Abstract
Escherichia coli sequence type 131 (ST131) is an extensively antimicrobial-resistant E. coli clonal group that has spread explosively throughout the world. Recent molecular epidemiologic and whole-genome phylogenetic studies have elucidated the fine clonal structure of ST131, which comprises multiple ST131 subclones with distinctive resistance profiles, including the (nested) H30, H30-R, and H30-Rx subclones. The most prevalent ST131 subclone, H30, arose from a single common fluoroquinolone (FQ)-susceptible ancestor containing allele 30 of fimH (type 1 fimbrial adhesin gene). An early H30 subclone member acquired FQ resistance and launched the rapid expansion of the resulting FQ-resistant subclone, H30-R. Subsequently, a member of H30-R acquired the CTX-M-15 extended-spectrum beta-lactamase and launched the rapid expansion of the CTX-M-15-containing subclone within H30-R, H30-Rx. Clonal expansion clearly is now the dominant mechanism for the rising prevalence of both FQ resistance and CTX-M-15 production in ST131 and in E. coli generally. Reasons for the successful dissemination and expansion of the key ST131 subclones remain undefined but may include increased transmissibility, greater ability to colonize and/or persist in the intestine or urinary tract, enhanced virulence, and more-extensive antimicrobial resistance compared to other E. coli. Here we discuss the epidemiology and molecular phylogeny of ST131 and its key subclones, possible mechanisms for their ecological success, implications of their widespread dissemination, and future research needs.
INTRODUCTION
Escherichia coli sequence type 131 (ST131) is a recently emerged, extensively antimicrobial-resistant E. coli clonal group that has spread explosively throughout the world, driving the rapid increase in prevalence of antimicrobial resistance in E. coli (1–5). Such widespread expansion of a single clonal group is unprecedented in E. coli populations, although it has been seen in other antimicrobial-resistant pathogens such as methicillin-resistant Staphylococcus aureus, penicillin-resistant Streptococcus pneumoniae, and the NAP1 strain of Clostridium difficile (6, 7), and was foreshadowed by the notorious but less extensive expansion of E. coli “clonal group A” (8–10). Despite ST131 being recognized as a pandemic clonal group that threatens public health, ST131 has received comparatively less attention in the United States than have other antimicrobial-resistant pathogens. Here we review the epidemiology and molecular phylogeny of ST131, possible mechanisms for its ecological success, and implications of its widespread dissemination.
EARLY STUDIES OF ST131 EPIDEMIOLOGY AND RESISTANCE
ST131, which is defined by the sequences of the 7 housekeeping genes that are commonly used for multilocus sequence typing (MLST) in E. coli according to the Achtman MLST scheme (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli), was first reported in 2008 by two research groups that were studying CTX-M-type extended-spectrum-beta-lactamase (ESBL)-producing E. coli (2, 11). By 2009, ST131 had been identified in 9 countries spanning 3 continents (3–5). ST131 belongs to (virulence-associated) E. coli phylogenetic group B2, and most isolates exhibit serotype O25b:H4, except for a small subset that are serotype O16:H5 (12) and rare variants with other serotypes (13). ST131 is associated with distinct combinations of extraintestinal virulence factors, compared with non-ST131 E. coli (4, 14–17), and exhibits diverse pulsed-field gel electrophoresis (PFGE) profiles, several of which predominate globally (18).
According to recent studies, the prevalence of ST131 among human clinical E. coli isolates varies by geographic region and host population, ranging overall from 12.5% (19) to nearly 30% (15, 20, 21) of E. coli clinical isolates. Like other extraintestinal pathogenic E. coli (ExPEC) strains, ST131 causes a variety of extraintestinal infections, including bacteremia, pneumonia, and urinary tract, intra-abdominal, and wound infections. Whether ST131 is associated with worse clinical outcomes than other E. coli strains is unclear, since some studies suggest that ST131 is more likely to cause persistent or recurrent urinary tract infections (20) or a higher frequency of sepsis (22), whereas others have found no difference in outcomes of infections with ST131 versus other E. coli strains (23, 24). In one study, patients infected with ST131 had persistent or recurrent symptoms in part because they received empirical therapy with fluoroquinolones (FQs), which are ineffective against most members of this clonal group (20).
Regarding epidemiological correlates, it is not clear if ST131 is more strongly associated with community settings or with health care settings. Most of the studies that found a high prevalence of ST131 among community-associated isolates have utilized convenience samples of ESBL-producing E. coli or have not taken into account the extent of health care contact among community dwellers (25–27). In contrast, in a population-based cohort study that evaluated consecutively collected isolates from Olmsted County, MN, and recorded several host characteristics, including health care exposure, ST131 was significantly associated with more health care-associated infections than community-associated infections (20, 28).
Other epidemiologic and ecologic associations include ST131's predilection for elderly hosts and its high prevalence among long-term-care-facility (LTCF) residents in both the United States (20) and Europe (29–34). Additionally, infections due to ESBL-positive ST131 have been associated with international travel (26, 35–37). ST131 is also broadly disseminated among diverse nonhuman sources, including companion animals, other animals, food sources, and the environment (38–41). However, the prevalence of ST131 colonization and infection is substantially greater among humans than among nonhuman hosts (42, 43), evidence suggesting that the ST131 pandemic is primarily a human-based phenomenon.
Although ST131 first came to attention because of its association with ESBL-producing E. coli strains, in particular, those expressing CTX-M-15, most ST131 isolates in many locales are ESBL negative, but resistant to FQs, and often are coresistant to aminoglycosides and/or trimethoprim-sulfamethoxazole (4, 19, 21). Among diverse patient groups from a variety of locales, ST131 consistently accounts for approximately 70% to 80% of FQ-resistant isolates (4, 15, 19–21, 44–46), and for nearly two-thirds of ESBL-producing isolates (4, 15), but for only 0% to 7% of FQ-susceptible isolates (4, 15, 20). Much of the diversity of susceptibility profiles among ST131 isolates, and possibly some of the epidemiologic and host group associations, is explained by the fine clonal structure of ST131, as discussed below.
H30 AND H30-Rx SUBCLONES OF ST131
Until recently, ST131 has been regarded primarily as a unitary entity, despite clear evidence of genetic diversity at the level of virulence gene content and pulsed-field gel electrophoresis profiles. The fine clonal structure of ST131 was recently elucidated by using multiple molecular typing methods (15, 47, 48). This led to the identification of multiple ST131 subclones with distinctive resistance profiles and provided insights into the relative importance of horizontal gene transfer or mutation, versus clonal expansion, in the pandemic emergence of ST131 and its characteristic resistance traits.
The most prevalent subclone of ST131, called H30 because it contains the H30 variant of the type 1 fimbrial adhesin gene fimH, was identified initially through sub-ST analysis of over 1,000 historical and recent E. coli isolates (both ST131 and non-ST131) using a combination of typing strategies, including sequencing of fimH, gyrA, and parC, multilocus sequence typing (MLST), and PFGE (44). Investigators observed that the H30 ST131 subclone comprised approximately half of all recent FQ-resistant E. coli isolates from diverse locales and sources, compared with <1% of FQ-susceptible isolates, and that the H30 lineage first appeared in 2000 and expanded abruptly thereafter. The close genetic similarity of most H30 strains suggested that they all arose from a single fimH30-carrying ancestor. This indicated that the dramatic emergence of FQ-resistant ST131 strains has been driven by clonal expansion and dissemination rather than by independent acquisition of FQ resistance genes in heterogeneous strains.
Further support for this conclusion was provided by the finding of a tight linkage between the H30 ST131 subclone and a single FQ-resistance-conferring gyrA-plus-parC allele combination, despite evidence for widespread horizontal transfer of gyrA and parC alleles among other lineages (44). The H30 ST131 subclone was also found to be associated with combined resistance to ≥3 antibiotic classes and with CTX-M-15 (44). A novel and rapid form of sequence typing based on sequencing of the fumC and fimH loci (called CH typing) reliably identifies H30 ST131 as clonotype CH40-30 (22, 47) and has been used in subsequent studies to detect the H30 subclone, as has a PCR-based assay that detects fimH30-specific single-nucleotide polymorphisms (SNPs) (15).
More recently, an important sublineage within H30, called H30-Rx because of its more extensive antimicrobial resistance profile, was identified by Price et al. using whole-genome phylogenetic analysis (48). Those investigators first analyzed 524 ST131 isolates collected between 1967 and 2011 using PFGE and found intermixing of FQ-resistant, FQ-susceptible, CTX-M-positive, and CTX-M-negative strains within the PFGE tree, suggesting frequent horizontal acquisition of FQ resistance and ESBL genes. However, when 105 of these isolates underwent whole-genome sequencing and SNP analysis to reconstruct the phylogeny of ST131, both FQ resistance and CTX-M-15 were shown to be almost entirely confined to the H30 subclone, which formed the most highly derived portion of the tree. Moreover, within H30, those H30 strains that carried CTX-M-15 formed a discrete sublineage, H30-Rx, that was separated from ESBL-negative H30 strains by 3 core genome SNPs.
These data indicate that the H30 ST131 lineage comprises a series of nested subclones, all derived from a single common FQ-susceptible H30 ancestor. Within H30, the FQ-resistant subclone, i.e., H30-R, encompasses nearly all FQ-resistant ST131 isolates, whereas within H30-R, the H30-Rx subclone encompasses the vast majority of CTX-M-15-containing ST131 isolates (Fig. 1). This nested clonal structure results in a continuum of antimicrobial resistance among H30-associated ST131 subclones, from the most susceptible, H30 (non-R, non-Rx), to the more resistant H30-R, to the most extensively resistant, H30-Rx (17, 48). The study by Price et al. confirmed at the whole-genome level that clonal expansion is indeed the dominant mechanism for the rising prevalence of CTX-M-15 production and FQ resistance, both in ST131 specifically and, given the prominence of ST131 among resistant E. coli strains, in E. coli generally. Petty et al. independently reported similar findings shortly thereafter (49).
FIG 1.
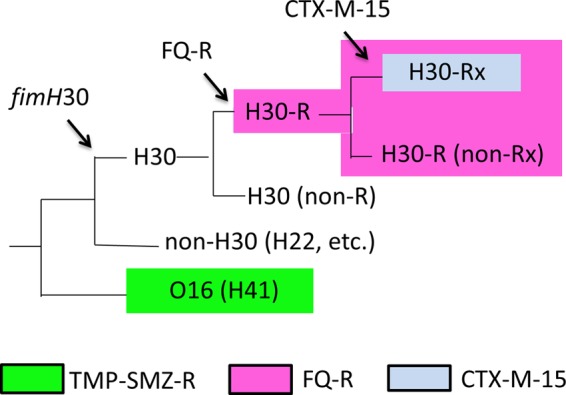
Schematic dendrogram of ST131 phylogeny reconstructed using whole-genome single nucleotide polymorphism analysis. Arrows indicate emergence of fimH30 allele, FQ resistance, and CTX-M-15 ESBL. Colors indicate resistance traits significantly associated with (although not confined to) specific lineages. FQ, fluoroquinolone; TMP-SMZ, trimethoprim-sulfamethoxazole; R, resistance.
The prevalence and resistance phenotypes of the H30, H30-R, and H30-Rx ST131 subclones have thus far been characterized in only a few populations. Among E. coli clinical isolates from U.S. veterans in 2011, ST131 accounted for 78% and 64%, respectively, of FQ-resistant and ESBL-producing isolates but only 7% of FQ-susceptible isolates. Among these ST131 isolates, the H30 ST131 subclone (encompassing all H30, H30-R, and H30-Rx isolates) accounted for 95% to 98% of FQ-resistant or ESBL-producing isolates but only 12.5% of FQ-susceptible isolates (15). Similarly, in a case-control study conducted in the Chicago region, approximately half of ESBL-producing E. coli isolates were ST131, and of those ST131 isolates, 98% were H30 and 92% were H30-Rx (17). In that study, aggregate resistance scores were higher among H30 than non-H30 ST131 isolates.
Few studies have evaluated host factors associated with these important ST131 subclones. In a population-based study of consecutively collected E. coli isolates in Olmsted County, MN, the prevalence of ST131 subgroups (classified broadly as H30 versus non-H30) varied with patient age and type of infection. Specifically, non-H30 subclone ST131 isolates (encompassing isolates with fimH alleles 41 and 22 and others) were more common among young patients, whereas H30 isolates predominated among patients older than 50 years (28). Additionally, H30 ST131 was also significantly more prevalent among health-care-associated (43%) than among community-associated (11%) isolates. In a multicenter study utilizing CH typing to analyze >1,600 extraintestinal E. coli isolates, CH clonotype 40-30, corresponding to the H30 subclone of ST131, was the most prevalent clonotype overall and was statistically associated with recurrent or persistent urinary tract infections and sepsis (22). Price et al., who further divided the H30 subclone into H30-Rx and H30-non-Rx isolates, observed that sepsis was significantly associated specifically with H30-Rx, whereas the (non-Rx) H30 and the non-H30 ST131 isolates had associated sepsis rates no different from those of non-ST131 isolates (48). Whether these differential outcomes classified by ST131 subclone are related to pathogen or host factors is not clear.
O16 SUBCLONE OF ST131
A small subset of ST131 isolates exhibits serotype O16:H5. Such strains derive from the most basal clade with the ST131 phylogeny, well separated from the more prominent O25b:H4 ST131 clade (Fig. 1). The O16 and O25b subsets within ST131 are identified as representing distinct STs according to the Pasteur Institute (but not the Achtman) MLST schema (50, 51). The O16 ST131 isolates uniformly contain the fimH41 allele, whereas most O25b:H4 ST131 isolates have the fimH30 allele, a minority have fimH22, and the remainder have one of several rarer fimH alleles (12, 52). O16 ST131 isolates also have distinct combinations of gyrA and parC alleles compared with other ST131 isolates (12, 52). The O16 ST131 subclone is not detected by a commonly used PCR screening assay for ST131 that targets the O25 rfb variant and pabB (12). In contrast, it is detected both by PCR amplification of a subclone-specific allele of trpA and by combined detection of the O16 rfb variant and ST131-specific alleles of mdh and gyrB (12).
Characteristics of the O16 ST131 clade were recently described within two large clinical E. coli collections (total n = 4,239). The O16 ST131 isolates accounted for 1% to 5% of the E. coli isolates from each contributing center. Additionally, comparison with a large private reference library identified O16 ST131 isolates collected from humans and pets in diverse geographic regions. O16 subclone isolates had a higher prevalence of resistance to trimethoprim-sulfamethoxazole and gentamicin than the H30 ST131 subclone isolates but a lower prevalence of resistance to FQs and ceftriaxone (Table 1). The O16 ST131 isolates broadly resembled O25b:H4 ST131 isolates for virulence genotype and experimental virulence in mouse models, although in a separate study O16 ST131 isolates killed mice less rapidly than did O25b:H4 ST131 isolates (12, 52).
TABLE 1.
Prevalence of antimicrobial resistance shown by ST131 subclone
| Drug | % prevalence of resistance, range |
|||||
|---|---|---|---|---|---|---|
| ST131 |
Non-ST131a,f | |||||
| O16a | O25b |
|||||
| Non-H30b | H30 |
|||||
| All H30c | H30-R |
|||||
| H30-R, non-Rxd | H30-Rxd,e | |||||
| Ampicillin | 87–94 | 61–80 | 76–88 | 98 | 34–48 | |
| Gentamicin | 31–44 | 6–20 | 20–30 | 32 | 3–6 | |
| TMP-SMZg | 57–66 | 23–40 | 42–51 | 45 | 13–26 | |
| Ciprofloxacin | 7–19 | 0–36 | 84–100 | 100 | 98–100 | 9–14 |
| Ceftriaxone | 0–13 | 3–29 | 4–25 | 6 | 77–89 | 2–8 |
BASIS FOR ST131 EMERGENCE
It is remarkable that the pandemic emergence of ST131, and specifically its H30-R and H30-Rx subclones, occurred over less than 10 years (4, 5, 44). Reasons for the successful dissemination and expansion of ST131 remain undefined but may include higher transmissibility, a greater ability to colonize and/or persist in the intestine or urinary tract, enhanced virulence (i.e., ability to cause disease), and more-extensive antimicrobial resistance compared to other E. coli. Evidence pertaining to each of these mechanisms is discussed below.
Transmission.
Multiple case reports describe seemingly efficient person-to-person transmission of ST131. Intrafamilial transmission of ST131 is suggested by the occurrence in an 8-month-old girl of septic arthritis and osteomyelitis caused by an ST131 isolate with the same PFGE profile as a FQ-resistant ST131 isolate from her mother's feces (53). Similarly, nearly identical ST131 CTX-M-15-producing strains caused severe infections in 2 adult sisters who resided temporarily in the same home (48, 54, 55). Likewise, in a reported father-daughter pair, the adult daughter, who had limited contact with her elderly father, visited him once while he was hospitalized for treatment of E. coli pyelonephritis and renal abscesses and used his hospital room lavatory. She subsequently developed septic shock, E. coli bacteremia, and emphysematous pyelonephritis. The two patients' infections were caused by ST131 strains with highly similar PFGE profiles, suggesting possible host-to-host transmission during the patients' brief hospital room encounter (56). ST131 strain sharing has even been documented between 2 dogs and 3 cats within a household (57).
Regarding within-institution transmission, recent evidence suggests spread of a CTX-M-15-producing ST131 strain within a French day care center, where 7 children had intestinal colonization with the same strain (58). At the other extreme of the age spectrum, several residents of LTCFs in Olmsted County, MN, had intestinal colonization with ST131 isolates that exhibited similar PFGE profiles, which clustered by LTCF, suggesting within-facility horizontal transmission (R. Banerjee, unpublished data). Although close person-to-person contact is known to be associated with multihost sharing of extraintestinal E. coli strains (59–62), whether ST131 is more efficiently transmitted than other E. coli strains is unknown and deserves systematic evaluation.
Colonization.
Because intestinal colonization with extraintestinal E. coli is believed to be a prerequisite for extraintestinal infection (63), it also is possible that superior intestinal colonizing ability underlies the widespread dissemination of ST131. In a mouse model of intestinal colonization, an ST131 strain outcompeted the several tested commensal strains when it was mixed with them in a 1:1 ratio and administered enterally into streptomycin-pretreated mice. Although that study was limited by its evaluation of only a single ST131 strain, it provided evidence that at least some ST131 strains can efficiently colonize the intestine (64).
It is unclear whether the prevalence or duration of intestinal colonization in humans is different for ST131 compared to other E. coli strains or for the more successful ST131 subclones (i.e., H30, H30-R, and H30-Rx) compared to other ST131 subclones (e.g., H41 and H22). In studies of the prevalence of ST131 carriage among individuals colonized specifically with ESBL-producing E. coli, ST131 colonization rates have varied greatly by patient population, from nearly 10% among healthy adults (65) and healthy mother-newborn pairs (66) to 38% among hospitalized international patients (36) and 44% among children in French day care centers (58).
However, fewer studies have evaluated the prevalence of ST131 among “all-comer” fecal E. coli isolates, without selection for ESBL production. Kudinha et al. noted ST131 prevalence values of 0% to 4% among fecal E. coli isolates from 580 healthy children, men, and women in Australia (67–69). In contrast, among 133 elderly residents of LTCFs in Olmsted County, MN, 24% subjects had fecal colonization with ST131 (Banerjee, unpublished). Similarly, among adult inpatients admitted to two urban hospitals in the United States who had intestinal colonization with FQ-resistant E. coli, approximately 25% were colonized with ST131 (J. R. Johnson, unpublished data). Taken together, those studies suggest that ST131 intestinal colonization rates are likely to vary by ESBL status, host characteristics, and geographic region. Comparisons with other E. coli lineages are needed to help interpret these findings.
Virulence.
It is also possible that the globally increasing prevalence of ST131 among clinical isolates is occurring because ST131 strains are more virulent than other E. coli strains, giving it a fitness advantage in the pathogenic niche. However, the available molecular epidemiological, conventional epidemiological, and in vivo experimental data that bear on this question are conflicted, pointing to no clear answer. In molecular epidemiological studies, ST131 is consistently found to have a greater number of virulence factor genes (i.e., higher aggregate virulence scores) than other comparably antimicrobial-resistant E. coli strains (4, 15, 16). Moreover, within ST131, the H30, non-H30, and H30-Rx subclones have characteristic virulence profiles that are distinct from those of non-ST131 E. coli, conceivably conferring enhanced virulence (17). In that regard, Blanco et al. have recently used the presence of specific virulence genes to define five main virotypes within ST131 that appear to have distinct epidemiologic correlates (70) and virulence phenotypes in a mouse model, as discussed below (52). ST131 isolates have also been noted to have a high prevalence of biofilm production, which may be an important virulence-promoting trait (67).
Conventional epidemiological studies provide some support for the prediction of enhanced virulence of ST131 by demonstrating that ST131 is more prevalent among invasive than noninvasive E. coli isolates. In a series of studies from Australia, ST131 accounted for 30% of pyelonephritis isolates among E. coli isolates from women, versus only 13% of cystitis isolates and 4% of fecal isolates, and similar prevalence trends were seen among men and children (67–69). Likewise, in a study from the United Kingdom, ST131 prevalence was 21% among bacteremia isolates compared to only 7% among urinary isolates (71). Additionally, as noted above, H30 (and, specifically, H30-Rx) ST131 strains have exhibited epidemiological associations with sepsis (22, 48).
However, all of these studies lack detailed patient-level clinical information, which is an important limitation, since host factors and portal of entry impact disease severity and outcomes of invasive E. coli infections (72). Indeed, when investigators adjusted for host factors, no differences in cure or mortality were found between patients with infections due to ST131 and those with infections due to other E. coli strains (20).
Studies using experimental animal models likewise do not clearly support the hypothesis that ST131 is more virulent than other E. coli strains. For example, Lavigne et al. assessed the virulence of 3 ST131 and 6 non-ST131 E. coli strains in infection models involving Caenorhabditis elegans (a nematode) and zebrafish and concluded that ST131 strains were no more virulent than non-ST131 E. coli strains (73). Furthermore, virulence levels differed among the 3 tested ST131 strains, and a given strain's virulence was not consistent across the nematode and zebrafish models, raising questions about which, if either, model best predicts virulence in humans (73).
Similarly, 4 different studies using mouse models to investigate ST131 virulence yielded discordant results. Using a mouse sepsis model, Clermont et al. demonstrated that the 4 studied ST131 strains were highly virulent, each killing all 10 challenged mice (74), although this might have reflected in part the nonphysiological nature of the model, which involves the subcutaneous injection of 109 CFU/ml bacteria into the abdominal wall (64). Using a similar mouse subcutaneous sepsis model, Johnson et al. assessed 61 ST131 and non-ST131 isolates and found a wide range of lethality overall, with no correlation between mouse lethality or illness severity and either ST131 status or FQ resistance (75). Mora et al. used a similar mouse subcutaneous sepsis model to assess the virulence of 23 ST131 clinical isolates and found that two-thirds of the strains were highly lethal, killing >90% of inoculated mice. Virulence levels differed by virotype, with virotypes A, B, and C (all H30 associated) being most lethal and virotypes D (H22 associated) and E (rare; H30 associated) being least lethal (52). Lastly, Vimont et al. evaluated an ST131 strain in a mouse model of ascending urinary tract infection (64) and observed higher bladder and kidney concentrations with the ST131 strain than with the two non-ST131 comparison strains, suggesting that at least some ST131 strains may have an enhanced ability to acutely colonize the urinary tract (64, 76).
The discrepant results from the various mouse model studies possibly can be explained by the use of different models, experimental conditions, and strains or by chance. It also is possible that current models that involve subcutaneous or transurethral inoculation, and that assess only short-term endpoints, bypass critical steps in the pathogenesis pathways where ST131 may have a fitness advantage in nature, such as vaginal or periurethral colonization, entry into or persistence within the urinary tract, or penetration into tissue or the bloodstream from a primary urinary tract source.
Resistance.
Widespread use (including much misuse) of antimicrobials has likely contributed to the emergence and spread of ST131. The strong association between H30 ST131 and FQ resistance suggests that use of FQs, the most commonly prescribed antibiotics in many parts of the world, likely has driven the expansion of H30 ST131. Likewise, use of extended-spectrum cephalosporins may have driven the expansion of H30-Rx, which is strongly associated with CTX-M-15. A few in vivo studies suggest that FQ-resistant E. coli stains may not be less fit than FQ-susceptible strains and that they are able to become effective intestinal colonizers during FQ selection pressure and even after FQ withdrawal (77, 78).
Still, why extensive antimicrobial use should select specifically for ST131 over other antimicrobial-resistant E. coli strains is not immediately apparent. Notably, compared to other resistant E. coli strains, although H30 ST131 strains have similarly (or even slightly less) extensive antimicrobial resistance profiles overall, they are more likely to be resistant specifically to first-line agents such as FQs and extended-spectrum cephalosporins. Additionally, compared with other FQ-resistant isolates, FQ-resistant H30 ST131 strains may have higher FQ MICs, thereby potentially having an advantage in the presence of FQ selection (Johnson, unpublished).
Epidemiological evidence supports the idea of an association between antibiotic exposure and ST131 infection and colonization. For example, in a population-based cohort study, after multivariable adjustment, antecedent use of FQs and extended-spectrum cephalosporins was significantly associated with development of an ST131 infection (20). Similarly, in several case-control studies, recent use of antibiotics was a risk factor for ESBL-producing E. coli infections (26, 31). The high prevalence of ST131 in environments with extensive antimicrobial use, such as health care settings and LTCFs, indirectly supports the idea that antimicrobial use facilitates ST131 propagation.
CONCLUSIONS
Recent molecular epidemiologic studies utilizing advanced technologies such as whole-genome phylogenetic analysis have demonstrated that the H30 ST131 lineage emerged in 2000, followed by rapid expansion of its subclones H30-R and H30-Rx, thereby dramatically changing the population structure of E. coli. Most current FQ-resistant E. coli strains originated from a common H30-R ST131 ancestor, which gave rise to what are now the most prevalent and among the most extensively antimicrobial-resistant human-associated E. coli lineages in the world. With its combination of multidrug resistance and ecological success, H30 ST131 counters the hypothesis that increased antimicrobial resistance entails a significant fitness cost.
The explosive expansion of ST131 has important clinical implications. This expansion has been greatest in health care settings and LTCFs, which are characterized by frequently suboptimal infection control practices, numerous direct interactions among patients/occupants, and extensive antimicrobial use. This emphasizes the necessity of implementing in these settings rigorous antimicrobial stewardship programs (especially those that focus on reducing use of FQs and extended-spectrum cephalosporins) and effective infection control interventions (including hand hygiene campaigns and environmental disinfection). Because LTCF residents move frequently between community and hospital settings and are reservoirs for ST131, as well as for other multidrug-resistant organisms, more research should focus on the role of LTCFs in facilitating spread of ST131 (79). Notably, however, there is currently no evidence to suggest that active surveillance and contact isolation for ST131 among hospitalized LTCF residents would prevent ST131 spread in hospitals or be cost-effective.
To prescribe more-effective empirical antimicrobial therapy, clinicians require increased awareness about ST131 and its high prevalence and extensive antimicrobial resistance capabilities and which patients are at risk for it. However, because reliance on clinical risk factors alone may fail to identify many patients with ST131 (80), rapid diagnostic tests that can quickly detect H30 and H30-Rx are also needed (22, 81) and may lead to more-timely, better-targeted antimicrobial therapy and improved clinical outcomes. Additionally, vaccines that target H30-associated antigens, and other H30-specific interventions directed toward the intestinal tract (or, in women, vaginal colonization), may reduce the reservoir of asymptomatic individuals colonized with ST131, in turn reducing the number of infected patients.
In summary, the current global epidemic of antimicrobial-resistant E. coli infections has an important clonal basis, being driven largely by the dissemination and expansion of ST131 and specifically by its nested H30-R and H30-Rx subclones. Like the proverbial new broom that sweeps clean, this cluster of newly emerged E. coli lineages has made a clean worldwide clonal sweep. The basis for the tremendous epidemiological success of these ST131 subclones is poorly understood but may include one or more of the following factors: increased transmission and/or colonization capacity, more-extensive or intense antimicrobial resistance, and enhanced virulence. Future research should address these hypotheses, through animal and human studies that assess overall transmissibility and prevalence and duration of intestinal colonization of ST131 compared to other E. coli strains and in vitro and animal studies that evaluate the molecular basis for differences in the transmission, colonization capacity, and virulence of ST131 subclones. Clinical studies are also needed to investigate the impact of specific infection control and antimicrobial stewardship interventions on ST131 prevalence (as both a commensal and a pathogen) in hospitals and LTCFs. It is only by better understanding ST131's secrets of success that we can hope to develop strategies to interrupt its continued emergence and spread.
ACKNOWLEDGMENTS
R.B. has no conflicts of interest to report. J.R.J. has received research grants or contracts from Merck, Rochester Medical, Syntiron, and Crucell and has patent applications for diagnostic tests for E. coli strains.
This work was supported by the National Institutes of Health 2KL2RR024151-07 (R.B.) and by the Office of Research and Development, Medical Research Service, Department of Veterans Affairs, grant no. 1 I01 CX000192 01 (J.R.J.).
ADDENDUM IN PROOF
A recently published review of ST131 phylogeny and epidemiology also highlights the remarkable global dissemination of the ST131 clonal group (M. H. Nicolas-Chanoine, X. Bertrand, and J. Y. Madec, Clin. Microbiol. Rev. 27:543-574, 2014, doi:10.1128/CMR.00125-13).
Footnotes
Published ahead of print 27 May 2014
REFERENCES
- 1.Coque TM, Baquero F, Canton R. 2008. Increasing prevalence of ESBL-producing enterobacteriaceae in Europe. Euro Surveill. 13:19044 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19044 [PubMed] [Google Scholar]
- 2.Nicolas-Chanoine M-H, Blanco J, Leflon-Guibout V, Demarty R, Alonso MP, Canica MM, Park Y-J, Lavigne J-P, Pitout J, Johnson JR. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61:273–281. 10.1093/jac/dkm464 [DOI] [PubMed] [Google Scholar]
- 3.Peirano G, Pitout JDD. 2010. Molecular epidemiology of Escherichia coli producing CTX-M-beta lactamases: the worldwide emergence of clone ST131 O25:H4. Int. J. Antimicrob. Agents 35:316–321. 10.1016/j.ijantimicag.2009.11.003 [DOI] [PubMed] [Google Scholar]
- 4.Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. 2010. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin. Infect. Dis. 51:286–294. 10.1086/653932 [DOI] [PubMed] [Google Scholar]
- 5.Rogers BA, Sidjabat HE, Paterson DL. 2011. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 66:1–14. 10.1093/jac/dkq415 [DOI] [PubMed] [Google Scholar]
- 6.McDonald LC, Killgore GE, Thompson A, Owens RC, Jr, Kazakova SV, Sambol SP, Johnson S, Gerding DN. 2005. An epidemic, toxin-gene-variant strain of Clostridium difficile. N. Engl. J. Med. 353:2433–2441. 10.1056/NEJMoa051590 [DOI] [PubMed] [Google Scholar]
- 7.Loo V, Poirier L, Miller MA, Oughton M, Libman MD, Michaud S, Bourgault A-M, Nguyen T, Frenette C, Kelly M, Vibien A, Brassard P, Fenn S, Dewar K, Hudson TJ, Horn R, Rene P, Monczak Y, Dascal A. 2005. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 353:2442–2449. 10.1056/NEJMoa051639 [DOI] [PubMed] [Google Scholar]
- 8.Manges AR, Johnson JR, Foxman B, O'Bryan TT, Fullerton KE, Riley LW. 2001. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N. Engl. J. Med. 345:1007–1013. 10.1056/NEJMoa011265 [DOI] [PubMed] [Google Scholar]
- 9.Johnson JR, Manges AR, O'Bryan TT, Riley LW. 2002. A disseminated multidrug-resistant clonal group of uropathogenic Escherichia coli in pyelonephritis. Lancet 359:2249–2251. 10.1016/S0140-6736(02)09264-4 [DOI] [PubMed] [Google Scholar]
- 10.Johnson JR, Menard ME, Lauderdale TL, Kosmidis C, Gordon D, Collignon P, Maslow JN, Andrasević AT, Kuskowski MA, Trans-Global Initiative for Antimicrobial Resistance Analysis Investigators 2011. Global distribution and epidemiologic associations of Escherichia coli clonal group A, 1998–2007. Emerg. Infect. Dis. 17:2001–2009. 10.3201/eid1711.110488 [DOI] [PubMed] [Google Scholar]
- 11.Coque TM, Novais A, Carattoli A, Poirel L, Pitout J, Peixe L, Baquero F, Canton R, Nordmann P. 2008. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg. Infect. Dis. 14:195–200. 10.3201/eid1402.070350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson JR, Clermont O, Johnston B, Clabots C, Tchesnokova V, Sokurenko E, Junka AF, Maczynska B, Denamur E. 2014. Rapid and specific detection, molecular epidemiology, and experimental virulence of the O16 subgroup within Escherichia coli sequence type 131. J. Clin. Microbiol. 52:1358–1365. 10.1128/JCM.03502-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.López-Cerero L, Navarro MD, Bellido M, Martín-Peña A, Viñas L, Cisneros JM, Gómez-Langley SL, Sánchez-Monteseirín H, Morales I, Pascual A, Rodríguez-Baño J. 2014. Escherichia coli belonging to the worldwide emerging epidemic clonal group O25b/ST131: risk factors and clinical implications. J. Antimicrob. Chemother. 69:809–814. 10.1093/jac/dkt405 [DOI] [PubMed] [Google Scholar]
- 14.Johnson JR, Urban C, Weissman SJ, Jorgensen JH, Lewis JS, II, Hansen G, Edelstein PH, Robicsek A, Cleary T, Adachi J, Paterson D, Quinn J, Hanson ND, Johnston BD, Clabots C, Kuskowski MA, AMERECUS Investigators 2012. Molecular epidemiological analysis of Escherichia coli sequence type ST131 (O25:H4) and blaCTX-M-15 among extended-spectrum-beta-lactamase-producing E. coli from the United States, 2000 to 2009. Antimicrob. Agents Chemother. 56:2364–2370. 10.1128/AAC.05824-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colpan A, Porter S, Johnston B, Clabots C, Anway R, Thao L, Kuskowski MA, Tchesnokova V, Sokurenko EV, Johnson JR. 2013. Escherichia coli sequence type 131 (ST131) as an emergent multidrug-resistant pathogen among U.S. veterans. Clin. Infect. Dis. 57:1256–1265. 10.1093/cid/cit503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olesen B, Hansen DS, Nilsson F, Frimodt-Møller J, Leihof RF, Struve C, Scheutz F, Johnston B, Krogfelt KA, Johnson JR. 2013. Prevalence and characteristics of the epidemic multiresistant Escherichia coli ST131 clonal group among extended-spectrum beta-lactamase-producing E. coli isolates in Copenhagen, Denmark. J. Clin. Microbiol. 51:1779–1785. 10.1128/JCM.00346-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banerjee R, Robicsek A, Kuskowski MA, Porter S, Johnston BD, Sokurenko E, Tchesnokova V, Price LB, Johnson JR. 2013. Molecular epidemiology of Escherichia coli sequence type 131 and its H30 and H30-Rx subclones among extended-spectrum-beta-lactamase-positive and -negative E. coli clinical isolates from the Chicago region, 2007–2010. Antimicrob. Agents Chemother. 57:6385–6388. 10.1128/AAC.01604-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson JR, Nicolas-Chanoine M-H, DebRoy C, Castanheira M, Robicsek A, Hansen G, Weissman S, Urban C, Platell J, Trott D, Zhanel G, Clabots C, Johnston BD, Kuskowski MA. 2012. Comparison of Escherichia coli ST131 pulsotypes, by epidemiologic traits, 1967–2009. Emerg. Infect. Dis. 18:598–607. 10.3201/eid1804.111627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.López-Cerero L, Bellido Mdel M, Serrano L, Liró J, Cisneros JM, Rodríguez-Baño J, Pascual A. 2013. Escherichia coli O25b:H4/ST131 are prevalent in Spain and are often not associated with ESBL or quinolone resistance. Enferm. Infecc. Microbiol. Clin. 31:385–388. 10.1016/j.eimc.2012.09.005 [DOI] [PubMed] [Google Scholar]
- 20.Banerjee R, Johnston B, Lohse C, Porter SB, Clabots C, Johnson JR. 2013. Escherichia coli sequence type ST131 is a dominant, antimicrobial-resistant clonal group associated with healthcare and elderly hosts. Infect. Control Hosp. Epidemiol. 34:361–369. 10.1086/669865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams-Sapper S, Diep BA, Perdreau-Remington F, Riley LW. 2013. Clonal composition and community clustering of drug-susceptible and -resistant Escherichia coli isolates from bloodstream infections. Antimicrob. Agents Chemother. 57:490–497. 10.1128/AAC.01025-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tchesnokova V, Billig M, Chattopadhyay S, Linardopoulou E, Aprikian P, Roberts PL, Skrivankova V, Johnston B, Gileva A, Igusheva I, Toland A, Riddell K, Rogers P, Qin X, Butler-Wu S, Cookson BT, Fang FC, Kahl B, Price LB, Weissman SJ, Limaye A, Scholes D, Johnson JR, Sokurenko EV. 2013. Predictive diagnostics of Escherichia coli infections based on the clonal association of antimicrobial resistance and clinical outcome. J. Clin. Microbiol. 51:2991–2999. 10.1128/JCM.00984-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williamson DA, Freeman JT, Porter S, Roberts S, Wiles S, Paterson DL, Johnson JR. 2013. Clinical and molecular correlates of virulence in Escherichia coli causing bloodstream infection following transrectal ultrasound-guided (TRUS) prostate biopsy. J. Antimicrob. Chemother. 68:2898–2906. 10.1093/jac/dkt276 [DOI] [PubMed] [Google Scholar]
- 24.Chung H-C, Lai C-H, Lin J-N, Huang C-K, Liang S-H, Chen W-F, Shih Y-C, Lin H-H, Wang J-L. 2012. Bacteremia caused by extended-spectrum beta-lactamase-producing Escherichia coli sequence type ST131 and non-ST131 clones: comparison of demographic data, clinical features, and mortality. Antimicrob. Agents Chemother. 56:618–622. 10.1128/AAC.05753-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doi Y, Park YS, Rivera JI, Adams-Haduch JM, Hingwe A, Sordillo EM, Lewis JS, Howard WJ, Johnson LE, Polsky B, Jorgensen JH, Richter SS, Shutt KA, Paterson DL. 2013. Community-associated extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli infection in the United States. Clin. Infect. Dis. 56:641–648. 10.1093/cid/cis942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banerjee R, Strahilevitz J, Johnson JR, Nagwekar PP, Schora DM, Shevrin I, Du H, Peterson LR, Robicsek A. 2013. Predictors and molecular epidemiology of community-onset extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli infection in a Midwestern community. Infect. Control Hosp. Epidemiol. 34:947–953. 10.1086/671725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cagnacci S, Gualco L, Debbia E, Schito GC, Marchese A. 2008. European emergence of ciprofloxacin-resistant Escherichia coli clonal groups O25:H4-ST131 and O15:K52:H1 causing community-acquired uncomplicated cystitis. J. Clin. Microbiol. 46:2605–2612. 10.1128/JCM.00640-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banerjee R, Johnston B, Lohse C, Chattopadhyay S, Tchesnokova V, Sokurenko EV, Johnson JR. 2013. The clonal distribution and diversity of extraintestinal Escherichia coli isolates vary according to patient characteristics. Antimicrob. Agents Chemother. 57:5912–5917. 10.1128/AAC.01065-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burke L, Humphreys H, Fitzgerald-Hughes D. 2012. The revolving door between hospital and community: extended-spectrum beta-lactamase-producing Escherichia coli in Dublin. J. Hosp. Infect. 81:192–198. 10.1016/j.jhin.2012.04.021 [DOI] [PubMed] [Google Scholar]
- 30.Rooney PJ, O'Leary MC, Loughrey AC, McCalmont M, Smyth B, Donaghy P, Badri M, Woodford N, Karisik E, Livermore DM. 2009. Nursing homes as a reservoir of extended-spectrum beta-lactamase (ESBL)-producing ciprofloxacin-resistant Escherichia coli. J. Antimicrob. Chemother. 64:635–641. 10.1093/jac/dkp220 [DOI] [PubMed] [Google Scholar]
- 31.Nicolas-Chanoine MH, Jarlier V, Robert J, Arlet G, Drieux L, Leflon-Guibout V, Laouénan C, Larroque B, Caro V, Mentré F, Group Coli β 2012. Patient's origin and lifestyle associated with CTX-M-producing Escherichia coli: a case-control-control study. PLoS One 7:e30498. 10.1371/journal.pone.0030498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Donk CF, Schols JM, Driessen CJ, Hagenouw RG, Meulendijks A, Stobberingh EE. 2013. Prevalence and spread of multidrug resistant Escherichia coli isolates among nursing home residents in the southern part of The Netherlands. J. Am. Med. Dir. Assoc. 14:199–203. 10.1016/j.jamda.2012.09.026 [DOI] [PubMed] [Google Scholar]
- 33.Nicolas-Chanoine M-H, Robert J, Vigan M, Laoenan C, Brisse S, Mentre F, Jarlier V. 2013. Different factors associated with CTX-M-producing ST131 and non-ST131 Escherichia coli clinical isolates. PLoS One 8:e72191. 10.1371/journal.pone.0072191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brisse S, Diancourt L, Laouenan C, Vigan M, Caro V, Arlet G, Drieux L, Leflon-Guibout V, Mentre F, Jarlier V, Nicolas-Chanoine MH. 2012. Phylogenetic distribution of CTX-M and non-extended-spectrum beta-lactamase-producing Escherichia coli isolates: group B2 isolates, except clone ST131, rarely produce CTX-M enzymes. J. Clin. Microbiol. 50:2974–2981. 10.1128/JCM.00919-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pitout JDD, Campbell L, Church DL, Gregson DB, Laupland KB. 2009. Molecular characteristics of travel-related extended-spectrum-beta-lactamase-producing Escherichia coli isolates from the Calgary Health Region. Antimicrob. Agents Chemother. 53:2539–2543. 10.1128/AAC.00061-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vasoo S, Madigan T, Cunningham SA, Mandrekar JN, Porter SB, Johnston B, Sampathkumar P, Tosh PK, Johnson JR, Patel R, Banerjee R. 2014. Prevalence of rectal colonization with multidrug-resistant Enterobacteriaceae among international patients hospitalized at Mayo Clinic, Rochester, Minnesota. Infect. Control Hosp. Epidemiol. 35:182–186. 10.1086/674853 [DOI] [PubMed] [Google Scholar]
- 37.Freeman JT, McBride SJ, Heffernan H, Bathgate T, Pope C, Ellis-Pegler RB. 2008. Community-onset genitourinary tract infection due to CTX-M-15-producing Escherichia coli among travelers to the Indian subcontinent in New Zealand. Clin. Infect. Dis. 47:689–692. 10.1086/590941 [DOI] [PubMed] [Google Scholar]
- 38.Vignaroli C, Luna GM, Pasquaroli S, Cesare Di A, Petruzzella R, Paroncini P, Biavasco F. 2013. Epidemic Escherichia coli ST131 and Enterococcus faecium ST17 in coastal marine sediments from an Italian beach. Environ. Sci. Technol. 47:13772–13780. 10.1021/es4019139 [DOI] [PubMed] [Google Scholar]
- 39.Colomer-Lluch M, Mora A, López C, Mamani R, Dahbi G, Marzoa J, Herrera A, Viso S, Blanco JE, Blanco M, Alonso MP, Jofre J, Muniesa M, Blanco J. 2013. Detection of quinolone-resistant Escherichia coli isolates belonging to clonal groups O25b:H4-B2-ST131 and O25b:H4-D-ST69 in raw sewage and river water in Barcelona, Spain. J. Antimicrob. Chemother. 68:758–765. 10.1093/jac/dks477 [DOI] [PubMed] [Google Scholar]
- 40.Vredenburg J, Varela AR, Hasan B, Bertilsson S, Olsen B, Narciso-da-Rocha C, Bonnedahl J, Stedt J, Da Costa PM, Manaia CM. 27 August 2013. Quinolone-resistant Escherichia coli isolated from birds of prey in Portugal are genetically distinct from those isolated from water environments and gulls in Portugal, Spain, and Sweden. Environ. Microbiol. 10.1111/1462-2920.12231 [DOI] [PubMed] [Google Scholar]
- 41.Zurfluh K, Hachler H, Nuesch-Inderbinen M, Stephan R. 2013. Characteristics of extended-spectrum beta-lactamase- and carbapenemase-producing Enterobacteriaceae isolates from rivers and lakes in Switzerland. Appl. Environ. Microbiol. 79:3021–3026. 10.1128/AEM.00054-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Platell JL, Johnson JR, Cobbold RN, Trott DJ. 2011. Multidrug-resistant extraintestinal pathogenic Escherichia coli of sequence type ST131 in animals and foods. Vet. Microbiol. 153:99–108. 10.1016/j.vetmic.2011.05.007 [DOI] [PubMed] [Google Scholar]
- 43.Manges AR, Johnson JR. 2012. Food-borne origins of Escherichia coli causing extraintestinal infections. Clin. Infect. Dis. 55:712–719. 10.1093/cid/cis502 [DOI] [PubMed] [Google Scholar]
- 44.Johnson JR, Tchesnokova V, Johnston B, Clabots C, Roberts PL, Billig M, Riddell K, Rogers P, Qin X, Butler-Wu S, Price LB, Aziz M, Nicolas-Chanoine MH, Debroy C, Robicsek A, Hansen G, Urban C, Platell J, Trott DJ, Zhanel G, Weissman SJ, Cookson BT, Fang FC, Limaye AP, Scholes D, Chattopadhyay S, Hooper DC, Sokurenko EV. 2013. Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J. Infect. Dis. 207:919–928. 10.1093/infdis/jis933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Croxall G, Hale J, Weston V, Manning G, Cheetham P, Achtman M, McNally A. 2011. Molecular epidemiology of extraintestinal pathogenic Escherichia coli isolates from a regional cohort of elderly patients highlights the prevalence of ST131 strains with increased antimicrobial resistance in both community and hospital care settings. J. Antimicrob. Chemother. 66:2501–2508. 10.1093/jac/dkr349 [DOI] [PubMed] [Google Scholar]
- 46.Liss MR, Peterson EM, Johnston B, Osann K, Johnson JR. 2013. Prevalence of ST131 among fluoroquinolone-resistant Escherichia coli obtained from rectal swabs before transrectal prostate biopsy. Urology 81:548–556. 10.1016/j.urology.2012.10.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weissman SJ, Johnson JR, Tchesnokova V, Billig M, Dykhuizen D, Riddell K, Rogers P, Qin X, Butler-Wu S, Cookson BT, Fang FC, Scholes D, Chattopadhyay S, Sokurenko E. 2012. High resolution two-locus clonal typing of extraintestinal pathogenic Escherichia coli. Appl. Environ. Microbiol. 78:1353–1360. 10.1128/AEM.06663-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Price LB, Johnson JR, Aziz M, Clabots C, Johnston B, Tchesnokova V, Nordstrom L, Pearson T, Keim P, Sokurenko EV. 2013. Epidemic clonal expansion of CTX-M-15-producing Escherichia coli ST131. mBio 4:e00377-13. 10.1128/mBio.00377-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petty NK, Zakour Ben NL, Stanton-Cook M, Skippington E, Totsika M, Forde BM, Phan MD, Moriel Gomes D, Peters KM, Davies M, Rogers BA, Dougan G, Rodriguez-Bano J, Pascual A, Pitout JD, Upton M, Paterson DL, Walsh TR, Schembri MA, Beatson SA. 2014. Global dissemination of a multidrug resistant Escherichia coli clone. Proc. Natl. Acad. Sci. U. S. A. 111:5694–5699. 10.1073/pnas.1322678111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsumura Y, Yamamoto M, Nagao M, Hotta G, Matsushima A, Ito Y, Takakura S, Ichiyama S. 2012. Emergence and spread of B2-ST131-O25b, B2-ST131-O16, and D-ST405 clonal groups among extended-spectrum beta-lactamase-producing Escherichia coli in Japan. J. Antimicrob. Chemother. 67:2612–2620. 10.1093/jac/dks278 [DOI] [PubMed] [Google Scholar]
- 51.Matsumura Y, Yamamoto M, Nagao M, Ito Y, Takakura S, Ichiyama S. 2013. Association of fluoroquinolone resistance, virulence genes, and IncF plasmids with extended-spectrum-beta-lactamase-producing Escherichia coli sequence type 131 (ST131) and ST405 clonal groups. Antimicrob. Agents Chemother. 57:4736–4742. 10.1128/AAC.00641-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mora A, Dahbi G, López C, Mamani R, Marzoa J, Dion S, Picard B, Blanco M, Alonso MP, Denamur E, Blanco J. 30 January 2014. Virulence patterns in a murine sepsis model of ST131 Escherichia coli clinical isolates belonging to serotypes O25b:H4 and O16:H5 are associated to specific virotypes. PLoS One 10.1371/journal.pone.0087025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson JR, Anderson JT, Clabots C, Johnston B, Cooperstock M. 2010. Within-household sharing of a fluoroquinolone-resistant Escherichia coli sequence type ST131 strain causing pediatric osteoarticular infection. Pediatr. Infect. Dis. J. 29:473–475. 10.1097/INF.0b013e3181c89bd7 [DOI] [PubMed] [Google Scholar]
- 54.Owens RC, Johnson JR, Stogsdill P, Yarmus L, Lolans K, Quinn J. 2011. Community transmission in the United States of a CTX-M-15-producing sequence type ST131 Escherichia coli strain resulting in death. J. Clin. Microbiol. 49:3406–3408. 10.1128/JCM.00993-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andersen PS, Stegger M, Aziz M, Contente-Cuomo T, Gibbons HS, Keim P, Sokurenko EV, Johnson JR, Price LB. 2013. Complete genome sequence of the epidemic and highly virulent CTX-M-15 producing H30-Rx subclone of Escherichia coli ST131. Genome Announc. 1:e00988-13. 10.1128/genomeA.00988-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ender PT, Gajanana D, Johnston B, Clabots C, Tamarkin FJ, Johnson JR. 2009. Transmission of an extended-spectrum beta-lactamase-producing Escherichia coli (sequence type ST131) strain between a father and daughter resulting in septic shock and emphysematous pyelonephritis. J. Clin. Microbiol. 47:3780–3782. 10.1128/JCM.01361-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnson JR, Miller S, Johnston B, Clabots C, DebRoy C. 2009. Sharing of Escherichia coli sequence type ST131 and other multidrug-resistant and urovirulent E. coli strains among dogs and cats within a household. J. Clin. Microbiol. 47:3721–3725. 10.1128/JCM.01581-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blanc V, Leflon-Guibout V, Blanco J, Haenni M, Madec J-Y, Rafignon G, Bruno P, Mora A, Lopez C, Dahbi G, Dunais B, Anastay M, Branger C, Moreau R, Pradier C, Nicolas-Chanoine M-H. 8 January 2014. Prevalence of day-care centre children (France) with faecal CTX-M-producing Escherichia coli comprising O25b:H4 and O16:H5 ST131 strains. J. Antimicrob. Chemother. 10.1093/jac/dkt519 [DOI] [PubMed] [Google Scholar]
- 59.Reves RR, Fong M, Pickering LK, Bartlett A, III, Alvarez M, Murray BE. 1990. Risk factors for fecal colonization with trimethoprim-resistant and multiresistant Escherichia coli among children in day-care centers in Houston, Texas. Antimicrob. Agents Chemother. 34:1429–1434. 10.1128/AAC.34.7.1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lietzau S, Raum E, von Baum H, Marre R, Brenner H. 2006. Clustering of antibiotic resistance of E. coli in couples: suggestion for a major role of conjugal transmission. BMC Infect. Dis. 6:119. 10.1186/1471-2334-6-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lietzau S, Raum E, von Baum H, Marre R, Brenner H. 2007. Household contacts were key factor for children's colonization with resistant Escherichia coli in community setting. J. Clin. Epidemiol. 60:1149–1155. 10.1016/j.jclinepi.2007.01.016 [DOI] [PubMed] [Google Scholar]
- 62.Hannah EL, Angulo FJ, Johnson JR, Haddadin B, Williamson J, Samore MH. 2005. Drug-resistant Escherichia coli, rural Idaho. Emerg. Infect. Dis. 11:1614–1617. 10.3201/eid1110.050140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamamoto S, Tsukamoto T, Terai A, Kurazono H, Takeda Y, Yoshida O. 1997. Genetic evidence supporting the fecal-perineal-urethral hypothesis in cystitis caused by Escherichia coli. J. Urol. 157:1127–1129. 10.1016/S0022-5347(01)65154-1 [DOI] [PubMed] [Google Scholar]
- 64.Vimont S, Boyd A, Bleibtreu A, Bens M, Goujon J-M, Garry L, Clermont O, Denamur E, Vandewalle A. 2012. The CTX-M-15-producing Escherichia coli clone O25b:H4-ST131 has high intestine colonization and urinary tract infection abilities. PLoS One 7:e46547. 10.1371/journal.pone.0046547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nicolas-Chanoine M-H, Gruson C, Bialek-Davenet S, Bertrand X, Thomas-Jean F, Berg F, Moyat M, Meiller E, Marcon E, Danchin N, Noussair L, Moreau R, Leflon-Guibout V. 2013. 10-Fold increase (2006–2011) in the rate of healthy subjects with extended-spectrum beta-lactamase-producing Escherichia coli faecal carriage in a Parisian check-up centre. J. Antimicrob. Chemother. 68:562–568. 10.1093/jac/dks429 [DOI] [PubMed] [Google Scholar]
- 66.Birgy A, Mariani-Kurkdjian P, Bidet P, Doit C, Genel N, Courroux C, Arlet G, Bingen E. 2013. Characterization of extended-spectrum-beta-lactamase-producing Escherichia coli strains involved in maternal-fetal colonization: prevalence of E. coli ST131. J. Clin. Microbiol. 51:1727–1732. 10.1128/JCM.03255-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kudinha T, Johnson JR, Andrew SC, Kong F, Anderson P, Gilbert GL. 2013. Escherichia coli sequence type 131 as a prominent cause of antibiotic resistance among urinary Escherichia coli isolates from reproductive-age women. J. Clin. Microbiol. 51:3270–3276. 10.1128/JCM.01315-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kudinha T, Johnson JR, Andrew SD, Kong F, Anderson P, Gilbert GL. 2013. Genotypic and phenotypic characterization of Escherichia coli isolates from children with urinary tract infection and from healthy carriers. Pediatr. Infect. Dis. J. 32:543–548. 10.1097/INF.0b013e31828ba3f1 [DOI] [PubMed] [Google Scholar]
- 69.Kudinha T, Johnson JR, Andrew SD, Kong F, Anderson P, Gilbert GL. 2013. Distribution of phylogenetic groups, sequence type ST131, and virulence-associated traits among Escherichia coli isolates from men with pyelonephritis or cystitis and healthy controls. Clin. Microbiol. Infect. 19:E173–E180. 10.1111/1469-0691.12123 [DOI] [PubMed] [Google Scholar]
- 70.Blanco J, Mora A, Mamani R, López C, Blanco M, Dahbi G, Herrera A, Marzoa J, Fernández V, de la Cruz F, Martínez-Martínez L, Alonso MP, Nicolas-Chanoine MH, Johnson JR, Johnston B, López-Cerero L, Pascual A, Rodríguez-Baño J, Spanish Group for Nosocomial Infections (GEIH) 2013. Four main virotypes among extended-spectrum-β-lactamase-producing isolates of Escherichia coli O25b:H4-B2-ST131: bacterial, epidemiological, and clinical characteristics. J. Clin. Microbiol. 51:3358–3357. 10.1128/JCM.01555-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alhashash F, Weston V, Diggle M, McNally A. 2013. Multidrug-resistant Escherichia coli bacteremia. Emerg. Infect. Dis. 19:1699–1701. 10.3201/eid1910.130309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lefort A, Panhard X, Clermont O, Woerther P-L, Branger C, Mentre F, Fantiin B, Wolff M, Denamur E. 2011. Host factors and portal of entry outweigh bacterial determinants to predict the severity of Escherichia coli bacteremia. J. Clin. Microbiol. 49:777–783. 10.1128/JCM.01902-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lavigne J-P, Bergunst AC, Goret L, Sotto A, Combescure C, Blanco J, O'Callaghan D, Nicholas-Chanoine M-H. 2012. Virulence potential and genomic mapping of the worldwide clone Escherichia coli ST131. PLoS One 7:e34294. 10.1371/journal.pone.0034294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Clermont O, Lavollay M, Vimont S, Deschamps C, Forestier C, Branger C, Denamur E, Arlet G. 2008. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J. Antimicrob. Chemother. 61:1024–1028. 10.1093/jac/dkn084 [DOI] [PubMed] [Google Scholar]
- 75.Johnson JR, Porter SB, Zhanel G, Kuskowski MA, Denamur E. 2012. Virulence of Escherichia coli clinical isolates in a murine sepsis model in relation to sequence type ST131 status, fluoroquinolone resistance, and virulence genotype. Infect. Immun. 80:1554–1562. 10.1128/IAI.06388-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chassin C, Vimont S, Cluzeaud F, Bens M, Goujon J-M, Fernandez B, Hertig A, Rondeau E, Arlet G, Hornet MW, Vandewalle A. 2008. TLR4 facilitates translocation of bacteria across renal collecting duct cells. J. Am. Soc. Nephrol. 19:2364–2374. 10.1681/ASN.2007121273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fantin B, Duval X, Massias L, Alavoine L, Chau F, Retout S, Andremont A, Mentre F. 2009. Ciprofloxacin dosage and emergence of resistance in human commensal bacteria. J. Infect. Dis. 200:390–398. 10.1086/600122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Andraud M, Rose N, Laurentie M, Sanders P, LeRoux A, Cariolet R, Chauvin C, Jouy E. 2011. Estimation of transmission parameters of a fluoroquinolone-resistant Escherichia coli strain between pigs in experimental conditions. Vet. Res. 42:44. 10.1186/1297-9716-42-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lautenbach E. 2013. Flying under the radar: the stealth pandemic of Escherichia coli sequence type 131. Clin. Infect. Dis. 57:1266–1269. 10.1093/cid/cit505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dumford D, Suwantarat N, Bhasker V, Kundrapu S, Zabarsky TF, Drawz P, Zhu H, Donskey CJ. 2013. Outbreak of fluoroquinolone-resistant Escherichia coli infections after transrectal ultrasound-guided biopsy of the prostate. Infect. Control Hosp. Epidemiol. 34:269–273. 10.1086/669512 [DOI] [PubMed] [Google Scholar]
- 81.Chattopadhyay S, Taub F, Paul S, Weissman SJ, Sokurenko EV. 2013. Microbial variome database: point mutations, adaptive or not, in bacterial core genomes. Mol. Biol. Evol. 30:1465–1470. 10.1093/molbev/mst048 [DOI] [PMC free article] [PubMed] [Google Scholar]


