Abstract
Recently, the newly emerged hypervirulent Klebsiella pneumoniae strain (hvKP) has caused great concern globally, but the clinical features and molecular characteristics of bacteremia caused by hvKP are rarely reported in mainland China. Seventy patients with K. pneumoniae bacteremia were investigated to study the clinical features of hvKP infection from 2008 till 2012 in Beijing Chao-Yang Hospital. The molecular characteristics of the hvKP strains were also studied using PCR, multilocus sequence typing, and pulsed-field gel electrophoresis (PFGE) methods. hvKP was identified in 31.4% of the patients with K. pneumoniae bacteremia, which displayed 4 serotypes (K1, K2, K20, and K57). Patients with hvKP infection tended to have no underlying diseases compared to those with classic K. pneumoniae (cKP). More hvKP-positive patients (95.5%) had community-acquired infection than did cKP-infected patients (35.4%) (P < 0.001). The 30-day mortality rate was lower in hvKP-infected patients than in cKP-infected patients (4.5% compared to 16.7%). Resistance to tested antimicrobials was significantly greater in cKP- than in hvKP-infected patients. Two extended-spectrum-beta-lactamase (ESBL)-producing hvKP strains were found. Seven novel sequence types (STs) and 4 new alleles of K. pneumoniae were revealed. A strong correlation was found between two STs (ST23, ST1265) and the K1 serotype. The hvKP isolates (n = 22) had 14 different PFGE patterns, and among them 10 K1 isolates shared similar PFGE patterns. The emerging hvKP strain was prevalent in patients with severe community-acquired infections in healthy individuals in China. Identification of ESBL-producing hvKP strains in hvKP-infected patients will facilitate clinical management of hvKP infection.
INTRODUCTION
The classic Klebsiella pneumoniae strain (cKP) is one of the major pathogens causing hospital-acquired (HA) infection, particularly in immunocompromised patients. It is the second-commonest cause of Gram-negative bacteremia (1, 2). A new hypervirulent variant of K. pneumoniae (hvKP) was identified 20 years ago in Taiwan, and it has been reported frequently in Asia ever since (3, 4, 5). This new variant is different from the classic strain in that the appearance of colonies grown on an agar plate is hypermucoviscous. Therefore, it is also called hypermucoviscous K. pneumoniae. In contrast to the cKP strain, the hypervirulent variant is not only able to cause nosocomial infection in immunocompromised patients, but, more importantly, it is able to cause life-threatening community-acquired (CA) infection in healthy individuals, which has caused a great concern worldwide (6). In addition, this new strain also has the ability to spread from the primary site of infection to other parts of the body, which is an unusual feature for enteric Gram-negative bacilli in nonimmunocompromised hosts (7). Recently, hvKP infection is increasingly reported from Europe (8), South America (9), Australia (10), and North America (11). Although such cases have been extensively described worldwide, particularly in Asian patients, to date, there have been few reports on the clinical features of hvKP in mainland China. In this report, we investigate the clinical features and molecular characteristics of K. pneumoniae bacteremia in 70 patients over a 3-year period.
MATERIALS AND METHODS
Hospital setting and case definition.
The Beijing Chao-Yang Hospital is a 1,500-bed tertiary care teaching hospital with 6 intensive-care-unit (ICU) wards and approximately 25,000 hospital admissions per year. We conducted a retrospective cohort study of patients treated in the hospital for K. pneumoniae bloodstream infections between June 2008 and April 2012. The patients were identified according to the records from the clinical microbiology department. Patient data were obtained from medical records, including demographic characteristics, clinical features, duration of hospital stay, antimicrobial therapy administration, mechanical ventilation, and use of invasive devices. Each case was differentiated between CA infection and HA infection. The CA bloodstream infection was defined as detection of K. pneumoniae in blood cultures taken within 48 h after admission. Conversely, HA bloodstream infection was defined as development of bacteremia >48 h into inpatient admission, including infections related to the presence of medical devices.
Study samples.
Isolates from 70 patients who were diagnosed to have had K. pneumoniae bloodstream infection were available for analysis and were identified as K. pneumoniae by using conventional microbiologic methods and further confirmed as K. pneumoniae by 16S rRNA sequencing. The “string test” was performed on all isolates. The string test is positive when a bacteriology inoculation loop is able to generate a viscous string of >5 mm in length by stretching bacterial colonies on an agar plate. K. pneumoniae strains with a positive string test were designated hvKP.
Antimicrobial susceptibility testing.
Antimicrobial susceptibility testing for isolated K. pneumonia was carried out by means of a Kirby-Bauer disk diffusion test on Mueller-Hinton agar, which was performed and interpreted according to the guidelines established by the Clinical and Laboratory Standards Institute (CLSI), USA (12). A panel of 23 antimicrobial agents was tested, including piperacillin, ampicillin-sulbactam, amoxicillin-clavulanic acid, ticarcillin-clavulanate, cefazolin, piperacillin-tazobactam, cefoxitin, cefuroxime, ceftriaxone, ceftazidime, cefotaxime, cefepime, aztreonam, imipenem, meropenem, ciprofloxacin, levofloxacin, tetracycline, minocycline, gentamicin, tobramycin, amikacin, and trimethoprim-sulfamethoxazole. All of the K. pneumoniae isolates were screened and confirmed by a double-disk synergy test for produced extended-spectrum beta-lactamases (ESBLs). K. pneumoniae ATCC 700603 and Staphylococcus aureus ATCC 25923 were included in each experiment as controls.
PCR for capsular polysaccharide synthesis (CPS) genotyping.
K1, K2, K5, K20, K54, and K57 serotypes were identified by detection of K serotype-specific wzy and wzx alleles using the PCR method as previously described (13). The PCR products were visualized by 1% agarose gel electrophoresis and sequenced commercially. The BLAST program at http://www.ncbi.nlm.nih.gov was used for final serotype identification.
MLST.
Multilocus sequence typing (MLST) was performed according to the protocol described on the K. pneumonia MLST website (www.pasteur.fr/mlst) (14). Internal fragments of seven housekeeping genes for K. pneumonia (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) were amplified, sequenced, and analyzed. Alleles and sequence types (STs) were determined according to the MLST database (www.pasteur.fr/mlst/Kpneumoniae.html). Alleles and STs that had not been previously described were submitted to the curator of the database and were assigned as new designations. The program eBURST version 3.0 software (based upon related sequence types) was used to analyze the clustering of related STs, which were classified as clonal complexes (CCs) (15). Clonal complexes were defined as groups of two or more independent isolates that shared identical alleles at six or more loci. Each complex was named after the putative founder ST.
PFGE.
Pulsed-field gel electrophoresis (PFGE) typing was conducted as previously described (16). Whole-cell genomic DNA representing each isolate was digested with the restriction enzyme XbaI (TaKaRa Biotechnology, Dalian, China) and separated by electrophoresis through 1% pulsed-field certified agarose (Bio-Rad, Richmond, CA, USA) by using a CHEF-Mapper (Bio-Rad). Electrophoretic switch times of 5 to 35 s were used with a 6-V/cm current and a switch angle of 120° under a constant temperature of 14°C. PFGE patterns were interpreted by using the criteria proposed by Tenover et al. (17).
Statistical analysis.
Data were analyzed using the statistical package SPSS for windows version 17.0. For categorical data, different groups were compared using the chi-square test to analyze the quantitative variables. A P value of ≤0.05 was considered to be statistically significant. All susceptibility data were analyzed using WHONET, version 5.6.
Ethics statement.
Permission for using the information in the medical records of the patients and the K. pneumoniae isolates for research purposes was granted by the ethical committee of Beijing Chao-Yang Hospital.
RESULTS
hvKP identification.
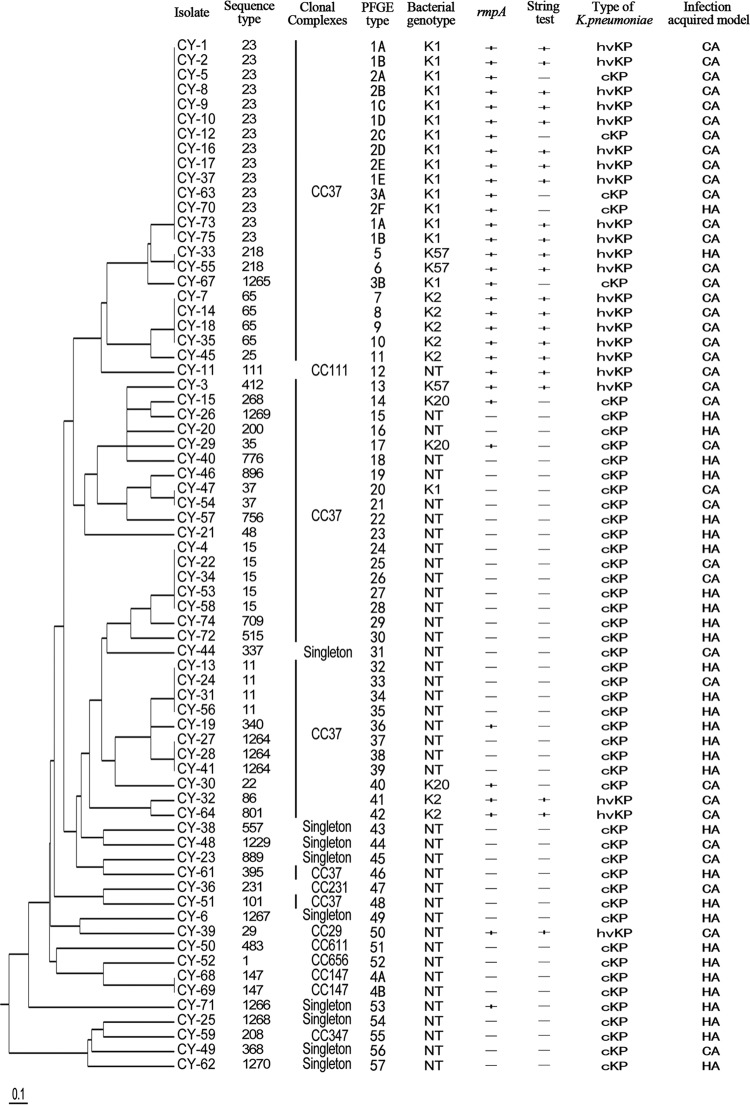
The string test was used to differentiate between hvKP and cKP among 70 K. pneumoniae isolates. Twenty-two isolates were found to be string test positive (Fig. 1, “String test” column).
FIG 1.
Graphic summary of molecular characteristic of 70 K. pneumoniae isolates.
Serotype identification.
To identify the serotypes of K. pneumoniae, PCR was performed on 70 K. pneumoniae isolates using primers for K1, K2, K5, K20, K54, and K57 serotypes as described in Materials and Methods. A summary of the results is shown in Fig. 1. A total of 29 (41.4%) isolates tested positive for K1, K2, K20, or K57 serotypes. Sixteen (55.2%) of them displayed the K1 serotype, 7 (24.1%) isolates showed the K2 serotype, while 3 (10.3%) isolates were K20 serotype positive and 3 (10.3%) were K57 serotype positive. However, no K5 and K54 serotypes were found.
Relationship between serotypes and site of infections.
K1 and K2 serotypes were more commonly found than other serotypes in patients with community-acquired infections (P < 0.001 and P = 0.013, respectively) (Table 1). We also found that patients with primary liver abscess more commonly tested positive for K1 and K2 (Table 2). In addition, K1 and K2 serotypes were also correlated with primary bacteremia, urological infection, and community-acquired pneumonia. In contrast, more classic K. pneumoniae-positive patients than hvKP-positive patients presented with hospital-acquired pneumonia (P = 0.004) (Table 2).
TABLE 1.
Different serotypes of K. pneumoniae bacteremic isolates from community-acquired or hospital-acquired infections
| Serotype | No. (%) of isolates |
P value (CA vs HA) | |
|---|---|---|---|
| Community-acquired infections (n = 38) | Hospital-acquired infections (n = 32) | ||
| K1 | 15 (39.5) | 1 (3.1) | <0.001b |
| K2 | 7 (18.4) | 0 | 0.013b |
| K20 | 3 (7.9) | 0 | 0.245 |
| K57 | 2 (5.3) | 1 (3.1) | 1.000 |
| K5 | 0 | 0 | |
| K54 | 0 | 0 | |
| NTa | 11 (28.9) | 30 (93.8) | <0.001b |
NT, not belonging to the K1, K2, K5, K20, K54, or K57 serotype.
A P value of ≤0.05 was considered to be statistically significant.
TABLE 2.
Difference between hypervirulent and classic K. pneumoniae-induced infections
| Infection type | No. of hvKP-induced infections (n = 22) |
No. of cKP-induced infections (n = 48) |
P value (hvKP vs cKP) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| K1 | K2 | K20 | K57 | NTa | K1 | K2 | K20 | K57 | NTa | ||
| Liver abscess | 6 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | <0.001b |
| Bacteremia | 1 | 1 | 0 | 2 | 0 | 2 | 0 | 1 | 0 | 6 | 0.195 |
| Catheter-related bloodstream infection | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0.301 |
| Hospital-acquired pneumoniae | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13 | 0.004b |
| Community-acquired pneumoniae | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 1.000 |
| Ventilator-associated pneumoniae | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1.000 |
| Biliary tract infection | 0 | 0 | 0 | 1 | 2 | 1 | 0 | 1 | 0 | 7 | 0.741 |
| Urological infections | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0.700 |
| Abdominal infection | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1.000 |
| Peritonitis | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.314 |
| Acute enteritis | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1.000 |
| Pyelonephritis | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1.000 |
| Perianal abscess | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1.000 |
NT, not belonging to the K1, K2, K5, K20, K54, or K57 serotype.
A P value of ≤0.05 was considered to be statistically significant.
Comparison of demographic and clinical characteristics of patients infected with hvKP and cKP.
Among the 70 K. pneumonia isolates analyzed, 22 (31.4%) of them were found to be hvKP and 48 (68.6%) of them were cKP. Overall, there was a trend of more male patients with K. pneumonia bloodstream infections. But the male predominance was more significant in hvKP-infected patients (90.9%) than in cKP-infected patients (62.5%, P = 0.021). There was no difference in age distribution between the two groups. More hvKP-positive patients (21/22, 95.5%) than cKP-infected patients (17/48, 35.4%) had CA infections (P < 0.001) (Table 3). It was also noted that 40.9% (9/22) of the hvKP-infected patients had no reported underlying diseases at the time of infection. In contrast, 95.8% (46/48) of the cKP-infected patients had one or more underlying illnesses. The 30-day mortality rate was lower in the hvKP group (4.5%) than in the cKP group (Table 3).
TABLE 3.
Demographic and clinical characteristics of patients with K. pneumoniae bacteremia
| Characteristic | No. (%) of patients |
P value (hvKP vs cKP) | ||
|---|---|---|---|---|
| Total (n = 70) | With hvKP infection (n = 22) | With cKP infection (n = 48) | ||
| Gender | ||||
| Male | 50 (71.4) | 20 (90.9) | 30 (62.5) | 0.021a |
| Female | 20 (28.6) | 2 (9.1) | 18 (37.5) | 0.021a |
| Age | ||||
| >60 yrs | 38 (54.2) | 9 (40.9) | 29 (60.4) | 0.196 |
| Acquired infection model | ||||
| Community-acquired infection | 38 (54.2) | 21 (95.5) | 17 (35.4) | <0.001a |
| Hospital-acquired infection | 32 (45.7) | 1 (4.5) | 31 (64.6) | <0.001a |
| Underlying diseases | ||||
| None | 11 (15.7) | 9 (40.9) | 2 (4.17) | <0.001a |
| Diabetes mellitus | 16 (22.9) | 6 (27.3) | 10 (20.8) | 0.760 |
| Biliary tract diseases | 9 (12.9) | 3 (13.6) | 6 (12.5) | 1.000 |
| Heart disease | 4 (5.7) | 0 (0) | 4 (8.3) | 0.301 |
| Central nervous system diseases | 5 (7.1) | 0 (0) | 5 (10.4) | 0.173 |
| Alcoholic cirrhosis | 3 (4.3) | 1 (4.5) | 2 (4.17) | 1.000 |
| Hematologic diseases | 3 (4.3) | 0 (0) | 3 (6.3) | 0.547 |
| Pulmonary infection | 5 (5.7) | 1 (4.5) | 4 (8.3) | 1.000 |
| Cancer | 14 (20) | 1 (4.5) | 13 (27.1) | 0.050a |
| Other | 9 (12.9) | 1 (4.5) | 8 (16.7) | 0.255 |
| Use of invasive devices | 36 (51.4) | 5 (22.7) | 31 (64.6) | 0.001a |
| Metastatic spread | 2 (2.9) | 2 (9.1) | 0 (0) | 0.096 |
| Outcome | ||||
| Discharged | 53 (75.7) | 16 (72.7) | 37 (77.1) | 0.767 |
| Died | 9 (14.5) | 1 (4.5) | 8 (16.7) | 0.255 |
| Lost to follow up | 8 (11.4) | 5 (22.7) | 3 (4.9) | 0.098 |
A P value of ≤0.05 was considered to be statistically significant.
Antimicrobial susceptibility of K. pneumoniae isolates.
Notably, the prevalence of cKP strains exhibiting resistance to the tested antimicrobials was higher than that of the hvKP strains (Table 4). Twenty-six (26/70, 37.1%) of the isolates produced ESBLs. The percentage of ESBL-producing cKP strains was significantly higher than that of ESBL-producing hvKP strains (50% compared to 9.09%, P = 0.001). Among all of the isolates, one of them (1/70, 1.4%) was resistant to carbapenem and produced K. pneumoniae carbapenemase 2 (KPC-2).
TABLE 4.
Differences of the antimicrobial susceptibility between hypervirulent and classic K. pneumoniae
| Drug | No. (%) of patients susceptible |
P value (hvKP vs cKP) | ||
|---|---|---|---|---|
| Total (n = 70) | hvKP infected (n = 22) | cKP infected (n = 48) | ||
| Piperacillin | 29 (41.4) | 11 (50.0) | 18 (37.5) | 0.434 |
| Ampicillin-sulbactam | 36 (51.4) | 16 (72.7) | 20 (41.7) | 0.021a |
| Amoxicillin-clavulanic acid | 48 (68.6) | 20 (90.9) | 28 (58.3) | 0.011a |
| Ticarcillin-clavulanate | 43 (61.4) | 20 (90.9) | 23 (47.9) | 0.001a |
| Piperacillin-tazobactam | 55 (78.6) | 21 (95.5) | 34 (70.8) | 0.026a |
| Cefazolin | 25 (35.7) | 9 (40.9) | 16 (33.3) | 0.597 |
| Cefoxitin | 47 (67.1) | 18 (81.8) | 29 (60.4) | 0.102 |
| Cefuroxime | 37 (52.9) | 18 (81.8) | 19 (39.6) | 0.002a |
| Ceftazidime | 45 (64.2) | 20 (90.9) | 25 (52.1) | 0.003a |
| Ceftriaxone | 45 (64.2) | 20 (90.9) | 25 (52.1) | 0.003a |
| Cefotaxime | 49 (70.0) | 20 (90.9) | 29 (60.4) | 0.011a |
| Cefepime | 61 (87.1) | 22 (100) | 39 (81.3) | 0.049a |
| Aztreonam | 49 (70.0) | 21 (95.5) | 28 (58.3) | 0.002a |
| Meropenem | 69 (98.6) | 22 (100) | 47 (97.9) | 1.000 |
| Imipenem | 69 (98.6) | 22 (100) | 47 (97.9) | 1.000 |
| Ciprofloxacin | 44 (62.9) | 20 (90.9) | 24 (50.0) | 0.001a |
| Levofloxacin | 46 (65.7) | 20 (90.9) | 26 (54.2) | 0.003a |
| Tetracycline | 44 (62.9) | 19 (86.4) | 25 (52.1) | 0.007a |
| Minocycline | 39 (55.7) | 16 (72.7) | 23 (47.9) | 0.071 |
| Gentamicin | 55 (78.6) | 22 (100) | 33 (68.8) | 0.002a |
| Tobramycin | 54 (77.1) | 22 (100) | 32 (66.7) | 0.001a |
| Amikacin | 63 (90.0) | 22 (100) | 41 (85.4) | 0.089 |
| Trimethoprim-sulfamethoxazole | 47 (67.1) | 20 (90.9) | 27 (56.3) | 0.005a |
| ESBLs | 26 (37.1) | 2 (9.09) | 24 (50.0) | 0.001a |
A P value of ≤0.05 was considered to be statistically significant.
MLST and PFGE analysis of K. pneumoniae isolates and identification of clonal complexes.
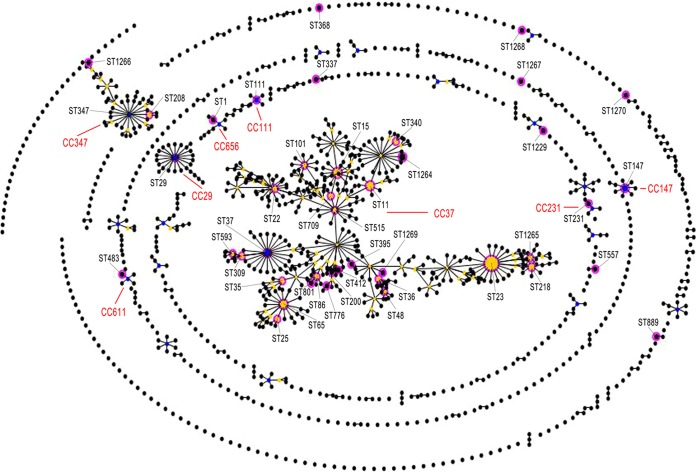
MLST analysis identified 42 STs among the 70 K. pneumoniae isolates, including 7 newly identified STs (ST1264, ST1265, ST1266, ST1267, ST1268, ST1269, ST1270) and 4 novel alleles (rpoB88, tonB225, tonB226, pgi107), as shown in Fig. 1. The most prevalent ST in K. pneumoniae isolates was ST23 (n = 14; 20%), followed by ST15 (n = 5; 7.1%), ST65 (n = 4; 5.7%), and ST11 (n = 4; 5.7%). These 4 STs accounted for 38.6% (27/70) of the total K. pneumoniae isolates. Further, 34 STs were represented by 34 isolates, respectively. Clustering analysis by eBURST showed that these 42 STs could be grouped into 8 clonal complexes (CCs) and 9 singletons with the stringent default definition of sharing the same sequences at at least 6 of 7 loci. CC37 was found to be the predominant type, because 57.1% (24/42) of the isolates belonged to this group, as shown in Fig. 2.
FIG 2.
Comparative eBURST analyses. The population snapshot shows the clonal assignment of the STs presented in this study compared to that of the STs in the entire K. pneumonia MLST isolate database. Each black dot represents one ST in the database. The blue dots indicate individual founders, while yellow spots are subfounders. The pink circle highlights the matching ST type in this study with the database. The different clonal complexes are shown in red.
We found a strong correlation between ST23 and the K1 serotype. Interestingly, one of the novel STs, ST1265, was also associated with the K1 serotype. This new sequence type shares 6 alleles with ST23, indicating a new addition to the K1 serotype.
PFGE analysis showed that the 70 isolates revealed 57 different PFGE profiles. The banding patterns were designated PFGE types 1 to 57 (Fig. 1, “PFGE type” column), and a gel image is shown in Fig. 3. In addition, cKP isolates (n = 48) showed 43 different PFGE patterns, suggesting a polyclonal origin. The hvKP isolates (n = 22) had 14 different PFGE patterns, also indicating a polyclonal origin. However, among the 10 K1 serotype isolates in the hvKP group, we found two groups of isolates (n = 7 and n = 3, respectively) that shared similar PFGE patterns, suggesting a clonal origin.
FIG 3.
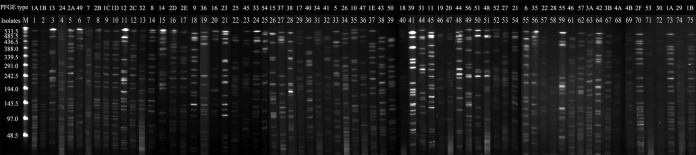
Gel image of PFGE result. Genomic DNA was digested using XbaI enzyme and subjected to pulsed-field gel electrophoresis.
Association between serotype and presence of the rmpA gene.
rmpA-carrying strains are often associated with the hypermucoviscous phenotype (18). As summarized in Fig. 1, we found that all of the string test-positive isolates were also rmpA positive. But 10 rmpA-positive strains were string test negative. Two rmpA and string test-positive isolates were K1, K2, K5, K20, K54, and K57 serotype negative.
DISCUSSION
In this study, we identified the hypervirulent K. pneumoniae in 31.4% (22/70) of patients with K. pneumoniae bacteremia. In addition, we found that the hvKP was more commonly encountered in community-acquired infections, including liver abscess, urological infection, and pneumonia complicated with bacteremia, especially among patients without known underlying diseases. This suggests that this strain may play an important role in community-acquired infections. Furthermore, we identified seven novel STs and four new alleles of K. pneumoniae, indicating the uniqueness of this bacterial population in China.
To date, hvKP infection has been reported mainly in the Asian population. Although recently there was a similar study reported from China (19), unlike the present report, there was little information on the characteristics of hvKP bloodstream infection. More importantly, we found two ESBL-producing hvKP strains. To date, antimicrobial resistance in hvKP strains has been rarely reported outside mainland China. Documenting the hvKP isolates would be useful not only for clinical doctors in Asia but also for clinicians in the Western countries where hvKP infection is being reported more and more frequently.
K. pneumoniae is encapsulated, with at least 78 capsular polysaccharide serotypes existing to date (20). The capsule is an important virulence factor, and some capsular serotypes, particularly K1, K2, K5, K16, K20, K54, K57, and KN1, are recognized as hypervirulent variants of K. pneumoniae (20, 21, 22). In this study, we identified 4 hvKP serotypes (K1, K2, K20, and K57), which agrees with previous reports from Taiwan (20, 23). However, none of our isolates was positive for K5 or K54, which has been reported previously (24, 25). In addition, we also noted that the K1 serotype was correlated with not only liver abscess but also urological infection, community-acquired pneumonia, and primary bloodstream infection, which agrees with previous studies (26, 27). Most importantly, we found that hvKP was able to cause serious infection in nonimmunocompromised hosts in the current study, because 60% (6/10) of the liver abscess patients had no underlying diseases.
Previously, it was documented that the ST23 strain was dominant in liver abscesses and clonally related in Taiwan patients (28, 29). However, some studies have shown that K. pneumoniae-related liver abscesses are not caused by a clonally spread strain (30). In the current study, we found that the six strains of ST23 K1 isolates that were associated with primary liver abscess also shared similar PFGE patterns, suggesting that they are clonally related, which supports previous findings from Taiwan (30). We also noticed that several isolates (n = 4) from patients with liver abscess had different PFGE patterns, demonstrating that genetic diversity also existed in liver abscess-causing hvKP (see Fig. 1).
In summary, we have shown clinical and molecular differences between hypervirulent K. pneumonia infections and classic K. pneumonia infections. We hope our study will draw attention from clinicians, which may lead to prompt recognition and successful management of hvKP infections.
ACKNOWLEDGMENTS
We thank the team of curators from the Institut Pasteur MLST system (Paris, France) for importing novel alleles, profiles, and/or isolates at http:/www.pasteur.fr/mlst. We thank Yongfei Hu, Chunxia Yang, Chunlei Wang, Shoushan Qu, Fang Li, Shanshan Wang, Jiuxin QU, Zhengjia Liu, and Peng Wang for their assistance.
We have no transparency declarations to make.
This work was financially supported by Beijing Natural Science Fund Project—Beijing City Board of Education Science and Technology Key Project (KZ201210025025) and National Natural Science Foundation of China (81070005/H0104) for Bin Cao.
Footnotes
Published ahead of print 30 June 2014
REFERENCES
- 1.Podschun R, Ullmann U. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11:589–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donnenberg MS. 2010. Enterobacteriaceae, p 2815–2833 In Mandell GL, Bennett JE, Dolin R. (ed), Principles and practice of infectious diseases, 7th ed. Churchill-Livingstone, Philadelphia, PA [Google Scholar]
- 3.Liu YC, Cheng DL, Lin CL. 1986. Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch. Intern. Med. 146:1913–1916. 10.1001/archinte.1986.00360220057011 [DOI] [PubMed] [Google Scholar]
- 4.Yeh KM, Kurup A, Siu LK, Koh YL, Fung CP, Lin JC, Chen TL, Chang FY, Koh TH. 2007. Capsular serotype K1 or K2, rather than magA and rmpA, is a major virulence determinant for Klebsiella pneumoniae liver abscess in Singapore and Taiwan. J. Clin. Microbiol. 45:466–471. 10.1128/JCM.01150-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung DR, Lee SS, Lee HR, Kim HB, Choi HJ, Eom JS, Kim JS, Choi YH, Lee JS, Chung MH, Kim YS, Lee H, Lee MS, Park CK, Korean Study Group for Liver Abscess 2007. Emerging invasive liver abscess caused by K1 serotype Klebsiella pneumoniae in Korea. J. Infect. 54:578–583. 10.1016/j.jinf.2006.11.008 [DOI] [PubMed] [Google Scholar]
- 6.Shon AS, Bajwa RP, Russo TA. 2013. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 15:107–118. 10.4161/viru.22718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng DL, Liu YC, Yen MY, Liu CY, Wang RS. 1991. Septicmetastatic lesions of pyogenic liver abscess. Their association with Klebsiella pneumoniae bacteraemia in diabetic patients. Arch. Intern. Med. 151:1557–1559 [PubMed] [Google Scholar]
- 8.Decré D, Verdet C, Emirian A, Le Gourrierec T, Petit JC, Offenstadt G, Maury E, Brisse S, Arlet G. 2011. Emerging severe and fatal infections due to Klebsiella pneumoniae in two university hospitals in France. J. Clin. Microbiol. 49:3012–3014. 10.1128/JCM.00676-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vila A, Cassata A, Pagella H, Amadio C, Yeh KM, Chang FY, Siu LK. 2011. Appearance of Klebsiella pneumoniae liver abscess syndrome in Argentina: case report and review of molecular mechanisms of pathogenesis. Open Microbiol. J. 5:107–113. 10.2174/1874285801105010107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang L, Bastian I, Warner M. 2013. Survey of Klebsiella pneumoniae bacteraemia in two South Australian hospitals and detection of hypermucoviscous phenotype and magA/rmpA genotypes in K. pneumoniae isolates. Infection 41:559–563. 10.1007/s15010-012-0374-y [DOI] [PubMed] [Google Scholar]
- 11.McCabe R, Lambert L, Frazee B. 2010. Invasive Klebsiella pneumoniae infections, California, USA. Emerg. Infect. Dis. 16:1490–1491. 10.3201/eid1609.100386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. 2012. Performance standards for antimicrobial susceptibility testing; 22nd informational supplement. CLSI M100-S22. CLSI, Wayne, PA [Google Scholar]
- 13.Fang CT, Lai SY, Yi WC, Hsueh PR, Liu KL, Chang SC. 2007. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin. Infect. Dis. 45:284–293. 10.1086/519262 [DOI] [PubMed] [Google Scholar]
- 14.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43:4178–4182. 10.1128/JCM.43.8.4178-4182.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518–1530. 10.1128/JB.186.5.1518-1530.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu VL, Hansen DS, Ko WC, Sagnimeni A, Klugman KP, von Gottberg A, Goossens H, Wagener MM, Benedi VJ, International Klebsiella Study Group 2007. Virulence characteristics of Klebsiella and clinical manifestations of K. pneumoniae bloodstream infections, Emerg. Infect. Dis. 13:986–993. 10.3201/eid1307.070187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1989. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nassif X, Honoré N, Vasselon T, Cole ST, Sansonetti PJ. 1989. Positive control of colanic acid synthesis in Escherichia coli by rmpA and rmpB, two virulence-plasmid genes of Klebsiella pneumoniae. Mol. Microbiol. 3:1349–1359. 10.1111/j.1365-2958.1989.tb00116.x [DOI] [PubMed] [Google Scholar]
- 19.Li W, Sun G, Yu Y, Li N, Chen M, Jin R, Jiao Y, Wu H. 2014. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin. Infect. Dis. 58:225–232. 10.1093/cid/cit675 [DOI] [PubMed] [Google Scholar]
- 20.Pan YJ, Fang HC, Yang HC, Lin TL, Hsieh PF, Tsai FC, Keynan Y, Wang JT. 2008. Capsular polysaccharide synthesis regions in Klebsiella pneumoniae serotype K57 and a new capsular serotype. J. Clin. Microbiol. 46:2231–2240. 10.1128/JCM.01716-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chuang YP, Fang CT, Lai SY, Chang SC, Wang JT. 2006. Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 193:645–654. 10.1086/499968 [DOI] [PubMed] [Google Scholar]
- 22.Cheng NC, Yu YC, Tai HC, Hsueh PR, Chang SC, Lai SY, Yi WC, Fang CT. 2012. Recent trend of necrotizing fasciitis in Taiwan: focus on monomicrobial Klebsiella pneumoniae necrotizing fasciitis. Clin. Infect. Dis. 55:930–939. 10.1093/cid/cis565 [DOI] [PubMed] [Google Scholar]
- 23.Lin CJ, Lin CY, Li WY, Hsiue HC, Huang YT, Ruan SY, Wang JT, Hsueh PR. 2009. Repeated bacteraemia with subsequent septic arthritis caused by Klebsiella pneumoniae capsular serotype K57 in a patient with diabetes. Clin. Infect. Dis. 49:1284–1286. 10.1086/605689 [DOI] [PubMed] [Google Scholar]
- 24.Chen YJ, Chen SY, Wang JT, Hsueh PR. 2009. Mycotic aneurysm caused by gas-forming serotype K5 Klebsiella pneumoniae. Int. J. Infect. Dis. 13:e47–e48. 10.1016/j.ijid.2008.06.008 [DOI] [PubMed] [Google Scholar]
- 25.Hsu CR, Lin TL, Chen YC, Chou HC, Wang JT. 2011. The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiology 157:3446–3457. 10.1099/mic.0.050336-0 [DOI] [PubMed] [Google Scholar]
- 26.Ko WC, Paterson DL, Sagnimeni AJ, Hansen DS, Von Gottberg A, Mohapatra S, Casellas JM, Goossens H, Mulazimoglu L, Trenholme G, Klugman KP, McCormack JG, Yu VL. 2002. Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg. Infect. Dis. 8:160–166. 10.3201/eid0802.010025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee HC, Chuang YC, Yu WL, Lee NY, Chang CM, Ko NY, Wang LR, Ko WC. 2006. Clinical implications of hypermucoviscosity phenotype in Klebsiella pneumoniae isolates: association with invasive syndrome in patients with community-acquired bacteraemia. J. Intern. Med. 259:606–614. 10.1111/j.1365-2796.2006.01641.x [DOI] [PubMed] [Google Scholar]
- 28.Turton JF, Englender H, Gabriel SN, Turton SE, Kaufmann ME, Pitt TL. 2007. Genetically similar isolates of Klebsiella pneumoniae serotype K1 causing liver abscesses in three continents. J. Med. Microbiol. 56:593–597. 10.1099/jmm.0.46964-0 [DOI] [PubMed] [Google Scholar]
- 29.Lau YJ, Hu BS, Wu WL, Lin YH, Chang HY, Shi ZY. 2000. Identification of a major cluster of Klebsiella pneumoniae isolates from patients with liver abscess in Taiwan. J. Clin. Microbiol. 38:412–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng HP, Chang FY, Fung CP, Siu LK. 2002. Klebsiella pneumoniae liver abscess in Taiwan is not caused by a clonal spread strain. J. Microbiol. Immunol. Infect. 35:85–88 [PubMed] [Google Scholar]