Abstract
Antibiotic-resistant Campylobacter constitutes a serious threat to public health, and resistance to macrolides is of particular concern, as this class of antibiotics is the drug of choice for clinical therapy of campylobacteriosis. Very recently, a horizontally transferrable macrolide resistance mediated by the rRNA methylase gene erm(B) was reported in a Campylobacter coli isolate, but little is known about the dissemination of erm(B) among Campylobacter isolates and the association of erm(B)-carrying isolates with clinical disease. To address this question and facilitate the control of antibiotic-resistant Campylobacter, we determined the distribution of erm(B) in 1,554 C. coli and Campylobacter jejuni isolates derived from food-producing animals and clinically confirmed human diarrheal cases. The results revealed that 58 of the examined isolates harbored erm(B) and exhibited high-level resistance to macrolides, and most were recent isolates, derived in 2011-2012. In addition, the erm(B)-positive isolates were all resistant to fluoroquinolones, another clinically important antibiotic used for treating campylobacteriosis. The erm(B) gene is found to be associated with chromosomal multidrug resistance genomic islands (MDRGIs) of Gram-positive origin or with plasmids of various sizes. All MDRGIs were transferrable to macrolide-susceptible C. jejuni by natural transformation under laboratory conditions. Molecular typing of the erm(B)-carrying isolates by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) identified diverse genotypes and outbreak-associated diarrheal isolates. Molecular typing also suggested zoonotic transmission of erm(B)-positive Campylobacter. These findings reveal an emerging and alarming trend of dissemination of erm(B) and MDRGIs in Campylobacter and underscore the need for heightened efforts to control their further spread.
INTRODUCTION
Campylobacter is the leading bacterial cause of food-borne illnesses worldwide and primarily causes gastroenteritis (1). For clinical treatment of campylobacteriosis, fluoroquinolones and macrolides are often prescribed (2); however, the resistance of Campylobacter to clinically important antibiotics is increasingly reported. Indeed, the 2013 CDC report identified antibiotic-resistant Campylobacter as a serious antibiotic resistance threat in the U.S. (http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf). Due to the increasing prevalence of fluoroquinolone-resistant Campylobacter worldwide (3–5), macrolides are now considered the drug of choice for therapeutic purposes (2, 6). Thus, development and spread of macrolide resistance in Campylobacter will significantly limit the options for clinical treatment.
Historically, macrolide resistance in Campylobacter has been at a low level of incidence and is mainly mediated by point mutations in the 23S rRNA or in the rplD and rplV genes encoding 50S ribosomal subunit proteins L4 and L22. The mutations occur at low frequencies and incur fitness costs in Campylobacter (6–9), partly explaining the low prevalence of macrolide resistance in Campylobacter. rRNA methylases, which represent major mechanisms for macrolide resistance in other bacterial organisms (10), were not identified in Campylobacter until recently. A recent work by our group reported the identification of a Gram-positive erm(B) gene in a single porcine Campylobacter coli isolate (11), which carried no mutations in the 23S rRNA or in the rplD and rplV genes. erm(B) encodes a rRNA methylase and mediates high-level macrolide resistance (MIC = 512 μg/ml) in Campylobacter, and it can be transferred between Campylobacter jejuni and Campylobacter coli via natural transformation (11). This was the first identification of a horizontally transferrable macrolide resistance mechanism in thermophilic Campylobacter spp.
The identification of a horizontally transferrable erm(B) in Campylobacter is alarming, as the gene confers high-level resistance to macrolides and can be disseminated by horizontal gene transfer. However, nothing is known about the distribution of erm(B) in Campylobacter and the association of erm(B)-carrying Campylobacter strains with clinical disease. To answer this question and facilitate the control of antibiotic-resistant Campylobacter, we investigated the incidence of erm(B) in Campylobacter isolates derived from both human diarrheal cases and food-producing animals, determined the gene environments surrounding erm(B), and characterized the erm(B)-positive isolates by molecular typing methods.
MATERIALS AND METHODS
Campylobacter strains, growth conditions, and antimicrobial susceptibility testing.
A total of 1,554 Campylobacter isolates (1,157 C. coli and 397 C. jejuni isolates) were analyzed in this study. Detailed information on isolates is listed in Table S1 in the supplemental material. These include 75 human isolates, 789 swine isolates, 433 chicken isolates, 227 duck isolates, and 30 isolates from chicken carcasses. The previously identified erm(B)-carrying C. coli ZC113 of swine origin was included as a reference in this study (11). The 75 human C. coli isolates were from the retrospective collection of Chinese Centers for Disease Control and Prevention (CDC, Beijing, China) and were obtained during 2001–2011 from clinically confirmed gastroenteritis in four different provinces or cities. Due to the lack of a surveillance system for the Campylobacter spp. in patients with diarrhea or gastrointestinal disease in hospitals in China, clinical Campylobacter isolates were available from only a few hospitals, which had a collaborative relationship with China CDC. All patients have not been administered macrolides, but whether these patients had taken macrolides by themselves at home was unknown. The swine isolates were cultured from feces, and the chicken and duck isolates were cultured from cloacal swabs during 2008–2012 from Shandong, Ningxia, and Guangdong provinces under the routine program of surveillance for antimicrobial resistance in bacteria of animal origin (12, 13). Although information on antimicrobial usage for the herds/flocks from which the isolates were obtained was not available, the antibiotic usage records for some of the farms in which the isolates was collected indicated that macrolides, such as tylosin, spiramycin, and erythromycin, had been commonly used for curing or preventing bacterial infections (14). In addition, 30 Campylobacter isolates were cultured from whole chicken carcasses, which were collected after processing but before distribution in two slaughterhouses located in geographically separated regions of Shandong province in May 2011. All of the above-mentioned isolates were obtained based on one isolate per animal. In laboratory, all these Campylobacter isolates were grown on Mueller-Hinton agar (MHA; Sigma-Aldrich, MO, USA) at 42°C under microaerobic conditions (5% O2, 10% CO2, and 85% N2). The Campylobacter-specific growth supplements and selective agents (Oxoid, Hampshire, United Kingdom) were added to the media when needed. The isolates were further confirmed by PCR as C. coli or C. jejuni as previously described (15). The standard agar dilution method recommended by Clinical and Laboratory Standards Institute (CLSI; 2008) was used to determine the MICs of various antibiotics in the Campylobacter isolates (16). C. jejuni ATCC 33560 was used as the quality control strain.
PCR identification of erm(B) in various Campylobacter isolates, and determination of its location and genetic environments.
The erm(B) was identified in various Campylobacter isolates by PCR and sequencing. An 421-bp amplicon of erm(B) was produced by using the primers erm(B)-F (5′-GGGCATTTAACGACGAAACTGG) and erm(B)-R (5′-CTGTGGTATGGCGGGTAAGT), which were designed according to the conserved regions of the erm(B) genes found in C. coli ZC113, Enterococcus faecium (accession no. JN899585), Streptococcus pneumoniae (X52632), and Lactobacillus reuteri (AY082384). A modified random primer-walking strategy as previously described was performed using primers which were complementary to locations inside erm(B) for the sequence of entire open reading frame (ORF) of this gene (17). To determine whether erm(B) was localized in plasmids or on the chromosome, we analyzed the erm(B)-positive Campylobacter isolates by S1 nuclease pulsed-field gel electrophoresis (PFGE) and Southern blotting. Whole cells of the isolates were embedded in agarose gel plugs and digested with S1 nuclease (TaKaRa, Dalian, China), and the DNA was separated by PFGE as described previously (18). For Southern blotting, the 421-bp PCR product of erm(B) was used as the probe for hybridization. The probe was labeled with a digoxigenin (DIG) High Prime I DNA labeling and detection starter kit (Roche Diagnostics, Mannheim, Germany). Hybridization was performed at 44°C for 14 h. Membranes were washed twice at room temperature (22 to 25°C) with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% SDS for 5 min and twice at 68°C with 0.5× SSC and 0.1% SDS for 15 min. DIG was detected with specific antibodies using a DIG High Prime I DNA labeling and detection starter kit according to the protocol of the supplier. The genetic environments of erm(B) in 26 C. coli isolates that harbored a chromosomally carried erm(B) were determined by using a long-range PCR with primers cadf-F2 and pfo-R2 or a modified random primer-walking sequencing strategy as described previously (15, 17).
Genotyping.
Multilocus sequence typing (MLST) for Campylobacter was performed following the method described previously (19). The allelic profiles and the sequence types were generated by blasting the Campylobacter sequences in the MLST database (http://www.pubmlst.org/campylobacter). PFGE was performed using a CHEF-DR III apparatus (Bio-Rad Laboratories, Hercules, CA, USA), according to the rapid protocol for Campylobacter (20), and Salmonella H9812 was used as the reference marker (digested with XbaI), while all Campylobacter isolates were digested with SmaI. The dendrograms were constructed from the PFGE data by the unweighted pair group method with arithmetic average (UPGMA) with the Dice coefficient using InfoQuest FP software, version 4.5 (Bio-Rad Laboratories, USA).
Plasmid typing.
Plasmid typing was performed for the C. coli isolates that harbored a plasmid-borne erm(B) gene (Table 1). Since no Campylobacter-specific plasmid typing methods are available (21, 22), the incompatibility groups were characterized using the replicon typing methods for Enterobacteriaceae as previously described (23, 24).
TABLE 1.
Characteristics of the 58 erm(B)-positive Campylobacter isolates identified in this study
| Isolatea | Hostb | Province/cityc | Sample | Yr of isolation | Mutation in 23S rRNAg | Location of erm(B)d | MDRGI typee | MIC (μg/ml)f |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CIP (4) | ERY (32) | CLI (8) | GEN (8) | TET (16) | CHL (32) | ||||||||
| ZC113 (reference strain) | S | SD | Feces | 2008 | — | C | I | 32 | 512 | 256 | 128 | 128 | 4 |
| C179b | C | GD | Feces | 2012 | — | C | — | 32 | >512 | 256 | 1 | 256 | 4 |
| YC74 | S | NX | Feces | 2009 | A2075G | P | 64 | 128 | 16 | >256 | 128 | 4 | |
| ZP-GX-1 | C | SD | Carcass | 2011 | — | C | III | 16 | 128 | >256 | 256 | 512 | 8 |
| ZP-GX-5 | C | SD | Carcass | 2011 | — | C | III | 32 | 512 | 128 | 128 | 512 | 8 |
| ZP-GX-8 | C | SD | Carcass | 2011 | — | C | III | 32 | 512 | 128 | 128 | 512 | 8 |
| ZP-GX-12 | C | SD | Carcass | 2011 | — | C | III | 32 | 512 | 128 | 128 | 512 | 8 |
| ZP-GX-14 | C | SD | Carcass | 2011 | — | C | III | 64 | 512 | 128 | 128 | 512 | 8 |
| ZP-GX-20 | C | SD | Carcass | 2011 | — | C | III | 32 | 512 | 128 | 128 | 512 | 8 |
| ZP-GX-26 | C | SD | Carcass | 2011 | — | C | III | 64 | 512 | 128 | 128 | 256 | 8 |
| ZP-GX-28 | C | SD | Carcass | 2011 | — | C | III | 16 | 128 | >256 | 128 | 512 | 8 |
| ZP-GX-35 | C | SD | Carcass | 2011 | — | C | III | 32 | 512 | 128 | 128 | 512 | 8 |
| ZP-GX-36 | C | SD | Carcass | 2011 | — | C | III | 64 | 512 | 128 | 128 | 512 | 8 |
| ZP-GX-37 | C | SD | Carcass | 2011 | — | C | III | 64 | 512 | 128 | 128 | 256 | 8 |
| LH-GX-14 | C | SD | Carcass | 2011 | — | C | III | 64 | 256 | 128 | 128 | 256 | 4 |
| DH46a | D | SD | Feces | 2012 | — | C | — | 64 | 512 | 256 | >256 | 128 | 16 |
| DH67 | D | SD | Feces | 2008 | — | C | III | 64 | 512 | 256 | 256 | 128 | 64 |
| DZ5 | S | SD | Feces | 2012 | A2075G | C | — | 32 | 512 | 32 | 1 | 512 | 8 |
| DZ50 | S | SD | Feces | 2008 | A2075G | C, P | — | 32 | 512 | 8 | 256 | 256 | 4 |
| DZ14a | S | SD | Feces | 2012 | A2075G | P | 16 | 512 | 32 | >256 | 128 | 4 | |
| DZB1 | S | SD | Feces | 2012 | A2075G | P | 32 | 512 | 16 | 1 | 512 | 4 | |
| DZB4 | S | SD | Feces | 2012 | — | C | II | 32 | 256 | 256 | >256 | 128 | 4 |
| DZB5 | S | SD | Feces | 2012 | A2075G | P | 32 | 512 | 32 | >256 | 512 | 8 | |
| DZB7 | S | SD | Feces | 2012 | — | C | — | 64 | 64 | 16 | 2 | 64 | 4 |
| DZB12 | S | SD | Feces | 2012 | — | C | I | 32 | 512 | 256 | >256 | 128 | 8 |
| DZB41 | S | SD | Feces | 2012 | A2075G | P | 16 | 512 | 32 | 1 | 512 | 4 | |
| 158 | C | SD | Feces | 2008 | — | C | III | 32 | 32 | >256 | 128 | 128 | 2 |
| TH34 | S | GD | Feces | 2012 | A2075G | P | 32 | 128 | 32 | 1 | 128 | 16 | |
| TH45 | S | GD | Feces | 2012 | — | P | 32 | >512 | 32 | >256 | 512 | 16 | |
| TH48 | S | GD | Feces | 2012 | A2075G | P | 16 | >512 | 64 | 256 | 128 | 32 | |
| TH62 | S | GD | Feces | 2012 | A2075G | P | 16 | >512 | 32 | 1 | 512 | 8 | |
| TH74 | S | GD | Feces | 2012 | A2075G | P | 16 | >512 | 64 | 1 | 512 | 32 | |
| TH80 | S | GD | Feces | 2012 | A2075G | P | 32 | >512 | 32 | >256 | 32 | 32 | |
| TH96 | S | GD | Feces | 2012 | A2075G | P | 8 | >512 | 64 | 4 | 128 | 32 | |
| TH117 | S | GD | Feces | 2012 | A2075G | C | — | 32 | >512 | 128 | 256 | 512 | 8 |
| TH119 | S | GD | Feces | 2012 | A2075G | P | 16 | >512 | 64 | 1 | 32 | 4 | |
| 52-3b | S | GD | Feces | 2012 | A2075G | P | 32 | >512 | 32 | 1 | 128 | 32 | |
| 84a | S | GD | Feces | 2012 | A2075G | C | — | 16 | 512 | 32 | 1 | 128 | 4 |
| 08-23 | S | GD | Feces | 2012 | A2075G | C | — | 32 | >512 | 32 | >256 | 64 | 16 |
| 08-120 | S | GD | Feces | 2012 | A2075G | P | 8 | >512 | 32 | 2 | 128 | 4 | |
| 10-5-18 | S | GD | Feces | 2012 | A2075G | P | 8 | >512 | 64 | 256 | 128 | 32 | |
| 10-7-1a | S | GD | Feces | 2012 | — | P | 8 | >512 | 64 | 1 | 512 | 4 | |
| 10-7-1c | S | GD | Feces | 2012 | — | P | 8 | >512 | 32 | 1 | 512 | 4 | |
| 10-13-6a | S | GD | Feces | 2012 | A2075G | P | 32 | >512 | 32 | 256 | 128 | 8 | |
| 10-33-66 | S | GD | Feces | 2012 | A2075G | P | 32 | >512 | 32 | 256 | 128 | 64 | |
| JW16 | S | NX | Feces | 2012 | — | P | 32 | 32 | 32 | 4 | 64 | 4 | |
| LWC2 | S | NX | Feces | 2012 | — | P | 8 | 512 | 32 | 1 | 128 | 2 | |
| LWD21 | S | NX | Feces | 2012 | — | P | 16 | 32 | 16 | 0.5 | 32 | 4 | |
| LWD84 | S | NX | Feces | 2012 | — | P | 32 | 64 | 8 | 2 | 128 | 4 | |
| HN-CCD07046 | H | HN | Stool | 2007 | — | C | IV | 128 | 128 | 256 | 256 | 128 | 32 |
| SH-CCD11C073 | H | SH | Stool | 2011 | — | C | V | 32 | 128 | 256 | 128 | 512 | 32 |
| SH-CCD11C226 | H | SH | Stool | 2011 | — | C | III | 64 | 512 | 256 | >256 | 128 | 64 |
| SH-CCD11C287 | H | SH | Stool | 2011 | — | C | VI | 256 | 512 | 256 | 256 | 128 | 64 |
| SH-CCD11C365 | H | SH | Stool | 2011 | — | C | VI | 256 | 512 | 256 | 128 | 512 | 16 |
| SH-CCD11C416 | H | SH | Stool | 2011 | — | C | VI | 64 | 128 | 256 | 128 | 512 | 16 |
| SH-CCD11C419 | H | SH | Stool | 2011 | — | C | III | 64 | 512 | 256 | 128 | 128 | 32 |
| SH-CCD11C490 | H | SH | Stool | 2011 | — | C | VI | 128 | 128 | 256 | 256 | 128 | 64 |
| SH-CCD11C518 | H | SH | Stool | 2011 | — | C | VI | 256 | 512 | 256 | 256 | 512 | 64 |
| SH-CCD11C682 | H | SH | Stool | 2011 | — | C | VI | 64 | 128 | 256 | 128 | 128 | 16 |
All but one are C. coli, and C179b is C. jejuni. The erm(B)-carrying isolate C. coli ZC113 identified in the previous study (11) is included as a reference.
S, swine; C, chicken; D, duck; H, human.
SD, Shandong; NX, Ningxia; GD, Guangdong; HN, Henan; SH, Shanghai.
erm(B) is on the chromosome (C), the plasmid (P), or both (C, P).
—, the genetic environment of erm(B) in the chromosome could not be determined by the primer walking strategy used in this study.
CIP, ciprofloxacin; ERY, erythromycin; CLI, clindamycin; GEN, gentamicin; TET, tetracycline; CHL, chloramphenicol. Numbers in parentheses are MIC breakpoints. The MICs over the breakpoint are in bold.
—, no mutations detected in 23S rRNA gene.
Transfer of macrolide resistance by natural transformation or electrotransformation.
Macrolide resistance in C. coli isolates was transferred to the macrolide-susceptible C. jejuni strain NCTC 11168 or 81-176 by natural transformation, which was performed according to the method described by Wang and Taylor (25). We also determined if these erm(B) gene-carrying plasmids in C. coli isolates could be transferred to C. jejuni NCTC 11168 or 81-176 and C. coli ATCC 33559 using both the natural transformation and electroporation methods (26). The transformants were selected on MH agar plates containing erythromycin (10 μg/ml). The transformation without donor DNA was used as a negative control. All copies of the 23S rRNA gene of the transformants were sequenced to confirm lack of mutations in this gene in the transformants (27). Moreover, to confirm the transfer of the erm(B) gene and other resistance genes to the transformants, long-range PCR assays were conducted in the transformants using primers cadf-F2 and pfo-R2 to amplify the region between cadF and cj1476c genes (15), and primers rdxA-F (5′-GGTATTTTGGCTCGTGAGCTTG) and ACE-R (5′-GCATATGAAGACAAGGGAGCT) to amplify the region between the nfsB and Δaac genes.
Nucleotide sequence accession numbers.
The erm(B)-carrying segments in various isolates have been deposited in GenBank, and their accession numbers are KC876748 (ZP-GX-1), KC876749 (DZB4), KC876750 (HN-CCD07046), KC876751 (SH-CCD11C073), and KC876752 (SH-CCD11C365).
RESULTS AND DISCUSSION
Presence of erm(B) in Campylobacter isolates.
In total, 1,554 Campylobacter isolates (1,157 C. coli and 397 C. jejuni isolates) were examined in this study, including 75 isolates from human diarrheal cases and the rest from swine, chicken, and duck species (see Table S1 in the supplemental material). Among the isolates examined, 58 were positive for erm(B), including 57 C. coli and 1 C. jejuni isolates (Table 1). Ten of the erm(B)-positive isolates were from humans, 32 from swine, 14 from chicken, and 2 from duck, and the percentage of erm(B)-positive Campylobacter isolates in each of the sources is indicated in Table S2 in the supplemental material. The MIC results indicated that all 58 isolates were resistant to erythromycin, clindamycin, ciprofloxacin, and tetracycline, and 39 of them (67%) were also resistant to gentamicin (Table 1). Most of the 58 isolates showed an erythromycin MIC of ≥512 μg/ml. Twenty-two of the 58 (38%) erm(B)-positive Campylobacter strains harbored the A2075G (equivalent to A2059G in E. coli) mutation in 23S rRNA, and these isolates exhibited no significant difference (P > 0.05) in the MICs of erythromycin compared with the other 36 isolates that had no mutation in the target gene (Table 1). Sequencing of the PCR products revealed three variants of erm(B) among the 58 isolates with lengths of 738 bp (n = 34), 753 bp (n = 21), and 765 bp (n = 3). PFGE and Southern blotting revealed that 33 (57%) isolates, including the C. jejuni isolate C179b, carried erm(B) on the chromosome, while 24 (41%) isolates harbored erm(B) on plasmids of various sizes (Table 1; representative results are shown in Fig. S1 in the supplemental material).
Notably, most (53/58) of the erm(B)-positive Campylobacter isolates were obtained in recent years (2011-2012) (see Table S1 in the supplemental material). Among the 720 isolates collected from 2007 to 2009, 5 (0.7%) were positive with erm(B), while 53 (6.4%) of the 829 isolates collected during 2011-2012 were positive for erm(B). This finding suggests the recent emergence and a rising trend for erm(B)-carrying Campylobacter isolates in China.
Genetic environments of the erm(B) gene on the chromosome of C. coli isolates.
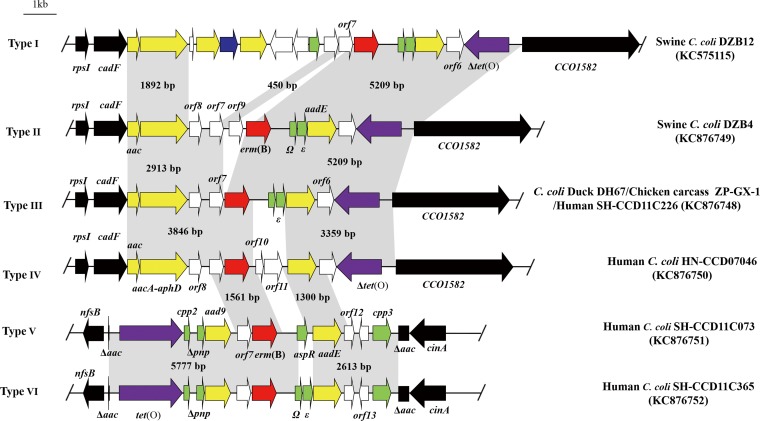
We further determined the genetic environments of erm(B) in 26 C. coli isolates that harbored a chromosomally carried erm(B) gene. In all cases, erm(B) was associated with multidrug resistance genomic islands (MDRGIs), which were inserted between cadF and CCO1582 (19 isolates) or in the aac gene, located between nfsB and cinA (7 isolates) (Fig. 1). The MDRGIs were classified into six types (designated I to VI) according to their gene contents and insertion sites on chromosome, and the previously identified MDRGI in ZC113 was designated type I (Table 1; Fig. 1). Types I to IV were bordered by cadF and CCO1582, while types V and VI were inserted in the aac gene. The GC contents of the MDRGIs ranged from 34.3% to 37.3%, higher than that (31.4%) of the genome of C. coli RM2228. Among them, type III was the most common and was observed in 16 isolates originated from both human and other animal species. Types IV, V, and VI were detected only in the human isolates, while types I and II were observed only in the swine isolates (Table 1).
FIG 1.
Chromosomal organization and comparison of six different types (I to VI) of MDRGIs in erm(B)-positive C. coli isolates. erm(B) is in red, aminoglycoside resistance genes are in yellow, the streptothricin resistance gene (sat4) is in blue, the tetracycline resistance gene [tet(O)] is in purple, genes with predicted functions are in green, and genes coding hypothetical proteins are in white. The tet(O) gene is intact in types V and VI but is truncated in other types. The border genes of the MDRGIs are depicted by black box arrows. The gray shading indicates regions sharing more than 98% DNA identity. A representative strain for each type of MDRGIs is indicated on the right of the panel.
Most of the ORFs in the MDRGIs are either identical or >90% identical to their orthologs in Gram-positive bacteria (see Table S3 in the supplemental material), implying that all erm(B)-carrying MDRGIs in Campylobacter were derived from Gram-positive origins. The orf7 gene encodes a conserved hypothetical protein that shares 95.8 to 97.1% amino acid (aa) identity to a hypothetical ORF in Clostridium difficile NAP08 (ZP_06892641) and is located immediately upstream of erm(B) in all but type II MDRGIs (Fig. 2). For type II, orf9 is inserted between orf7 and erm(B). The deduced amino acid sequence of orf9 showed 64.7% amino acid (aa) identity (86/133) to the fosfomycin resistance protein FosX of Listeria monocytogenes (YP_005962817). The two ORFs (Ω and ε) immediately downstream of erm(B) were conserved in types I, II, III, and VI. The Ω gene encodes 71 aa and shares 97.2% (69/71) aa identity to the Ω gene in Streptococcus agalactiae (ZP_22245542). The ε gene encodes 95 aa and shows 98.8% (89/90) aa identity to the corresponding region of ε in Streptococcus pyogenes (YP_232758). Regarding the other resistance genes, the aminoglycoside resistance gene aadE, located downstream of erm(B), was highly conserved (99.9% nucleotide identity) in all MDRGIs, while the aac and aacA-aphD genes in types I to IV exhibited nucleotide identity of 99.9%. The aad9 gene in types V and VI also showed 99.8% nucleotide identity to the aad9 gene on pCG8245 in C. jejuni (AY701528) (28). The tetracycline resistance gene tet(O) was intact in types V and VI but was truncated in the other types of MDRGIs. Detailed information on the ORFs in types I to VI is presented in Table S3 in the supplemental material. During the investigation of the genetic environment of erm(B) in C179b and in other seven C. coli isolates, in which the erm(B) gene was chromosome borne but was not inserted between cadF and CCO1582 or in the aac gene located between nfsB and cinA, the primer-walking strategy used in this study was not successful despite several attempts. Thus, the flanking regions of erm(B) in these isolates are unknown. Whole-genome sequence analysis will be needed to determine the exact location of the erm(B) insertion in these isolates.
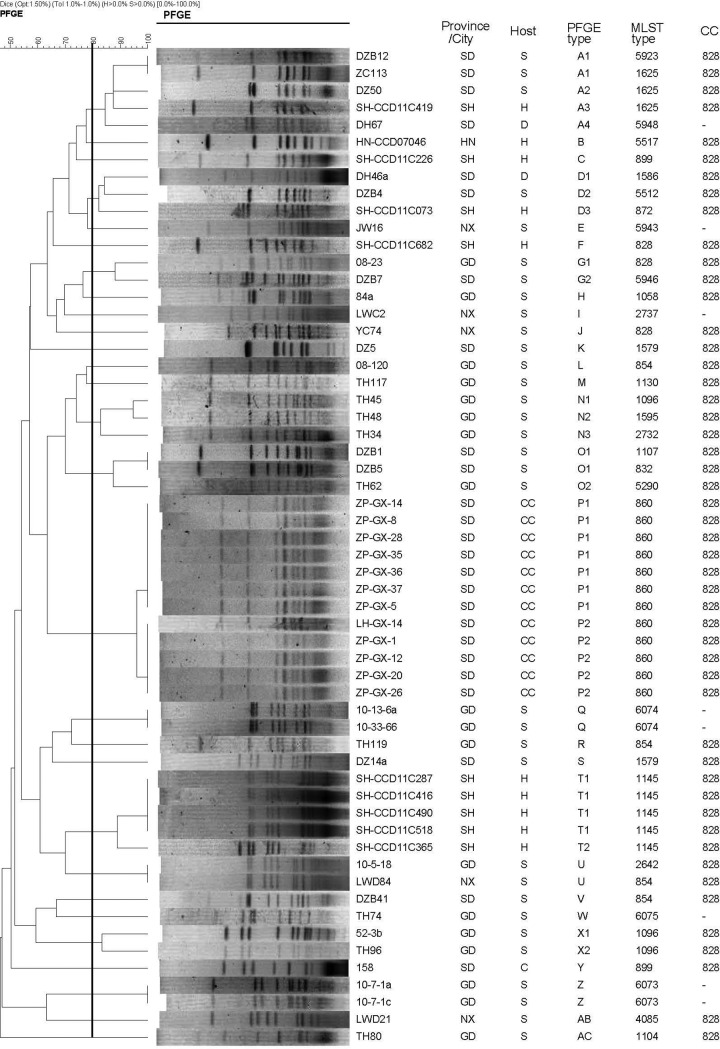
FIG 2.
PFGE and MLST typing of 58 erm(B)-positive C. coli isolates. STs 5923, 5943, 5946, 5948, 6073, 6074, and 6075 were newly designated in this study. SmaI was used for PFGE. Provinces and cities include Shandong (SD), Ningxia (NX), Guangdong (GD), Henan (HN), and Shanghai (SH). Sources and host species include swine (S), chicken (C), chicken carcasses (CC), duck (D), and human (H). −, a clonal complex cannot be assigned.
Although the identified MDGRIs vary in size, gene content, and insertion sites on the chromosome (Fig. 1), they shared a common origin of Gram-positive bacteria. The erm(B) gene and its neighboring genes in the MDRGIs are either identical or almost identical to their orthologs in Gram-positive bacteria, including Streptococcus, Enterococcus, Clostridium, Listeria, and Lactobacillus (see Table S3 in the supplemental material), implying that all erm(B)-carrying MDRGIs in Campylobacter were derived from Gram-positive origins. How Campylobacter acquired the MDGRIs from Gram-positive bacteria is unknown and remains to be determined.
Molecular typing and phylogenetic analysis of the erm(B)-carrying C. coli isolates.
PFGE and MLST were used to type the 57 erm(B)-positive C. coli isolates plus the previously reported erm(B)-carrying C. coli isolate ZC113. MLST generated 30 sequence types (STs), seven of which were newly designated in this study (Fig. 2). All 30 STs except for eight singletons were clustered into one clonal complex, CC828. Using a cutoff of 80% pattern similarity, the erm(B)-positive C. coli isolates were clustered into 28 PFGE subtypes (Fig. 2). The PFGE results generally corresponded to the MLST typing data, as most isolates of the same STs also appeared in the same PFGE subtypes. For instance, 4 isolates of human origin had the same ST (1145) and PFGE type (T1). Likewise, the 12 isolates from chicken carcasses, all belonged to ST 860 and could be clustered together at >95% (P1 and P2) according to the PFGE types (Fig. 2). Among the 75 human C. coli isolates examined in this study, 10 of them harbored the erm(B) gene and all of them were on chromosomal MDRGIs. These 10 isolates were from hospitals in Shanghai (n = 9, collected in 2011) and Henan province (n = 1, collected in 2007). Six of the 10 isolates were from pediatric patients (50 days to 6 years of age), while the others were from adults (24 to 54 years of age). Notably, 5 of the human isolates (4 from children and 1 from an adult) shared the same PFGE type and MLST type (Fig. 2), and were all from Shanghai, suggesting that this particular genotype was associated with an outbreak in the area. It should be acknowledged that nine out of the 10 human erm(B)-positive isolates were from Shanghai, suggesting that the true extent of erm(B) gene dissemination among humans in China cannot be determined in this study. Interestingly, a human isolate (SH-CCD11C419) from Shanghai and swine isolates (ZC113 and DZ50) from Shandong were clustered together by PFGE and MLST, all of which belong to ST1625 and PFGE subtype (A) using a cutoff of 80% genetic similarity for PFGE (Fig. 2), suggesting that they are clonal and suggesting the possibility of zoonotic transmission of this clone.
Transfer of erm(B)-carrying MDRGIs between C. coli and C. jejuni.
Our previous study showed that the type I MDRGI from ZC113 could be transferred to C. jejuni NCTC 11168 via natural transformation (11). In this study, we examined five other types of MDRGIs and found that they were also transferable from C. coli isolates to C. jejuni NCTC 11168 or 81-176 by natural transformation. Transfer of the MDRGIs to the transformants was confirmed by long-range PCR. Six different amplicons of expected sizes, including 12,210 bp, 8,927 bp, 8,281 bp, 8,384 bp, 9,660 bp, and 9,684 bp, were obtained from transformants 11168-type I, 81-176-type II, 11168-type III, 11168-type IV, 81-176-type V, and 81-176-type VI, respectively (Table 2). The sequencing results confirmed that the MDRGIs in the transformants were identical to those in the six types of donors. All transformants exhibited high-level resistance to erythromycin and clindamycin (Table 2), consistent with the known function of erm(B) in conferring resistance to macrolides, lincosamides, and streptogramin B (10, 29). Although the transformation was done under laboratory conditions, this finding is alarming, as Campylobacter is naturally transformable, and transfer of erm(B) between C. coli and C. jejuni could conceivably occur in natural environments, which would pose a great concern for public health, as a large portion of human campylobacteriosis is associated with C. jejuni (30, 31).
TABLE 2.
Antimicrobial susceptibility of Campylobacter transformants carrying the six types of MDRGIs
| Straina | MIC (μg/ml)b |
||||
|---|---|---|---|---|---|
| CIP | ERY | CLI | GEN | TET | |
| NCTC 11168 | <0.25 | 1 | 0.125 | 0.5 | 0.5 |
| 81–176 | <0.25 | 0.5 | 0.25 | 0.25 | 128 |
| 11168-type I (ZC113) | <0.25 | 512 | 256 | >64 | 0.5 |
| 81–176-type II (DZB4) | <0.25 | 64 | 128 | 64 | 128 |
| 11168-type III (ZP-GX-8) | <0.25 | 128 | 512 | >64 | 0.25 |
| 11168-type IV (HN-CCD07046) | <0.25 | 128 | 256 | >64 | 0.25 |
| 81–176-type V (SH-CCD11C073) | <0.25 | 128 | 128 | 1 | 128 |
| 81–176-type VI (SH-CCD11C365) | <0.25 | 256 | 128 | 1 | 128 |
The strains in parentheses indicate the donors carrying MDRGIs.
CIP, ciprofloxacin; ERY, erythromycin; CLI, clindamycin; GEN, gentamicin; TET, tetracycline.
We also tried to transfer some of the erm(B)-carrying plasmids by natural transformation and electroporation. However, both methods failed to move the plasmids to recipient Campylobacter strains (C. coli ATCC 33559 and C. jejuni NCTC 11168 and 81-176). This could be due to the difficulty of genetic manipulation of Campylobacter, as transformation of Campylobacter with plasmid DNA is less efficient than that with chromosomal DNA (32). Additionally, most of the erm(B)-carrying plasmids were above 70 kb in size (data not shown), which may limit the efficiency of transformation. At present, it is unknown if these plasmids can be transferred by conjugation. Despite the difficulty in transferring the plasmids, we performed plasmid typing with the 24 C. coli isolates harboring a plasmid-borne erm(B). The PCR and sequencing results revealed that 14 of them were typeable and belonged to four incompatibility (Inc) groups, including IncA/C (isolates JW16, 10-13-6a, 10-33-66, TH119, and TH74), IncW (isolates LWC2, LWD21, TH34, and DZ50), IncY (isolates DZB5, 10-5-18, and TH96), and IncFIA (isolates 10-7-1a and 10-7-1c). Ten of the 24 isolates were not typeable with the methods used in this study. However, it should be noted that it is unknown whether these identified plasmid types belonged to the erm(B)-carrying plasmids or to other plasmids, as a given C. coli isolate may contain more than one plasmid. The detailed genetic features of the erm(B)-carrying plasmids and their transfer mechanisms remain to be determined in future studies.
The identification of multiple erm(B)-positive isolates in this study reinforced the findings in our recent report (11) and indicates that erm(B) has emerged and is disseminating in Campylobacter. This finding is significant, as macrolides are the key antibiotic for clinical therapy of Campylobacter infections and resistance mediated by rRNA methylase was not identified until recently (11). Previously identified macrolide resistance mechanisms in Campylobacter involved 23S rRNA mutations in conjunction with the function of the CmeABC efflux pump (33, 34). The 23S rRNA mutations are well known for conferring high-level macrolide resistance in Campylobacter (7), but resistance-conferring mutations in 23S rRNA occur at low frequencies and incur a significant fitness cost in the absence of antibiotic selection pressure (35), contributing to the low prevalence of macrolide-resistant Campylobacter. The contribution of erm(B) to macrolide resistance in Campylobacter was shown in our recent report (11) and in this study using the transformants, in which 23S rRNA mutations were absent and erm(B) alone elevated the MICs up to 512 μg/ml (Table 2). The data from Table 2 and part of the data in Table 1 (for isolates with no 23S rRNA mutations) clearly show the significance of erm(B) in conferring high-level macrolide resistance in Campylobacter. Interestingly, the presence of both 23S rRNA mutation and erm(B) did not appear to show an additive effect on resistance, as macrolide MICs of the isolates harboring both mechanisms did not differ significantly (P > 0.05) from those of the isolates having only erm(B) (Table 1). The emergence of erm(B) in Campylobacter will likely change the landscape of macrolide resistance in Campylobacter, as this resistance determinant is horizontally transferable and may not cause a fitness burden in the organism. This possibility warrants further investigation.
In summary, this study reveals the presence of a horizontally transferable erm(B) in multiple Campylobacter isolates from various sources. In 59% of the cases, erm(B) was carried on chromosomal MDRGIs, along with other antibiotic resistance determinants. The MDRGIs thus confer resistance to multiple classes of antibiotics, including macrolides, lincosamides, and aminoglycosides. Additionally, all erm(B)-carrying isolates exhibited resistance to fluoroquinolones and tetracycline (Table 1). Thus, the erm(B)-positive isolates, regardless of the location of erm(B) (on the chromosome or a plasmid) or their origin of isolation (from human patients or farm animals), are resistant to all antibiotics that are important for clinical treatment of campylobacteriosis. The multidrug resistance phenotype of the erm(B)-positive isolates underlines the need for enhanced efforts to curb their further spread, as there will be limited options for clinical treatment of human patients infected by these isolates.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by grants from National Natural Science Foundation of China (U1031004), National Basic Research Program of China (2013CB127200), and the Key Projects in the National Science & Technology Pillar Program during the Twelfth Five-year Plan Period (2012BAK01B02).
Footnotes
Published ahead of print 30 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.03039-14.
REFERENCES
- 1.Ruiz-Palacios GM. 2007. The health burden of Campylobacter infection and the impact of antimicrobial resistance: playing chicken. Clin. Infect. Dis. 44:701–703. 10.1086/509936 [DOI] [PubMed] [Google Scholar]
- 2.Blaser MJ, Engberg J. 2008. Clinical aspects of Campylobacter jejuni and Campylobacter coli infections, p 99–121 In Nachamkin I, Szymanski CM, Blaser MJ. (ed), Campylobacter, 3rd ed. ASM Press, Washington, DC [Google Scholar]
- 3.Nelson JM, Chiller TM, Powers JH, Angulo FJ. 2007. Fluoroquinolone-resistant Campylobacter species and the withdrawal of fluoroquinolones from use in poultry: a public health success story. Clin. Infect. Dis. 44:977–980. 10.1086/512369 [DOI] [PubMed] [Google Scholar]
- 4.Gupta A, Nelson JM, Barrett TJ, Tauxe RV, Rossiter SP, Friedman CR, Joyce KW, Smith KE, Jones TF, Hawkins MA, Shiferaw B, Beebe JL, Vugia DJ, Rabatsky-Ehr T, Benson JA, Root TP, Angulo FJ, NARMS Working Group 2004. Antimicrobial resistance among Campylobacter strains, United States, 1997–2001. Emerg. Infect. Dis. 10:1102–1109. 10.3201/eid1006.030635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo N, Pereira S, Sahin O, Lin J, Huang S, Michel L, Zhang Q. 2005. Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc. Natl. Acad. Sci. U. S. A. 102:541–546. 10.1073/pnas.0408966102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luangtongkum T, Jeon B, Han J, Plummer P, Logue CM, Zhang Q. 2009. Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Future Microbiol. 4:189–200. 10.2217/17460913.4.2.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibreel A, Taylor DE. 2006. Macrolide resistance in Campylobacter jejuni and Campylobacter coli. J. Antimicrob. Chemother. 58:243–255. 10.1093/jac/dkl210 [DOI] [PubMed] [Google Scholar]
- 8.Luangtongkum T, Shen Z, Seng V, Sahin O, Jeon B, Liu P, Zhang Q. 2012. Impaired fitness and transmission of macrolide-resistant Campylobacter jejuni in its natural host. Antimicrob. Agents Chemother. 56:1300–1308. 10.1128/AAC.05516-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han F, Pu S, Wang F, Meng J, Ge B. 2009. Fitness cost of macrolide resistance in Campylobacter jejuni. Int. J. Antimicrob. Agents 34:462–466. 10.1016/j.ijantimicag.2009.06.019 [DOI] [PubMed] [Google Scholar]
- 10.Roberts MC. 2008. Update on macrolide-lincosamide-streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol. Lett. 282:147–159. 10.1111/j.1574-6968.2008.01145.x [DOI] [PubMed] [Google Scholar]
- 11.Qin S, Wang Y, Zhang Q, Deng F, Shen Z, Wu C, Wang S, Zhang J, Shen J. 2014. Report of ribosomal RNA methylase gene erm(B) in multidrug resistant Campylobacter coli. J. Antimicrob. Chemother. 69:964–968. 10.1093/jac/dkt492 [DOI] [PubMed] [Google Scholar]
- 12.Qin S, Wu C, Wang Y, Jeon B, Shen Z, Wang Y, Zhang Q, Shen J. 2011. Antimicrobial resistance in Campylobacter coli isolated from pigs in two provinces of China. Int. J. Food Microbiol. 146:94–98. 10.1016/j.ijfoodmicro.2011.01.035 [DOI] [PubMed] [Google Scholar]
- 13.Chen X, Naren GW, Wu C, Wang Y, Dai L, Xia LN, Luo P, Zhang Q, Shen J. 2010. Prevalence and antimicrobial resistance of Campylobacter isolates in broilers from China. Vet. Microbiol. 144:133–139. 10.1016/j.vetmic.2009.12.035 [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, He T, Schwarz S, Zhao Q, Shen Z, Wu C, Shen J. 2013. Multidrug resistance gene cfr in methicillin-resistant coagulase-negative staphylococci from chickens, ducks, and pigs in China. Int. J. Med. Microbiol. 303:84–87. 10.1016/j.ijmm.2012.12.004 [DOI] [PubMed] [Google Scholar]
- 15.Qin S, Wang Y, Zhang Q, Chen X, Shen Z, Deng F, Wu C, Shen J. 2012. Identification of a novel genomic island conferring resistance to multiple aminoglycoside antibiotics in Campylobacter coli. Antimicrob. Agents Chemother. 56:5332–5339. 10.1128/AAC.00809-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; informational supplement. CLSI document M31-A3. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 17.Zhang K, McClure JA, Elsayed S, Conly JM. 2009. Novel staphylococcal cassette chromosome mec type, tentatively designated type VIII, harboring class A mec and type 4 ccr gene complexes in a Canadian epidemic strain of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53:531–540. 10.1128/AAC.01118-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235–240. 10.1006/abio.1995.1220 [DOI] [PubMed] [Google Scholar]
- 19.Dingle KE, Colles FM, Falush D, Maiden MC. 2005. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J. Clin. Microbiol. 43:340–347. 10.1128/JCM.43.1.340-347.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ribot EM, Fitzgerald C, Kubota K, Swaminathan B, Barrett TJ. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39:1889–1894. 10.1128/JCM.39.5.1889-1894.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tenover FC, Williams S, Gordon KP, Nolan C, Plorde JJ. 1985. Survey of plasmids and resistance factors in Campylobacter jejuni and Campylobacter coli. Antimicrob. Agents Chemother. 27:37–41. 10.1128/AAC.27.1.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazi W, Senok A, Al-Mahmeed A, Arzese A, Bindayna K, Botta G. 2008. Trends in antibiotic sensitivity pattern and molecular detection of tet(O)-mediated tetracycline resistance in Campylobacter jejuni isolates from human and poultry sources. Jpn. J. Infect. Dis. 61:82–84 [PubMed] [Google Scholar]
- 23.Johnson TJ, Wannemuehler YM, Johnson SJ, Logue CM, White DG, Doetkott C, Nolan LK. 2007. Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl. Environ. Microbiol. 73:1976–1983. 10.1128/AEM.02171-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228. 10.1016/j.mimet.2005.03.018 [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Taylor DE. 1990. Natural transformation in Campylobacter species. J. Bacteriol. 172:949–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van V, Wood AC, Henderson J, Wooldridge KG, Ketley JM. 1998. Genetic manipulation of enteric Campylobacter species. Methods Microbiol. 27:407–419. 10.1016/S0580-9517(08)70301-5 [DOI] [Google Scholar]
- 27.Alonso R, Mateo E, Churruca E, Martinez I, Girbau C, Fernández-Astorga A. 2005. MAMA-PCR assay for the detection of point mutations associated with high-level erythromycin resistance in Campylobacter jejuni and Campylobacter coli strains. J. Microbiol. Methods 63:99–103. 10.1016/j.mimet.2005.03.013 [DOI] [PubMed] [Google Scholar]
- 28.Nirdnoy W, Mason CJ, Guerry P. 2005. Mosaic structure of a multiple-drug-resistant, conjugative plasmid from Campylobacter jejuni. Antimicrob. Agents Chemother. 49:2454–2459. 10.1128/AAC.49.6.2454-2459.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weisblum B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577–585. 10.1128/AAC.39.3.577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States- major pathogens. Emerg. Infect. Dis. 17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Humphrey T, O'Brien S, Madsen M. 2007. Campylobacters as zoonotic pathogens: a food production perspective. Int. J. Food Microbiol. 117:237–257. 10.1016/j.ijfoodmicro.2007.01.006 [DOI] [PubMed] [Google Scholar]
- 32.Taylor DE. 1992. Genetics of Campylobacter and Helicobacter. Annu. Rev. Microbiol. 46:35–64. 10.1146/annurev.mi.46.100192.000343 [DOI] [PubMed] [Google Scholar]
- 33.Cagliero C, Mouline C, Payot S, Cloeckaert A. 2005. Involvement of the CmeABC efflux pump in the macrolide resistance of Campylobacter coli. J. Antimicrob. Chemother. 56:948–950. 10.1093/jac/dki292 [DOI] [PubMed] [Google Scholar]
- 34.Payot S, Bolla JM, Corcoran D, Fanning S, Mégraud F, Zhang Q. 2006. Mechanisms of fluoroquinolone and macrolide resistance in Campylobacter spp. Microbes Infect. 8:1967–1971. 10.1016/j.micinf.2005.12.032 [DOI] [PubMed] [Google Scholar]
- 35.Luangtongkum T, Shen Z, Seng VW, Sahin O, Jeon B, Liu P, Zhang Q. 2012. Impaired fitness and transmission of macrolide-resistant Campylobacter jejuni in its natural host. Antimicrob. Agent Chemother. 56:1300–1308. 10.1128/AAC.05516-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.