Abstract
Candida kefyr is an increasingly reported pathogen in patients with hematologic malignancies. We studied a series of bloodstream isolates that exhibited reduced echinocandin susceptibilities (RES). Clinical and surveillance isolates were tested for susceptibilities to all three echinocandins, and those isolates displaying RES to one or more echinocandins were selected for molecular and biochemical studies. The isolates were analyzed for genetic similarities, and a subset was analyzed for mutations in the echinocandin target gene FKS1 and glucan synthase echinocandin sensitivities using biochemical methods. The molecular typing did not indicate strong genetic relatedness among the isolates except for a series of strains recovered from a single patient. Two unrelated isolates with RES had previously uncharacterized FKS1 mutations: R647G and deletion of amino acid 641 (F641Δ). Biochemical analysis of the semipurified R647G glucan synthase generated differential echinocandin sensitivity (resistance to micafungin only), while the deletion of F641 resulted in a glucan synthase highly insensitive to all three echinocandins. The consecutive isolates from a single patient with RES all harbored the common S645P mutation, which conferred resistance to all three echinocandins. The MIC values paralleled the glucan synthase inhibition kinetic data, although the S645P isolates displayed relatively higher susceptibility to caspofungin (2 μg/ml) than the other two echinocandins (>8 μg/ml). These findings highlight novel and common FKS1 mutations in C. kefyr isolates. The observation of differential susceptibilities to echinocandins may provide important mechanistic insights for echinocandin antifungals.
INTRODUCTION
Candida kefyr (teleomorph: Kluyveromyces marxianus) has been reported as a rare but potentially increasing cause of invasive candidiasis (IC) (1–3), especially in patients with hematologic malignancies (2, 4–6). Isolates of this species have been noted to develop reduced susceptibility to echinocandins, although the majority examined in population analyses remain susceptible (7).
Decreased susceptibility to echinocandins is associated with mutations and polymorphic changes in FKS1, the gene that encodes the target enzyme, β-1,3-d-glucan synthase (Fks1p) (8–14). Fks1p is a plasma membrane protein with several regions exposed to the environment on the outer leaflet of the membrane phospholipid layer (13, 15). The catalytic Fks subunits (Fks1p, Fks2p, and Fks3p) together with the regulatory subunit, Rho1p, compose the glucan synthase (GS) complex (16, 17). Reduced echinocandin susceptibilities (RES) are associated with amino acid alterations at mainly two regions or hot spots (HS) of Fks1p (Fksp2 in Candida glabrata). Both of these regions (HS1 and HS2) are highly conserved and thought to be important for the interaction of the enzyme with echinocandins, although the mechanism of resistance remains unclear (13, 18). A third region of FKS1, HS3, has also been shown to affect the susceptibility to echinocandins (15, 18). HS3 maps downstream and near HS1 and also codes for the amino acids predicted to reside in an environment-exposed region of Fks1p.
We recently observed an increase in the number of C. kefyr IC cases in patients with hematologic malignancies at the John Hopkins Hospital (JHH). This observation prompted a retrospective review of all C. kefyr infections in two major hematologic wards between 2004 and 2010 (19). Notable findings of that study included a striking seasonality in isolate recovery, with high rates during summer months. The observation of reduced echinocandin antifungal susceptibilities prompted this detailed study of mechanisms.
MATERIALS AND METHODS
C. kefyr clinical isolates.
The study was approved by the JHH institutional review board (IRB). All available C. kefyr clinical isolates from patients between 1 January 2009 and 31 December 2012 were recovered from the JHH mycology laboratory. Patients with hematologic malignancies receiving intensive chemotherapy at JHH have fungal surveillance cultures (FSC) of throat and rectal swabs or stool specimens performed systematically on admission and weekly thereafter until their discharge. Clinical data, including demographics, the type of the underlying hematologic malignancy, the treatment regimen, and the administration of antifungal agents within 30 days prior to the first positive C. kefyr isolate, were summarized.
Other strains and media.
C. kefyr type strain ATCC 4922 (American Type Culture Collection, Manassas, VA), isolated from buttermilk (20), was used as an unrelated control for genotyping and echinocandin MIC determinations. Candida albicans clinical isolate SC5314 (21) was used as an outlier control. All yeasts were routinely maintained or propagated on yeast extract-peptone-dextrose (YPD) liquid or agar plate medium (1% yeast extract, 2% peptone, and 2% dextrose) and grown at 30°C.
Echinocandin susceptibility testing.
All strains were grown on Sabouraud dextrose agar at 35°C for 24 h prior to testing for echinocandin susceptibilities by the broth microdilution assay as described by Clinical and Laboratory Standards Institute (CLSI) document M27-A3 (22) (concentration range, 0.015 to 8.0 μg/ml). Echinocandins were obtained from their respective manufacturers (caspofungin, Merck & Co., Rahway, NJ; micafungin, Astellas Pharma Inc., Deerfield, IL; anidulafungin, Pfizer, Inc., New York, NY) and suspended in dimethyl sulfoxide (DMSO) (Sigma, St. Louis, MO) prior to dilution into RPMI medium for MIC assays. The MIC value was determined as the concentration that produced a prominent reduction in turbidity (≥50% reduction in growth) at 24 h. The strains were tested at least twice on different days. Since we repeatedly observed elevated MICs above the published epidemiological cutoff values for C. kefyr (23), the modal MICs were calculated using strains ATCC 4922, C115, C117, and C130 as references for the remaining clinical isolates. The strains were chosen because these harbored FKS HS amino acid sequences (see below) of known susceptible C. albicans (12, 13) and C. kefyr (14) strains. The modal MIC values were derived from 2 to 10 measurements per isolate.
C. kefyr genotyping.
A repetitive sequence-based PCR method developed for Candida rugosa (24) was adapted to fingerprint the C. kefyr clinical isolates. Genomic DNA was prepared, using a MasterPure yeast DNA purification kit (Epicentre Biotechnologies, Madison, WI), from Candida spp. grown for ∼18 h at 30°C in YPD broth cultures with shaking. Oligonucleotides Ca-21 (5′-CATCTGTGGTGGAAAGTAAAC-3′) and Ca-22 (5′-ATAATGCTCAAAGGTGGTAAG-3′) (24) were used at 1.0 μM in a PCR volume of 25 μl containing 100 ng of genomic DNA, using a TaKaRa Ex Taq kit (Clontech, Mountain View, CA) with the cold start method as per the manufacturer's recommendation. PCR conditions were initial denaturation at 94°C for 5 min, followed by 35 cycles with a ramping temperature rate of 1.5°C/s for denaturation at 94°C for 15 s, annealing at 51°C for 30 s, and extension at 72°C for 30 s, and a final extension step at 72°C for 5 min. The amplicons were resolved in 6% acrylamide Tris-borate-EDTA (TBE) gels (Invitrogen/Life Technologies Corporation, Grand Island, NY), stained with ethidium bromide, and visualized under UV light. DNA amplicon banding images were captured with a UVP GelDoc-It imaging system, and the data were imported into PyElph, an open-source software tool (sourceforge.net/projects/pyelph/files/releases/) (25) to generate dendrograms, using the unweighted-pair group method of arithmetic mean (UPGMA).
FKS1 HS sequencing.
HS1 and HS2 were amplified from C. kefyr genomic DNA using C. albicans FKS1 oligonucleotides FKS1-HS1F (5′-AATGGGCTGGTGCTCAACAT-3′), FKS1-HS1R (5′-CCTTCAATTTCAGATGGAACTTGATG-3′), FKS1-HS2F (5′-AAGATTGGTGCTGGTATGGG-3′), and FKS1-HS2R (5′-TAATGGTGCTTGCCAATGAG-3′), as described by Garcia-Effron et al. (26). Each sample reaction mixture contained 0.5 μM primers and 200 ng of genomic DNA in a 25-μl volume and was performed using a JumpStart REDTaq kit (Sigma, St. Louis, MO). The PCR conditions were initial denaturation at 94°C for 2 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 2 min, extension at 72°C for 3 min, and final extension at 72°C for 5 min. The amplicons were column purified (Qiagen Sciences Inc., Germantown, MD) and sequenced using an AB3730XL DNA analyzer (AB Biosystems, Fremont, CA) at the DNA Analysis Facility of Johns Hopkins University (JHU). Multiple sequence alignments were conducted using ClustalW and T-Coffee (MacVector v. 12.6.0; MacVector, Inc., Cary, NC), while pairwise sequence alignments were done with EMBOSS Matcher (www.ebi.ac.uk).
Glucan synthase echinocandin inhibition assays.
The strains were grown with vigorous shaking at 37°C to the early stationary phase in modified YPD (2% yeast extract, 4% Bacto Peptone, and 4% dextrose) broth, and cells were collected by centrifugation. Cell disruption, membrane protein extraction, and partial 1,3-β-d-glucan synthase purification by-product entrapment were performed as previously described (27). The sensitivity to echinocandin drugs was measured in a polymerization assay using a 96-well multiscreen HTS filtration system (Millipore Corporation, Bedford, MA) in a final volume of 100 μl, as previously described (11). Serial dilutions of the three echinocandin drugs (0.01 to 10,000 ng/ml) were used to determine the inhibition kinetics yielding 50% inhibitory concentration (IC50) values. Control reactions were performed in the presence of 1% DMSO. The reactions were initiated by the addition of partially purified glucan synthase. Inhibition profiles and IC50s were determined using a sigmoidal response (variable-slope) curve-fitting algorithm or two-site competition fitting algorithm with GraphPad Prism software (v. 4.0; GraphPad Software, Irvine, CA).
Nucleotide sequence accession numbers.
The partial FKS nucleotide sequence data for each isolate were deposited at GenBank (www.ncbi.nlm.nih.gov/GenBank/) and assigned the following accession numbers (in parentheses, HS1 and HS2, respectively): ATCC 4922 (KJ685779 and KJ685792), C113 (KJ685780 and KJ685793), C114 (KJ685781 and KJ685794), C115 (KJ685782 and KJ685795), C116 (KJ685783 and KJ685796), C117 (KJ685784 and KJ685797), C130 (KJ685785 and KJ685798), C131 (KJ685786 and KJ685799), C132 (KJ685787 and KJ685800), C133 (KJ685788 and KJ685801), C134 (KJ685789 and KJ685802), C135 (KJ685790 and KJ685803), and C136 (KJ685791 and KJ685804).
RESULTS
Reduced echinocandin susceptibilities in C. kefyr isolates.
Twenty-five C. kefyr isolates were recovered from 17 patients (Table 1): 11 blood and 14 FSC isolates from 7 and 14 patients, respectively. One patient (number 17) had 7 sequential isolates retrieved from blood (n = 4) and stool (n = 3) cultures. All patients with C. kefyr IC had an underlying diagnosis of acute myelogenous leukemia (AML) and had received prior treatment with multiple antifungal agents. In 6 of 7 (86%) patients with candidemia, C. kefyr isolates had elevated MICs to one or multiple echinocandins (relative to susceptible stains; see modal MIC values in Table 1). In 2 patients, bloodstream isolates were resistant to all echinocandins, whereas 1 patient had a C. kefyr isolate (C113) that displayed differential susceptibilities to the echinocandins (micafungin MIC, 4.0 μg/ml; caspofungin MIC, 0.125 μg/ml; and anidulafungin MIC, 0.25 μg/ml). All 3 patients had received micafungin (100 mg/day) as the primary prophylaxis or empirical treatment within 30 days prior to a positive culture (median, 14 days; range, 8 to 20 days). In contrast, only 2 of the remaining 4 patients with IC who had low echinocandin MICs had been preexposed to micafungin.
TABLE 1.
JHH Candida kefyr isolates and echinocandin susceptibilities
| Patient no. | Hematologic malignancya | Chemotherapyb | Isolate | Source | Date of culture (mo and yr) | Treatment with antifungals ≤30 days prior to positive culture (days)c |
MICd |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MICA | FLU | VOR | LAMB | ANID | CAS | MICA | ||||||
| 1 | AML (new) | FLAM | C113 | Blood | Jul 2009 | 8 | No | No | No | 0.25 | 0.125 | 4 |
| 2 | AML (new) | AcDVP16 | C124 | Mucosa | Jul 2009 | 20 | No | No | No | 0.5 | 0.125 | 2 |
| 3 | MM (relapse) | Othere | C125 | Mucosa | Jul 2009 | No | No | No | No | 0.06 | 0.06 | 0.03 |
| 4 | AML (new) | AcDVP16 | C126 | Mucosa | Jul 2009 | 11 | No | No | No | 0.06 | 0.06 | 0.25 |
| 5 | AML (new) | FLAM | C127 | Mucosa | Jul 2009 | 1 | No | No | 9 | 0.06 | 0.03 | 0.25 |
| 6 | Pre-B cell ALL | Otherf | C128 | Mucosa | Jul 2009 | 4 | No | No | No | 0.06 | 0.06 | 0.25 |
| 7 | AML (new) | None | C129 | Mucosa | Jul 2009 | 6 | No | No | No | 0.06 | 0.06 | 0.25 |
| 8 | AML (relapse) | Otherg | C112 | Blood | Aug 2009 | No | No | No | No | 0.03 | 0.06 | 0.25 |
| C115 | Blood | Jul 2010 | No | No | No | No | 0.25 | 0.25 | 0.5 | |||
| 9 | AML (new) | FLAM | C123 | Mucosa | Aug 2009 | 10 | No | 7 | No | 0.5 | 0.125 | 1 |
| 10 | AML (relapse) | FLAM | C117 | Blood | Oct 2009 | No | 10 | No | No | 0.06 | 0.06 | 0.25 |
| 11 | AML (new) | FLAM | C122 | Mucosa | Dec 2009 | No | No | 2 | 14 | 0.06 | 0.06 | 0.125 |
| 12 | AML (new) | AcDVP16 | C116 | Blood | Aug 2010 | 20 | 5 | 2 | No | >8.0 | 4 | >8.0 |
| 13 | AML (relapse) | FLAM | C119 | Mucosa | Sep 2010 | No | No | 8 | 21 | 0.06 | 0.06 | 0.125 |
| C120 | Mucosa | Sep 2010 | 8 | No | 3 | 30 | 0.06 | 0.06 | 0.125 | |||
| 14 | GS (relapse) | Otherg | C121 | Mucosa | Sep 2010 | 12 | No | 11 | No | 0.06 | 0.06 | 0.125 |
| 15 | AML (relapse) | AcDVP16 | C118 | Blood | May 2011 | 14 | No | No | 23 | 0.25 | 0.25 | 0.5 |
| 16 | AML (relapse) | Otherh | C114 | Blood | Feb 2010 | 18 | 7 | No | 9 | 0.5 | 0.25 | 1 |
| 17 | AML (relapse) | AcDVP16 | C130 | Stool | Oct 2012 | 1 | No | No | No | 0.03 | 0.06 | 0.25 |
| C131 | Stool | Oct 2012 | 8 | No | No | No | 1 | 0.25 | 1 | |||
| C132 | Blood | Oct 2012 | 14 | No | No | No | >8.0 | 2 | >8.0 | |||
| C133 | Blood | Oct 2012 | 14 | No | No | No | >8.0 | 2 | >8.0 | |||
| C134 | Blood | Oct 2012 | 15 | No | No | No | >8.0 | 2 | >8.0 | |||
| C135 | Blood | Oct 2012 | 16 | No | No | No | >8.0 | 2 | >8.0 | |||
| C136 | Stool | Oct 2012 | 17 | No | No | No | >8.0 | 2 | >8.0 | |||
| C. kefyri | 0.125 | 0.25 | 0.125 | |||||||||
| Wild-type MICsj | 0.06 | 0.125 | 0.25 | |||||||||
AML, acute myelogenous leukemia; MM, multiple myeloma; ALL, acute lymphocytic leukemia; GS, granulocytic sarcoma.
FLAM, flavopiridol, cytarabine, and mitoxantrone; AcDVP16, cytarabine, daunorubicin, and etoposide.
MICA, micafungin; FLU, fluconazole; VOR, voriconazole; LAMB, liposomal amphotericin B.
ANID, anidulafungin; CAS, caspofungin.
Other: bortezomib, cyclophosphamide, and lenolidomide.
Other: polyethylene glycol (PEG)-asparaginase.
Other: Investigational agent, topotecan, and carboplatin.
Other: Chk-1 inhibitor and clofarabine.
C. kefyr ATCC-4922.
Modal values were calculated using MICs observed for C. kefyr isolates ATCC-4922, C115, C117, and C130. See Materials and Methods.
There were 10 patients with C. kefyr-positive FSC with no evidence of IC. All but 2 of these isolates displayed MICs within one dilution of the epidemiological cutoff values for all echinocandins (0.25 μg/ml, 0.03 μg/ml, and 0.125 μg/ml for anidulafungin, caspofungin, and micafungin, respectively) (7). Notably, two patients (numbers 1 and 2), had C. kefyr isolates that had relatively high MICs to micafungin while those to anidulafungin and caspofungin remained relatively low. Patient number 17 had multiple stool isolates positive for C. kefyr: the first two were susceptible to all echinocandins, while the last one, recovered late during his treatment course, had high echinocandin MICs (Table 1 and details below).
Isolate genotyping and C. kefyr FKS1 analysis.
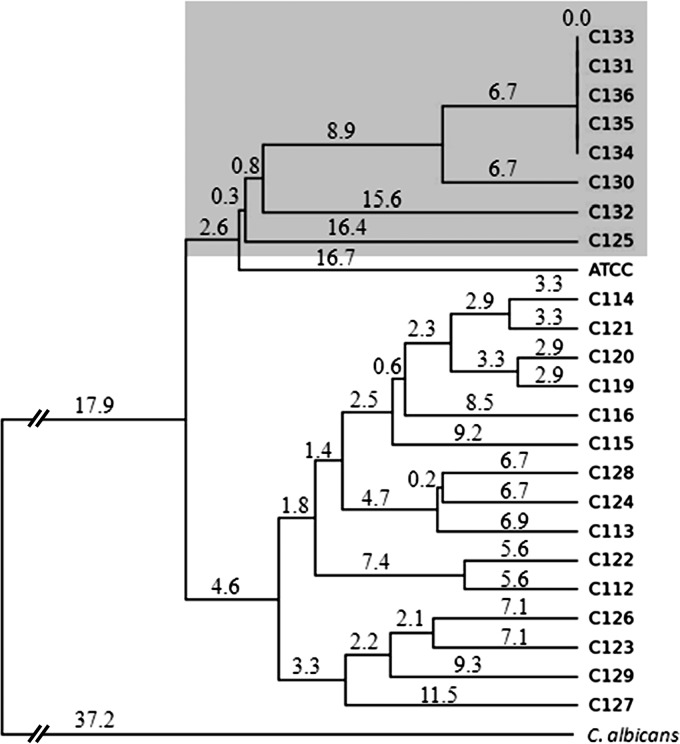
All isolates from different patients were DNA typed and found to share little genetic similarity (Fig. 1). However, sequential isolates recovered from stool and bloodstream cultures from patient 17 shared strong genetic relatedness. Individual bloodstream isolates with RES (C113 and C116), and sequential related isolates recovered from patient 17 were chosen for detailed study.
FIG 1.
Genetic relatedness of C. kefyr isolates shown by a dendrogram and tree analysis of all C. kefyr isolates. Genetic relatedness was assessed from the Rep-PCR amplicon patterns using the unweighted-pair group method with arithmetic mean (UPGMA) (35) generated by PyElph software. The genetic distances are shown above the branches. The serial isolates recovered from a single patient are highlighted within a gray shaded box. ATCC, C. kefyr ATCC 4922.
The C. kefyr FKS1 homolog (CkFKS1) HS1, HS2, and HS3 regions were amplified and sequenced, using primers to C. albicans FKS1 sequences (26). Alignment of the translated sequences against the corresponding regions of echinocandin-sensitive C. albicans (SC5314) (21), two other echinocandin-sensitive C. kefyr blood isolates, and a reference C. kefyr ATCC strain (ATCC 4922) revealed amino acid changes in HS1. Isolate C113 had an amino acid change at position 647 (R647G; amino acid positions are relative to C. albicans Fksp1), while C116 was missing the codon for amino acid F641 (F641Δ) (Fig. 2). Both DNA strands of the C116 HS1 were sequenced and were in 100% agreement. C113 and C116 had amino acid sequences identical to those of echinocandin-sensitive C. kefyr strains at HS2 and HS3 (18; data not shown).
FIG 2.

Sequence analysis of C. kefyr FKS1 HS1 from two unrelated blood isolates. Two echinocandin-sensitive isolates, C115 and C117, were used as controls. The sequences were aligned (36, 37) to the homologous FKS1 region from an irrelevant C. kefyr (ATCC 4922) strain and to C. albicans (SC5314). The amino acid changes in C113 and C116 are shaded in gray. The most common mutation to confer echinocandin resistance occurs at S645 (11, 13), indicated by a number sign (#) for reference. The amino acid residue numbering (above) is relative to C. albicans Fks1p (Gsc1p/Orf19.2929p) (Candida Genome Database, http://www.candidagenome.org/).
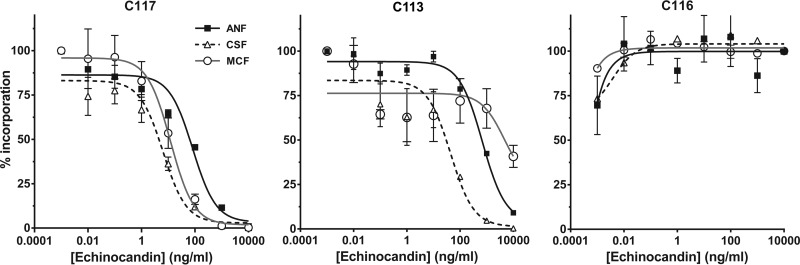
Isolates C113 and C116 were evaluated for in vitro GS inhibition with all three echinocandins (11) relative to that for the echinocandin-sensitive C117 isolate (Fig. 3). The GS from the control echinocandin sensitive C117 isolate showed characteristic inhibitory kinetic profiles with 50% inhibitory concentrations (IC50s) of 107.1, 5.4, and 11.8 ng/ml for anidulafungin, caspofungin, and micafungin, respectively. Resistant isolate C113 showed a differential GS inhibition profile with IC50s of 790.9, 33.11, and >10,000 ng/ml for anidulafungin, caspofungin, and micafungin, respectively. These IC50s corresponded to the relatively low MICs to anidulafungin and caspofungin but high MIC to micafungin (Table 1). The GS from the F641Δ strain C116, which exhibited high MICs to all echinocandins, appeared insensitive to inhibition at the highest level of drug (10,000 ng/ml) tested.
FIG 3.
Echinocandin inhibition profiles of enriched GS complexes from the susceptible and resistant C. kefyr isolates. The in vitro inhibition of product-entrapped 1,3-β-d-glucan synthase (GS) complexes isolated from three C. kefyr strains was performed to determine the 50% inhibitory concentration (IC50). Incorporation of [3H]glucose into the polymerized product was measured in GS isolated from one sensitive strain (C117) and two RES strains (C113 [R647G] and C116 [F641Δ]). The GS complexes prepared from isolates C116 and C133 were insensitive to echinocandins at up to 10,000 ng/ml. Error bars represent standard errors of the means (SEM). ANF, anidulafungin; CSF, caspofungin; MCF, micafungin.
Analysis of sequential FSC and bloodstream isolates.
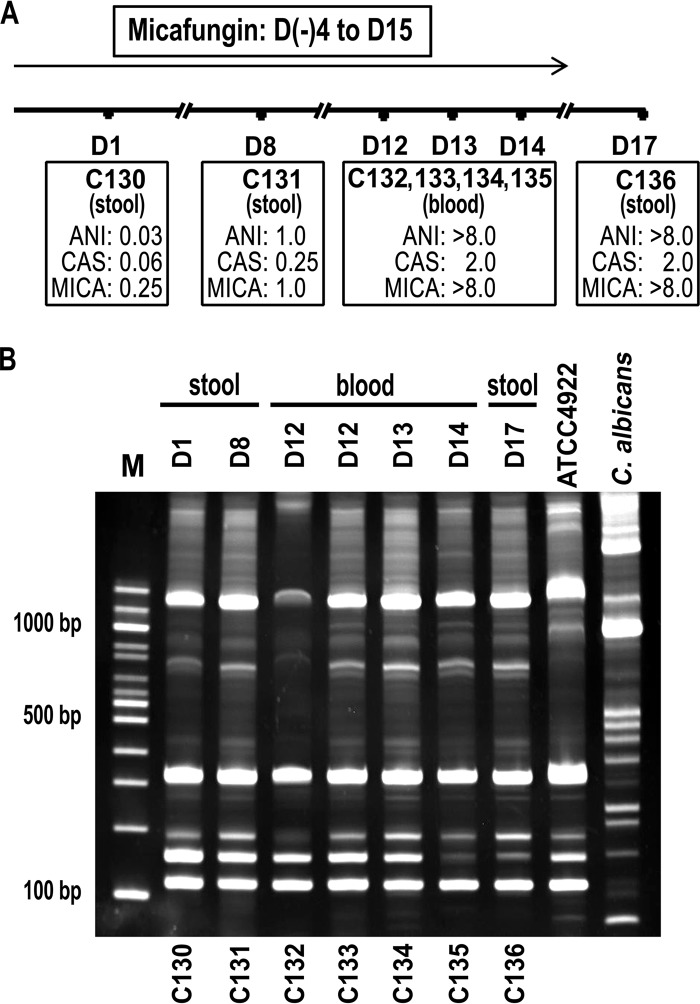
Isolates C130 to C136 recovered sequentially from stool and bloodstream cultures from patient 17 were further investigated. Isolates C130 and C133 were first recovered in the patient's stool and blood cultures 4 and 12 days, respectively, after initiation of micafungin (Fig. 4A). The repetitive PCR (Rep-PCR) results suggested that all isolates but one were closely related to the original stool isolate C130 (Fig. 1 and 4B). This dominant genotype (C130) strain that originated from the gastrointestinal tract demonstrated elevated MICs to all echinocandins within a week and was found in the bloodstream in 12 days. The same dominant genotype was repeatedly recovered from the patient's blood cultures on days 12, 13, and 14 and from his stool culture on day 17, 2 days after stopping micafungin. A second unrelated genotype (C132) was recovered from the same blood culture on the same day as the dominant strain (C133), and displayed RES to all echinocandin drugs as well.
FIG 4.
Genetic analysis of C. kefyr FKS1 HS1 from a series of isolates recovered from a single patient. (A) Time line of sequential C. kefyr isolates. Micafungin was stopped 4 days prior to the last culture. (B) The amplicons generated by Rep-PCR with primers Ca-21 and Ca-22 were separated in a 6% TBE acrylamide gel. The day and source of culture are shown above; strain designations are shown below each lane. A study-independent C. kefyr (ATCC 4922), was included for comparison as was C. albicans (right two lanes, respectively). Isolate C132 produced the same banding pattern from independent Rep-PCRs of genomic DNA replicates. Molecular size markers are shown on the left (lane M). ANI, anidulafungin; CAS, caspofungin; MICA, micafungin.
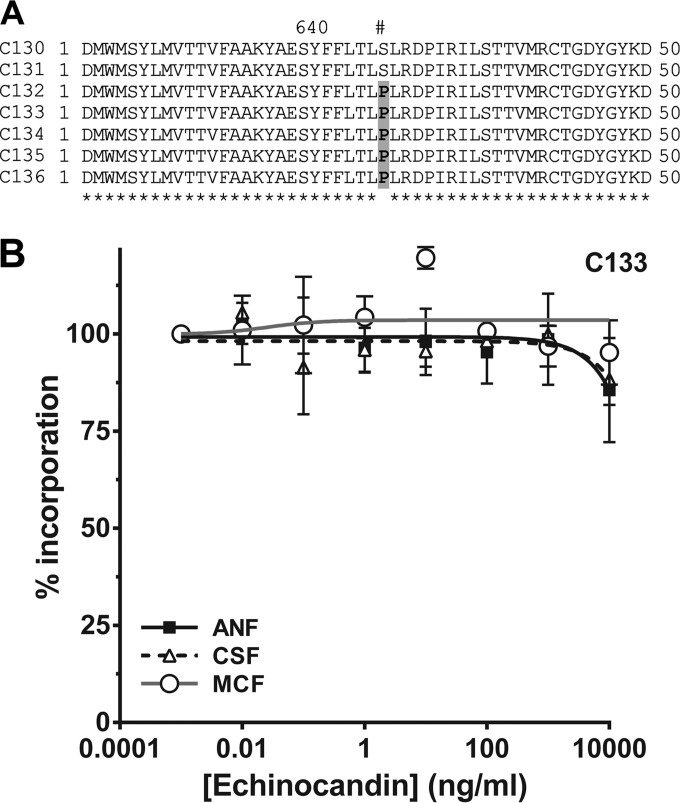
Hot spot regions (HS1, HS2, and HS3) of the CkFKS1 gene from all of these isolates were evaluated by DNA sequencing. A mutation encoding a S645P amino acid change in HS1 of the RES isolates first recovered from the blood was noted and persisted through isolates C132 to C136 (Fig. 5A). No other mutations were noted in HS1, HS2, or HS3 (data not shown). S645P corresponds as well with the most frequent substitution associated with RES in C. albicans and C. glabrata (13). Although species-specific breakpoints are not clear for C. kefyr, the S645P amino acid substitution appeared to generate a GS enzyme complex that is highly insensitive to all three echinocandins (Fig. 5B). However, it was not possible to obtain in-range IC50s, as the sigmoidal dose-response curves did not converge similarly to that observed for a true susceptible isolate (C117) (Fig. 3). Thus, high concentrations of all three echinocandins affected the extracted GS enzyme complex similarly, suggesting that differential susceptibilities measured in in vitro testing may reflect a cumulative cellular phenotype.
FIG 5.
Molecular and biochemical analysis of the CkFKS1 and GS complex from a series of isolates recovered from a single patient. (A) DNA sequence analysis of HS1 of isolates C130 to C136. RES isolates C132 to C136 harbor the most common amino acid substitution within HS1 leading to echinocandin resistance (S645P; shaded in gray). Amino acid residue numbers (above) are relative to the C. albicans Fks1p (Gsc1p/Orf19.2929p) (Candida Genome Database, http://www.candidagenome.org/). (B) Echinocandin inhibition assay of the enriched GS complex isolated from the representative isolate C133. The incorporation of [3H]glucose into the polymerized product was measured as a function of the echinocandin concentration. The GS complex from this isolate was insensitive to all three echinocandins up to 10,000 ng/ml. Error bars represent standard errors of the means (SEM).
DISCUSSION
Candida kefyr has been reported as a pathogen of increased concern, especially among people with hematologic malignancies (2, 3). Our center's experience over the last 5 years confirmed an increase in isolation of this species from patients with severe mucositis and neutropenia (19). Data generated in this analysis of FSC and bloodstream isolates reveal two important findings: (i) FKS1 HS1 mutations in C. kefyr are consistent with those in other Candida species that generate high-level echinocandin resistance, and these mutations render the GS enzyme complex as relatively resistant to drug inhibition in vitro; and (ii) a novel HS1 mutation confers relative resistance of C. kefyr to micafungin but preserves activity of the other echinocandin drugs. Testing of enzyme inhibition confirmed that this mutation reduces susceptibility at the level of the GS enzyme complex.
A survey of non-albicans Candida collected between 2001 and 2010, as part of the ARTEMIS Global Antifungal Susceptibility Program and the SENTRY Antimicrobial Surveillance Program, a multicenter and multicountry repository of fungal species, found C. kefyr isolates susceptible to all three echinocandins (23). However, echinocandin resistance within 10 days of treatment with caspofungin was recently reported for an echinocandin-susceptible C. kefyr blood isolate from a patient with AML (14). In our series, echinocandin resistance was observed in 86% of patients who developed IC, all during treatment for AML. Analysis of sequential isolates suggests that the mechanism of IC in these patients is via translocation through the gastrointestinal tract. It is likely that these patients are at particularly high risk due to prolonged neutropenia and severe mucositis, with extensive exposure to antifungals administered in a preventative or empirical fashion. Development of colonization resistance as a prerequisite for bloodstream invasion is suggested by the observation that bloodstream but not mucosal isolates exhibit high MICs in patients who had prolonged exposure to micafungin.
All of our RES C. kefyr isolates had amino acid changes in the echinocandin target Fks1p in the region encoded by HS1. Echinocandins exert their action by inhibiting the biosynthesis of the major fungal cell wall component, β-1,3-d-glucan, by interfering with the activity of the catalytic subunit of GS, Fks1p. How these antifungals interact and inhibit Fks1p activity is not entirely understood (13, 15). Although other cellular factors unrelated to Fks1p have been implicated in reduced susceptibility in some Candida species, in some Aspergillus isolates (28–31), clinical isolates of otherwise susceptible strains such as C. albicans and C. glabrata have been noted to harbor amino acid changes in Fks1p and Fks2p, respectively. All of the Fks1p homologous regions encompassed by HS1, HS2, and HS3 from different fungi are predicted to reside in a conserved domain of the enzyme embedded in the outer leaflet of the plasma membrane (15).
The molecular and biochemical analyses of C. kefyr isolates demonstrate similarities and differences in the way that echinocandins inhibit β-glucan synthesis. Substitution of F641, which is conserved in diverse yeasts and molds, has been reported in up to 33% of RES C. albicans strains (13, 26), implicating this residue as an important amino acid for Fks1p activity and echinocandin resistance. Candida kefyr isolate C116, which demonstrated reduced susceptibilities to all three echinocandins, had lost this amino acid, with a GS enzyme complex that demonstrated reduced binding to all three drugs. This is the first report to associate loss of F641 (F641Δ) in Fks1p or its orthologous amino acid with echinocandin resistance. A homologous deletion in the C. glabrata Fks2p (F659Δ) also confers RES in that species (27, 32). The relative fitness of C116 has not been studied in detail, but no obvious in vitro growth phenotype was observed. Recovery of the isolate from a patient's blood culture suggested little loss of infectivity.
The detailed study of sequential C. kefyr isolates recovered from one patient confirms that mucosal isolates can become resistant to all three echinocandins after prolonged administration of micafungin. Isolate C133 had particularly high MICs to all three echinocandins but displayed a lower MIC to caspofungin, repeatedly measured at 2 μg/ml (although a value above the susceptible range). This isolate harbored the most common HS1 amino acid substitution associated with RES in C. albicans and C. glabrata at amino acid 645 (S645P) (13). The extracted GS enzyme complex was relatively resistant to all three drugs, suggesting that other cellular factors impacted the apparent differential MIC.
One particularly interesting bloodstream isolate (C113) exhibited an unusual in vitro MIC pattern with relatively reduced susceptibility to micafungin compared to that for the other echinocandins. This isolate exhibited a novel substitution in HS1, R647G. GS inhibition studies revealed an enzyme much less susceptible to micafungin inhibition than to inhibition by either anidulafungin or caspofungin. This phenotype is similar to that observed in C. albicans isolates that harbor P649H (33). These data suggest that the amino acids R647 and P649 are particularly important for GS inhibition by micafungin. Definitive confirmation of this finding requires mutagenesis and reintroduction of the gene into an echinocandin-sensitive isolate.
An unexpected finding was the high level of echinocandin resistance (IC50 of >10,000 ng/ml) of some of the C. kefyr GS-bearing amino acid changes in HS1, and this correlated with MIC values of >8.0 μg/ml. This level of resistance has not been documented previously in the literature, and its significance is not yet understood. Our findings suggest that mutations in CkFKS1 HS1 are readily generated in vivo, do not appear to affect strain fitness, and at times produce highly resistant echinocandin strains. Similarly, Fekkar et al. (14) reported on an RES C. kefyr harboring an HS1 amino acid change (F641Y) from the bloodstream of a patient with leukemia.
In conclusion, C. kefyr is a rare (34) but emerging cause of IC in vulnerable patient populations, such as patients with hematologic malignancies (14, 15). Our data suggest high rates of echinocandin resistance among C. kefyr blood isolates in AML patients with prior drug exposure, associated with specific mutations in the CkFKS1 HS1. In addition, analysis of these isolates suggests differential echinocandin binding to the GS enzyme complex. Further study of this emerging species may provide more information on how these drugs interact with the GS enzyme complex to impart antifungal activity.
ACKNOWLEDGMENTS
We thank Kausik Datta for data analysis and manuscript preparation.
This work was supported by Merck and Company, Inc. (grant MISP 506052 to K.A.M.), Pfizer, Inc. (D.S.P.), and the National Institutes of Health (grants AI085118 to K.A.M. and AI069397 to D.S.P.).
Footnotes
Published ahead of print 30 June 2014
REFERENCES
- 1.Corpus K, Hegeman-Dingle R, Bajjoka I. 2004. Candida kefyr, an uncommon but emerging fungal pathogen: report of two cases. Pharmacotherapy 24:1084–1088. 10.1592/phco.24.11.1084.36140 [DOI] [PubMed] [Google Scholar]
- 2.Reuter CW, Morgan MA, Bange FC, Gunzer F, Eder M, Hertenstein B, Ganser A. 2005. Candida kefyr as an emerging pathogen causing nosocomial bloodstream infections in neutropenic leukemia patients. Clin. Infect. Dis. 41:1365–1366. 10.1086/497079 [DOI] [PubMed] [Google Scholar]
- 3.Sendid B, Lacroix C, Bougnoux ME. 2006. Is Candida kefyr an emerging pathogen in patients with oncohematological diseases? Clin. Infect. Dis. 43:666–667. 10.1086/506573 [DOI] [PubMed] [Google Scholar]
- 4.Borg-von Zepelin M, Kunz L, Ruchel R, Reichard U, Weig M, Gross U. 2007. Epidemiology and antifungal susceptibilities of Candida spp. to six antifungal agents: results from a surveillance study on fungaemia in Germany from July 2004 to August 2005. J. Antimicrob. Chemother. 60:424–428. 10.1093/jac/dkm145 [DOI] [PubMed] [Google Scholar]
- 5.Gomez-Lopez A, Pan D, Cuesta I, Alastruey-Izquierdo A, Rodriguez-Tudela JL, Cuenca-Estrella M. 2010. Molecular identification and susceptibility profile in vitro of the emerging pathogen Candida kefyr. Diagn. Microbiol. Infect. Dis. 66:116–119. 10.1016/j.diagmicrobio.2009.06.007 [DOI] [PubMed] [Google Scholar]
- 6.Pfaller MA, Diekema DJ. 2010. Epidemiology of invasive mycoses in North America. Crit. Rev. Microbiol. 36:1–53. 10.3109/10408410903241444 [DOI] [PubMed] [Google Scholar]
- 7.Pfaller MA, Boyken L, Hollis RJ, Kroeger J, Messer SA, Tendolkar S, Jones RN, Turnidge J, Diekema DJ. 2010. Wild-type MIC distributions and epidemiological cutoff values for the echinocandins and Candida spp. J. Clin. Microbiol. 48:52–56. 10.1128/JCM.01590-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balashov SV, Park S, Perlin DS. 2006. Assessing resistance to the echinocandin antifungal drug caspofungin in Candida albicans by profiling mutations in FKS1. Antimicrob. Agents Chemother. 50:2058–2063. 10.1128/AAC.01653-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douglas CM, D'Ippolito JA, Shei GJ, Meinz M, Onishi J, Marrinan JA, Li W, Abruzzo GK, Flattery A, Bartizal K, Mitchell A, Kurtz MB. 1997. Identification of the FKS1 gene of Candida albicans as the essential target of 1,3-beta-d-glucan synthase inhibitors. Antimicrob. Agents Chemother. 41:2471–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katiyar S, Pfaller M, Edlind T. 2006. Candida albicans and Candida glabrata clinical isolates exhibiting reduced echinocandin susceptibility. Antimicrob. Agents Chemother. 50:2892–2894. 10.1128/AAC.00349-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park S, Kelly R, Kahn JN, Robles J, Hsu MJ, Register E, Li W, Vyas V, Fan H, Abruzzo G, Flattery A, Gill C, Chrebet G, Parent SA, Kurtz M, Teppler H, Douglas CM, Perlin DS. 2005. Specific substitutions in the echinocandin target Fks1p account for reduced susceptibility of rare laboratory and clinical Candida sp. isolates. Antimicrob. Agents Chemother. 49:3264–3273. 10.1128/AAC.49.8.3264-3273.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perlin DS. 2007. Resistance to echinocandin-class antifungal drugs. Drug Resist. Updat. 10:121–130. 10.1016/j.drup.2007.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perlin DS. 2011. Current perspectives on echinocandin class drugs. Future Microbiol. 6:441–457. 10.2217/fmb.11.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fekkar A, Meyer I, Brossas JY, Dannaoui E, Palous M, Uzunov M, Nguyen S, Leblond V, Mazier D, Datry A. 2013. Rapid emergence of echinocandin resistance during Candida kefyr fungemia treated with caspofungin. Antimicrob. Agents Chemother. 57:2380–2382. 10.1128/AAC.02037-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson ME, Edlind TD. 2012. Topological and mutational analysis of Saccharomyces cerevisiae Fks1. Eukaryot. Cell 11:952–960. 10.1128/EC.00082-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delley PA, Hall MN. 1999. Cell wall stress depolarizes cell growth via hyperactivation of RHO1. J. Cell Biol. 147:163–174. 10.1083/jcb.147.1.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazur P, Baginsky W. 1996. In vitro activity of 1,3-beta-d-glucan synthase requires the GTP-binding protein Rho1. J. Biol. Chem. 271:14604–14609. 10.1074/jbc.271.24.14604 [DOI] [PubMed] [Google Scholar]
- 18.Johnson ME, Katiyar SK, Edlind TD. 2011. New Fks hot spot for acquired echinocandin resistance in Saccharomyces cerevisiae and its contribution to intrinsic resistance of Scedosporium species. Antimicrob. Agents Chemother. 55:3774–3781. 10.1128/AAC.01811-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dufresne SF, Marr KA, Sydnor E, Staab JF, Karp JE, Lu K, Zhang SX, Lavallee C, Perl TM, Neofytos D. 2014. Epidemiology of Candida kefyr in patients with hematologic malignancies. J. Clin. Microbiol. 52:1830–1837. 10.1128/JCM.00131-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrison FC. 1928. A systematic study of some Torulas. Trans. R. Soc. Can. 22:187–225 [Google Scholar]
- 21.Gillum AM, Tsay EY, Kirsch DR. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179–182. 10.1007/BF00328721 [DOI] [PubMed] [Google Scholar]
- 22.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard—3rd ed. CLSI document M27-A3 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 23.Pfaller MA, Castanheira M, Diekema DJ, Messer SA, Jones RN. 2011. Triazole and echinocandin MIC distributions with epidemiological cutoff values for differentiation of wild-type strains from non-wild-type strains of six uncommon species of Candida. J. Clin. Microbiol. 49:3800–3804. 10.1128/JCM.05047-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Redkar RJ, Dube MP, McCleskey FK, Rinaldi MG, Del Vecchio VG. 1996. DNA fingerprinting of Candida rugosa via repetitive sequence-based PCR. J. Clin. Microbiol. 34:1677–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pavel AB, Vasile CI. 2012. PyElph—a software tool for gel images analysis and phylogenetics. BMC Bioinformatics 13:9. 10.1186/1471-2105-13-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia-Effron G, Katiyar SK, Park S, Edlind TD, Perlin DS. 2008. A naturally occurring proline-to-alanine amino acid change in Fks1p in Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis accounts for reduced echinocandin susceptibility. Antimicrob. Agents Chemother. 52:2305–2312. 10.1128/AAC.00262-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia-Effron G, Lee S, Park S, Cleary JD, Perlin DS. 2009. Effect of Candida glabrata FKS1 and FKS2 mutations on echinocandin sensitivity and kinetics of 1,3-beta-d-glucan synthase: implication for the existing susceptibility breakpoint. Antimicrob. Agents Chemother. 53:3690–3699. 10.1128/AAC.00443-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staab JF, Kahn JN, Marr KA. 2010. Differential Aspergillus lentulus echinocandin susceptibilities are Fksp independent. Antimicrob. Agents Chemother. 54:4992–4998. 10.1128/AAC.00774-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stevens DA. 2009. Frequency of paradoxical effect with caspofungin in Candida albicans. Eur. J. Clin. Microbiol. Infect. Dis. 28:717. 10.1007/s10096-008-0688-y [DOI] [PubMed] [Google Scholar]
- 30.Stevens DA, White TC, Perlin DS, Selitrennikoff CP. 2005. Studies of the paradoxical effect of caspofungin at high drug concentrations. Diagn. Microbiol. Infect. Dis. 51:173–178. 10.1016/j.diagmicrobio.2004.10.006 [DOI] [PubMed] [Google Scholar]
- 31.Walker LA, Munro CA, de Bruijn I, Lenardon MD, McKinnon A, Gow NA. 2008. Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 4:e1000040. 10.1371/journal.ppat.1000040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Costa-de-Oliveira S, Marcos Miranda I, Silva RM, Pinto ESA, Rocha R, Amorim A, Goncalves Rodrigues A, Pina-Vaz C. 2011. FKS2 mutations associated with decreased echinocandin susceptibility of Candida glabrata following anidulafungin therapy. Antimicrob. Agents Chemother. 55:1312–1314. 10.1128/AAC.00589-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Effron G, Park S, Perlin DS. 2009. Correlating echinocandin MIC and kinetic inhibition of fks1 mutant glucan synthases for Candida albicans: implications for interpretive breakpoints. Antimicrob. Agents Chemother. 53:112–122. 10.1128/AAC.01162-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parmeland L, Gazon M, Guerin C, Argaud L, Lehot JJ, Bastien O, Allaouchiche B, Michallet M, Picot S, Bienvenu AL. 2013. Candida albicans and non-Candida albicans fungemia in an institutional hospital during a decade. Med. Mycol. 51:33–37. 10.3109/13693786.2012.686673 [DOI] [PubMed] [Google Scholar]
- 35.Murtagh F. 1984. Complexities of hierarchic clustering algorithms: the state of the art. Comput. Stat. 1:101–113 [Google Scholar]
- 36.Di Tommaso P, Moretti S, Xenarios I, Orobitg M, Montanyola A, Chang JM, Taly JF, Notredame C. 2011. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39:W13–W17. 10.1093/nar/gkr245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taly JF, Magis C, Bussotti G, Chang JM, Di Tommaso P, Erb I, Espinosa-Carrasco J, Kemena C, Notredame C. 2011. Using the T-Coffee package to build multiple sequence alignments of protein, RNA, DNA sequences and 3D structures. Nat. Protoc. 6:1669–1682. 10.1038/nprot.2011.393 [DOI] [PubMed] [Google Scholar]