Abstract
Biofilm-related infections account for at least 65% of all human infections, but there are no available antimicrobials that specifically target biofilms. Their elimination by available treatments is inefficient since biofilm cells are between 10- and 1,000-fold more resistant to conventional antibiotics than planktonic cells. Here we describe the synergistic interactions, with different classes of antibiotics, of a recently characterized antibiofilm peptide, 1018, to potently prevent and eradicate bacterial biofilms formed by multidrug-resistant ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) pathogens. Combinations of peptide 1018 and the antibiotic ceftazidime, ciprofloxacin, imipenem, or tobramycin were synergistic in 50% of assessments and decreased by 2- to 64-fold the concentration of antibiotic required to treat biofilms formed by Pseudomonas aeruginosa, Escherichia coli, Acinetobacter baumannii, Klebsiella pneumoniae, Salmonella enterica, and methicillin-resistant Staphylococcus aureus. Furthermore, in flow cell biofilm studies, combinations of low, subinhibitory levels of the peptide (0.8 μg/ml) and ciprofloxacin (40 ng/ml) decreased dispersal and triggered cell death in mature P. aeruginosa biofilms. In addition, short-term treatments with the peptide in combination with ciprofloxacin prevented biofilm formation and reduced P. aeruginosa PA14 preexisting biofilms. PCR studies indicated that the peptide suppressed the expression of various antibiotic targets in biofilm cells. Thus, treatment with the peptide represents a novel strategy to potentiate antibiotic activity against biofilms formed by multidrug-resistant pathogens.
INTRODUCTION
Biofilms are structured, surface-associated multicellular communities of microbes embedded in a biopolymeric matrix rich in macromolecular components (1–3). Microbes form biofilms under several circumstances, including in response to nutritional cues or starvation, attachment to surfaces, or stresses, such as subinhibitory antibiotics. Biofilms colonize indwelling medical devices, prosthetics, and body surfaces (e.g., skin, bladder, lung, heart) and give rise to difficult-to-treat chronic infections, such as lung infections in patients with cystic fibrosis (CF) or endocarditis (1, 2). Biofilms represent a physiologically distinct state of bacteria, with hundreds of genes changing expression compared to their expression in free-living (planktonic) cells.
The most alarming aspect of biofilm-related infections is that they are highly (10- to 1,000-fold) resistant to conventional antibiotics that were developed to kill planktonic cells (4, 5). As a consequence, there is a lack of antibiofilm agents in the antibiotic development pipeline. The increased antibiotic resistance of biofilm cells is transient, reverting upon conversion to the planktonic state, and thus is adaptive. There are a variety of explanations that have been provided for adaptive resistance in biofilms, including limited diffusion of antibiotics, binding to matrix components like DNA, limited growth and metabolism of cells deep within the biofilm, the presence of persisters, and the upregulation of resistance mechanisms as part of the altered transcriptional profile of biofilms (5). In addition, the frequency of mutations in part is due to responses to stress, and therefore, the emergence of resistance increases in biofilms, and horizontal transfer of resistance genes is also increased by the close proximity of cells (6, 7). As a result of this and other factors, in the clinic, chronic infections are often treated with combinations of different antibiotics, which can lead to the emergence of multidrug-resistant bacteria (3). Currently, the most recalcitrant bacteria in our society are termed the ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) pathogens.
We recently demonstrated that peptide 1018 is a broad-spectrum antibiofilm peptide that acts by binding to and causing the degradation of the widespread signaling nucleotide (p)ppGpp, which is involved in stress responses and biofilm formation (8). Peptide 1018 exhibited modest antimicrobial activity against planktonic cells, but at concentrations well below the MIC, it completely prevented biofilm formation and eradicated preformed mature biofilms caused by both Gram-negative and Gram-positive bacterial pathogens (8). Dispersal of biofilms was observed after treatment with 0.8 μg/ml of peptide, while biofilm cells were almost completely killed at higher concentrations of peptide (10 μg/ml). Here, we have investigated the synergistic interactions against biofilms of antibiofilm peptide 1018 in conjunction with different classes of antibiotics commonly used in the clinic.
MATERIALS AND METHODS
Bacterial strains.
The strains utilized included Pseudomonas aeruginosa wild-type strains PAO1 (strain H103) and PA14 and clinical isolates of Escherichia coli O157, Salmonella enterica serovar Typhimurium (clinical isolate 14028S), methicillin-resistant Staphylococcus aureus (MRSA; clinical isolate SAP0017), Klebsiella pneumoniae (ATTC 13883; a colistin-heteroresistant reference strain from the American Type Culture Collection, Manassas, VA), Acinetobacter baumannii (SENTRY 8; a polymyxin B-resistant blood clinical isolate from the United States obtained through the SENTRY surveillance system), and Burkholderia cenocepacia genomovar IIIa (Vancouver Children's Hospital clinical isolate 4813). P. aeruginosa PAO1 (p)ppGpp mutant ΔrelA spoT (with the deletion of nucleotides 181 to 2019 in relA and nucleotides 200 to 1948 in spoT) was a kind gift from D. Nguyen (9).
MIC assay and MBIC checkerboard assay.
The broth microdilution method (10) was used for measuring the MIC of ciprofloxacin. The broth microdilution method with minor modifications for cationic peptides (8) was used for measuring the minimal biofilm inhibitory concentration (MBIC) of peptide 1018 and the antibiotics. The peptide 1018 (VRLIVAVRIWRR-NH2) used in this study was synthesized by CPC Scientific using solid-phase 9-fluorenylmethoxy carbonyl (Fmoc) chemistry and purified to a purity of ∼95% using reversed-phase high-performance liquid chromatography (HPLC). The peptide was dissolved in water and stored in glass vials. MBIC assays were performed in sterile 96-well polypropylene microtiter plates (catalog no. 3790; Costar). The peptide and antibiotics were added to the plate at the desired concentrations, and the bacteria were inoculated to a final concentration of 5 × 105 CFU/ml per well. The plates were incubated at 37°C for 24 h before being washed to remove unattached cells. Bound cells (biofilm) were stained with crystal violet, which was subsequently extracted with 95% ethanol. The number of attached cells was analyzed by measuring the absorbance at 595 nm. The MBIC was defined as the concentration in the well containing the minimal amount of compounds leading to an absorbance equivalent to that of the background control.
Biofilm cultivation in flow cell chambers and microscopy.
Biofilms were grown for 72 h in the absence or presence of the desired concentration of peptide 1018 and the antibiotics (ciprofloxacin, ceftazidime, imipenem, and tobramycin) at 37°C in flow chambers with channel dimensions of 1 by 4 by 40 mm. The medium used was always BM2 minimal medium [62 mM potassium phosphate buffer, pH 7.0, 7 mM (NH4)2SO4, 2 mM MgSO4, 10 μM FeSO4] containing 0.4% (wt/vol) glucose as a carbon source. Silicone tubing (inner diameter, 0.062 in.; outer diameter, 0.125 in.; wall thickness, 0.032-in.; VWR) was autoclaved, and the system was assembled and sterilized by pumping a 0.5% hypochlorite solution through the system at 6 rpm for 30 min using a Watson Marlow 205S multichannel peristaltic pump. The system was then rinsed at 6 rpm with sterile water and medium for 30 min each. Flow chambers were inoculated by injecting 400 μl of an overnight culture diluted to an optical density at 600 nm of 0.05. After inoculation, the chambers were left without flow for 2 h, after which medium (with or without a subinhibitory concentration of 1018) was pumped through the system at a constant rate of 0.5 rpm (2.4 ml/h). Biofilm cells were stained using a LIVE/DEAD BacLight bacterial viability kit (Molecular Probes, Eugene, OR) or Syto-9 alone prior to the microscopy experiments. A ratio of Syto-9 (green fluorescence, live cells) to propidium iodide (PI; red fluorescence, dead cells) of 1:5 was used. Microscopy was done using a confocal laser scanning microscope (Fluoview FV1000; Olympus), and three-dimensional reconstructions were generated using the Imaris software package (Bitplane AG). Biofilm biovolume (μm3) was calculated using Imaris software.
For inhibition of biofilm growth, the treatment was applied from the beginning of the experiment. For treatment of preformed biofilms, bacteria were allowed to develop structured 2-day-old biofilms prior to peptide and antibiotic treatment for the following 24 h. For shot experiments, injection of peptide and antibiotic diluted in medium was done directly in the flow cell chamber. The flow was stopped during application of the treatment (2 h for inhibition and 1 h for eradication).
Evaluation of cells dispersed from biofilms.
Dispersion experiments were performed using P. aeruginosa PA14 in BM2 medium with 0.4% glucose. Strains were grown in the flow cell system to form biofilm on a plastic surface. P. aeruginosa PA14 biofilms were treated with 0.8 μg/ml of peptide or 40 ng/ml of ciprofloxacin, or both. To assay for dispersed cells, aliquots of 30-min flow rate effluent cells were sampled at the designated times (0, 3, and 24 h), 1 ml was taken and centrifuged, the pellet was resuspended in 1 ml of LB and serially diluted 10-fold, and 100-μl portions from serial dilutions of these aliquots were plated onto LB agar plates. The plates were incubated at 37°C overnight, and colony counts were performed to obtain the numbers of CFU/ml at each time point. The experiment was repeated at least 3 times.
Cell survival determination.
The amount of planktonic cells surviving after a treatment was determined using P. aeruginosa PA14. Strains were grown in BM2 medium in the presence or absence of serine hydroxamate (SHX; 500 μM) to induce amino acid starvation. After 3 h, P. aeruginosa PA14 cultures were treated with 40 ng/ml of ciprofloxacin or 0.8 μg/ml of peptide 1018, or both. To assay for survivors, aliquots were withdrawn after 23 h and serially diluted 10-fold, and 100-μl portions from serial dilutions of these aliquots were plated onto LB agar plates. The plates were incubated at 37°C overnight, and colony counts were performed to obtain the numbers of CFU/ml.
qRT-PCR.
Flow cell biofilms were grown for 2 days as described above and treated with peptide 1018 at 0.8 μg/ml for 24 h or not treated. Then, cells were scraped off the flow cell chamber and harvested. Total RNA was then isolated, using RNeasy minicolumns (Qiagen, Mississauga, ON, Canada). DNase treatment of RNA samples, cDNA synthesis, and quantitative real-time reverse-transcription PCR (qRT-PCR) were carried out as described previously (11). All reactions were normalized to the rpsL gene, encoding the 30S ribosomal protein S12. The stability of the rpsL gene under different conditions was confirmed by comparing the respective cycle thresholds (CTs): 16.8 ± 0.1 for untreated biofilm and 17.0 ± 0.4 for 1018-treated biofilm. The difference was not statistically significant (Student's t test).
RESULTS
Synergy between antibiofilm peptide 1018 and antibiotics to inhibit biofilm formation.
Synergy between antibiofilm peptide 1018 and conventional antibiotics (ciprofloxacin, ceftazidime, imipenem, or tobramycin) was determined using a checkerboard titration assay, as previously described (12), except that we measured the effect of peptide diluted in one dimension and antibiotic in another on biofilms formed by bacterial adherence to the surface of microtiter wells (13). The lowest concentration of the peptide-antibiotic combination that led to the complete inhibition of biofilm growth (minimal biofilm inhibitory concentration [MBIC]) was determined, and the fractional inhibitory concentration (FIC) was calculated on the basis of the MBIC of each compound alone and in combination (12); the MBIC represents the minimal concentration required to fully prevent biofilm formation. The FIC provides a measure of the degree of synergy between two antimicrobial agents against a particular microorganism. FIC values below 0.5 indicate synergy, values just above 0.5 indicate a 2-fold change in the MBIC for one agent and a very large change in the MBIC for the other, and those above 4.0 indicate antagonistic interactions (8, 12).
Antibiofilm peptide 1018 exhibited synergistic interactions with all of the antibiotics tested in this study against biofilms formed by at least one of the 6 tested bacterial species (Table 1), with 50% of all combinations giving synergy (FIC ≤ 0.5) and 16.7% giving close to synergy (FIC < 0.53). Addition of the peptide in 20 of 24 cases reduced by 4- to 64-fold the levels of antibiotic needed to completely inhibit biofilm formation (Table 1).
TABLE 1.
Antibiofilm peptide 1018 showed synergy with conventional antibiotics in a broad-spectrum mannera
| Strain | FIC |
Fold decrease in antibiotic concn |
||||||
|---|---|---|---|---|---|---|---|---|
| CTZ | CIP | IMI | TOB | CTZ | CIP | IMI | TOB | |
| P. aeruginosa PA14 | 0.38 | 0.14 | 0.5 | 0.5 | 16 | 16 | 4 | 2 |
| E. coli O157 | 1 | 0.69 | 0.75 | 0.38 | 4 | 4 | 4 | 4 |
| A. baumannii | 0.37 | 0.52 | 0.53 | 0.38 | 16 | 32 | 8 | 4 |
| K. pneumoniae | 0.75 | 0.63 | 0.53 | 0.31 | 4 | 2 | 32 | 16 |
| S. enterica | 0.31 | 1 | 1 | 0.75 | 4 | 2 | 2 | 4 |
| MRSA | 0.16 | 0.25 | 0.25 | 0.52 | 32 | 64 | 4 | 64 |
Checkerboard titrations were performed to assess synergistic interactions between peptide 1018 and conventional antibiotics to prevent biofilm formation. Briefly, one compound (e.g., the peptide) was diluted along the rows of a microtiter plate, and the other compound (e.g., an antibiotic) was diluted along the columns. In this method, one is looking for a reduction in the minimal biofilm inhibitory concentration (MBIC) of each compound in the presence of the other. The result is expressed as the fractional inhibitory concentration (FIC) index, as described in Results. In all cases, peptide 1018, when combined with antibiotics, reduced the antibiotic MBIC when used alone, here depicted as the fold decrease in antibiotic concentration at the FIC. Values that indicate synergy are shown in boldface. CTZ, ceftazidime; CIP, ciprofloxacin; IMI, imipenem; TOB, tobramycin.
Some variability was observed for different species and different antibiotics. Sub-MBICs of the peptide in combination with all antibiotics tested led to synergistic (FIC = 0.14 to 0.5) inhibition of P. aeruginosa PA14 biofilm formation. Particularly significant was the case of ciprofloxacin (FIC = 0.14), in which a 16-fold decrease in the concentration of ciprofloxacin required to inhibit biofilm growth when used in combination with 1018 was shown (Table 1). Synergy (FIC = 0.16 to 0.25) was also observed between peptide 1018 and three of the antibiotics tested against Gram-positive methicillin-resistant S. aureus (MRSA) biofilms, while tobramycin showed near synergy with peptides due to a 64-fold reduction in the effective concentration of this antibiotic (Table 1). Combinations of the peptide with ceftazidime (FIC = 0.37) and tobramycin (FIC = 0.38) were synergistic against biofilms formed by A. baumannii SENTRY 8, while the other two antibiotics were 8- to 32-fold more effective in the presence of peptide. Peptide 1018 was also synergistic with the aminoglycoside antibiotic tobramycin (FIC = 0.38) in inhibiting biofilm formation by E. coli O157 (Table 1). Conversely, with the related species S. enterica serovar Typhimurium, synergy was observed only when combining 1018 with ceftazidime (FIC = 0.31). For K. pneumoniae ATCC 13883, synergy was observed when using peptide 1018 in combination with tobramycin (FIC = 0.31), while effects were also observed for the other three antibiotics tested, with the MBIC for imipenem being notably reduced by 32-fold (Table 1). Thus, synergy was highly dependent on both the particular species being tested and the antibiotic used in combination.
Synergy of antibiofilm peptide 1018 with conventional antibiotics assessed in flow cells.
The best combination obtained in the checkerboard microtiter plate assay for each bacterial species (the peptide-antibiotic combinations that led to the lowest FIC values) was used to confirm the synergistic interactions using the more sophisticated flow cell system. Biofilms were allowed to develop for 3 days in flow cell chambers with a constant flow of BM2 glucose minimal medium across the developing biofilms. Peptide and/or the different antibiotics tested were added to the flow medium during the entire 3 days of the experiment, after which the resulting surface-adherent cells or biofilm was stained for total cells (stained green with the fluorescent dye Syto-9) and dead cells (stained red with normally impermeant propidium iodide) and visualized using confocal laser scanning microscopy, as previously described (8, 12).
Synergy was confirmed using this system, as in all cases the combination of peptide plus antibiotic significantly increased the biofilm inhibitory activity of each compound when used alone (on the basis of a lack of live adherent biofilm colonies; Fig. 1). For example, P. aeruginosa biofilm formation was inhibited by 0.8 μg/ml of peptide 1018 combined with 40 ng/ml ciprofloxacin, leading to a significant reduction in biofilm thickness, while the few small microcolonies observed were mostly composed of dead cells (Fig. 1). A few cells remained attached to the surface of the flow cell chambers after combination treatment of E. coli O157 (1.6 μg/ml tobramycin plus 8 μg/ml 1018), K. pneumoniae (0.1 μg/ml tobramycin plus 2 μg/ml 1018), S. aureus (8 μg/ml ceftazidime plus 8 μg/ml 1018), and A. baumannii (32 μg/ml ceftazidime plus 32 μg/ml 1018) (Fig. 1). However, these cells appeared to be dead, as determined by propidium iodide uptake (a change of the color from yellow to red when merged with Syto-9). No attached S. enterica cells were observed after the application of 2 μg/ml peptide and 62.5 ng/ml ceftazidime (Fig. 1). These results confirmed the results obtained using the checkerboard assay and clearly showed that antibiofilm peptide 1018 acted in synergy with conventional antibiotics, thus markedly reducing the concentration of antibiotic needed to prophylactically treat bacterial biofilm formation.
FIG 1.
Antibiofilm peptide 1018 synergized with conventional antibiotics to inhibit biofilm formation. Biofilms were grown in a flow cell system (see Materials and Methods). Treatment (antibiotic-peptide combination) was added to the flowthrough medium at the beginning of the experiment and maintained for the entire 3 days of the experiment. After 3 days, bacteria were stained green with the all-bacteria stain Syto-9 and red with the dead-bacteria stain propidium iodide (merge shows as a color change from yellow to red) prior to confocal imaging. Each panel shows reconstructions from the top in the large panel and sides in the right and bottom panels (x-y, y-z, and x-z dimensions, respectively). The FIC values provided were those calculated using checkerboard assays (Table 1) and are provided to the right of each image (synergy is an FIC of <0.5).
Eradication of preformed biofilms by combination of peptide 1018 with antibiotics.
The same combinations of peptide and antibiotics used in the inhibition studies (and first determined to show activity in the checkerboard assays) were also used to treat preexisting biofilms. Biofilms were grown for 2 days in flow cells, after which they had already formed biofilm colonies, and were then treated for the last 24 h of the experiment. As a comparative control, we first tested the susceptibility of P. aeruginosa PA14 biofilms to ciprofloxacin (Fig. 2). Biofilm resistance to ciprofloxacin was observed, as treatment with the antibiotic at its MIC, 10× MIC, or 100× MIC did not eradicate 2-day-old P. aeruginosa PA14 biofilms (Fig. 2), and treatment with 10× MIC and 100× MIC triggered only some cell death of inner biofilm clusters (Fig. 2). Next, a combination of peptide and antibiotic or each compound individually was tested. In all cases, treatment with conventional antibiotics alone did not clear preformed biofilms and did not significantly induce biofilm cell death (Fig. 3). Treatment with low levels of the antibiofilm peptide 1018 alone led to decreased biofilm thickness, disrupted the overall biofilm structure, and triggered some cell death (Fig. 3), as previously observed with peptide 1018 (8). These effects were significantly increased in the presence of low concentrations of antibiotics that by themselves did not affect preformed biofilms (Fig. 3). For example, combined treatment with ceftazidime and peptide 1018 completely cleared mature biofilms formed by A. baumannii (Fig. 3). The same combination of antimicrobials at lower concentrations disrupted S. aureus MRSA mature biofilms and led to cell death (Fig. 3). Peptide 1018 in combination with tobramycin led to biofilm clearance for K. pneumoniae and killed biofilm cells of E. coli O157 (Fig. 3). Similarly, treatment of P. aeruginosa PA14 mature biofilms with 1018 and ciprofloxacin led to very small microcolonies composed of dead cells (Fig. 3). On the other hand, treatment with the peptide and ceftazidime did not clear mature biofilms formed by S. enterica; however, it did reduce biofilm thickness and led to substantial cell death (Fig. 3). Therefore, we can conclude that, when used in combination with conventional antibiotics, peptide 1018 potently enhanced antibiotic action to both prevent biofilm formation and treat mature biofilms formed by multidrug-resistant pathogens.
FIG 2.

Pseudomonas aeruginosa PA14 preformed biofilms were highly resistant to ciprofloxacin. Biofilms were grown in a flow cell system. Ciprofloxacin at its MIC (160 ng/ml), 10× MIC, and 100× MIC was applied on a 2-day-old biofilm for 24 h. After 3 days, bacteria were stained green with the all-bacteria stain Syto-9 and red with the dead-bacteria stain propidium iodide (merge shows as a color change from yellow to red) prior to confocal imaging. Each panel shows reconstructions from the top in the large panel and sides in the right and bottom panels (x-y, y-z, and x-z dimensions, respectively).
FIG 3.
Synergy between antibiofilm peptide 1018 with conventional antibiotics in eradicating preformed biofilms. Bacteria were grown as biofilms in a flow cell system. Treatments (antibiotic, peptide, or combinations, as specified in Fig. 1) were added after 2 days of biofilm growth for a subsequent 24 h. After 3 days, bacteria were stained green with the all-bacteria stain Syto-9 and red with the dead-bacteria stain propidium iodide (merge shows as a color change from yellow to red) prior to confocal imaging. Each panel shows reconstructions from the top in the large panel and sides in the right and bottom panels (x-y, y-z, and x-z dimensions, respectively).
Combinations of peptide and ciprofloxacin significantly reduced dispersal of viable cells from P. aeruginosa mature biofilms.
The lowest FIC obtained in checkerboard assays corresponded to the combined use of peptide 1018 with ciprofloxacin against P. aeruginosa PA14 (FIC = 0.14 at 40 ng/ml of ciprofloxacin and 0.8 μg/ml of 1018; Table 1). To further assess the effect of this combined therapy on the dispersal of cells and cell death in P. aeruginosa PA14 mature biofilms, we performed cell dispersal assays, measuring the numbers of cells collected from the flowthrough medium. Briefly, biofilms were allowed to grow untreated for 2 days and subsequently treated with ciprofloxacin alone or in combination with 1018 (Fig. 4). Cells that dispersed from the biofilms in the flow cell chambers were collected in tubes and plated on LB agar for assessment of viable cell counts. Treatment with 40 ng/ml ciprofloxacin led to no increase in the number of live dispersed cells (P > 0.05; Fig. 4). When the same concentration of ciprofloxacin was used together with 0.8 μg/ml 1018, the number of dispersed cells was significantly reduced after both 3 and 23 h (Fig. 4). This can be contrasted with observations of the cells remaining attached to the flow cell, which formed very small microcolonies composed largely of dead cells (>61%, in contrast to 2.5% ± 2% dead cells for the control; P < 0.001) (Fig. 3), while biofilms treated only with ciprofloxacin remained virtually unaffected (P > 0.05). Moreover, the ciprofloxacin, ceftazidime, and tobramycin susceptibilities of dispersed cells from P. aeruginosa PA14 biofilms treated with 0.8 μg/ml of 1018 were identical to those of dispersed cells from untreated biofilms (MICs, 0.16, 3.2, and 0.8 μg/ml for ciprofloxacin, ceftazidime, and tobramycin, respectively), indicating that live dispersed cells from samples treated with low levels of the peptide did not develop adaptive resistance to any of these antibiotics.
FIG 4.
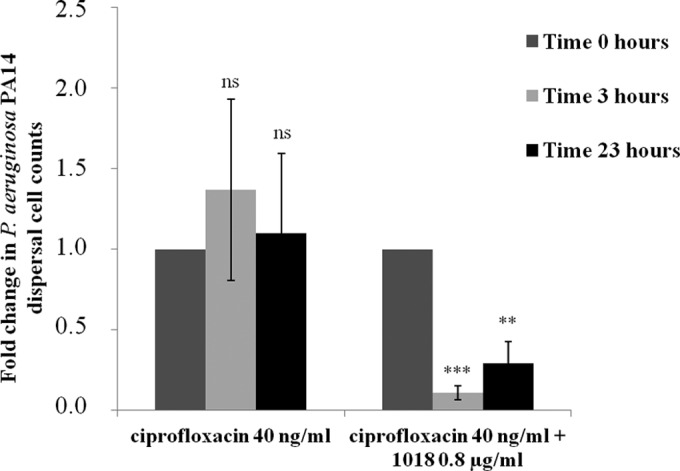
Low levels of 1018 in combination with ciprofloxacin decreased the dispersal of viable cells from P. aeruginosa PA14 biofilms. Dispersed cells from 2-day-old biofilms grown in the flow cell system were collected after 0, 3, and 23 h of treatment with 40 ng/ml of ciprofloxacin or 40 ng/ml of ciprofloxacin and 0.8 μg/ml of peptide 1018 and enumerated by the use of plate counts. Student's t test was done to compare the counts at different time points to those at time zero and determine the statistical significance (ns, not significant; **, P < 0.01; ***, P < 0.001).
Short-term therapy using combinations to treat P. aeruginosa PA14 mature biofilms.
To further evaluate the antibiofilm potential of peptide 1018, we performed short-term combined treatments using 1018 and ciprofloxacin on P. aeruginosa PA14 biofilms (Fig. 5). Treatment with both compounds for only 2 h at the beginning of biofilm growth (inhibition conditions) substantially inhibited biofilm formation, leaving only a few aggregated cells posttreatment (Fig. 5). Eradication conditions involved growing P. aeruginosa PA14 biofilms untreated for 2 days before treating them with 1018 and ciprofloxacin. Treatment of 2-day-old biofilms once with 1018-ciprofloxacin for 1 h demonstrated modest effects, with a decreased biofilm thickness and an altered overall biofilm structure (Fig. 5), while treatment of the biofilms twice on days 2 and 3 for 1 h each time virtually eradicated the Pseudomonas biofilms (Fig. 5).
FIG 5.
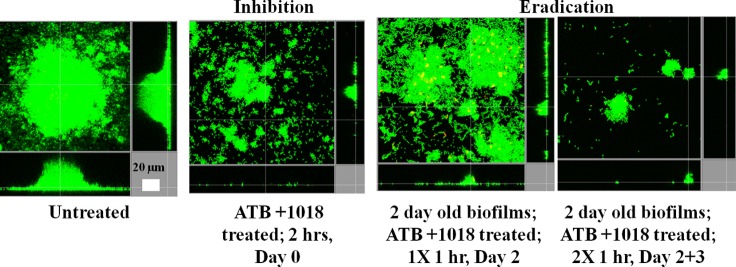
Short-term treatments with combinations of low concentrations of peptide 1018 and ciprofloxacin still inhibited biofilm formation. Peptide 1018 (0.8 μg/ml) and ciprofloxacin (40 ng/ml) were injected for 2 h at the beginning of P. aeruginosa PA14 biofilm formation or into 2-day-old P. aeruginosa PA14 biofilms for 1 h on day 2 or for 1 h on day 2 plus 1 h on day 3. After 3 days, bacteria were stained green with the all-bacteria stain Syto-9 and red with the dead-bacteria stain propidium iodide (merge shows as a color change from yellow to red) prior to confocal imaging. Each panel shows reconstructions from the top in the large panel and sides in the right and bottom panels (x-y, y-z, and x-z dimensions, respectively). ATB, antibiotic (ciprofloxacin).
Low concentrations of peptide 1018 increased the susceptibility of otherwise resistant, SHX-treated P. aeruginosa PA14 planktonic cells and led to the dysregulation of genes involved in antibiotic resistance.
We previously demonstrated that peptide 1018 acts against biofilms by binding to the signaling nucleotide ppGpp, marking it for degradation (8). This explains the antibiofilm activity of the peptide, since (p)ppGpp has been shown to be involved in biofilm formation and maintenance (8). Multiple studies have also demonstrated that (p)ppGpp overproduction leads to adaptive antibiotic resistance (9, 14). Conversely, bacteria unable to produce (p)ppGpp exhibit increased susceptibility to antibiotics (9, 15), which was confirmed here in MIC assays using a P. aeruginosa ΔrelA spoT mutant that lacks the ability to produce (p)ppGpp (Table 2). Indeed, the (p)ppGpp mutant was at least 2- to 4-fold more susceptible to ciprofloxacin, tobramycin, imipenem, and ceftazidime than its parent strain under the tested medium conditions (Table 2).
TABLE 2.
The PAO1 ΔrelA spoT mutant was more susceptible to antibiotics than its parent straina
| Strain | MIC (μg/ml) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ciprofloxacin |
Tobramycin |
Imipenem |
Ceftazidime |
|||||||||
| LB | BM2 + CAA | BM2 − CAA | LB | BM2 + CAA | BM2 − CAA | LB | BM2 + CAA | BM2 − CAA | LB | BM2 + CAA | BM2 − CAA | |
| PAO1 | 0.64 | 0.64 | 1.28 | 0.64 | 1.28 | 1.28 | 10.24 | 2.56 | 0.64 | 1.28 | 0.64 | 0.64 |
| PAO1 ΔrelA spoT | 0.32 | 0.32 | 0.32 | 0.32 | 0.64 | 0.64 | 5.12 | 1.28 | 0.32 | 0.64 | 0.32 | 0.32 |
The MICs of ciprofloxacin, tobramycin, imipenem, and ceftazidime for which we observed 100% inhibition of planktonic growth were determined for Pseudomonas aeruginosa PAO1 and its (p)ppGpp mutant, PAO1 ΔrelA spoT. The media used were rich medium (LB), BM2 glucose minimal medium with 0.5% Casamino Acids (BM2 + CAA), and BM2 glucose minimal medium without additional CAA (BM2 − CAA).
Here, we assessed with planktonic cells if synergy between peptide 1018 and ciprofloxacin occurred only under conditions where (p)ppGpp overproduction occurred due to the serine limitation created by the addition of the serine analogue SHX. In the absence of SHX, the combination of peptide plus ciprofloxacin had absolutely no effect on the viability of planktonic P. aeruginosa PA14 cells compared to that of ciprofloxacin alone (Fig. 6). In contrast, in SHX-treated samples, the combination of 0.8 μg/ml of 1018 with 40 ng/ml of ciprofloxacin led to significantly increased susceptibility compared to that to ciprofloxacin alone (Fig. 6). Furthermore, SHX moderately decreased susceptibility to ciprofloxacin by about 2-fold (Fig. 6).
FIG 6.

Low concentrations of peptide 1018 synergized with ciprofloxacin against SHX-treated P. aeruginosa PA14 planktonic cells. Liquid cultures of P. aeruginosa PA14 were prepared in the presence or absence of SHX and treated with 40 ng/ml of ciprofloxacin or 40 ng/ml of ciprofloxacin combined with 0.8 μg/ml of 1018. Cell counts were performed after 23 h of treatment. Statistical significance was calculated using Student's t test comparing the SHX-treated and SHX-untreated groups (*, P < 0.05).
To better understand the synergy between antibiofilm peptide 1018 and the antibiotics used in this study, we extracted RNA from cells harvested from P. aeruginosa PA14 mature biofilms that were either untreated or treated with 0.8 μg/ml of 1018. Next, we performed qRT-PCR to assess the effect of low levels of the peptide on the expression of genes that are well-known antibiotic targets and/or involved in antibiotic resistance mechanisms (Table 3). The expression of the genes for the ciprofloxacin target gyrase gyrA and the other gyrase subunit, gyrB (16), was significantly downregulated by about 5-fold in the presence of the peptide (Table 3). Imipenem and ceftazidime are known to bind penicillin-binding proteins (PBPs) (17, 18), including PBP 7 (encoded by the pbpG gene) and PBP 3 (encoded by the ftsI and pbpB genes). The genes for these two proteins were also downregulated in peptide-treated biofilm samples compared to their regulation in untreated samples (Table 3). Treatment with peptide 1018 also led to an ∼5-fold decrease in the expression of pelB, a gene involved in Pel matrix polysaccharide biosynthesis, biofilm formation, and aminoglycoside resistance (19) (Table 3). Moreover, the nirS gene, which is responsible for nitric oxide synthesis and is involved in biofilm formation (20), was downregulated by 4.5-fold in peptide-treated biofilms (Table 3).
TABLE 3.
Downregulation of antibiotic targets in biofilms treated with peptide 1018a
| Gene | Mean ± SD fold change | Function | Biological role |
|---|---|---|---|
| gyrA | −4.74 ± 2.2 | DNA gyrase subunit A | DNA replication |
| gyrB | −5.00 ± 0.45 | DNA gyrase subunit B | DNA replication |
| pbpG | −4.53 ± 2 | d-Alanyl-d-alanine endopeptidase | Cell wall |
| ftsI | −2.63 ± 1.0 | Penicillin-binding protein 3 | Cell wall |
| pelB | −4.83 ± 0.43 | Biofilm matrix protein | Exopolysaccharide synthesis |
| nirS | −4.5 ± 2 | Nitrite reductase precursor | Denitrification |
Results of qRT-PCR performed on RNA extracted from flow cell biofilms. We evaluated the dysregulation of genes involved in antibiotic resistance: gyrA and gyrB are known targets of ciprofloxacin, pbpG and ftsI are involved in ceftazidime and imipenem resistance, and pelB is involved in tobramycin resistance. The gene nirS is involved in biofilm formation. We determined the fold decrease in gene expression by mature biofilms treated with 0.8 μg/ml of peptide 1018 for 24 h compared to the level of expression by untreated mature biofilms.
DISCUSSION
The increase in antibiotic resistance during biofilm infections is a substantial problem in public health, and solutions need to be provided. Several strategies have been proposed over the years in an attempt to efficiently treat bacterial biofilms, including prevention, weakening, disruption, or killing (21). We recently demonstrated that antibiofilm peptide 1018 inhibited biofilm formation and eradicated preformed biofilms formed by Gram-negative and Gram-positive multidrug-resistant pathogens (8). In addition, 1018 was shown to disperse mature biofilms at low concentrations (0.8 μg/ml) and kill biofilm cells at higher concentrations (10 μg/ml) (8). To evaluate the clinical potential of this broad-spectrum antibiofilm peptide, we tested here its potential synergy with conventional antibiotics.
Using checkerboard assays and performing confirmatory studies using the flow cell methodology, we demonstrated that peptide 1018, in combination with 4 conventional antibiotics used in the clinic, prevented biofilm formation and eradicated preformed biofilms formed by bacterial pathogens that are notorious for their high antibiotic resistance. Interactions between the peptide and antibiotics led to synergy in half of the tested combinations and substantial reductions (up to 64-fold) in the concentrations of antibiotic required to inhibit biofilm formation (Table 1). This might have significant potential in treating chronic infections that usually involve biofilms and are highly recalcitrant to treatment. Indeed, to treat chronic infections, the antibiotic dose is often increased but can still fail to clear the infection, thus leading to high-level antibiotic resistance (3). The use of a peptide like 1018 may provide a solution to this problem due to its ability to potentiate the antibiofilm activity of conventional antibiotics that are otherwise largely effective only against planktonic bacteria.
We previously observed that low concentrations of peptide 1018 (0.8 μg/ml) increased biofilm dispersal after 23 h of treatment (8). Dispersal of cells from biofilms represents a potential danger in the clinical setting, as this may lead to infection at other sites or even septic shock (2, 22). Critically when the peptide was combined with ciprofloxacin at sub-MICs, the number of live dispersed cells dropped dramatically by ∼20-fold compared to the normal level of cells dispersed from wild-type biofilms (Fig. 4). This may be due to the increase in cell death in biofilms. These results indicate that combination treatment with peptide 1018 and an antibiotic (ciprofloxacin) may be a good strategy to treat biofilm infections and avoid potential future infections derived from cells dispersed from biofilms.
Mechanistic studies indicated that overproduction of (p)ppGpp led to the increased susceptibility of planktonic cells to the combination of peptide and ciprofloxacin, indicating that it is the action of the peptide on (p)ppGpp production in biofilms that mediates the synergistic effect (8). In addition, qRT-PCR studies revealed that low levels of peptide 1018 (0.8 μg/ml) led to the downregulation of genes involved in the mechanism of action of antibiotics as well as in the formation of biofilms, likely making biofilms more susceptible to antibiotics (Table 3).
In conclusion, we have demonstrated the ability of antibiofilm peptide 1018 to synergize with conventional antibiotics to inhibit biofilm formation and disrupt preformed biofilms in a broad-spectrum manner. Mechanistically, we propose that the ability of the peptide to target (p)ppGpp leads to increased bacterial susceptibility to the antibiotic ciprofloxacin and further postulate that this may extend to other antibiotics.
ACKNOWLEDGMENTS
We thank Dao Nguyen for providing the PAO1relAspoT mutant and its parent strain.
The research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R21AI098701 and by grant MOP-74493 from the Canadian Institutes for Health Research. R.E.W.H. holds a Canada Research Chair in Health and Genomics. C.D.L.F.-N. received a scholarship from the Fundación la Caixa and Fundación Canadá (Spain).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Additionally, we have filed a provisional patent application on the use of cationic antibiofilm peptides (U.S. patent application 61/870,655).
Footnotes
Published ahead of print 30 June 2014
REFERENCES
- 1.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. 10.1126/science.284.5418.1318 [DOI] [PubMed] [Google Scholar]
- 2.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95–108. 10.1038/nrmicro821 [DOI] [PubMed] [Google Scholar]
- 3.Høiby N, Ciofu O, Johansen HK, Song ZJ, Moser C, Jensen PØ, Molin S, Givskov M, Tolker-Nielsen T, Bjarnsholt T. 2011. The clinical impact of bacterial biofilms. Int. J. Oral Sci. 3:55–65. 10.4248/IJOS11026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Römling U, Balsalobre C. 2012. Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med. 272:541–561. 10.1111/joim.12004 [DOI] [PubMed] [Google Scholar]
- 5.de la Fuente-Núñez C, Reffuveille F, Fernández L, Hancock REW. 2013. Bacterial biofilm development as a multicelular adaptation: antibiotic resistance and new therapeutic strategies. Curr. Opin. Microbiol. 16:580–589. 10.1016/j.mib.2013.06.013 [DOI] [PubMed] [Google Scholar]
- 6.Driffield K, Miller K, Bostock JM, O'Neill AJ, Chopra I. 2008. Increased mutability of Pseudomonas aeruginosa in biofilms. J. Antimicrob. Chemother. 61:1053–1056. 10.1093/jac/dkn044 [DOI] [PubMed] [Google Scholar]
- 7.Hausner M, Wuertz S. 1999. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl. Environ. Microbiol. 65:3710–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de la Fuente-Núñez C, Reffuveille F, Haney EF, Strauss SK, Hancock REW. 2014. Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog. 10:e1004152. 10.1371/journal.ppat.1004152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986. 10.1126/science.1211037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiegand I, Hilpert K, Hancock REW. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3:163–175. 10.1038/nprot.2007.521 [DOI] [PubMed] [Google Scholar]
- 11.Fernández L, Gooderham WJ, Bains M, McPhee JB, Wiegand I, Hancock REW. 2010. Adaptive resistance to the “last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob. Agents Chemother. 54:3372–3382. 10.1128/AAC.00242-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall MJ, Middleton RF, Westmacot D. 1983. The fractional inhibitory concentration (FIC) index as a measure of synergy. J. Antimicrob. Chemother. 11:427–433. 10.1093/jac/11.5.427 [DOI] [PubMed] [Google Scholar]
- 13.de la Fuente-Núñez C, Korolik V, Bains M, Nguyen U, Breidenstein EBM, Horsman S, Lewenza S, Burrows L, Hancock REW. 2012. Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrob. Agents Chemother. 56:2696–2704. 10.1128/AAC.00064-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert P, Collier PJ, Brown MR. 1990. Influence of growth rate on susceptibility to antimicrobial agents: biofilms, cell cycle, dormancy, and stringent response. Antimicrob. Agents Chemother. 34:1865–1868. 10.1128/AAC.34.10.1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geiger T, Goerke C, Fritz M, Schäfer T, Ohlsen K, Liebeke M, Lalk M, Wolz C. 2010. Role of the (p)ppGpp synthase RSH, a RelA/SpoT homolog, in stringent response and virulence of Staphylococcus aureus. Infect. Immun. 78:1873–1883. 10.1128/IAI.01439-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levine C, Hiasa H, Marians KJ. 1998. DNA gyrase and topoisomerase IV: biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochim. Biophys. Acta 1400:29–43. 10.1016/S0167-4781(98)00126-2 [DOI] [PubMed] [Google Scholar]
- 17.Hashizume T, Ishino F, Nakagawa J, Tamaki S, Matsuhashi M. 1984. Studies on the mechanism of action of imipenem (N-formimidoylthienamycin) in vitro: binding to the penicillin-binding proteins (PBPs) in Escherichia coli and Pseudomonas aeruginosa, and inhibition of enzyme activities due to the PBPs in E. coli. J. Antibiot. (Tokyo) 37:394–400. 10.7164/antibiotics.37.394 [DOI] [PubMed] [Google Scholar]
- 18.Hayes MV, Orr DC. 1983. Mode of action of ceftazidime: affinity for the penicillin-binding proteins of Escherichia coli K12, Pseudomonas aeruginosa and Staphylococcus aureus. J. Antimicrob. Chemother. 12:119–126. 10.1093/jac/12.2.119 [DOI] [PubMed] [Google Scholar]
- 19.Colvin KM, Gordon VD, Murakami K, Borlee BR, Wozniak DJ, Wong GC, Parsek MR. 2011. The Pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 7:e1001264. 10.1371/journal.ppat.1001264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de la Fuente-Núñez C, Reffuveille F, Fairfull-Smith KE, Hancock REW. 2013. Effect of nitroxides on swarming motility and biofilm formation, multicellular behaviors in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 57:4877–4881. 10.1128/AAC.01381-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bjarnsholt T, Ciofu O, Molin S, Givskov M, Høiby N. 2013. Applying insights from biofilm biology to drug development—can a new approach be developed? Nat. Rev. Drug Discov. 12:791–808. 10.1038/nrd4000 [DOI] [PubMed] [Google Scholar]
- 22.Hall-Stoodley L, Stoodley P. 2005. Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 13:7–10. 10.1016/j.tim.2004.11.004 [DOI] [PubMed] [Google Scholar]




