Abstract
Glycopeptides and β-lactams inhibit bacterial peptidoglycan synthesis in Gram-positive bacteria; resistance to these antibiotics is studied intensively in enterococci and staphylococci because of their relevance to infectious disease. Much less is known about antibiotic resistance in glycopeptide-producing actinomycetes that are likely to represent the evolutionary source of resistance determinants found in bacterial pathogens. Nonomuraea sp. ATCC 39727, the producer of A40926 (the precursor for the semisynthetic dalbavancin), does not harbor the canonical vanHAX genes. Consequently, we investigated the role of the β-lactam-sensitive d,d-peptidase/d,d-carboxypeptidase encoded by vanYn, the only van-like gene found in the A40926 biosynthetic gene cluster, in conferring immunity to the antibiotic in Nonomuraea sp. ATCC 39727. Taking advantage of the tools developed recently to genetically manipulate this uncommon actinomycete, we varied vanYn gene dosage and expressed vanHatAatXat from the teicoplanin producer Actinoplanes teichomyceticus in Nonomuraea sp. ATCC 39727. Knocking out vanYn, complementing a vanYn mutant, or duplicating vanYn had no effect on growth but influenced antibiotic resistance and, in the cases of complementation and duplication, antibiotic production. Nonomuraea sp. ATCC 39727 was found to be resistant to penicillins, but its glycopeptide resistance was diminished in the presence of penicillin G, which inhibits VanYn activity. The heterologous expression of vanHatAatXat increased A40926 resistance in Nonomuraea sp. ATCC 39727 but did not increase antibiotic production, indicating that the level of antibiotic production is not directly determined by the level of resistance. The vanYn-based self-resistance in Nonomuraea sp. ATCC 39727 resembles the glycopeptide resistance mechanism described recently in mutants of Enterococcus faecium selected in vitro for high-level resistance to glycopeptides and penicillins.
INTRODUCTION
Glycopeptide antibiotics are an important class of natural products composed of glycosylated nonribosomal peptides produced by a diverse group of filamentous actinomycetes (1). Vancomycin, made by Amycolatopsis orientalis, and teicoplanin, produced by Actinoplanes teichomyceticus, are currently used for the treatment of life-threatening infections of pathogens, such as β-lactam-resistant Staphylococcus aureus, Enterococci spp., and Clostridium difficile (2–4). They arrest bacterial cell wall biosynthesis in Gram-positive bacteria by binding to the d-alanyl-d-alanine termini (d-Ala-d-Ala) of peptidoglycan (PG) precursors at the outer surface of the bacterial cell membrane, and they block PG polymerization by steric hindrance (5). The complex is stabilized by an array of hydrophobic interactions and five hydrogen bonds (H-bonds) (5).
High-level resistance to glycopeptides by target modification is encoded by the van genes that remodel cell wall biosynthesis, reducing the affinity of the antibiotics for their targets (6, 7). The most studied mechanism occurs in clinical isolates of enterococci and staphylococci that have acquired transposon Tn1546 (8); this results in the production of PG precursors ending in d-lactate (d-Lac) instead of d-Ala, as well as high levels of vancomycin and teicoplanin resistance (9, 10). The substitution of the amide moiety in d-Ala-d-Ala by an ester linkage in d-Ala-d-Lac results in the loss of one H-bond, reducing binding affinity to vancomycin 1,000-fold and rendering the antibiotic therapeutically ineffective (6, 11). The synthesis of the alternative PG precursors requires the coordinated action of enzymes encoded by vanHAX (7). vanH codes for a dehydrogenase (α-keto acid reductase) that reduces pyruvate to d-lactate, vanA encodes a ligase that catalyzes the formation of the d-Ala-d-Lac depsipeptide that replaces the dipeptide d-Ala-d-Ala in PG synthesis, and the d,d-peptidase VanX selectively removes the intracellular pool of d-Ala-d-Ala produced by the native d-Ala-d-Ala ligase, ensuring that the d-Ala-d-Lac depsipeptide is incorporated into the PG precursor. The production of an accessory protein, the d,d-carboxypeptidase VanY, acts as a subsidiary mechanism by removing the C-terminal d-Ala residue of any unmodified PG precursors inserted into the inner leaflet of the membrane (12, 13). These d-Ala-d-Ala ending precursors originate from incomplete elimination by VanX of the dipeptide d-Ala-d-Ala produced by the host ligase (13).
Variants of vanHAX (which confers the VanA phenotype) that determine glycopeptide resistance phenotypes with various levels of resistance to vancomycin and susceptibility or resistance to teicoplanin (e.g., VanB, VanC, VanD, VanE, VanG, VanL, VanM, and VanN) continue to be discovered in enterococci (6, 14). Close homologues (54 to 64% amino acid sequence identities) and similarly organized vanHAX gene clusters occur in actinomycetes that produce glycopeptides and that use them for immunity during antibiotic production (15–18) or in taxonomically related strains that share the same antibiotic-containing ecological niche, such as Streptomyces coelicolor (19). vanHAX genes are present in Amycolatopsis spp., which produce vancomycin and the vancomycin-like molecules balhimycin, chloroeremomycin, ristocetin, and avoparcin (15, 18), in Streptomyces toyocaensis strain NRRL 15009, the producer of the teicoplanin-like A47934 (16), and in the teicoplanin producer A. teichomyceticus strain ATCC 31121 (17). In these last two actinomycetes, van-like genes are located within the gene clusters dedicated to the biosynthesis of A47934 (the sta biosynthetic cluster) and teicoplanin (the tcp biosynthetic cluster). It has been suggested that the van-like glycopeptide resistance determinants found in bacterial pathogens may have originated from antibiotic-producing actinomycetes (15).
Nonomuraea sp. ATCC 39727 produces the teicoplanin-like glycopeptide antibiotic A40926 (20). A40926 is the precursor of the semisynthetic derivative dalbavancin, which is a promising second-generation glycopeptide currently completing phase III of clinical development (second-generation glycopeptides [telavancin, oritavancin, and dalbavancin] are semisynthetic derivatives of natural vancomycin- and teicoplanin-like glycopeptides [2]). Dalbavancin shows improved activity, pharmacokinetics, and pharmacodynamics compared to those of teicoplanin (2). Nonomuraea sp. ATCC 39727 does not possess a vanHAX-like gene cluster (21). We demonstrated recently that the biosynthetic gene cluster (dbv) dedicated to A40926 production contains a vanYn gene encoding a metallo-d,d-carboxypeptidase that hydrolyzes the C-terminal d-Ala residue of PG pentapeptide precursors (22). An analysis of UDP-linked PG precursors in Nonomuraea sp. ATCC 39727 identified the predominant presence of the tetrapeptide UDP-MurNAc-l-Ala-d-Glu-meso-Dap-d-Ala, which binds glycopeptides poorly, suggesting a role for VanYn in conferring some level of resistance to teicoplanin and vancomycin. Consistent with that, a vanYn-null mutant of Nonomuraea sp. ATCC 39727 was more sensitive to glycopeptides (21), and heterologous expression of vanYn in sensitive Streptomyces species increased the level of glycopeptide resistance (23).
In this study, we took advantage of tools developed recently to genetically manipulate Nonomuraea sp. ATCC 39727 (24, 25) in order to investigate how altering vanYn gene dosage and inserting vanHatAatXat from A. teichomyceticus ATCC 31121 into Nonomuraea sp. ATCC 39727 affect glycopeptide and β-lactam resistance, as well as A40926 production.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli strain DH5α (26) (Invitrogen) was used for routine cloning. E. coli strain ET12567 (27) was the nonmethylating strain from which DNA was extracted for protoplast transformation. E. coli ET12567/pUZ8002 (28) was the nonmethylating plasmid donor strain for intergeneric conjugation with Nonomuraea sp. ATCC 39727. E. coli strains were grown in Luria-Bertani (LB) liquid medium (29), and antibiotics (all from Sigma-Aldrich) were added at the following concentrations to appropriate cultures to maintain plasmids: 100 μg/ml ampicillin, 50 μg/ml kanamycin, 25 μg/ml chloramphenicol, and 50 μg/ml apramycin.
Nonomuraea sp. ATCC 39727 and its recombinant strains (listed in Table 1) were maintained as lyophilized master cell banks (MCBs). The mycelium from the MCBs was streaked on slants of salt medium (SM) (30) solidified with agar (15 g/liter). After growth, the mycelium from a slant was homogenized in 10 ml of 0.9% (wt/vol) NaCl, inoculated into liquid SM, grown for 96 h at 28°C with aeration, and stored as a working cell bank (WCB) in 1.5-ml cryovials at −80°C. One vial was used to inoculate each 300-ml Erlenmeyer flask containing 50 ml cultivation medium, and the flasks were incubated at 28°C, with shaking at 200 rpm. Unless otherwise stated, Nonomuraea sp. recombinant strains were cultivated in VSP medium (24), and antibiotics were added to the cultures when required to maintain plasmids. Biomass was measured as dry weight after the mycelium was harvested by centrifugation for 10 min at 4,000 × g and the pellet was dehydrated for 24 h in a 50°C oven. Glucose consumption and broth pH were estimated by using Diastix sticks (Bayer) and pH test strips (Sigma-Aldrich), respectively. Surface cultures were grown on V0.1 agar (24) containing 2.4 g/liter soluble starch (Difco), 0.1 g/liter dextrose (A.D.E.A), 0.3 g/liter malt extract (Costantino), 0.5 g/liter yeast extract (Costantino), 0.5 g/liter tryptose (Difco), and deionized water to 1 liter (pH 7.2) and solidified with agar (15 g/liter).
TABLE 1.
Strains and plasmids
| Plasmid or strain | Genotypea | Reference or source |
|---|---|---|
| Plasmids | ||
| pSET152 | E. coli-Streptomyces shuttle vector; lacZα oriT; pUC replicon, ΦC31 attB-int; Aprr | 32 |
| pST30 | Integrative plasmid pSET152 containing the resistance genes vanHatAatXat from Actinoplanes teichomyceticus | 34 |
| pSET152Y | Integrative plasmid pSET152 containing the resistance gene vanYn | This work |
| pRT802 | pRT801 derived, ΦBT1 attB-int; Kanr | 33 |
| pRT802Y | Integrative plasmid pRT802 containing the resistance gene vanYn | This work |
| pIJ86 | oriT RK2, ermEp*, rep pIJ101, Aprr | 21 |
| pIJ86Y | Multicopy plasmid pIJ86 containing the resistance gene vanYn under the constitutive ermE* promoter | 21 |
| Escherichia coli strains | ||
| DH5α | F− ϕ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) phoA supE44 thi-1 gyrA96 relA1 λ− | 26 |
| ET12567 | DNA methylation-deficient host, dam dcm hsdM hsdS hsdR cat tet | 27 |
| ET12567/pUZ8002 | Derived from the methylation-defective strain ET12567 carrying plasmid pUZ8002 | 28 |
| Nonomuraea sp. strains | ||
| ATCC 39727 (wild type) | Producer of the glycopeptide A40926; vanYn is located in the A40926 biosynthetic cluster dbv | 20 |
| Strain carrying pSET152 | Wild-type strain with pSET152 inserted in the ΦC31 attB site; Aprr | This work |
| Strain carrying pST30 | Wild-type strain with the integrative pST30 containing the resistance genes vanHatAatXat from A. teichomyceticus | This work |
| Strain carrying pSET152Y | Wild-type strain with pST152Y inserted in the ΦC31 attB site; Aprr | This work |
| Strain carrying pRT802 | Wild-type strain with pRT802 integrated in the ΦBT1 attB site; Kanr | This work |
| Strain carrying pRT802Y | Wild-type strain with pRT802Y integrated in the ΦBT1 attB site; Kanr | This work |
| Strain carrying pIJ86 | Wild-type strain with the multicopy pIJ86; Aprr | This work |
| Strain carrying pIJ86Y | Wild-type strain with pIJ86Y; Aprr | This work |
| ΔvanYn strain | vanYn-null mutant, sensitive to glycopeptides; Aprr | 21 |
| ΔvanYn-pRT802 strain | ΔvanYn mutant with pRT802; Aprr Kanr | This work |
| ΔvanYn-pRT802Y strain | ΔvanYn mutant with pRT802Y to complement the deletion; Aprr Kanr | This work |
Apr, apramycin; Kan, kanamycin.
Recombinant DNA techniques.
Plasmid DNA isolation, digestion with restriction endonucleases (Roche), ligation with T4 DNA ligase (Roche), and transformation of E. coli DH5α with recombinant plasmid DNA were performed by standard methods (31).
Plasmid construction and Nonomuraea sp. recombinant strains.
The plasmids, recombinant strains, and oligonucleotides used in this study are listed in Tables 1 and 2. To generate pSET152Y, the vanYn gene (with its promoter and terminator regions) was amplified from the genomic DNA of Nonomuraea sp. ATCC 39727 using the primers FB_137F and FB_137R. pSET152Y was constructed by first cloning the resulting PCR product into the TOPO pCRII vector (Invitrogen) and then excising it as a 1.2-kb EcoRI fragment (containing vanYn with its own promoter, ribosome binding site, and transcriptional terminator). This fragment was then cloned into pSET152 (32). pRT802Y was constructed by cloning a 869-bp fragment containing vanYn (amplified from Nonomuraea sp. ATCC 39727 genomic DNA with oligonucleotides FB_137F and FB_137R) with its natural promoter, ribosome binding site, and transcriptional terminator into EcoRV-digested pRT802 (33). pSET152Y and pRT802Y were then used to transform the nonmethylating strain E. coli ET12567.
TABLE 2.
Oligonucleotides used in this study
| Name | Sequence (5′ to 3′)a |
|---|---|
| FB_137F | GTGATCGAAACCGTCCCG |
| FB_137R | CTGGAGGAGCTTCCGCAG |
| FB_138F | TTCGCGTTCGTGGACGAG |
| FB_138R | CTACACGATCGGAAAATCAAA |
| LM_005F | ATGGATCCCAGACTGGAGGAGAGGGATG |
| LM_005R | GATAAGCTTCGATCCTGGAGTTCGTCTTC |
| LM_001F | AGAGCGGTCACACGTTCTCT |
| LM_001R | CGATCCTGGAGTTCGTCTTC |
| LM_002F | GCTAACTAGTAGTTCCTTCGTCACC |
| LM_002R | CCGGCTCGTATGTTGTGTG |
| LM_006F | TCGCCATTCAGGCTGC |
| LM_006R | CTCATTAGGCACCCCAGG |
Introduced restriction sites for BamHI and HindIII are in bold type.
To construct pIJ86Y, the vanYn protein-coding sequence was amplified from Nonomuraea sp. ATCC 39727 genomic DNA using Expand high-fidelity polymerase (Roche) and primers LM_005F and LM_005R, which introduced the BamHI and HindIII restriction sites (Table 2, bold type), respectively, into the PCR product. The PCR product was purified, digested with BamHI and HindIII, and ligated with pIJ86 that had been cleaved with the same enzymes. pIJ86Y, with vanYn transcribed from the strong constitutive ermE* promoter (30), was used to transform E. coli ET12567/pUZ8002 to yield ET12567/pUZ8002/pIJ86Y. All of the above constructs were confirmed by DNA sequencing. pST30 (34) was used to introduce vanHatAatXat into Nonomuraea sp. ATCC 39727, which lacks vanHAX homologues (21).
The Nonomuraea sp. strains carrying pSET152Y and pRT802Y were obtained by protoplast transformation of the wild-type strain ATCC 39727, as previously described (24), using plasmid DNA extracted from E. coli ET12567 and selecting for apramycin and kanamycin resistance, respectively. Nonomuraea sp. strains carrying pIJ86Y and pST30 were obtained by conjugation (25) using E. coli ET12567/pUZ8002/pIJ86Y and E. coli ET12567/pUZ8002/pST30, respectively, as donors and Nonomuraea sp. ATCC 39727 as the recipient and selecting for apramycin resistance. Strains resulting from conjugation or protoplast transformation were confirmed to carry the appropriate plasmid by colony PCR using the following primers: LM_002F and LM_002R for pRT802Y, LM_006F and LM_006R for pSET152Y, LM_005F and LM_005R for pIJ86Y, and FB_138F and FB_138R for pST30.
The Nonomuraea sp. ΔvanYn strain was constructed by PCR targeting (21). Nonomuraea sp. strain ΔvanYn protoplasts were generated and transformed with pRT802Y as described for Nonomuraea sp. ATCC 39727 (24, 25). Clones of the Nonomuraea sp. ΔvanYn-pRT802Y strain (complemented mutant) were selected as apramycin and kanamycin resistant and confirmed by PCR as above.
Determination of MICs.
Cultures of Nonomuraea sp. ATCC 39727 and the recombinant strains listed in Table 1 were treated as follows to determine the MICs of glycopeptides, β-lactams, and bacitracin. Cryovials of WCBs were thawed at room temperature and used to inoculate VSP medium. The strains were incubated to exponential phase (approximately 72 h) at 28°C with shaking. Mycelium was harvested by centrifugation, suspended in 0.9% (wt/vol) NaCl, and fragmented by sonication with a Vibracell Albra sonicator 400 W (21). A suspension of sonicated hyphae (corresponding to 107 CFU) was seeded onto V0.1 agar plates supplemented with increasing concentrations of the following antibiotics (all from Sigma-Aldrich): 0 to 60 μg/ml vancomycin in 10-μg/ml increments; 0 to 1 μg/ml teicoplanin in 0.1-μg/ml increments; 0 to 15 μg/ml or 0 to 50 μg/ml A40926, depending on the strain, in 2.5-μg/ml or 10-μg/ml increments, respectively; 0 to 100 μg/ml ampicillin in 20-μg/ml increments; 0 to 100 μg/ml penicillin G in 20-μg/ml increments; and 0 to 100 μg/ml bacitracin in 20-μg/ml increments. The plates were allowed to dry and then incubated at 28°C. The MIC values were determined as the lowest antibiotic concentrations that inhibited visible growth after 10 days of incubation. When glycopeptide MICs were determined in the presence of penicillin G or ampicillin, 50 μg/ml of the β-lactam antibiotic was added to the agar medium.
d,d-Peptidase and d,d-carboxypeptidase assays.
Cells were harvested by centrifugation at 3,600 × g for 20 min at 4°C and suspended in 2 ml of 0.9% (wt/vol) NaCl per g of cells (wet weight). All of the following manipulations were carried out at 0 to 4°C. Mycelium was fragmented by sonication with a Sonics Vibracell VCX 130. Sonication was carried out for ≥5 min on ice, with cycles of 30 s with an amplitude of 90% (90% of 60 Hz), with breaks of 10 s. The samples were then centrifuged at 39,000 × g for 15 min, and the supernatants (cytoplasmic fractions) were collected. Alkaline extraction of the pellets (cell debris and membrane fractions) was carried out by adapting a protocol developed previously for extracting membrane-bound proteins in enterococci (23, 35). The sedimented pellets were resuspended in ice-cold distilled water containing proteinase inhibitors (0.19 mg/ml phenylmethanesulfonyl fluoride and 0.7 μg/ml pepstatin, both purchased from Sigma-Aldrich), and then, immediately before centrifugation (28,000 × g for 15 min at 4°C), the pH was adjusted to 12 by adding an appropriate volume of 2.5 N NaOH. Immediately after centrifugation, the supernatants were neutralized to pH 7 by adding 0.5 M sodium acetate (pH 5.4). Enzymatic activities in the supernatant and in the resuspended insoluble fraction were assayed as reported previously (22, 23) by measuring the release of d-Ala from commercially available dipeptide (d-Ala-d-Ala, 10 mM; Sigma-Aldrich) and tripeptide (Nε-acetyl-l-Lys-d-Ala-d-Ala, 10 mM; Sigma-Aldrich). d,d-Carboxypeptidase activity was detectable in the supernatant fraction, as previously reported (23, 35), and it was confirmed using 10 mM UDP-MurNAc-l-Ala-d-Glu-meso-Dap-d-Ala-d-Ala (UK-BaCWAN, University of Warwick) as a substrate. The release of d-Ala was followed spectrophotometrically with a d-amino acid oxidase-peroxidase coupled reaction that oxidized the colorimetric substrate 4-aminoantipyrine to chinonemine (22). To determine the inhibition of the activities of d,d-peptidase and d,d-carboxypeptidase, 0 mM to 100 mM penicillin G was added to the assay mixtures. Protein concentration was estimated by the biuret method (36). To compare the d,d-peptidase and d,d-carboxypeptidase activities in the cytoplasmic and membrane extracts, the activity was expressed as the number of nmol of d-Ala released from the dipeptide or tripeptide per min per mg of protein in the extract.
PG precursor extraction and analysis.
The extraction of UDP-linked PG precursors was performed as previously described (17, 21, 24). In brief, cells were grown to exponential phase (ca. 72 to 96 h) in VSP medium at 28°C with shaking at 200 rpm, bacitracin (Sigma-Aldrich) was added to 100 μg/ml, and the culture was incubated for a further 90 min to accumulate PG precursors. Mycelium was harvested by centrifugation, washed twice, suspended in distilled water (0.2 g of wet weight per ml), sonicated (with Sonics Vibracell VCX 130, as above), boiled for 20 min, and then centrifuged at 39,000 × g for 1 h. The supernatant was lyophilized and the residue dissolved in 0.1 volume of water adjusted to pH 3 with formic acid (Sigma-Aldrich). The samples were analyzed by reverse-phase high-performance liquid chromatography (HPLC) and electrospray ionization mass spectrometry (ESI-MS) on an LCQ Deca spectrometer equipped with an ion-trap analyzer (Thermo Finnigan) (17, 21, 24). UDP-MurNAc-l-Ala-d-Glu-meso-Dap-d-Ala-d-Ala was obtained from the UK-BaCWAN PG precursor synthesis facility (University of Warwick) and used as a standard. The results were expressed as a percentage of the total PG precursors (UDP-MurNAc-tetrapeptide, UDP-MurNAc-pentapeptide, and UDP-MurNAc-pentadepsipeptide) determined from the integrated peak areas.
A40926 production.
Vegetative cultures were obtained from one vial of the WCB inoculated into 50 ml of vegetative medium E26 (21) in 300-ml baffled flasks. The strains were grown for 72 to 96 h on a rotary shaker at 200 rpm and 28°C. Batch fermentations were conducted using the industrial production medium FM2 (8 g/liter Bacto-yeast extract [Costantino], 30 g/liter soybean flour [Sigma-Aldrich], 30 g/liter dextrose [A.D.E.A], 15 g/liter malt extract [Costantino], 4 g/liter CaCO3 [Sigma-Aldrich], 1 g/liter l-valine [Sigma-Aldrich], and deionized water to 1 liter [pH 7.4]). Fermentation runs were started by adding a 10% (vol/vol) inoculum from the vegetative cultures into FM2 medium in a 2-liter-working-volume P-100 Applikon glass reactor (height, 25 cm; diameter, 13 cm) equipped with an AD1030 Biocontroller and AD1032 motor (21, 37). The temperature was maintained at 30°C, stirring was at 500 rpm (corresponding to 1.17 m/s of tip speed), and the aeration rate was 2 liters/min. Foam production was controlled by the addition of antifoam (Sigma-Aldrich). Temperature, dissolved oxygen, and pH were measured throughout the experiments. Twenty-five milliliters of the cultures was sampled every day for a total of 10 days of fermentation to estimate biomass (dry weight, see above), glucose consumption (Diastix sticks, see above), and A40926 production (see below). The MICs during production were determined by sampling 10 ml of culture every 24 h and plating as described above.
HPLC analysis of culture extracts.
A40926 was extracted by mixing 1 volume of mycelium and 3 volumes of borate buffer (100 mM H3BO3, 100 mM NaOH [pH 12]). The samples were centrifuged (16,000 × g for 15 min) and incubated for 1 h at 50°C. The glycopeptide-containing supernatant was filtered through a Durapore membrane filter (0.45 μm) (Millipore). Chromatography was performed with a VWR Hitachi diode array L-2455 HPLC system with detection at 254 nm. The A40926 titers in the batch cultivations were estimated by injecting 50 μl of sample onto a 5-μm-particle-size Ultrasphere ODS (Beckman) HPLC column (4.6 by 250 mm) and eluting at a flow rate of 1 ml/min with a 26-min linear gradient from 25% to 37% of phase B. Phase A was 20 mM HCOONH4 (pH 4.5)-CH3CN (95:5 [vol/vol]), and phase B was 20 mM HCOONH4 (pH 4.5)-CH3CN (5:95 [vol/vol]) mixture. Ten microliters of a pure sample of 1 mg/ml A40926 (Sigma-Aldrich) was used as an internal standard.
RESULTS
Effect of vanYn gene dosage on growth and glycopeptide resistance.
Nonomuraea sp. strains harboring additional copies of vanYn were constructed using three plasmids described in Table 1. pSET152Y and pRT802Y integrate at a single copy into two different regions of the actinomycete genome, i.e., the ϕC31 attB and ϕBT1 attB sites, respectively (33). In each case, vanYn was expressed from its own promoter (21). The third plasmid used was the multicopy vector pIJ86, in which vanYn was expressed from the strong constitutive heterologous ermE* promoter (21, 23). Recombinant strains carrying empty vectors were used as negative controls.
Nonomuraea sp. ATCC 39727 growing exponentially in VSP medium, where no detectable A40926 production occurred, showed moderate resistance to glycopeptides, with MIC values of 4, 0.9, and 30 μg/ml for A40926, teicoplanin, and vancomycin, respectively (Table 3). The introduction of an additional copy of vanYn, carried by either pSET152Y or pRT802Y, did not significantly affect growth rate or biomass accumulation (Fig. 1), but it did increase the level of resistance to glycopeptides, particularly to A40926 (to 15 μg/ml) (Table 3). The overexpression of vanYn from pIJ86Y dramatically reduced the growth rate and biomass accumulation (Fig. 1), and it changed the morphology of the mycelium: the hyphae were thinner and did not clump like those of the wild-type strain and the control strain carrying the empty vector (data not shown). This might reflect a deleterious effect of d,d-carboxypeptidase overexpression on cell wall biosynthesis. The vanYn-null mutant grew similarly to the wild-type strain, but it was considerably more susceptible to glycopeptides (MICs, 2, 0.2, and 4 μg/ml for A40926, teicoplanin, and vancomycin, respectively). Complementation of the vanYn-null mutant with pRT802Y restored resistance, but surprisingly, the A40926 MIC was much higher than that of the wild-type strain and corresponded to that found in wild-type Nonomuraea spp. carrying pSET152Y, pRT802Y, or pIJ86Y (15 μg/ml, Table 3). This was not due to the vector itself, since the vanYn-null mutant transformed with the empty vector showed the same resistance profile as the vanYn-null mutant (data not shown). We speculate that the integration of vanYn outside the dbv cluster might compromise the regulatory mechanism that usually controls A40926 biosynthesis and self-resistance and result in levels of vanYn expression that exceed those of the wild-type strain (see below).
TABLE 3.
MICs of glycopeptides for Nonomuraea sp. ATCC 39727 and its recombinant strains, measured in the presence or absence of 50 μg/ml penicillin G
| Nonomuraea sp. strain genotype or plasmid | MIC (μg/ml) fora: |
|||||
|---|---|---|---|---|---|---|
| VAN | VAN + PENb | TEC | TEC + PENb | A40 | A40 + PENb | |
| Wild type | 30 | 17.5 | 0.9 | 0.5 | 4 | 3.5 |
| pSET152 | 30 | 17.5 | 0.9 | 0.5 | 4 | 3.5 |
| pSET152Y | 40 | 30 | 2 | 1.5 | 15 | 4 |
| pRT802 | 30 | 17.5 | 0.9 | 0.5 | 4 | 3.5 |
| pRT802Y | 30 | 20 | 1.25 | 0.75 | 15 | 10 |
| pIJ86 | 30 | 17.5 | 1 | 0.6 | 5 | 3.5 |
| pIJ86Y | 25 | 20 | 1.25 | 1 | 15 | 10 |
| ΔvanYn | 4 | 4 | 0.2 | 0.2 | 2 | 2 |
| ΔvanYn-pRT802 | 4 | 4 | 0.2 | 0.2 | 2 | 2 |
| ΔvanYn-pRT802Y | 25 | 15 | 1.25 | 0.5 | 15 | 10 |
| pST30 | 15 | 15 | 2.75 | 2.75 | 17.5 | 17.5 |
MICs were determined using strains grown in VSP medium for 72 h and the agar dilution method (21). The values represent the average of the data from three independent experiments. VAN, vancomycin; TEC, teicoplanin; A40, A40926; PEN, penicillin.
Penicillin G was added to the agar medium together with the different concentrations of glycopeptides.
FIG 1.
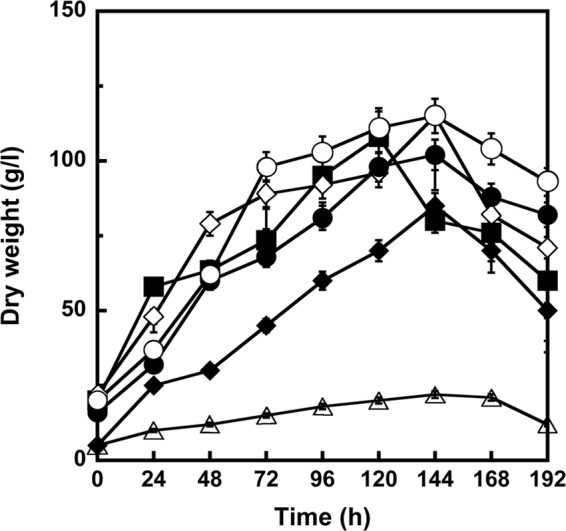
The effect of vanYn gene dosage and vanHatAatXat heterologous expression on the growth of Nonomuraea sp. The cells were grown in VSP medium, where production of A40926 was not detected by HPLC (limit of detection, 2 mg/liter). ■, Nonomuraea sp. ATCC 39727 wild type; ○, Nonomuraea sp. strain carrying pSET152Y; ●, Nonomuraea sp. strain carrying pRT802Y; ◇, Nonomuraea sp. ΔvanYn-pRT802Y strain; △, Nonomuraea sp. strain carrying pIJ86Y; ◆, Nonomuraea sp. strain carrying pST30. The values represent the averages and standard deviations from three independent experiments, with a standard deviation of <5%.
Expression of VanYn.
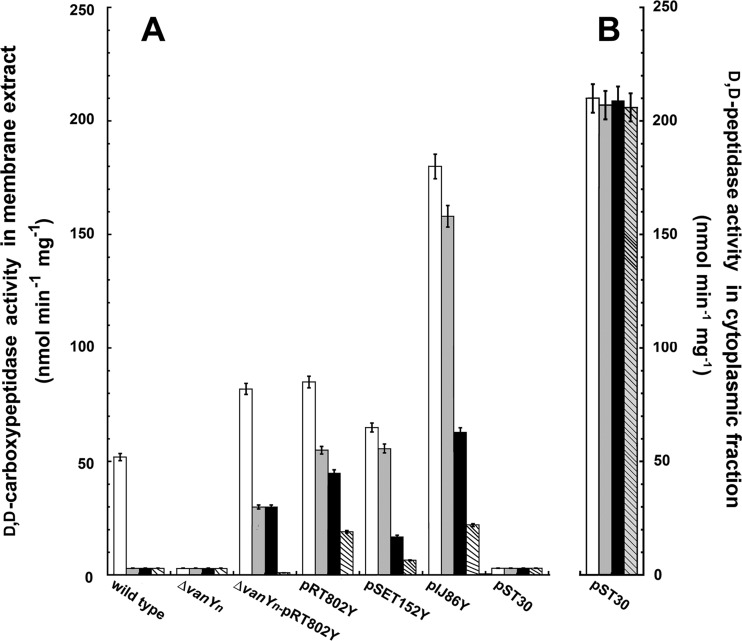
To confirm its predicted role in glycopeptide resistance, VanYn expression and cellular localization were studied in Nonomuraea sp. ATCC 39727 and in its recombinant strains by measuring d,d-peptidase and d,d-carboxypeptidase activities in their cytoplasmic and membrane fractions (Table 4). d,d-Peptidase and d,d-carboxypeptidase activities were measured by determining the amount of d-Ala released from the hydrolysis of the d-Ala-d-Ala dipeptide and the Nε-acetyl-l-Lys-d-Ala-d-Ala tripeptide using a d-amino acid oxidase coupled to a peroxidase (22). The d,d-carboxypeptidase activity was also confirmed by using the UDP-MurNAc-pentapeptide PG precursor of Nonomuraea sp. ATCC 39727, UDP-MurNAc-l-Ala-d-Glu-meso-Dap-d-Ala-d-Ala, as a substrate (data not shown). Both d,d-peptidase and d,d-carboxypeptidase activities were completely abolished in the vanYn-null mutant. In the complemented ΔvanYn-pRT802Y strain, both enzymatic activities were restored in the membrane fractions, confirming that VanYn is responsible for both the d,d-peptidase and d,d-carboxypeptidase activities. These data agree with our previous biochemical characterization of purified recombinant VanYn produced in E. coli as a bifunctional d,d-peptidase/d,d-carboxypeptidase: recombinant VanYn preferentially cleaved the last d-Ala from the acetyl-l-Lys-d-Ala-d-Ala tripeptide, but this activity was nearly halved if the substrate was d-Ala-d-Ala (22). In all of the strains harboring one, two, or more copies of vanYn, d,d-carboxypeptidase activity was higher than d,d-peptidase activity. Neither d,d-peptidase nor d,d-carboxypeptidase activities were detectable in the cytoplasmic fractions (Table 4), strongly suggesting that VanYn is associated with the membrane. This is consistent with the predicted VanYn structure, which contains a cytoplasmic domain at the N terminus (the first 20 amino acids) and a hydrophobic transmembrane portion (20 amino acids), followed by the active C-terminal domain exposed on the external face of the cytoplasmic membrane (22, 23). The lack of any cytoplasmic d,d-carboxypeptidase activity is also consistent with the finding that Nonomuraea sp. ATCC 39727 does not harbor any vanX-related genes.
TABLE 4.
d,d-Peptidase and d,d-carboxypeptidase activities in cytoplasmic and membrane extracts from Nonomuraea sp. ATCC 39727 and its recombinant strains grown in VSP medium
| Nonomuraea sp. strain genotype or plasmid | Enzyme activity (nmol min−1 mg−1)a |
|||
|---|---|---|---|---|
| Mean ± SD d,d-carboxypeptidase activityb |
Mean ± SD d,d-peptidase activityb |
|||
| Membrane | Cytoplasmic | Membrane | Cytoplasmic | |
| Wild type | 52 ± 3.4 | BDLc | 20 ± 2.6 | BDL |
| ΔvanYn | BDL | BDL | BDL | BDL |
| ΔvanYn-pRT802Yn | 82 ± 2.3 | BDL | 49 ± 3.5 | BDL |
| pSET152Yn | 65 ± 2.8 | BDL | 39 ± 3.9 | BDL |
| pRT802Yn | 85 ± 3.1 | BDL | 44 ± 2.7 | BDL |
| pIJ86Yn | 180 ± 4.2 | BDL | 88 ± 3.1 | BDL |
| pST30 | BDL | BDL | 18 ± 1.8 | 210 ± 4.6 |
Enzymatic activities were measured in the cytoplasmic and membrane fractions from strains grown in VSP medium for 72 h. The fractions were prepared as described in Materials and Methods. The results were obtained from a minimum of three independent bacterial extracts.
The hydrolysis of d-Ala from 10 mM d-Ala-d-Ala and 10 mM acetyl-l-Lys-d-Ala-d-Ala was determined by using a d-amino acid oxidase-peroxidase coupled assay (23).
BDL, below detection limit (5 nmol) of the spectrophotometric assay.
As expected, in those strains carrying an additional copy of vanYn, membrane-associated VanYn activity was higher than in the wild-type strain. VanYn activity was higher in the Nonomuraea sp. strain carrying pRT802Y than in the Nonomuraea sp. strain carrying pSET152Y, suggesting that a different genome integration site may influence the level of gene expression. Consistent with its elevated level of resistance (Table 3), the Nonomuraea sp. ΔvanYn-pRT802Y strain possessed a higher level of VanYn activity than the wild-type strain (Table 4). The overexpression of vanYn in the Nonomuraea sp. strain carrying pIJ86Y notably increased d,d-peptidase/d,d-carboxypeptidase activity in the membrane fractions, with high levels of activity observed early in growth, consistent with the expression of vanYn from the strong constitutive ermE* promoter.
Heterologous expression of A. teichomyceticus vanHatAatXat genes in Nonomuraea sp. ATCC 39727.
A40926 belongs to the lipoglycopeptide family and is structurally closely related to teicoplanin (2, 20). A. teichomyceticus, the producer of teicoplanin, possesses a constitutively expressed vanHatAatXat gene cluster that produces PG precursors ending in d-Lac (UDP-MurNAc-pentadepsipeptide), making the cell intrinsically resistant to glycopeptides (17). We investigated the consequences of introducing pST30 containing the vanHatAatXat gene cluster (34) into Nonomuraea sp. ATCC 39727. The growth rate and biomass accumulation of Nonomuraea sp. containing pST30 were slightly reduced in comparison to those of the wild-type strain (Fig. 1); the transformed strain was more resistant to teicoplanin and A40926 but not to vancomycin (Table 3). To confirm the expression of the heterologous van gene cluster, d,d-peptidase and d,d-carboxypeptidase activities were measured in the cytoplasmic and membrane extracts of the Nonomuraea sp. strain carrying pST30 (Table 4). A markedly high level of d,d-peptidase activity was measured in the cytoplasmic fraction compared to that in the other strains, presumably attributable to heterologous expression of VanXat (Table 4). Transient and membrane-bound d,d-peptidase/d,d-carboxypeptidase activity (42 nmol min−1 mg−1 protein), which was slightly weaker than in the wild-type strain, was detected in Nonomuraea sp. containing pST30 48 h after inoculation, but it rapidly decreased to an undetectable level after 72 h of growth (Table 4), presumably reflecting host VanYn activity.
β-Lactam inhibition of VanYn.
Nonomuraea sp. ATCC 39727 and its recombinant strains are resistant to >100 μg/ml of ampicillin and penicillin G when assessed using the agar dilution method, and the addition of 50 or 100 μg/ml ampicillin and penicillin G to liquid cultures did not influence growth rate or biomass accumulation (data not shown). However, the level of glycopeptide resistance of these strains generally decreased significantly in the presence of 50 μg/ml ampicillin (data not shown) or penicillin G (Table 3). The only cases in which the glycopeptide MICs remained unchanged in the presence of penicillin G were the vanYn-null mutant and the strain containing pST30 harboring vanHatAatXa from A. teichomyceticus. These results indicate that β-lactams may reduce the level of glycopeptide resistance in Nonomuraea sp. by targeting VanYn; however, this effect is overcome if β-lactam-insensitive vanHatAatXat-mediated resistance is introduced. These findings are in agreement with our previous unexpected observation of β-lactam inhibition of purified VanYn produced in E. coli (22). Purified VanYn was inhibited by increasing concentrations of penicillins, with a 50% inhibitory dose (ID50) of 1 mM for penicillin G (22); in contrast, VanX peptidases and VanY carboxypeptidases from enterococci are resistant to β-lactams (9, 12, 38). Consequently, the effect of penicillin G on d,d-peptidase and d,d-carboxypeptidase activities was measured in the cytoplasmic and membrane fractions from the different mutants. Ten millimolar penicillin G abolished d,d-carboxypeptidase activity in the membrane extracts of Nonomuraea sp. wild-type strains completely, and it was significantly reduced in strains with an increased vanYn gene copy number (Fig. 2A). The addition of increasing penicillin G concentrations led to complete inhibition of VanYn activity in those strains expressing d,d-carboxypeptidase activity higher than that expressed by the wild type. In contrast, the same penicillin G concentrations did not inhibit d,d-peptidase cytoplasmic activity in the strain carrying the vanHatAatXat gene cluster (Fig. 2B).
FIG 2.
Penicillin G effect on VanYn (A) and VanXat (B) activities. d,d-Carboxypeptidase and d,d-peptidase activities were determined by measuring the amount of d-Ala released by hydrolysis of the Nϵ-acetyl-l-Lys-d-Ala-d-Ala tripeptide and d-Ala-d-Ala dipeptide, respectively, using a d-amino acid oxidase coupled to a peroxidase (22). Different concentrations of penicillin G (PEN) (white bar, 0 mM; gray bar, 10 mM; black bar, 50 mM black bar; striped bar, 100 mM) were added to the assay mixture. (A) VanYn activity was measured in membrane extracts of Nonomuraea sp. ATCC 39727 and its recombinant strains (listed in Table 1) grown in VSP medium for 72 h. (B) VanXat activity was measured in the cytoplasmic fraction of the Nonomuraea sp. strain carrying pST30 grown in VSP for 72 h.
Analysis of cell wall precursors.
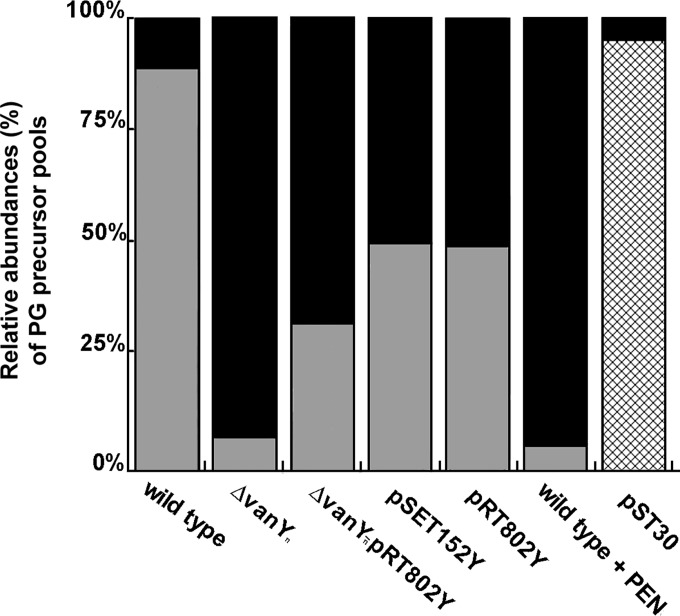
PG cytoplasmic precursors were extracted from exponentially growing cultures of Nonomuraea sp. ATCC 39727 and its recombinant strains following incubation with bacitracin, which blocks the dephosphorylation of undecaprenyl-phosphate, resulting in the accumulation of PG precursors (18). UDP-MurNAc-pentapeptide and UDP-MurNAc-tetrapeptide were then quantified by HPLC-tandem mass spectrometry (MS/MS) (17, 21, 24). Nonomuraea sp. ATCC 39727 produced mostly UDP-MurNAc-tetrapeptide (88%), with UDP-MurNAc-pentapeptide present in much smaller amounts (12%) (Fig. 3). The proportion was essentially reversed in the vanYn-null mutant (92% UDP-MurNAc-pentapeptide and 8% UDP-MurNAc-tetrapeptide), confirming the role of VanYn in removing the last amino acid from the PG precursor. The same UDP-MurNAc-pentapeptide-to-UDP-MurNAc-tetrapeptide ratio as in the vanYn-null mutant was found when 50 μg/ml penicillin G was added to the growth medium of the wild-type strain, suggesting that functional inactivation of VanYn by β-lactam inhibition gives the same phenotype as genetic inactivation of vanYn. However, for reasons we do not understand, the complemented ΔvanYn-pRT802Y mutant and the recombinant strains carrying an additional vanYn gene (Nonomuraea sp. strains carrying pSET152Y and pRT802Y) produced more pentapeptide and less tetrapeptide than expected. The expression of vanHatAatXat from A. teichomyceticus in Nonomuraea sp. resulted in the marked accumulation of the UDP-MurNAc-pentadepsipeptide (terminating in d-Lac). The complete replacement of UDP-MurNAc-tetrapeptide by the UDP-MurNAc-pentadepsipeptide in this last strain confirms that VanXat is capable of removing the intracellular pool of d-Ala-d-Ala. Since VanYn lacks d,d-carboxylesterase activity on a d-Ala-d-Lac-terminating peptide (22), in the presence of vanHatAatXat, this enzyme may act as only a subsidiary resistance mechanism, cleaving the C-terminal d-Ala residue from PG precursors that have escaped VanXat hydrolysis (13). Accordingly, vanY is considered to be an ancillary gene in most of the glycopeptide-resistant enterococci expressing vanHAX-mediated resistance (13).
FIG 3.
Analysis of PG precursor pools in Nonomuraea sp. ATCC 39727 and its derivatives. The Nonomuraea sp. strains listed in Table 1 were grown for 72 h in VSP medium. The wild-type strain was grown in the absence and presence of 50 μg/ml penicillin G. Cytoplasmic PG precursors were extracted from exponentially growing cultures treated with bacitracin and analyzed by HPLC-MS/MS. The relative abundances (%) of UDP-MurNAc-pentapeptide (black bar) and UDP-MurNAc-tetrapeptide (gray bar) were calculated by the integration of absorbance at 262 nm (mean value from a minimum of three independent cultures). In the Nonomuraea sp. strain carrying pST30 and expressing vanHatAatXa from A. teichomyceticus, the UDP-MurNAc-pentadepsipeptide terminated in d-Lac (cross-hatched bar).
A40926 biosynthesis and self-resistance.
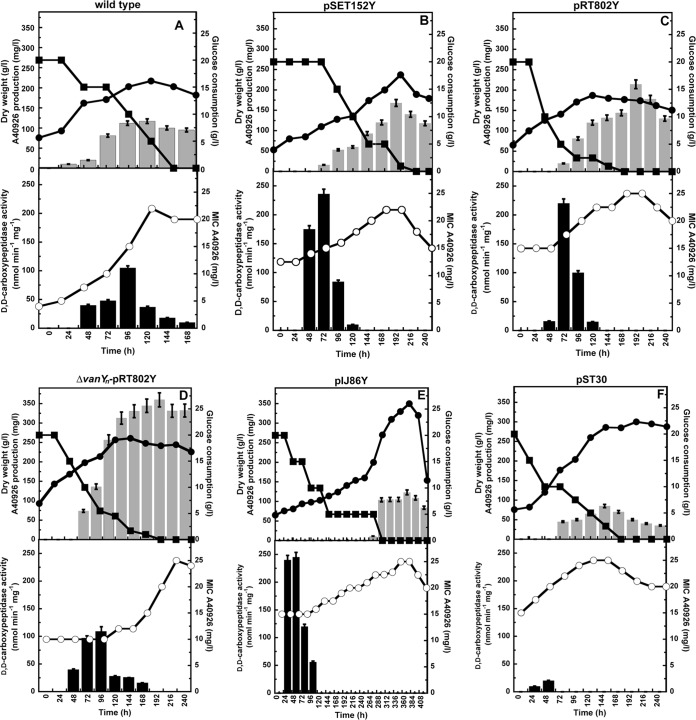
In our previous work in shake flasks, we reported that the growth and A40926 production curves were almost the same in the wild-type and the vanYn-null mutant, indicating that knocking out the resistance gene did not affect antibiotic production (21). The link between A40926 production, self-resistance, and VanYn expression was next studied at a bioreactor scale using FM2 production medium. The growth of Nonomuraea sp. ATCC 39727 in the presence of 50 μg/ml ampicillin or penicillin G had no effect on A40926 production (data not shown), which is consistent with our earlier work. In FM2 medium, the Nonomuraea sp. wild type reached a maximum A40926 productivity of 120 mg/liter after 120 h of fermentation, and resistance to A40926 increased from 4 to slightly >20 mg/liter from the inoculation to antibiotic production phase (Fig. 4A). While knocking out vanYn did not affect growth and A40926 production (21), resistance to A40926 increased from an initial 2 mg/liter (as in VSP medium) to >10 mg/liter during the production phase, indicating that resistance determinants other than vanYn may contribute to a moderate increase of resistance under antibiotic-producing conditions. As observed in VSP medium, the introduction of an additional copy of vanYn on either pSET152Y or pRT802Y (Fig. 4B and C) increased the initial level of resistance to A40926 from 4 mg/liter to 15 mg/liter; this then rose to 22.5 and 25 mg/liter during the production phase, respectively. This initial elevated level of resistance occurred concomitantly with increased A40926 productivity, reaching a maximum of 175 mg/liter and 225 mg/liter, respectively, although it was delayed in comparison to the wild-type strain (120 mg/liter). Interestingly, in the complemented Nonomuraea sp. ΔvanYn-pRT802Y strain, the initial level of resistance was 10 mg/liter, increasing to 25 mg/liter during production, while A40926 production reached 400 mg/liter after 216 h of fermentation (Fig. 4D), representing a 3.4-fold increase compared to the wild-type strain. The higher level of A40926 productivity in the complemented mutant than in the strains carrying an additional copy of vanYn is unexpected and might reflect a complex regulatory relationship between resistance and production. Nevertheless, a role for VanYn in antibiotic production was further suggested by the addition of 50 μg/ml ampicillin or penicillin G to the complemented mutant, which reverted its elevated production phenotype to that of the wild-type strain (data not shown). As in VSP medium, the growth of the Nonomuraea sp. strain carrying pIJ86Y was impaired by the overexpression of VanYn, and A40926 was not detectable until 288 h after inoculation (Fig. 4E). Maximum productivity was around 125 mg/liter, but it was much delayed compared to that of the wild-type strain. Glycopeptide resistance in the Nonomuraea sp. strain carrying pIJ86Y increased from 15 to 25 mg/liter and reached its maximum during the A40926 production phase. In Nonomuraea sp. harboring vanHatAatXat, the A40926 MIC was initially 15 mg/liter and increased during the production phase, reaching 25 mg/liter, but antibiotic productivity was lower than in the wild-type strain (75 mg/liter at 144 h of fermentation; Fig. 4F). When these results are taken together, glycopeptide resistance seems to be regulated in a growth-dependent manner, since it increased during culture growth and generally reached a maximum during A40926 production. A moderate growth-dependent increase in glycopeptide resistance during the production phase was observed even in the vanYn-null mutant, indicating that vanYn is not the only resistance determinant involved in this transition.
FIG 4.
Growth, resistance, and A40926 production in Nonomuraea sp. ATCC 39727 and its derivatives. The Nonomuraea spp. strains listed in Table 1 were grown in FM2 medium in 2-liter bioreactors. Growth parameters (glucose consumption, ■; dry weight, ●), levels of A40926 production (gray bars), the MIC of A40926 (○), and VanYn activity (black bars) were measured as described in Materials and Methods in Nonomuraea sp. ATCC 39727 (A), Nonomuraea sp. strain carrying pSET152Y (B), Nonomuraea sp. strain carrying pRT802Y (C), the Nonomuraea sp. ΔvanYn-pRT802Y strain (D), Nonomuraea sp. strain carrying pIJ86Y (E), and Nonomuraea sp. strain carrying pST30 (F). Data represent the averages from three independent experiments, and error bars indicate the standard deviation.
To gain insights into the role of VanYn during A40926 accumulation, its activity was studied in strains grown in FM2 at a bioreactor scale. As in VSP medium, VanYn activity was associated with the membrane fractions; maximum specific activities were always at least double those detected in VSP growth medium (Fig. 4 and Table 4), again suggesting a correlation between antibiotic production and resistance. In the wild-type strain, maximum VanYn activity was detectable 96 h after inoculation, immediately preceding the peak of A40926 production (Fig. 4A). In Nonomuraea sp. strains carrying pSET152Y or pRT802Y, maximum VanYn activity was 2-fold greater than in the wild-type strain and occurred long before maximum A40926 production compared to the wild-type strain (Fig. 4B and C), indicating again that the introduction of an additional copy of vanYn outside the dbv cluster may compromise the regulatory mechanism normally controlling A40926 resistance and production. In the complemented Nonomuraea sp. ΔvanYn-pRT802Y mutant, VanYn-specific activity was restored to the wild-type level but, as in the strains carrying pSET152Y and pRT802Y, its maximum expression markedly preceded A40926 production (Fig. 4D). As already observed in VSP medium, VanYn activity in the Nonomuraea sp. strain carrying pIJ86Y reached its maximum earlier, i.e., during the first 48 h of cultivation (Fig. 4E). In the Nonomuraea sp. strain carrying pST30, a low level of membrane-associated VanYn activity was detectable in the first 48 h (Fig. 4F), while ca. 70 nmol min−1 mg of protein−1 of a β-lactam-insensitive d,d-peptidase activity attributable to VanXat was found in the cytoplasmic fraction 72 h after inoculation (data not shown).
DISCUSSION
The vanYn-based self-resistance in Nonomuraea spp. described here resembles the glycopeptide resistance mechanism described recently in mutants of Enterococcus faecium selected in vitro for high-level resistance to glycopeptides and β-lactams (39, 40), providing further support for the notion that many clinically relevant resistance mechanisms originated in antibiotic-producing actinomycetes (41). Thus, understanding self-resistance mechanisms in glycopeptide-producing actinomycetes may prove useful in predicting and controlling the future emergence of mechanisms of cross-resistance to clinically used β-lactam and glycopeptide antibiotics. In E. faecium, as in Nonomuraea sp. ATCC 39727, the production of a metallo-d,d-carboxypeptidase, DdcY, eliminates the target of glycopeptides by hydrolysis of the C-terminal d-Ala residue of PG pentapeptide precursors. This modification, if complete, is usually lethal, since classical d,d-transpeptidases of the penicillin binding protein (PBP) family use precursors containing a pentapeptide stem as acyl donors; they catalyze the formation of 4→3 cross-links connecting the 4th amino acid residue of the acyl donor to the 3rd position of the acyl acceptor (42). The cross-linking of a tetrapeptide stems requires a PBP surrogate, l-d-transpeptidase (LDT), which is structurally unrelated to PBPs and catalyzes the formation of 3→3 linkages. In contrast to PBPs, this unusual PG cross-linking enzyme is resistant to penicillins (43). An examination of the PG composition of a range of bacteria has revealed that LDTs are more prevalent than expected, especially among Actinomycetales, which show the following percentages of 3→3 cross-links: 60 to 80% in mycobacteria, 38% in Corynebacterium jeikeium (41, 44), 57% in S. coelicolor, and 49% (exponential phase) to 31% (stationary phase) in Nonomuraea sp. ATCC 39727 (45).
Our results with the vanYn-null mutant and its complemented derivative demonstrate that VanYn contributes to glycopeptide resistance in Nonomuraea sp. ATCC 39727 by supplying tetrapeptide acyl donors that are essential substrates for the formation of the 3→3 PG cross-links by LDT. Such a transpeptidase has yet to be identified in Nonomuraea sp. ATCC 39727, but since the growth of the strain is completely unaffected by the addition of penicillin G and ampicillin to the culture medium, we assume that like the enterococcal LDT (43), it is resistant to these β-lactams.
The incomplete conversion (88%) of UDP-MurNAc-pentapeptide into the UDP-MurNAc-tetrapeptide in Nonomuraea sp. ATCC 39727 may account for its moderate resistance to glycopeptides. In enterococci, the levels of resistance to glycopeptides are determined by the extent of elimination of the PG precursors ending in d-Ala-d-Ala, in both the VanHAX (46) and LDT resistance pathways (39). Full elimination (<2%) of d-Ala-d-Ala-ending precursors is required for high-level vancomycin resistance, with teicoplanin resistance requiring an even more drastic elimination of the pentapeptide precursor (13).
A notable difference between Nonomuraea sp. VanYn and DdcY from E. faecium is that VanYn activity is inhibited by penicillin G and ampicillin. This inhibition was demonstrated initially with the purified recombinant enzyme produced in E. coli (22) and in this work on membrane extracts of Nonomuraea sp. ATCC 39727. We can speculate that the inhibition of VanYn activity might contribute to the reduction of the glycopeptide resistance level observed in Nonomuraea sp. ATCC 39727 in the presence of β-lactams, but this aspect needs to be further investigated.
VanYn is a membrane-bound bifunctional d,d-peptidase and d,d-carboxypeptidase containing conserved motifs (SxHxxGxAxD and ExxH) involved in the coordination of zinc and in the active site that are typical of VanY and VanX zinc-dependent d,d-carboxypeptidases and d,d-peptidases characterized in glycopeptide-resistant enterococci (12, 13, 15, 22, 38). Enterococcal VanY enzymes are typically insensitive to penicillins, which inhibit the activity of low-molecular-weight (LMW) membrane-bound PBPs involved in PG metabolism in many bacteria. Both VanY and LMW PBPs catalyze the same reaction but have a completely different protein domain architecture (41). Similar to classical VanY enzymes, VanYn lacks the canonical SxxK motif found in the active sites of PBPs; thus, determining the way in which β-lactams inhibit VanYn activity requires further analysis.
In enterococci and staphylococci, vanHAX are always present in the same order and are cotranscribed (6, 7, 9, 13), and the VanHAX proteins show high levels of amino acid sequence identity across different resistance genotypes (i.e., ca. 60% identity between enterococcal vanA and vanB gene clusters) and with those of actinomycetes (15, 41). The VanY d,d-carboxypeptidases in enterococci are more diverse than those in VanHAX (i.e., ca. 24% identity between enterococcal vanA and vanB gene clusters), and their genes occupy different positions in different resistance genotypes, i.e., upstream or downstream of vanHAX (41). This suggests that the van gene clusters in enterococci may have been generated by the recruitment of different genes from different glycopeptide producers.
The resistance of Nonomuraea sp. ATCC 39727 to A40926 increased as fermentation progressed, in contrast to A. teichomyceticus, in which resistance was expressed constitutively (17). This increase in resistance, albeit at a lower level, also occurred in the vanYn-null mutant, indicating that yet-undescribed growth-dependent resistance mechanisms (other than those involving vanYn) are involved in the transition from growth to antibiotic production. The integration of an additional copy of vanYn into the Nonomuraea sp. ATCC 39727 genome increased d,d-carboxypeptidase activity and resistance to A40926 during exponential growth, and it resulted in delayed but elevated A40926 production during the stationary phase. The introduction of an additional copy of the gene into the wild-type strain did not double enzyme activity, while complementation of the vanYn-null mutant gave a delayed increase in both resistance and A40926 productivity, the A40926 productivity being considerably improved compared to that of the wild-type and the other mutants. These results suggest that the two vanYn genes exhibit some level of coordinate regulation, perhaps mediated by the same growth-dependent signals that govern the increase in resistance observed in the vanYn-null mutant. While the heterologous expression of vanHatAatXat also increased the level of resistance, it did not enhance A40926 production. Thus, there is not a simple direct correlation between self-resistance and the level of antibiotic production. In a previous study (21), we reported that A40926 MICs increased in the presence of subinhibitory concentrations of A40926 but not of vancomycin and teicoplanin, suggesting a specific mechanism of induction by A40926 in the producing organism. Further investigation may clarify if A40926 has a role in controlling VanYn expression, self-resistance, and its own production. In empirical antibiotic strain improvement programs carried out in the pharmaceutical industry, the selection for increased resistance to a product often results in higher yields (47). This approach was also used during classical strain improvement programs (i.e., mutagenesis and selection of resistant colonies) with the A40926 producer (F. Marinelli, unpublished data). A thorough understanding of A40926 resistance in the producing microorganism may shed new light on the molecular mechanisms linking self-resistance to antibiotic production, thus enabling knowledge-based approaches to strain improvement for glycopeptide-producing actinomycetes.
ACKNOWLEDGMENTS
This work was supported by grants from the Fondo di Ateneo per la Ricerca to F. Marinelli, Progetto Cariplo:Promuovere Capitale Umano d'Eccellenza to G. L. Marcone and E. Binda, by MIUR fellowship to G. L. Marcone, and by the Consorzio Interuniversitario Biotecnologie fellowship to G. L. Marcone and E. Binda.
Footnotes
Published ahead of print 23 June 2014
REFERENCES
- 1.Yim G, Thaker MN, Koteva K, Wright G. 2014. Glycopeptide antibiotic biosynthesis. J. Antibiot. (Tokyo) 67:31–41. 10.1038/ja.2013.117 [DOI] [PubMed] [Google Scholar]
- 2.Jeya M, Moon HJ, Lee KM, Kim IW, Lee JK. 2011. Glycopeptide antibiotics and their novel semi-synthetic derivatives. Curr. Pharm. Biotechnol. 12:1194–1204. 10.2174/138920111796117382 [DOI] [PubMed] [Google Scholar]
- 3.Rossolini GM, Arena F, Pollini S. 2014. Novel infectious diseases and emerging Gram-positive multi-resistant pathogens in hospital and community acquired infections, p 11–28 In Marinelli F, Genilloud O. (ed), Antimicrobials—new and old molecules in the fight against multi-resistant bacteria. Springer Berlin Heidelberg, Berlin, Germany [Google Scholar]
- 4.Jovetic S, Zhu Y, Marcone GL, Marinelli F, Tramper J. 2010. β-Lactam and glycopeptide antibiotics: first and last line of defense? Trends Biotechnol. 28:596–604. 10.1016/j.tibtech.2010.09.004 [DOI] [PubMed] [Google Scholar]
- 5.Barna JC, Williams DH. 1984. The structure and mode of action of glycopeptide antibiotics of the vancomycin group. Annu. Rev. Microbiol. 38:339–357. 10.1146/annurev.mi.38.100184.002011 [DOI] [PubMed] [Google Scholar]
- 6.Courvalin P. 2006. Vancomycin resistance in Gram-positive cocci. Clin. Infect. Dis. 42(Suppl 1):S25–S34. 10.1086/491711 [DOI] [PubMed] [Google Scholar]
- 7.Arthur M, Reynolds PE, Depardieu F, Evers S, Dutka-Malen S, Quintiliani R, Jr, Courvalin P. 1996. Mechanisms of glycopeptide resistance in enterococci. J. Infect. 32:11–16. 10.1016/S0163-4453(96)80003-X [DOI] [PubMed] [Google Scholar]
- 8.Arthur M, Molinas C, Depardieu F, Courvalin P. 1993. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 175:117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reynolds PE, Depardieu F, Dutka-Malen S, Arthur M, Courvalin P. 1994. Glycopeptide resistance mediated by enterococcal transposon Tn1546 requires production of VanX for hydrolysis of d-alanyl-d-alanine. Mol. Microbiol. 13:1065–1070. 10.1111/j.1365-2958.1994.tb00497.x [DOI] [PubMed] [Google Scholar]
- 10.Walsh CT, Fisher SL, Park IS, Prahalad M, Wu Z. 1996. Bacterial resistance to vancomycin: five genes and one missing hydrogen bond tell the story. Chem. Biol. 3:21–28. 10.1016/S1074-5521(96)90079-4 [DOI] [PubMed] [Google Scholar]
- 11.Bugg TD, Wright GD, Dutka-Malen S, Arthur M, Courvalin P, Walsh CT. 1991. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry 30:10408–10415. 10.1021/bi00107a007 [DOI] [PubMed] [Google Scholar]
- 12.Wright GD, Molinas C, Arthur M, Courvalin P, Walsh CT. 1992. Characterization of VanY, a d,d-carboxypeptidase from vancomycin-resistant Enterococcus faecium BM4147. Antimicrob. Agents Chemother. 36:1514–1518. 10.1128/AAC.36.7.1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arthur M, Depardieu F, Cabanié L, Reynolds P, Courvalin P. 1998. Requirement of the VanY and VanX d,d-peptidases for glycopeptide resistance in enterococci. Mol. Microbiol. 30:819–830. 10.1046/j.1365-2958.1998.01114.x [DOI] [PubMed] [Google Scholar]
- 14.Lebreton F, Depardieu F, Bourdon N, Fines-Guyon M, Berger P, Camiade S, Leclercq R, Courvalin P, Cattoir V. 2011. d-Ala-d-Ser VanN-type transferable vancomycin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 55:4606–4612. 10.1128/AAC.00714-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall CG, Lessard IA, Park I, Wright GD. 1998. Glycopeptide antibiotic resistance genes in glycopeptide-producing organisms. Antimicrob. Agents Chemother. 42:2215–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pootoolal J, Thomas MG, Marshall CG, Neu JM, Hubbard BK, Walsh CT, Wright GD. 2002. Assembling the glycopeptide antibiotic scaffold: the biosynthesis of A47934 from Streptomyces toyocaensis NRRL 15009. Proc. Natl. Acad. Sci. U. S. A. 99:8962–8967. 10.1073/pnas.102285099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beltrametti F, Consolandi A, Carrano L, Bagatin F, Rossi R, Leoni L, Zennaro E, Selva E, Marinelli F. 2007. Resistance to glycopeptide antibiotics in the teicoplanin producer is mediated by van gene homologue expression directing the synthesis of a modified cell wall peptidoglycan. Antimicrob. Agents Chemother. 51:1135–1141. 10.1128/AAC.01071-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schäberle TF, Vollmer W, Frasch HJ, Hüttel S, Kulik A, Röttgen M, von Thaler AK, Wohlleben W, Stegmann E. 2011. Self-resistance and cell wall composition in the glycopeptide producer Amycolatopsis balhimycina. Antimicrob. Agents Chemother. 55:4283–4289. 10.1128/AAC.01372-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutchings MI, Hong HJ, Buttner MJ. 2006. The vancomycin resistance VanRS two-component signal transduction system of Streptomyces coelicolor. Mol. Microbiol. 59:923–935. 10.1111/j.1365-2958.2005.04953.x [DOI] [PubMed] [Google Scholar]
- 20.Goldstein BP, Selva E, Gastaldo L, Berti M, Pallanza R, Ripamonti F, Ferrari P, Denaro M, Arioli V, Cassani G. 1987. A40926, a new glycopeptide antibiotic with anti-Neisseria activity. Antimicrob. Agents Chemother. 31:1961–1966. 10.1128/AAC.31.12.1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marcone GL, Beltrametti F, Binda E, Carrano L, Foulston L, Hesketh A, Bibb M, Marinelli F. 2010. Novel mechanism of glycopeptide resistance in the A40926 producer Nonomuraea sp. ATCC 39727. Antimicrob. Agents Chemother. 54:2465–2472. 10.1128/AAC.00106-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Binda E, Marcone GL, Pollegioni L, Marinelli F. 2012. Characterization of VanYn, a novel d,d-peptidase/d,d-carboxypeptidase involved in glycopeptide antibiotic resistance in Nonomuraea sp. ATCC 39727. FEBS J. 279:3203–3213. 10.1111/j.1742-4658.2012.08706.x [DOI] [PubMed] [Google Scholar]
- 23.Binda E, Marcone GL, Berini F, Pollegioni L, Marinelli F. 2013. Streptomyces spp. as efficient expression system for a d,d-peptidase/d,d-carboxypeptidase involved in glycopeptide antibiotic resistance. BMC Biotechnol. 13:24. 10.1186/1472-6750-13-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcone GL, Carrano L, Marinelli F, Beltrametti F. 2010. Protoplast preparation and reversion to the normal filamentous growth in antibiotic-producing uncommon actinomycetes. J. Antibiot. (Tokyo) 63:83–88. 10.1038/ja.2009.127 [DOI] [PubMed] [Google Scholar]
- 25.Marcone GL, Foulston L, Binda E, Marinelli F, Bibb M, Beltrametti F. 2010. Methods for the genetic manipulation of Nonomuraea sp. ATCC 39727. J. Ind. Microbiol. Biotechnol. 37:1097–1103. 10.1007/s10295-010-0807-5 [DOI] [PubMed] [Google Scholar]
- 26.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580. 10.1016/S0022-2836(83)80284-8 [DOI] [PubMed] [Google Scholar]
- 27.MacNeil DJ, Klapko LM. 1987. Transformation of Streptomyces avermitilis by plasmid DNA. J. Ind. Microbiol. 2:209–218. 10.1007/BF01569542 [DOI] [Google Scholar]
- 28.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100:1541–1546. 10.1073/pnas.0337542100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 30.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 31.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 32.Bierman M, Logan R, O'Brien K, Seno ET, Rao RN, Schoner BE. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43–49. 10.1016/0378-1119(92)90627-2 [DOI] [PubMed] [Google Scholar]
- 33.Gregory MA, Till R, Smith MC. 2003. Integration site for Streptomyces phage phiBT1 and development of site-specific integrating vectors. J. Bacteriol. 185:5320–5323. 10.1128/JB.185.17.5320-5323.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Serina S, Radice F, Maffioli S, Donadio S, Sosio M. 2004. Glycopeptide resistance determinants from the teicoplanin producer Actinoplanes teichomyceticus. FEMS Microbiol. Lett. 240:69–74. 10.1016/j.femsle.2004.09.017 [DOI] [PubMed] [Google Scholar]
- 35.Kariyama R, Massidda O, Daneo-Moore L, Shockman GD. 1990. Properties of cell wall-associated d,d-carboxypeptidase of Enterococcus hirae (Streptococcus faecium) ATCC 9790 extracted with alkali. J. Bacteriol. 172:3718–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gornall AG, Bardawill CJ, David MM. 1949. Determination of serum proteins by means of the biuret reaction. J. Biol. Chem. 177:751–766 [PubMed] [Google Scholar]
- 37.Taurino C, Frattini L, Marcone GL, Gastaldo L, Marinelli F. 2011. Actinoplanes teichomyceticus ATCC 31121 as a cell factory for producing teicoplanin. Microb. Cell Fact. 10:82. 10.1186/1475-2859-10-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Z, Wright GD, Walsh CT. 1995. Overexpression, purification, and characterization of VanX, a d,d-dipeptidase which is essential for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry 34:2455–2463. 10.1021/bi00008a008 [DOI] [PubMed] [Google Scholar]
- 39.Cremniter J, Mainardi JL, Josseaume N, Quincampoix JC, Dubost L, Hugonnet JE, Marie A, Gutmann L, Rice LB, Arthur M. 2006. Novel mechanism of resistance to glycopeptide antibiotics in Enterococcus faecium. J. Biol. Chem. 281:32254–32262. 10.1074/jbc.M606920200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sacco E, Hugonnet JE, Josseaume N, Cremniter J, Dubost L, Marie A, Patin D, Blanot D, Rice LB, Mainardi JL, Arthur M. 2010. Activation of the l,d-transpeptidation peptidoglycan cross-linking pathway by a metallo-d,d-carboxypeptidase in Enterococcus faecium. Mol. Microbiol. 75:874–885. 10.1111/j.1365-2958.2009.07014.x [DOI] [PubMed] [Google Scholar]
- 41.Mainardi JL, Villet R, Bugg TD, Mayer C, Arthur M. 2008. Evolution of peptidoglycan biosynthesis under the selective pressure of antibiotics in Gram-positive bacteria. FEMS Microbiol. Rev. 32:386–408. 10.1111/j.1574-6976.2007.00097.x [DOI] [PubMed] [Google Scholar]
- 42.Cordillot M, Dubée V, Triboulet S, Dubost L, Marie A, Hugonnet JE, Arthur M, Mainardi JL. 2013. In vitro cross-linking of Mycobacterium tuberculosis peptidoglycan by l,d-transpeptidases and inactivation of these enzymes by carbapenems. Antimicrob. Agents Chemother. 57:5940–5945. 10.1128/AAC.01663-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mainardi JL, Fourgeaud M, Hugonnet JE, Dubost L, Brouard JP, Ouazzani J, Rice LB, Gutmann L, Arthur M. 2005. A novel peptidoglycan cross-linking enzyme for a beta-lactam-resistant transpeptidation pathway. J. Biol. Chem. 280:38146–38152. 10.1074/jbc.M507384200 [DOI] [PubMed] [Google Scholar]
- 44.Lavollay M, Fourgeaud M, Herrmann JL, Dubost L, Marie A, Gutmann L, Arthur M, Mainardi JL. 2011. The peptidoglycan of Mycobacterium abscessus is predominantly cross-linked by l,d-transpeptidases. J. Bacteriol. 193:778–782. 10.1128/JB.00606-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hugonnet JE, Haddache N, Veckerlé C, Dubost L, Marie A, Shikura N, Mainardi JL, Rice LB, Arthur M. 2014. Peptidoglycan cross-linking in glycopeptide resistant Actinomycetales. Antimicrob. Agents Chemother. 58:1749–1756. 10.1128/AAC.02329-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arthur M, Depardieu F, Reynolds P, Courvalin P. 1996. Quantitative analysis of the metabolism of soluble cytoplasmic peptidoglycan precursors of glycopeptide-resistant enterococci. Mol. Microbiol. 21:33–44. 10.1046/j.1365-2958.1996.00617.x [DOI] [PubMed] [Google Scholar]
- 47.Cundliffe E, Demain AL. 2010. Avoidance of suicide in antibiotic-producing microbes. J. Ind. Microbiol. Biotechnol. 37:643–672. 10.1007/s10295-010-0721-x [DOI] [PubMed] [Google Scholar]