Abstract
Silver in its ionic form (Ag+), but not the bulk metal (Ag0), is toxic to microbial life forms and has been used for many years in the treatment of wound infections. The prevalence of bacterial resistance to silver is considered low due to the nonspecific nature of its toxicity. However, the recent increased use of silver as an antimicrobial agent for medical, consumer, and industrial products has raised concern that widespread silver resistance may emerge. Pseudomonas aeruginosa is a common pathogen that produces pyocyanin, a redox toxin and a reductant for molecular oxygen and ferric (Fe3+) ions. The objective of this study was to determine whether pyocyanin reduces Ag+ to Ag0, which may contribute to silver resistance due to lower bioavailability of the cation. Using surface plasmon resonance spectroscopy and scanning electron microscopy, pyocyanin was confirmed to be a reductant for Ag+, forming Ag0 nanoparticles and reducing the bioavailability of free Ag+ by >95% within minutes. Similarly, a pyocyanin-producing strain of P. aeruginosa (PA14) reduced Ag+ but not a pyocyanin-deficient (ΔphzM) strain of the bacterium. Challenge of each strain with Ag+ (as AgNO3) gave MICs of 20 and 5 μg/ml for the PA14 and ΔphzM strains, respectively. Removal of pyocyanin from the medium strain PA14 was grown in or its addition to the medium that ΔphzM mutant was grown in gave MICs of 5 and 20 μg/ml, respectively. Clinical isolates demonstrated similar pyocyanin-dependent resistance to Ag+. We conclude that pseudomonal silver resistance exists independently of previously recognized intracellular mechanisms and may be more prevalent than previously considered.
INTRODUCTION
Silver serves no known biological function, and while the bulk metal (Ag0) is relatively inert, in the ionic state (Ag+), it is highly toxic to most microbial life forms. With its broad-spectrum antimicrobial properties, silver has been employed extensively in various forms over many years for the topical treatment of infected wounds and burns. Ag+ toxicity arises from nonspecific binding to DNA and the inactivation of enzymes and membrane proteins, including those associated with electron transport (1). This ability to target multiple cellular sites suggests that microbial resistance to Ag+ should be low (2), although its occurrence is well recognized (3–6). The current view of bacterial resistance to silver is that it is genetically determined and requires silver-binding proteins and associated efflux pumps to export the bound cation from the cell (4, 5). Recently, concern has been raised that the increased use of silver products for clinical, industrial, and domestic antimicrobial use may give rise to widespread silver resistance due to horizontal gene transfer (6).
Several reports have identified the reduction of soluble Ag+ to colloidal or nanoparticulate Ag0 by various bacterial species (7–10), including Pseudomonas aeruginosa (11), although in all cases the specific reductive mechanisms involved remain unknown (12). We postulated that bacterial reduction of Ag+ may represent a novel resistance mechanism, as it removes Ag+ from the bacterial environment before it enters the cell and decreases dependence on intracellular silver-binding proteins and efflux systems. P. aeruginosa is a common wound pathogen that produces the redox-active phenazine derivative pyocyanin, substantial amounts of which have been identified in burn wound exudates of infected patients (13). Pyocyanin acts as an extracellular electron carrier with NADH as the initial endogenous reductant and molecular oxygen acting as the terminal electron acceptor (14). Pyocyanin is also well recognized for its ability to reduce ferric (Fe3+) ions to the ferrous (Fe2+) state, a soluble form that can readily be taken up by P. aeruginosa for nutritional requirements (see Fig. S1 in the supplemental material) (15, 16). In aqueous solution (at 25°C), the standard electrode potential (Eo) for the iron redox couple (Fe3+/Fe2+) is 770 mV, whereas for the silver redox couple (Ag+/Ag0), it is 800 mV. We therefore further postulated that the very similar standard electrode potentials of these metal redox couples predict that Ag+ should be reducible by reduced pyocyanin (Eo = −34.0 mV) (17). Consequently, this study was undertaken to determine whether pyocyanin reduces Ag+ to Ag0, resulting in a lowering of the Ag+ concentration consistent with the survival of P. aeruginosa.
MATERIALS AND METHODS
Chemicals and reagents.
All chemicals and reagents were purchased from Sigma-Aldrich (Sydney, Australia). AgNO3 was used as the source of Ag+ for all of the studies described.
Bacterial strains and culture media.
Clinical isolates of P. aeruginosa were obtained from the Sydney South West Pathology Service (Liverpool Hospital, Liverpool, NSW, Australia), and their antibiotic sensitivities are presented in Table S1 in the supplemental material. In addition, we compared two strains of P. aeruginosa, strain PA14, a wild-type producer of pyocyanin, and an isogenic mutant, PA14 ΔphzM (ΔphzM), which is unable to produce the toxin due to a deficiency in methyltransferase production, which is required to methylate position 5 of the phenazine ring. However, the ΔphzM strain does produce substantial amounts of phenazine-1-carboxylic acid (PCA), a precursor in the biosynthesis of pyocyanin (see Fig. S2 in the supplemental material), which exhibits redox activity (Eo = −116 mV) (15). Although several PA14 pyocyanin-null mutants exist (e.g., ΔmvfR and ΔphnAB mutants), the ΔphzM mutant was chosen for this study, as the lesion is close to the final step in pyocyanin biosynthesis, whereas the lesions for other mutations are sufficiently distal as to cause additional phenotypic changes that may confound the interpretation of our results. The wild-type PA14 strain and isogenic mutant ΔphzM strain were maintained on Mueller-Hinton (II) agar (MHA) slopes, and all incubations were conducted in Mueller-Hinton (II) broth (MHB) at 32°C with agitation at 80 rpm for 24 h. Stock cultures of the P. aeruginosa strains and all subsequent incubations were undertaken in the dark, as pyocyanin is light sensitive (18). Preliminary experiments were undertaken to ensure that the growth characteristics of the two strains were equivalent. Initial cultures of each strain were set up by inoculating one loop (10 μl) of the PA14 or ΔphzM strain into MHB (20 ml) and incubating for 24 h. A 2% inoculum of each strain was then subcultured in MHB (20 ml) for a further 24 h. Before use, low-speed supernatants of these early stationary-phase cultures were prepared by centrifugation at 200 × g for 10 min to remove cell debris and aggregated material.
Extraction of phenazines.
Pyocyanin and PCA were extracted from the bacterial low-speed supernatants by liquid/liquid phase partitioning three times with equal volumes of chloroform followed by volume reduction of the organic phase under nitrogen. Quantitation of the phenazines was achieved by spectrophotometry in methanol at 690 nm for pyocyanin using an extinction coefficient of 5,816 and for PCA at 368 nm using an extinction coefficient of 3,020 (19). The identity of PCA was confirmed by tandem mass spectroscopy.
Synthesis of pyocyanin.
Chemically pure pyocyanin was prepared by photolysis of phenazine methosulfate as described previously (20). The crude material was purified by high-performance thin-layer chromatography (silica gel G; Merck, Sydney, Australia), and the structure and purity were confirmed by tandem mass spectroscopy (21).
Surface plasmon resonance (SPR) spectroscopy.
Mie theory describes the behavior of light interacting with particles of nanometer scale dimensions. On excitation by light, localized surface plasmons arise from the coherent oscillation of conduction band electrons at the interface of metal nanoparticles and the medium in which they are embedded (22). The extinction of light (QExt) by nanoparticles is due to absorption (QAbs) and scattering (QSca) effects of incident light, i.e.,
| (1) |
From Mie theory, for a particle of radius R (where 2R ≪ λ [λ is the wavelength of light]) and a frequency-dependent complex dielectric function such that
| (2) |
where ε′ and ε″ are the real and imaginary parts, respectively, of the dielectric function of the particle embedded in a medium of dielectric constant εm, the extinction cross section (CExt) can be expressed as follows:
| (3) |
If ε″ is small, when
| (4) |
the SPR condition is met, i.e., a localized surface plasmon resonance occurs and as
| (5) |
an extinction maximum can be detected spectroscopically. For Ag0, this condition occurs in the visible wavelength range. The theoretical SPR spectrum for a single silver nanosphere was calculated using the following parameters: wavelength range = 300 to 800 nm at a resolution of 1 nm for a nanoparticle of 60-nm diameter in water at 32°C. The theoretical QExt values are expressed in arbitrary units. Experimental SPR spectra due to pyocyanin-mediated reduction of Ag+ were determined by incubating AgNO3 (100 μM) with pyocyanin (5 μM) and NADPH (100 μM) as the source of reducing equivalents (23) in water (pH 7.0; 32°C; 18 MΩ · cm−1) followed by scanning spectrophotometry to observe the emergence of SPR bands associated with nanoparticulate Ag0 formation. The experimental QExt values are expressed in real units. To obtain SPR spectra in the presence of bacteria, low-speed supernatants (200 × g for 10 min) of the PA14 and ΔphzM strains were prepared and incubated with AgNO3 (100 μM) and monitored for the emergence of SPR bands at 32°C in MHB using a dual-beam spectrophotometer with the reference cell containing an identical suspension of bacteria in MHB in the absence of AgNO3.
Determination of Ag0 nanoparticle size and composition.
Following the reduction of Ag+ by the pyocyanin/NADPH system described above, drops (10 μl each) of the reaction mixture were placed on lacy carbon copper grids (300 mesh; ProSciTech, Australia) and air dried for transmission electron microscopy (TEM). Similarly, drops (10 μl each) of the reaction mixture were placed on aluminum stubs, air dried, and examined by scanning electron microscopy (SEM). The elemental composition of the nanoparticles was determined using energy dispersive X-ray (EDX) analysis of the nanoparticles in spot mode. All analyses were performed using a field emission scanning electron microscope (JEOL JSM-7001F; JEOL, Japan). To determine the nanoparticle size range, 300 particles were measured from 4 independent SEM fields using image analysis software (ImageJ, version 1.46; NIH).
Assay for free Ag+.
The concentration of free Ag+ was determined using the sensitive dimethylaminobenzylidene rhodanine method, as described previously (24). Ag0 nanoparticles were removed prior to assay of Ag+ by centrifugation at 5,000 rpm for 5 min using an Eppendorf microcentrifuge.
MIC and MBC assays.
MIC and minimum bactericidal concentration (MBC) assays were performed using low-speed supernatants diluted to an optical density at 600 nm (OD600) of 0.3 with MHB. This dilution ensured equivalent numbers of bacteria of each strain, verified by determining the number of CFU, while maintaining adequate pyocyanin concentrations. Bacterial suspensions of each strain (3 × 108 CFU/ml) were incubated at 32°C for 24 h with serial dilutions of AgNO3 (0 to 40 μg/ml). Prior to the addition of AgNO3, the suspensions were incubated for a minimum of 30 min to allow for bacterial reduction of pyocyanin, as it is readily oxidized by dissolved oxygen (21) during preparation procedures. Aliquots were subsequently plated onto MHA plates and incubated for a further 24 h at 37°C, the number of CFU was counted, and the MIC, MBC, and percent survival were determined.
Time course of bacterial killing by Ag+.
To determine the time course for killing of the PA14 strain by Ag+ in the absence of pyocyanin, the bacterial suspension (OD of 0.3) was centrifuged at 2,500 × g for 10 min, and the subsequent supernatant was discarded. The bacterial pellet was reconstituted in fresh MHB (1 ml) containing AgNO3 (10 μg/ml) and incubated at 32°C at 80 rpm. Aliquots were removed at intervals, and the number of CFU was determined by serial dilution.
Time course of pyocyanin production by the PA14 strain.
The time course for pyocyanin production by the PA14 strain was determined by centrifuging bacterial suspensions (OD of 0.3) at 2,500 × g for 10 min. The supernatants were discarded, and the bacterial pellets were reconstituted in fresh MHB (1 ml) and incubated at 32°C at 80 rpm. Pyocyanin was extracted and quantified at intervals as described above. Prior to extraction, aliquots were removed for determination of CFU.
Statistics.
Statistical analyses were performed using Prism 6 software (GraphPad Software, San Diego, CA, USA). Student's t test was used for comparison of Ag+ availability. Analysis of variance (ANOVA) with Dunnett's posttest comparison was used to determine significance values for the MIC studies. Spearman's correlation was used to determine the correlation coefficient between pyocyanin production by clinical isolates and Ag+ resistance.
RESULTS
Reduction of Ag+ by pyocyanin.
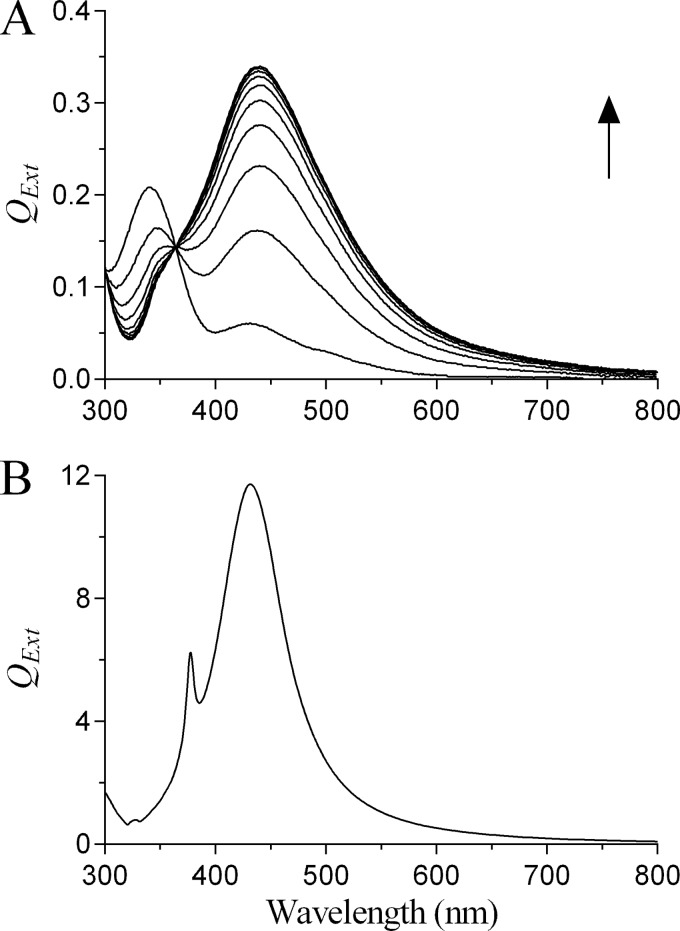
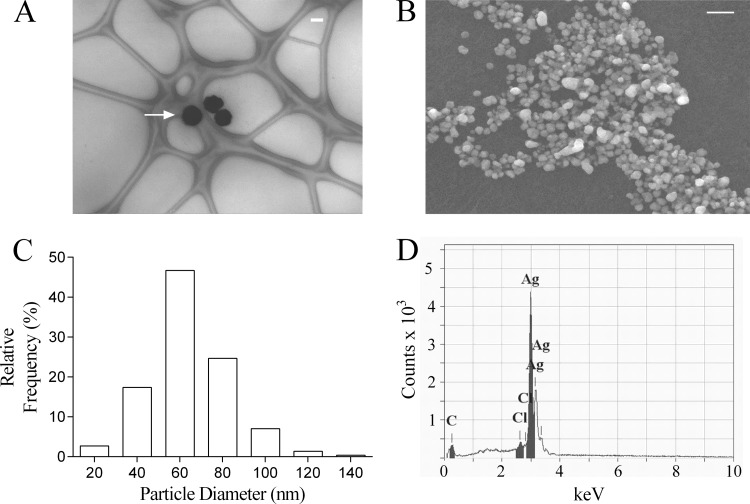
On incubation of Ag+ with pyocyanin in the presence of NADPH, SPR spectra indicating reduction of Ag+ to Ag0 were obtained (Fig. 1A). The experimental SPR spectra were consistent with the theoretical SPR spectrum for Ag0 generated using Mie theory (Fig. 1B). NADPH alone did not reduce Ag+ (data not shown). Ag0 nanoparticle formation was confirmed by TEM and SEM imaging (Fig. 2A and B) and, using image analysis, were found to be polydisperse with diameters ranging from approximately 10 to 140 nm with a mean diameter of 60 nm (Fig. 2C). Using energy-dispersive X-ray analysis, we observed emission peaks at approximately 3 keV, a characteristic of metallic silver, and determined the composition of the nanoparticles to be greater than 98% elemental silver (Fig. 2D).
FIG 1.
Reduction of Ag+ to Ag0 by pyocyanin. (A) SPR spectra were obtained by incubating Ag+ (as AgNO3; 100 μM) with pyocyanin (5 μM) and NADPH (100 μM) in water (pH 7.0; 32°C). Scans were repeated at 2-min intervals, and the arrow indicates the direction of increasing time (data are representative scans of experiments performed in triplicate with QExt values in real units). (B) Theoretical SPR spectrum of a single Ag0 nanoparticle of 60-nm diameter in water calculated using Mie theory with QExt values in arbitrary units.
FIG 2.
Ag0 nanoparticle analysis by electron microscopy. (A and B) Representative images of Ag0 nanoparticles (white arrow) by TEM (A) (on lacy carbon copper grid [300 mesh]) and by SEM (B). The Ag0 nanoparticles were prepared by the reduction of Ag+ (as AgNO3; 100 μM) by pyocyanin (5 μM) with NADPH (100 μM) in water (pH 7.0; 32°C). Bars, 100 nm (A) and 200 nm (B). (C) Histogram indicating the size distribution of the nanoparticles (300 particle measurements from four independent preparations with histogram bin width set at 20 nm). (D) Energy dispersive X-ray analysis of Ag0 nanoparticles to determine elemental composition. The analysis was undertaken in spot mode and is representative of four independent samples.
We next quantified the extent of reduction of Ag+ by the pyocyanin/NADPH system. The data from Fig. 1A indicated that the reaction was rapid, going to completion within 12 min of initiation. We therefore determined the Ag+ concentration at the initiation and termination of the reaction using the sensitive and specific dimethylaminobenzylidene rhodanine method after removal of the Ag0 nanoparticles by centrifugation. A reduction of approximately 95% in the available Ag+ concentration was achieved 12 min after initiating the reaction (Fig. 3).
FIG 3.
Bioavailability of Ag+ following reduction by pyocyanin. A solution of Ag+ (100 μM) was reduced in the presence of pyocyanin (5 μM) and NADPH (100 μM) in water (pH 7.0; 32°C), and the concentration of Ag+ was determined at the commencement (0 min) and conclusion (12 min) of the reaction (values are means plus standard deviations [error bars] from three independent experiments).
Reduction of Ag+ by P. aeruginosa is mediated by pyocyanin.
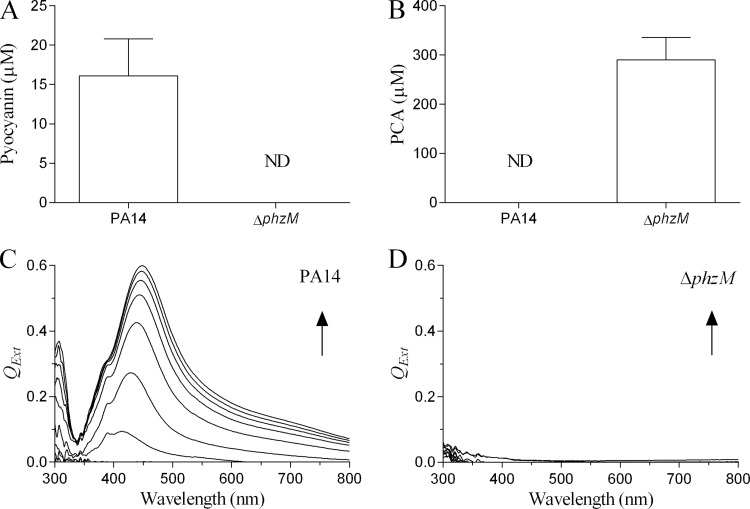
Although reduction of Ag+ to Ag0 by P. aeruginosa (11) and other bacterial species has been reported (7–10), the mechanism remains unexplained. To determine whether pyocyanin participated in the reduction of Ag+ by P. aeruginosa, we compared its reduction by the PA14 and ΔphzM strains. In preliminary experiments, we established that the growth characteristics of the two strains were identical in Mueller-Hinton (II) broth (data not shown). Using liquid/liquid phase partitioning of 24-h culture supernatants with chloroform, we quantified the amount of pyocyanin and PCA produced by the two strains. The mean concentration of pyocyanin produced by the PA14 culture was 16.1 ± 4.7 μM with no detectable PCA (Fig. 4A). The inability of the ΔphzM strain to produce pyocyanin was confirmed, although it secreted appreciable quantities of PCA (290.0 ± 45.3 μM). On incubation of low-speed bacterial culture supernatant with Ag+ (100 μM), we found that the cation was rapidly reduced to Ag0 within minutes by the PA14 strain (Fig. 4C), whereas reduction did not occur with the ΔphzM strain (Fig. 4D). Therefore, although PCA is redox active and is reduced by NADH (data not shown), this close analogue of pyocyanin is unable to reduce Ag+ under the conditions indicated. The SPR spectra shown in Fig. 4C are consistent with those produced by purified pyocyanin (Fig. 1A) and the theoretical SPR spectrum generated using Mie theory (Fig. 1B). These data demonstrate that P. aeruginosa can use Ag+ as a terminal electron acceptor. Moreover, the reduction of Ag+ by the PA14 strain occurred within a time frame comparable to that under cell-free conditions (Fig. 1A), indicating that the bacterium is able to reduce toxic levels of Ag+ within minutes and at pyocyanin concentrations found under clinical conditions (13).
FIG 4.
Pyocyanin and PCA production and reduction of Ag+ to Ag0 by P. aeruginosa strains. (A and B) Recovery of pyocyanin and PCA from 24-h low-speed supernatants (200 × g for 10 min) of the P. aeruginosa PA14 (A) and ΔphzM (B) strains after three sequential extractions with chloroform and quantitation by spectrophotometry (ND, not detected). (C and D) SPR spectra produced by incubating the low-speed supernatants of PA14 (C) and ΔphzM (D) strains with Ag+ (100 μM) at 32°C. Scans were repeated at 2-min intervals, and arrows indicate direction of increasing time (data are representative scans of experiments performed in triplicate).
Correlation of silver resistance with pyocyanin production by clinical isolates of P. aeruginosa.
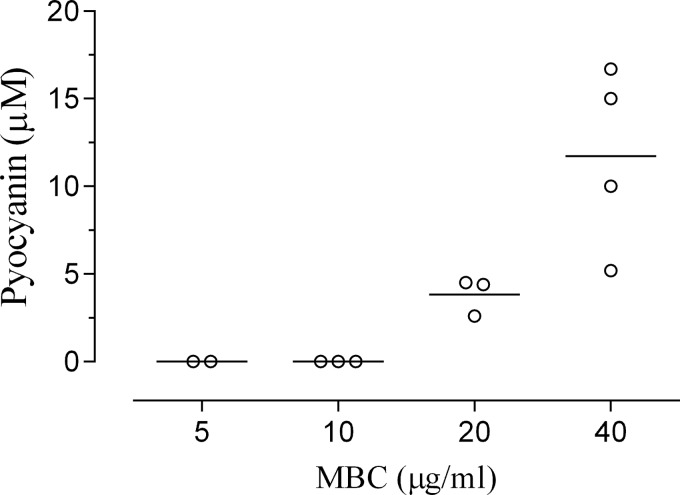
To determine whether pyocyanin production is an important determinant for avoiding Ag+ toxicity, we examined the survival of 12 clinical isolates of P. aeruginosa in the presence of AgNO3 (0 to 40 μg/ml) with differing capacities for pyocyanin production. We observed that the pyocyanin-deficient clinical isolates were more sensitive to AgNO3 with MBCs of 5 to 10 μg/ml, whereas those able to produce the toxin exhibited higher MBCs (20 to 40 μg/ml; Fig. 5). The correlation between pyocyanin production and resistance to Ag+ was positive and highly significant (P < 0.0001; correlation coefficient = 0.9456; n = 12) by Spearman's test.
FIG 5.
Correlation of pyocyanin production and Ag+ resistance by clinical isolates. Pyocyanin production by 12 clinical isolates of P. aeruginosa was determined by sequential chloroform extractions with quantitation by spectrophotometry. Suspensions of clinical isolates containing equivalent numbers of CFU were exposed to serial dilutions of AgNO3 (0 to 40 μg/ml) for 24 h. The surviving bacteria were transferred to MHA plates, the number of CFU was counted, and the minimum bactericidal concentration (MBC) was determined. The data were analyzed using the Spearman correlation method (P < 0.0001; correlation coefficient = 0.9456; n = 12). Each symbol represents the value for a clinical isolate of P. aeruginosa, and each bar represents the mean for a group of isolates.
Pyocyanin confers resistance by P. aeruginosa to Ag+.
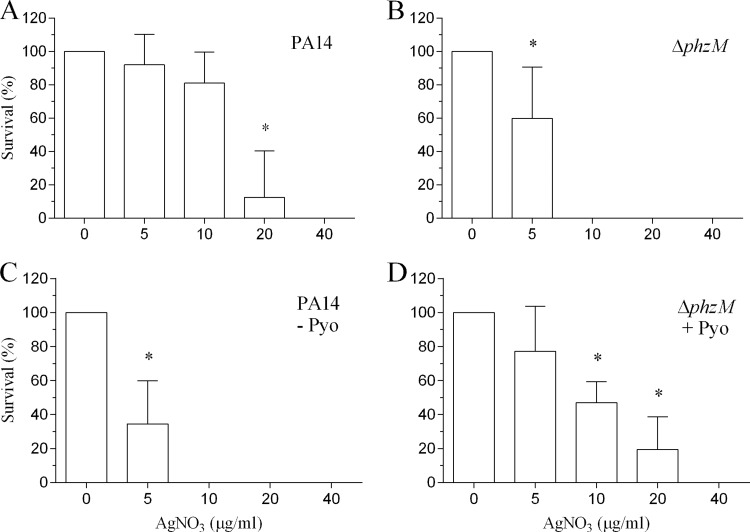
The conversion of toxic Ag+ to insoluble nontoxic Ag0 by pyocyanin effectively reduces the bioavailable concentration of Ag+ (Fig. 3), suggesting that the PA14 strain should be resistant to higher Ag+ concentrations compared to the ΔphzM strain. To demonstrate this, we exposed equivalent numbers of each strain (3 × 108 CFU/ml) to serial dilutions of AgNO3 (0 to 40 μg/ml) for 24 h. The MIC for the PA14 strain was 20 μg/ml (Fig. 6A), whereas the MIC for the ΔphzM strain was 5 μg/ml (Fig. 6B). The mean pyocyanin concentration of the control PA14 samples was 10.4 ± 1.1 μM (n = 5). To confirm that pyocyanin is essential for resistance, we centrifuged the PA14 culture, removed the supernatant, and resuspended the bacterial pellet in an equal volume of fresh medium devoid of pyocyanin. After the bacteria were challenged with AgNO3 for 24 h, we observed a 4-fold increase in sensitivity to Ag+ with a MIC comparable to that of the ΔphzM strain (Fig. 6C). Conversely, the addition of pyocyanin (final concentration of 20 μM) to the ΔphzM strain resulted in greater resistance to higher Ag+ concentrations with the MIC increasing to 20 μg/ml (Fig. 6D).
FIG 6.
Pyocyanin confers resistance to Ag+ toxicity by P. aeruginosa. Suspensions of the PA14 and ΔphzM strains containing equivalent numbers of CFU were exposed to serial dilutions of AgNO3 (0 to 40 μg/ml) for 24 h. The surviving bacteria were transferred to MHA plates, the number of CFU was counted, and the percent survival was determined. (A) The MIC for the PA14 strain was 20 μg/ml. (B) In contrast, the MIC for the ΔphzM strain was 5 μg/ml. (C) Removal of pyocyanin from the PA14 strain by centrifugation and resuspension in fresh MHB (devoid of pyocyanin [− Pyo]) resulted in increased sensitivity to Ag+. (D) The addition of pyocyanin (20 μM) to the ΔphzM strain increased the MIC to 20 μg/ml. Data represent means plus standard deviations from five independent experiments> Values that are significantly different (P < 0.001) from the value for no AgNO3 by ANOVA with Dunnett's multiple-comparison test are indicated by an asterisk.
Time course of pyocyanin production and bacterial killing by Ag+.
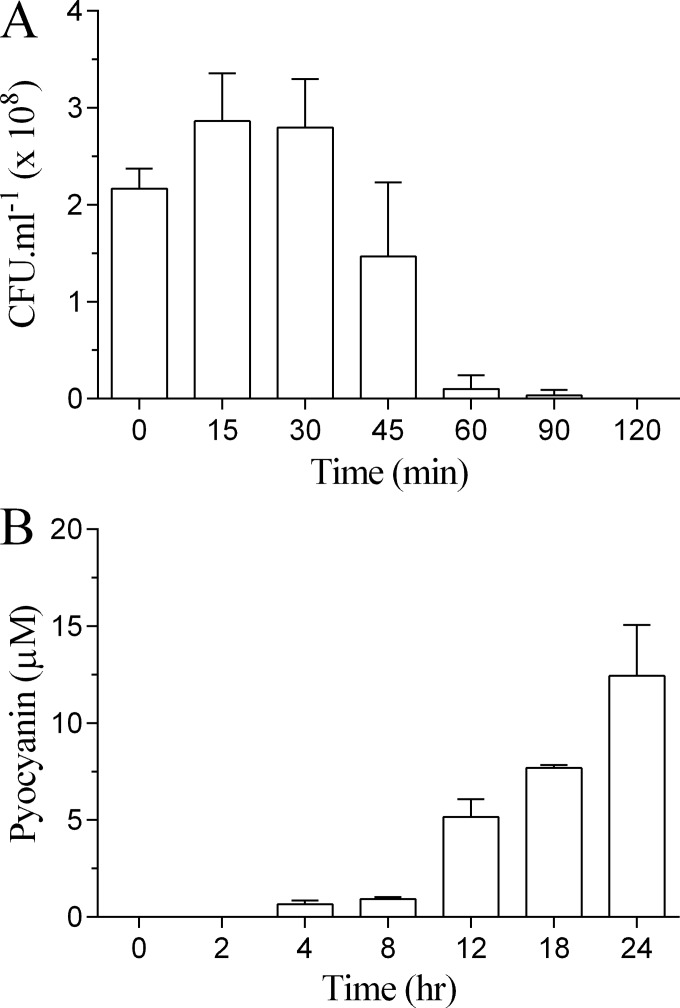
We further examined the relationship between de novo pyocyanin production by the PA14 strain following reconstitution in fresh medium and Ag+-dependent bacterial killing. In the absence of pyocyanin, the PA14 strain was susceptible to Ag+ (10 μg/ml) with no survival at 2 h (Fig. 7A). In contrast, and in the absence of AgNO3, detectable levels of pyocyanin were not observed until after 2 h of incubation following reconstitution in fresh medium (Fig. 7B).
FIG 7.
Time course for bacterial killing by Ag+ and pyocyanin production by strain PA14. (A) The time course for killing of the PA14 strain by Ag+, in the absence of pyocyanin, was determined by centrifugation of a 24-h culture, discarding the supernatant and resuspending the pellet in fresh MHB (1 ml; OD of 0.3) containing AgNO3 (10 μg/ml) and incubating at 32°C at 80 rpm. Aliquots were removed at intervals, and the number of CFU was determined by serial dilution. (B) The time course for pyocyanin production, in the absence of AgNO3, by the PA14 strain was determined by centrifuging a 24-h culture, discarding the supernatant, and resuspending the pellet in fresh MHB (1 ml; OD of 0.3) followed by incubation at 32°C at 80 rpm. Pyocyanin was extracted and quantified at the indicated intervals. Results represent means plus standard deviations from three independent experiments.
DISCUSSION
Within the clinical setting, the genes considered responsible for bacterial silver resistance (the sil genes) are widely viewed to be the key determinants for resistance, and several studies have considered their absence as sufficient evidence for Ag+ sensitivity (1–5). One study found sil-positive bacterial strains to have MIC values (for AgNO3) of ≥5 μg/ml, while for sil-negative strains, the MIC ranged from 1 to 2.5 μg/ml, at least a 2- to 5-fold difference in sensitivity (25). While differing test conditions do not permit direct comparisons, we identified a 4-fold difference between the MIC values for the PA14 strain and the same strain without pyocyanin, indicating that pyocyanin-mediated resistance provides a survival benefit against Ag+ comparable to that of the sil-based genetic mechanisms. Thus, our data demonstrate that further resistance mechanisms can be operative and independent of the sil genes and the silver-binding proteins and efflux pumps they encode. Moreover, our data indicate that P. aeruginosa does not need to acquire Ag+ resistance, as those clinical isolates that produce pyocyanin already possess intrinsic or constitutive resistance. It follows that the resistance of P. aeruginosa to Ag+ can be variable due to the greater or lesser availability of pyocyanin, as indicated in Fig. 5, and the extent that pyocyanin is in the reduced state in order to subsequently reduce Ag+. The equilibrium between the reduced and oxidized forms of the toxin will depend on the intrinsic reductive capacity of the bacterium and the availability of competing electron acceptors, such as molecular oxygen, in the extracellular environment. The sensitivity of P. aeruginosa strains to Ag+ (as AgNO3) is reported to range from MICs of 8 to 70 μg/ml (26, 27) with resistance considered to be unstable on repeated subculture (28), consistent with variable pyocyanin production.
We found a highly significant positive correlation between pyocyanin production by clinical isolates and resistance to Ag+. Furthermore, the MIC for the PA14 strain for Ag+ ranged from 5 to 20 μg/ml and was directly dependent on pyocyanin availability (Fig. 6). Therefore, we conclude that the sensitivity of a bacterial strain to Ag+ is determined not only by the bacterium but also by the immediate extracellular environment modified by the bacterium due to secretion of redox-active metabolites. At present, there remains a lack of consensus as to procedures for silver sensitivity testing. Current testing methods frequently require preparation of bacterial samples that result in loss of extracellular compounds, such as pyocyanin, that may confer resistance. By using these methods, P. aeruginosa would appear to exhibit greater sensitivity to silver than it otherwise would. Our data suggest silver testing protocols need to be reconsidered to take extracellular resistance mechanisms into account. SPR spectroscopy represents a simple yet effective screening methodology to identify bacterial species capable of the reductive inactivation of Ag+ and thus potentially for the identification of Ag+-resistant strains when used in conjunction with the MIC assay. However, the wavelength at which SPR maxima occur, and corresponding extinction coefficients, are variable and dependent on the dielectric constant of the incubation medium (εm) and the cross-sectional area of the resultant nanoparticles (equation 3). Indeed, our data demonstrate this to be the case with polydisperse nanoparticle formation (Fig. 2B and C), resulting in a substantial red-shift (approximately 10 to 20 nm) in the maximal wavelength over time (Fig. 1A and 4B), as predicted by equation 3, even though our value for εm remained constant.
The extracellular reductive mechanism adopted by P. aeruginosa for eliminating Ag+ from its immediate surroundings likely contributes to the environmental success of the organism. The biosynthesis and operation of specific silver-binding proteins and efflux pumps result in a high metabolic cost for bacteria. In contrast, pyocyanin is a low-molecular-weight and efficient redox cycling compound (23) that contributes to the maintenance of intracellular redox homeostasis by transferring reducing equivalents from the bacterium to molecular oxygen, or other acceptors, in the extracellular environment (14). The ability of pyocyanin to redox cycle using Ag+ as a terminal electron acceptor allows for small amounts of the compound to detoxify relatively high concentrations of the cation, as evidenced by the data shown in Fig. 3, and assist cellular respiration in an environment high in Ag+ where the cation is known to disrupt normal membrane-bound electron transport systems (1). Our data (Fig. 7) indicate that Ag+-dependent bacterial killing is a rapid process; therefore, the ability of pyocyanin to neutralize Ag+ within minutes would likely confer a survival advantage in an environment high in Ag+. Although our current data are specific to P. aeruginosa, other bacteria may similarly minimize Ag+ toxicity, and heavy metal ion toxicity in general, by the production of species-specific extracellular redox-active metabolites with suitable redox potentials, a view supported by the large number of bacterial species known to reduce Ag+ and other metal ions to nanoparticles (7–12). Indeed, the reduction of Ag+ by extracellular polymeric substances of Escherichia coli with reduced bactericidal activity has recently been demonstrated (29).
Our findings have important clinical implications. P. aeruginosa has been reported to account for greater than 50% of bacterial species isolated from chronic wounds (30, 31) where it typically exists in biofilm communities (32), and it is a major cause of infection for burn patients. Pyocyanin is considered to play an important regulatory role in pseudomonal biofilm organization and behavior (33), where it is actively maintained in the reduced state (34), the active form required for Ag+ reduction. It has been suggested that the concentration of free Ag+ available from current wound dressings is 1 to 10 orders of magnitude too low for a bactericidal effect against P. aeruginosa (35). Similarly, others found the concentrations of Ag+ produced by a range of silver preparations to be inadequate against the bacterium (36). It is apparent that different silver-containing products provide different levels of antibacterial activity and specificity for various bacteria (37) with considerable controversy as to the benefits of topical silver-containing creams (predominately silver sulfadiazine) compared to silver dressings (nanoparticulate silver) in their antibacterial activity (38, 39). Moreover, Ag+ bioavailability can be greatly influenced by various factors, including solutes in biological fluids such as chloride ion (3) and free sulfhydryl groups (40). Nevertheless, the efficacy of silver preparations is determined by the amount of bioavailable ionic silver they release, not the total amount of bound (sulfadiazine) or metallic (colloidal or nanoparticulate) silver present (41). The solubility product of silver sulfadiazine is 8.12 × 10−12 at 25°C (42), and in a simple nonbinding well-equilibrated medium at physiologic pH, the Ag+ concentration can be as low as 2.6 μM, although in the presence of human serum, which better approximates wound fluid, it can rise to 145 μM (43). Thus, the Ag+ concentrations used in the present study are relevant to the expected concentration of Ag+ available under clinical conditions. Similarly, the concentrations of pyocyanin shown to confer resistance in the present study are consistent with concentrations of the virulence factor demonstrated to be present in wound exudates (up to approximately 25 μM) from patients (13). Therefore, the production of pyocyanin by P. aeruginosa may enable the bacterium to survive various forms of silver therapy and contribute to delayed wound healing (3) with resultant suboptimal patient outcomes.
Conclusions.
In conclusion, while the current consensus is that the prevalence of bacterial resistance to silver is low, our study indicates that resistance may indeed be much more prevalent than previously thought and can exist independent of the presence of genes and plasmids generally considered necessary for resistance. Our findings further suggest that bacterial silver sensitivity testing protocols require reevaluation to accommodate extracellular redox-mediated resistance mechanisms. Moreover, our study identifies the specific extracellular reductive mechanism by which a common bacterium is able to biosynthesize high-purity metallic nanoparticles. Little is known of the diversity and function of bacterial redox-active compounds. The ability of bacteria to use these compounds for the extracellular reduction of toxic heavy metal ions in their environment may prove to be a vital survival strategy for many species with profound clinical implications.
Supplementary Material
ACKNOWLEDGMENTS
We thank F. Ausubel for the kind gift of the PA14 strains; M. Belkina of the Electron Microscope Unit, University of Western Sydney for technical assistance with the electron microscopy work; R. Pickford of the Mass Spectrometry Unit, University of Western Sydney for performing the mass spectroscopy analyses; and M. Maley and H. Ziochos (Liverpool Hospital, NSW, Australia) for the clinical isolates. We further thank Li Ching Ooi for technical assistance with the time course studies for pyocyanin production.
Footnotes
Published ahead of print 7 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.03069-14.
REFERENCES
- 1.Percival SL, Bowler PG, Russell D. 2005. Bacterial resistance to silver in wound care. J. Hosp. Infect. 60:1–7. 10.1016/j.jhin.2004.11.014 [DOI] [PubMed] [Google Scholar]
- 2.Percival SL, Woods E, Nutekpor M, Bowler P, Radford A, Cochrane C. 2008. Prevalence of silver resistance in bacteria isolated from diabetic foot ulcers and efficacy of silver-containing wound dressings. Ostomy Wound Manage. 54(3):30–40 [PubMed] [Google Scholar]
- 3.Haefeli C, Franklin C, Hardy K. 1984. Plasmid-determined silver resistance in Pseudomonas stutzeri isolated from a silver mine. J. Bacteriol. 158:389–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta A, Matsui K, Lo JF, Silver S. 1999. Molecular basis for resistance to silver cations in Salmonella. Nat. Med. 5:183–188. 10.1038/5545 [DOI] [PubMed] [Google Scholar]
- 5.Silver S. 2003. Bacterial silver resistance: molecular biology and uses and misuses of silver compounds. FEMS Microbiol. Rev. 27:341–353. 10.1016/S0168-6445(03)00047-0 [DOI] [PubMed] [Google Scholar]
- 6.Chopra I. 2007. The increasing use of silver-based products as antimicrobial agents: a useful development or a cause for concern? J. Antimicrob. Chemother. 59:587–590. 10.1093/jac/dkm006 [DOI] [PubMed] [Google Scholar]
- 7.Parikh RY, Singh S, Prasad BLV, Patole MS, Sastry M, Shouche YS. 2008. Extracellular synthesis of crystalline silver nanoparticles and molecular evidence of silver resistance from Morganella sp.: towards understanding biochemical synthesis mechanism. ChemBioChem 9:1415–1422. 10.1002/cbic.200700592 [DOI] [PubMed] [Google Scholar]
- 8.Klaus T, Joerger R, Olsson E, Granqvist C-G. 1999. Silver-based crystalline nanoparticles microbially fabricated. Proc. Natl. Acad. Sci. U. S. A. 96:13611–13614. 10.1073/pnas.96.24.13611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalimuthu K, Babu RS, Venkataraman D, Bilal M, Gurunathan S. 2008. Biosynthesis of silver nanocrystals by Bacillus licheniformis. Colloids Surf. B Biointerfaces 65:150–153. 10.1016/j.colsurfb.2008.02.018 [DOI] [PubMed] [Google Scholar]
- 10.Nanda A, Saravanan M. 2009. Biosynthesis of silver nanoparticles from Staphylococcus aureus and its antimicrobial activity against MRSA and MRSE. Nanomedicine 5:452–456. 10.1016/j.nano.2009.01.012 [DOI] [PubMed] [Google Scholar]
- 11.Kumar CG, Mamidyala SK. 2011. Extracellular synthesis of silver nanoparticles using culture supernatant of Pseudomonas aeruginosa. Colloids Surf. B Biointerfaces 84:462–466. 10.1016/j.colsurfb.2011.01.042 [DOI] [PubMed] [Google Scholar]
- 12.Thakkar KN, Mhatre SS, Parikh RY. 2010. Biological synthesis of metallic nanoparticles. Nanomedicine 6:257–262. 10.1016/j.nano.2009.07.002 [DOI] [PubMed] [Google Scholar]
- 13.Muller M, Li Z, Maitz PKM. 2009. Pseudomonas pyocyanin inhibits wound repair by inducing premature cellular senescence: role for p38 mitogen-activated protein kinase. Burns 35:500–508. 10.1016/j.burns.2008.11.010 [DOI] [PubMed] [Google Scholar]
- 14.Price-Whelan A, Dietrich LEP, Newman DK. 2007. Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. J. Bacteriol. 189:6372–6381. 10.1128/JB.00505-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Newman DK. 2008. Redox reactions of phenazine antibiotics with ferric (hydr)oxides and molecular oxygen. Environ. Sci. Technol. 42:2380–2386. 10.1021/es702290a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernandez ME, Kappler A, Newman DK. 2004. Phenazines and other redox-active antibiotics promote microbial mineral reduction. Appl. Environ. Microbiol. 70:921–928. 10.1128/AEM.70.2.921-928.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freidheim E, Michaelis L. 1931. Potentiometric study of pyocyanine. J. Biol. Chem. 91:355–368 [Google Scholar]
- 18.Propst C, Lubin L. 1979. Light-mediated changes in pigmentation of Pseudomonas aeruginosa cultures. J. Gen. Microbiol. 113:261–266. 10.1099/00221287-113-2-261 [DOI] [PubMed] [Google Scholar]
- 19.Jayatilake GS, Thornton MP, Leonard AC, Grimwade JE, Baker BJ. 1996. Metabolites from an Antarctic sponge-associated bacterium, Pseudomonas aeruginosa. J. Nat. Prod. 59:293–296. 10.1021/np960095b [DOI] [PubMed] [Google Scholar]
- 20.Knight M, Hartman PE, Hartman Z, Young VM. 1979. A new method of preparation of pyocyanin and demonstration of an unusual bacterial sensitivity. Anal. Biochem. 95:19–23. 10.1016/0003-2697(79)90179-9 [DOI] [PubMed] [Google Scholar]
- 21.Muller M. 2011. Glutathione modulates the toxicity of, but is not a biologically relevant reductant for, the Pseudomonas aeruginosa redox toxin pyocyanin. Free Radic. Biol. Med. 50:971–977. 10.1016/j.freeradbiomed.2011.01.012 [DOI] [PubMed] [Google Scholar]
- 22.Mulvaney P. 1996. Surface plasmon spectroscopy of nanosized metal particles. Langmuir 12:788–800. 10.1021/la9502711 [DOI] [Google Scholar]
- 23.Davis G, Thornalley PJ. 1983. Free radical production from the aerobic oxidation of reduced pyridine nucleotides catalysed by phenazine derivatives. Biochim. Biophys. Acta 724:456–464. 10.1016/0005-2728(83)90106-8 [DOI] [PubMed] [Google Scholar]
- 24.Ludlow LW, Guikema JA, Consigli RA. 1986. Use of 5-(4-dimethylamino-benzylidene) rhodanine in quantifying silver grains eluted from autoradiograms of biological material. Anal. Biochem. 154:104–109. 10.1016/0003-2697(86)90502-6 [DOI] [PubMed] [Google Scholar]
- 25.Woods EJ, Cochrane CA, Percival SL. 2009. Prevalence of silver resistance gene in bacteria isolated from human and horse wounds. Vet. Microbiol. 138:325–329. 10.1016/j.vetmic.2009.03.023 [DOI] [PubMed] [Google Scholar]
- 26.de Vicente A, Aviles M, Codina JC, Borrego JJ, Romero PR. 1990. Resistance to antibiotics and heavy metals of Pseudomonas aeruginosa isolated from natural waters. J. Appl. Bacteriol. 68:625–632. 10.1111/j.1365-2672.1990.tb05228.x [DOI] [PubMed] [Google Scholar]
- 27.Vasishta R, Chhibber S, Saxena M. 1989. Heavy metal resistance in clinical isolates of Pseudomonas aeruginosa. Folia Microbiol. (Praha) 34:448–452. 10.1007/BF02820752 [DOI] [PubMed] [Google Scholar]
- 28.Bridges K, Kidson A, Lowbury EJ, Wilkins MD. 1979. Gentamicin- and silver-resistant pseudomonas in a burns unit. Br. Med. J. 1:446–449. 10.1136/bmj.1.6161.446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang F, Alvarez PJ, Zhu D. 2014. Microbial extracellular polymeric substances reduce Ag+ to silver nanoparticles and antagonize bactericidal activity. Environ. Sci. Technol. 48:316–322. 10.1021/es403796x [DOI] [PubMed] [Google Scholar]
- 30.Gjødsbøl K, Christensen JJ, Karlsmark T, Jørgensen Klein BBM, Krogfelt KA. 2006. Multiple bacterial species reside in chronic wounds: a longitudinal study. Int. Wound J. 3:225–231. 10.1111/j.1742-481X.2006.00159.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madsen SM, Westh H, Danielsen L, Posdahl VT. 1996. Bacterial colonization and healing of venous leg ulcers. APMIS 104:895–899. 10.1111/j.1699-0463.1996.tb04955.x [DOI] [PubMed] [Google Scholar]
- 32.Bjarnsholt T, Kirketerp Møller, Jensen PØ K, Madsen KG, Phipps R, Krogfelt K, Høiby N, Givskov M. 2008. Why chronic wounds will not heal: a novel hypothesis. Wound Repair Regen. 16:2–10. 10.1111/j.1524-475X.2007.00283.x [DOI] [PubMed] [Google Scholar]
- 33.Dietrich LE, Teal TK, Price-Whelan A, Newman DK. 2008. Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science 321:1203–1206. 10.1126/science.1160619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koley D, Ramsey MM, Bard AJ, Whiteley M. 2011. Discovery of a biofilm electrocline using real-time 3D metabolite analysis. Proc. Natl. Acad. Sci. U. S. A. 108:19996–20001. 10.1073/pnas.1117298108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bjarnsholt T, Kirketerp Møller, Kristiansen K, Phipps S, Nielsen R, Jensen PØ AK, Høiby N, Givskov M. 2007. Silver against Pseudomonas aeruginosa biofilms. APMIS 115:921–928. 10.1111/j.1600-0463.2007.apm_646.x [DOI] [PubMed] [Google Scholar]
- 36.Santoro CM, Duchsherer NL, Grainger DW. 2007. Antimicrobial efficacy and ocular cell toxicity from silver nanoparticles. Nanobiotechnology 3:55–65. 10.1007/s12030-008-9007-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Percival SL, Bowler PG, Dolman J. 2007. Antimicrobial activity of silver-containing dressings on wound microorganisms using an in vitro model. Int. Wound J. 4:186–191. 10.1111/j.1742-481X.2007.00296.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castellano JJ, Shafii SM, Ko F, Donate G, Wright TE, Mannari RJ. 2007. Comparative evaluation of silver-containing antimicrobial dressings and drugs. Int. Wound J. 4:114–122. 10.1111/j.1742-481X.2007.00316.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wasiak J, Cleland H, Campbell F. 2008. Dressings for superficial and partial thickness burns. Cochrane Database Syst. Rev. 2008:CD002106. 10.1002/14651858.CD002106.pub3 [DOI] [PubMed] [Google Scholar]
- 40.Mulley G, Jenkins ATA, Waterfield NR. 2014. Inactivation of the antibacterial and cytotoxic properties of silver ions by biologically relevant compounds. PLoS One 9:e94409. 10.1371/journal.pone.0094409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor PL, Ussher AL, Burrell RE. 2005. Impact of heat on nanocrystalline silver dressings. Part I: Chemical and biological properties. Biomaterials 26:7221–7229. 10.1016/j.biomaterials.2005.05.040 [DOI] [PubMed] [Google Scholar]
- 42.Nesbitt RU, Jr, Sandmann BJ. 1977. Solubility studies of silver sulfadiazine. J. Pharm. Sci. 66:519–522. 10.1002/jps.2600660414 [DOI] [PubMed] [Google Scholar]
- 43.Tsipouras N, Rix CJ, Brady PH. 1995. Solubility of silver sulfadiazine in physiological media and relevance to treatment of thermal burns with silver sulfadiazine cream. Clin. Chem. 41:87–91 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.