Abstract
Here, we report the first autochthonous cases of infections caused by blaNDM-5 New Delhi metallo-β-lactamase-producing Escherichia coli strains recovered from urine and blood specimens of three patients from Algeria between January 2012 and February 2013. The three isolates belong to sequence type 2659 and they coexpress blaCTX-M-15 with the blaTEM-1 and blaaadA2 genes.
TEXT
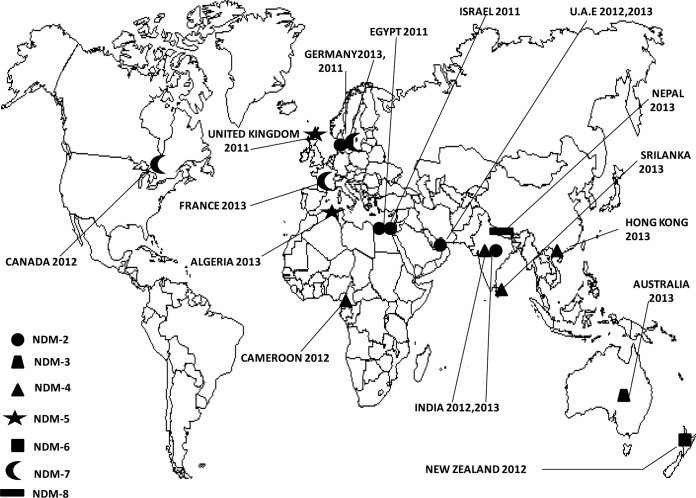
Escherichia coli is one of the most common causative agents of infection in humans, and the emergence of resistance to third-generation cephalosporins by extended-spectrum β-lactamases (ESBLs) has led to an increased use of carbapenem compounds (1). The growing incidence of resistance to carbapenems among Enterobacteriaceae is of major concern worldwide (1). Carbapenemase producers are mainly identified in Klebsiella pneumoniae and, to a lesser extent, in E. coli and other enterobacterial species. Carbapenemases are classified into three different classes (A, B, and D) and are now a serious problem due to their rapid spread in Enterobacteriaceae (1). Among the newly emerged β-lactamases in the world, New Delhi metallo-β-lactamase (NDM) represents the latest threat for public health (2). It was first reported from K. pneumoniae and E. coli isolates recovered from a Swedish patient previously hospitalized in India (2). Since then, seven additional NDM variants have been described worldwide (2) (Fig. 1). New Delhi metallo-β-lactamase 1 (NDM-1), which can be produced by different Enterobacteriaceae, has been reported worldwide, including recently in Acinetobacter baumannii clinical isolates in Algeria (3). In this report, we describe the first detection of blaNDM-5-containing New Delhi metallo-β-lactamase-producing E. coli clinical isolates in Algeria.
FIG 1.
Geographic distribution of NDM variants detected worldwide.
A total of 105 consecutive and nonduplicate E. coli clinical isolates were recovered from hospitalized and nonhospitalized patients at the University Hospital of Annaba, Algeria, and were screened for carbapenem resistance between January 2012 and February 2013. During the study period, out of the 105 isolates, a total of 3 isolates harbored the blaNDM-5 gene. Out of these 3 blaNDM-5-positive isolates, one was isolated from the blood of a 5-month-old child hospitalized in the pediatric ward, while two were isolated from urine samples from a 63-year-old man and a 75-year-old man. The isolates were identified using the Bruker Daltonics Microflex matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometer (Bremen, Germany), as previously described (3).
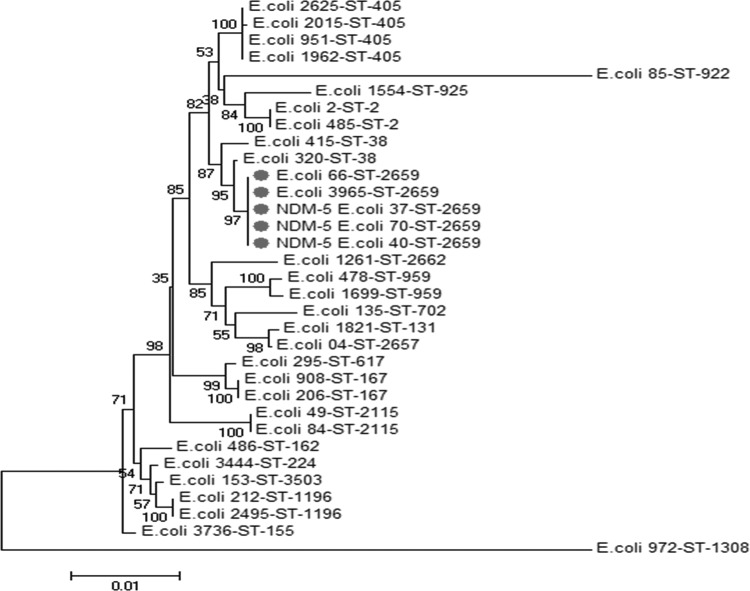
These three isolates were resistant to all β-lactams, with an imipenem MIC of >32 μg/ml and a high level of resistance to aminoglycosides and fluoroquinolones. They were susceptible only to tigecycline, fosfomycin, and colistin. Thus, NDM-5-harboring strains might be highly multidrug resistant, and a previous report on NDM-5-producing E. coli sequence type 648 (ST648) (GenBank accession no. JN104597) demonstrated that the strain was resistant to all available antimicrobials except tigecycline and colistin (4), and we observed the same phenotype for our three isolates. Carbapenemase activity was determined using the modified Hodge test and the disk approximation tests using EDTA (4) and was also confirmed using the recently described new MALDI-TOF MS carbapenemase assay, as published previously (5). Therefore, this method has emerged as a powerful and cost-effective tool for the rapid detection of carbapenem resistance (5). In our study, the presence of the blaNDM-5 gene in the three isolates was confirmed by real-time PCR and further verified by standard PCR and sequencing (5). In addition, the three isolates coexpressed the blaCTX-M-15 gene with the blaTEM-1 and blaaadA2 genes. In order to study the transferability of the resistance phenotype, a conjugation experiment was performed between our clinical donor isolates and azide-resistant E. coli strain J53 as a recipient. The transconjugants were selected on MacConkey agar plates containing 2 μg/ml imipenem and 100 μg/ml sodium azide, as described previously (2). PCR amplification of the plasmid DNA and susceptibility profiling showed that all transconjugants became resistant to all tested antibiotics, except aztreonam and ciprofloxacin (Table 1), and they acquired the blaNDM-5, blaTEM-1, and blaaadA2 genes. The result clearly revealed that these resistance genes were transferred via a plasmid that also confers resistance to most β-lactams, including imipenem and all aminoglycosides. Multilocus sequence typing (MLST) was performed to characterize the genetic relationship of the E. coli strains; it was carried out on 30 E. coli strains, in addition to the three NDM-5-positive isolates, using seven housekeeping genes (adk, fumC, icd, purA, gyrB, recA, and mdh), as described at the E. coli MLST Database (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli) (Fig. 2). The results revealed that the E. coli carbapenemase-positive strains belong to sequence type 2659, which was different from the NDM-5-producing ST648 E. coli strain (GenBank accession no. JN104597) isolated from the United Kingdom (6). This is the first reported ST2659 E. coli strain producing the NDM-5 carbapenemase-encoding gene.
TABLE 1.
Antimicrobial susceptibility of the three clinical E. coli isolates producing the blaNDM-5 gene and their transconjugants
| Antibiotic(s) | MIC (μg/ml) for: |
||
|---|---|---|---|
| E. coli J53 | E. coli NDM-5 | E. coli J53–NDM-5 transconjugants | |
| Ampicillin | 2 | >256 | >256 |
| Amoxicillin-clavulanate | 2 | >256 | >256 |
| Piperacillin-tazobactam | 1 | >256 | >256 |
| Cefoxitin | 4 | >256 | >256 |
| Cefotaxime | 2 | >256 | >256 |
| Cefuroxime | 4 | >256 | >256 |
| Ceftazidime | 0.064 | >256 | >256 |
| Aztreonam | 0.094 | >256 | 0.094 |
| Imipenem | 0.25 | >32 | >32 |
| Gentamicin | <1 | >512 | >512 |
| Amikacin | <4 | >512 | >512 |
| Tobramycin | <2 | >512 | >512 |
| Ciprofloxacin | 0.032 | >32 | 0.032 |
FIG 2.
Concatenated phylogenetic tree showing the molecular relationships of the seven genes analyzed (adk, fumC, icd, purA, gyrB, recA, and mdh) for 33 clinical E. coli isolates, including the three NDM-5-positive isolates. Gray circles indicate E. coli strains belonging to sequence type 2659. The number shown at each node indicates the bootstrap level from 500 replicates.
ST2659 has been reported only once, from a domesticated cat in Germany (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli). The reservoir of this gene in Algeria is unknown, but several contamination sources can be implicated. Various mobile genetic structures (insertion sequences, integrons, and transposons) can play an important role in the horizontal transfer of the blaNDM gene between different species of bacteria, such as from Acinetobacter spp. to E. coli (6). Travelers contribute significantly to the global movement of microbes and resistance genes (6). Although nosocomial transmission of the blaNDM gene has occurred in many countries (6), traveling to the Indian subcontinent is a significant risk factor for infection with an NDM-producing strain (6). The emergence of NDM-1-producing strains was linked to Asia and the Balkans (6). However, it has also been reported in autochthonous human cases worldwide (6). In contrast to other countries, where the blaNDM gene has been identified mainly in Enterobacteriaceae, the blaNDM gene has been reported only in A. baumannii clinical isolates from Algeria (3, 7). However, other types of carbapenemase-acquiring isolates have been reported in Algeria, such as blaVIM in Enterobacteriaceae (8) and P. aeruginosa (9). The blaOXA-24, blaOXA-23, and blaOXA-58 genes have been also reported in Annaba and Tlemcen, Algeria (3, 10, 11). However, no reports are available on isolates of NDM-producing E. coli from Algeria and, to the best of our knowledge, we report here the first blaNDM-5 gene in E. coli from Algeria and on the African continent.
We can conclude that the epidemiology of carbapenemase-encoding genes has changed in the African continent, and NDM gene variants efficiently disseminate worldwide. These cases should raise public concern once again over the increasing incidence of highly multidrug-resistant NDM-harboring strains.
Nucleotide sequence accession numbers.
The nucleotide sequences of the three blaNDM-5-containing E. coli strains have been deposited in the GenBank database under accession no. KF408072, KF408073, and KF408074.
ACKNOWLEDGMENTS
We thank Linda Hadjadj for technical assistance and Leanne McIlreavey for careful English corrections.
This work was partly funded by the Centre National de la Recherche Scientifique (CNRS) and IHU Méditerranée Infection.
We declare no conflicts of interest.
All authors read and approved the manuscript.
Footnotes
Published ahead of print 30 June 2014
REFERENCES
- 1.Walsh TR, Toleman MA, Poirel L, Nordmann P. 2005. Metallo-beta-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306–325. 10.1128/CMR.18.2.306-325.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054. 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mesli E, Berrazeg M, Drissi M, Bekkhoucha SN, Rolain JM. 2013. Prevalence of carbapenemase-encoding genes including New Delhi metallo-β-lactamase in Acinetobacter species, Algeria. Int. J. Infect. Dis. 17:739–743. 10.1016/j.ijid.2013.02.024 [DOI] [PubMed] [Google Scholar]
- 4.Hornsey M, Phee L, Wareham DW. 2011. A novel variant, NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob. Agents Chemother. 55:5952–5954. 10.1128/AAC.05108-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kempf M, Bakour S, Flaudrops C, Berrazeg M, Brunel JM, Drissi M, Mesli E, Touati A, Rolain JM. 2012. Rapid detection of carbapenem resistance in Acinetobacter baumannii using matrix-assisted laser desorption ionization–time of flight mass spectrometry. PLoS One 7:e31676. 10.1371/journal.pone.0031676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Wu C, Zhang Q, Qi J, Liu H, Wang Y, He T, Ma L, Lai J, Shen Z, Liu Y, Shen J. 2012. Identification of New Delhi metallo-β-lactamase 1 in Acinetobacter lwoffii of food animal origin. PLoS One 7:e37152. 10.1371/journal.pone.0037152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulanger A, Naas T, Fortineau N, Figueiredo S, Nordmann P. 2012. NDM-1-producing Acinetobacter baumannii from Algeria. Antimicrob. Agents Chemother. 10.1128/AAC.05653-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robin F, Aggoune-Khinache N, Delmas J, Naim M, Bonnet R. 2010. Novel VIM metallo-β-lactamase variant from clinical isolates of Enterobacteriaceae from Algeria. Antimicrob. Agents Chemother. 54:466–470. 10.1128/AAC.00017-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Touati M, Diene SM, Dekhil M, Djahoudi A, Racherache A, Rolain JM. 2013. Dissemination of a class I integron carrying VIM-2 carbapenemase in Pseudomonas aeruginosa clinical isolates from a hospital intensive care unit in Annaba, Algeria. Antimicrob. Agents Chemother. 57:2426–2427. 10.1128/AAC.00032-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Touati M, Diene SM, Racherache A, Dekhil M, Djahoudi A, Rolain JM. 2012. Emergence of blaOXA-23 and blaOXA-58 carbapenemase-encoding genes in multidrug-resistant Acinetobacter baumannii isolates from University Hospital of Annaba, Algeria. Int. J. Antimicrob. Agents 40:89–91. 10.1016/j.ijantimicag.2012.03.017 [DOI] [PubMed] [Google Scholar]
- 11.Bakour S, Kempf M, Touati A, Ait Ameur A, Haouchine D, Sahli F, Rolain JM. 2012. Carbapenemase-producing Acinetobacter baumannii in two university hospitals in Algeria. J. Med. Microbiol. 61:1341–1343. 10.1099/jmm.0.045807-0 [DOI] [PubMed] [Google Scholar]