 1.85 A resolution structure of the zinc (II) beta-lactamase from Bacillus cereus. Acta Crystallogr. D Biol. Crystallogr. 54:313–323. 10.1107/S0907444997010627 [DOI] [PubMed] [Google Scholar]
- 14.Zhang H, Hao Q. 2011. Crystal structure of NDM-1 reveals a common beta-lactam hydrolysis mechanism. FASEB J. 25:2574–2582. 10.1096/fj.11-184036 [DOI] [PubMed] [Google Scholar]
- 15.de Seny D, Prosperi-Meys C, Bebrone C, Rossolini GM, Page MI, Noel P, Frere JM, Galleni M. 2002. Mutational analysis of the two zinc-binding sites of the Bacillus cereus 569/H/9 metallo-beta-lactamase. Biochem. J. 363:687–696. 10.1042/0264-6021:3630687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z, Fast W, Benkovic SJ. 1999. On the mechanism of the metallo-beta-lactamase from Bacteroides fragilis. Biochemistry 38:10013–10023. 10.1021/bi990356r [DOI] [PubMed] [Google Scholar]
- 17.King DT, Worrall LJ, Gruninger R, Strynadka NC. 2012. New Delhi metallo-beta-lactamase: structural insights into beta-lactam recognition and inhibition. J. Am. Chem. Soc. 134:11362–11365. 10.1021/ja303579d [DOI] [PubMed] [Google Scholar]
- 18.Oelschlaeger P, Mayo SL, Pleiss J. 2005. Impact of remote mutations on metallo-beta-lactamase substrate specificity: implications for the evolution of antibiotic resistance. Protein Sci. 14:765–774. 10.1110/ps.041093405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moali C, Anne C, Lamotte-Brasseur J, Groslambert S, Devreese B, Van Beeumen J, Galleni M, Frere JM. 2003. Analysis of the importance of the metallo-beta-lactamase active site loop in substrate binding and catalysis. Chem. Biol. 10:319–329. 10.1016/S1074-5521(03)00070-X [DOI] [PubMed] [Google Scholar]
- 20.Yamaguchi Y, Jin W, Matsunaga K, Ikemizu S, Yamagata Y, Wachino J, Shibata N, Arakawa Y, Kurosaki H. 2007. Crystallographic investigation of the inhibition mode of a VIM-2 metallo-beta-lactamase from Pseudomonas aeruginosa by a mercaptocarboxylate inhibitor. J. Med. Chem. 50:6647–6653. 10.1021/jm701031n [DOI] [PubMed] [Google Scholar]
- 21.Borgianni L, Vandenameele J, Matagne A, Bini L, Bonomo RA, Frere JM, Rossolini GM, Docquier JD. 2010. Mutational analysis of VIM-2 reveals an essential determinant for metallo-beta-lactamase stability and folding. Antimicrob. Agents Chemother. 54:3197–3204. 10.1128/AAC.01336-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.CLSI. 2011. Performance standards for antimicrobial susceptibility testing; twenty-first informational supplement: Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 23.Trott O, Olson AJ. 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31:455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. 2012. ZINC: a free tool to discover chemistry for biology. J. Chem. Inf. Model. 52:1757–1768. 10.1021/ci3001277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas PW, Zheng M, Wu S, Guo H, Liu D, Xu D, Fast W. 2011. Characterization of purified New Delhi metallo-beta-lactamase-1. Biochemistry 50:10102–10113. 10.1021/bi201449r [DOI] [PubMed] [Google Scholar]
- 26.Kim Y, Tesar C, Mire J, Jedrzejczak R, Binkowski A, Babnigg G, Sacchettini J, Joachimiak A. 2011. Structure of apo- and monometalated forms of NDM-1—a highly potent carbapenem-hydrolyzing metallo-beta-lactamase. PLoS One 6:e24621. 10.1371/journal.pone.0024621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huntley JJ, Fast W, Benkovic SJ, Wright PE, Dyson HJ. 2003. Role of a solvent-exposed tryptophan in the recognition and binding of antibiotic substrates for a metallo-beta-lactamase. Protein Sci. 12:1368–1375. 10.1110/ps.0305303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown NG, Horton LB, Huang W, Vongpunsawad S, Palzkill T. 2011. Analysis of the functional contributions of Asn233 in metallo-beta-lactamase IMP-1. Antimicrob. Agents Chemother. 55:5696–5702. 10.1128/AAC.00340-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma NP, Hajdin C, Chandrasekar S, Bennett B, Yang KW, Crowder MW. 2006. Mechanistic studies on the mononuclear ZnII-containing metallo-beta-lactamase ImiS from Aeromonas sobria. Biochemistry 45:10729–10738. 10.1021/bi060893t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Materon IC, Palzkill T. 2001. Identification of residues critical for metallo-beta-lactamase function by codon randomization and selection. Protein Sci. 10:2556–2565. 10.1110/ps.ps.40884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang Z, Li L, Wang Y, Chen L, Kong X, Hong Y, Lan L, Zheng M, Guang-Yang C, Liu H, Shen X, Luo C, Li KK, Chen K, Jiang H. 2011. Molecular basis of NDM-1, a new antibiotic resistance determinant. PLoS One 6:e23606. 10.1371/journal.pone.0023606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garau G, Bebrone C, Anne C, Galleni M, Frere JM, Dideberg O. 2005. A metallo-beta-lactamase enzyme in action: crystal structures of the monozinc carbapenemase CphA and its complex with biapenem. J. Mol. Biol. 345:785–795. 10.1016/j.jmb.2004.10.070 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.