Abstract
VX-222, a thiophene-2-carboxylic acid derivative, is a selective nonnucleoside inhibitor of the hepatitis C virus (HCV) NS5B RNA-dependent RNA polymerase. In phase 1 and 2 clinical studies, VX-222 demonstrated effective antiviral efficacy, with substantial reductions in plasma HCV RNA in patients chronically infected with genotype 1 HCV. To characterize the potential for selection of VX-222-resistant variants in HCV-infected patients, the HCV NS5B gene was sequenced at baseline and during and after 3 days of VX-222 dosing (monotherapy) in a phase 1 study. Variants with the substitutions L419C/I/M/P/S/V, R422K, M423I/T/V, I482L/N/T, A486S/T/V, and V494A were selected during VX-222 dosing, and their levels declined over time after the end of dosing. Phenotypic analysis of these variants was conducted using HCV replicons carrying site-directed mutations. Of the 17 variants, 14 showed reduced susceptibility to VX-222 compared with the wild type, with the L419C/S and R422K variants having higher levels of resistance (>200-fold) than the rest of the variants (6.8- to 76-fold). The M423I and A486S variants remained susceptible to VX-222. The 50% effective concentration (EC50) for the L419P variant could not be obtained due to the poor replication of this replicon. The majority of the variants (15/17) were less fit than the wild type. A subset of the variants, predominately the L419S and R422K variants, were observed when the efficacy and safety of VX-222- and telaprevir-based regimens given for 12 weeks were investigated in genotype 1 HCV-infected patients in a phase 2 study. The NS3 and NS5B variants selected during the dual combination therapy showed reduced susceptibility to both telaprevir and VX-222 and had a lower replication capacity than the wild type. The phase 1b study has the ClinicalTrials.gov identifier NCT00911963, and the phase 2a study has ClinicalTrials.gov identifier NCT01080222.
INTRODUCTION
Approximately 170 million people worldwide are chronically infected with hepatitis C virus (HCV), which may lead to severe liver diseases, including fibrosis, cirrhosis, and hepatocellular carcinoma (1, 2). Treatment with peginterferon and ribavirin has a low success rate in patients infected with genotype 1 HCV and is associated with substantial adverse events (3, 4). In the last decade, the development of new classes of HCV therapy, the direct-acting antivirals (DAA), has been a major focus of drug discovery efforts.
Multiple DAAs are currently being marketed or in development, including inhibitors of the HCV NS3 protease, NS5A protein, and NS5B RNA-dependent RNA polymerase (5, 6). Two protease inhibitors, boceprevir and telaprevir, were the first to receive regulatory approval for use in combination with peginterferon and ribavirin, which marked the beginning of a new era in HCV therapy for genotype 1 HCV-infected patients. In phase 3 clinical studies, both boceprevir- and telaprevir-based therapies significantly improved sustained virologic response (SVR) rates for treatment-naive and previously treated patients compared with peginterferon plus ribavirin alone (7). However, adverse events, including severe rash and anemia, may occur in some patients receiving boceprevir or telaprevir treatment (7). Additionally, drug-resistant viral populations have been shown to emerge in patients who do not achieve an SVR with boceprevir or telaprevir treatment (8). Newer DAAs, simeprevir (a protease inhibitor) and sofosbuvir (a nucleoside NS5B polymerase inhibitor), showed improved tolerability and efficacy and were recently approved for registration (9–11). Future therapies for HCV infection will ideally be regimens that consist of combinations of DAAs and do not include peginterferon or even ribavirin, and the development of novel investigational DAAs for combinations is of great interest.
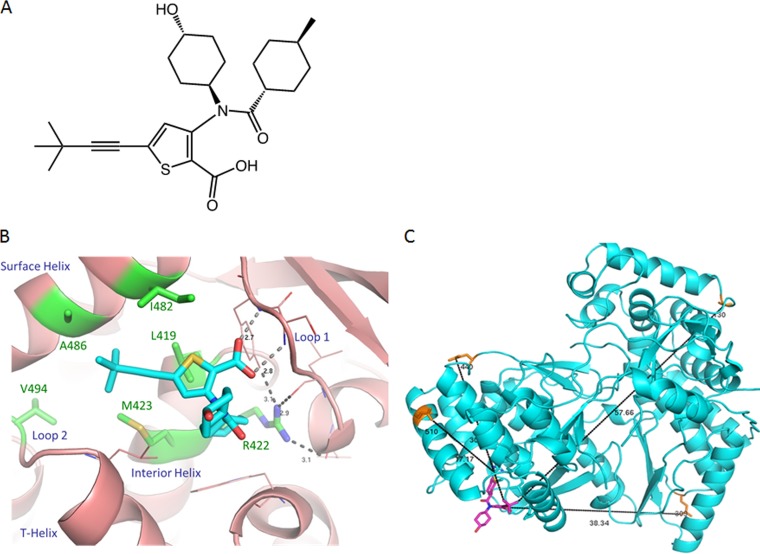
VX-222 (previously known as VCH-222), a thiophene-2-carboxylic acid derivative (Fig. 1A), is a selective nonnucleoside inhibitor (NNI) of the HCV NS5B polymerase that binds to an allosteric site located in the thumb domain (12–14). VX-222 is active against a purified NS5B polymerase with 50% inhibitory concentrations (IC50s) of 0.94 and 1.2 μM for genotypes 1a and 1b, respectively (15). It exhibits antiviral activity against genotypes 1a, 1b, and 2a in the HCV replicon, with 50% effective concentrations (EC50s) ranging from 4.6 to 22.3 nM (15). In a phase 1 clinical study, a reduction of more than 3 logs in HCV RNA was observed after 3 days of VX-222 monotherapy with a dose of 250, 500, or 750 mg twice daily or 1,500 mg once daily in treatment-naive patients infected with genotype 1 HCV (16). The combination of VX-222 and telaprevir in a phase 2 study resulted in a rapid initial decline in HCV RNA, with 23% of the patients having HCV RNA levels below the lower limit of quantitation (LLOQ) at week 2 and 43% achieving undetectable HCV RNA levels at week 4 of treatment. However, this regimen was discontinued due to high viral breakthrough rates prior to week 12 (17). The combination of VX-222 and telaprevir with ribavirin (a triple regimen) or with peginterferon and ribavirin (a quadruple regimen) led to significantly improved SVR rates of 67 to 90% (17).
FIG 1.
Structures of VX-222 and NS5B polymerase. (A) Chemical structure of VX-222 (full name: 5-(3,3-dimethylbut-1-ynyl)-3-[(4-hydroxycyclohexyl)-(4-methylcyclohexanecarbonyl)amino]thiophene-2-carboxylic acid). (B) Cocrystal structure of VX-222 and NS5B polymerase. The protein is in light magenta. The residues in Table 1 are labeled and highlighted in green; the side chains of these residues are shown by sticks. The key helices and loops are indicated. VX-222 is shown in stick representation, with carbon, nitrogen, oxygen, and sulfur atoms in cyan, blue, red, and yellow, respectively. The hydrogen bonds are shown by dashed lines, and the bond distances are indicated. (C) Position of exploratory variants in NS5B polymerase relative to VX-222. The protein is in cyan. The side chains of the residues with substitutions identified in the exploratory genotypic analysis are labeled and shown by orange sticks. VX-222 is shown in stick representation and in light purple. The distances between the positions of exploratory variants and VX-222 are indicated with black lines.
Resistance to thumb-binding NNIs has been previously reported. Substitutions at L419, M423, I482, and V494 in NS5B have been shown to affect the susceptibility to VCH-759, another thiophene-2-carboxylic acid derivative that also binds to the thumb domain of NS5B (18). In addition, NS5B substitutions at R422, M423, M426, I482, and V494 have been shown to be associated with resistance to a second class of thumb-binding NNIs (dihydropyrone class) in vitro or in patients (19–21). More recently, GS-9669, a thumb-binding NNI currently in clinical study in combination with sofosbuvir (22), has been shown to exhibit a decrease in potency against NS5B L419M, R422K, M423I/T/V, and I482L variants (23). VX-222 also selected for NS5B L419M/S, M423I/T/V, I482L, A486V, and V494A variants in replicon cells (our unpublished data). In this study, we report the sequence analysis of viral variants before, during, and after dosing with VX-222 for 3 days in a phase 1 study of treatment-naive patients infected with genotype 1 HCV (16). The phenotypic susceptibilities of these variants to VX-222 and other HCV inhibitors, as well as the replication capacity, are presented. Additionally, we report the phenotypic characteristics of the NS3 and NS5B variants observed in the 12-week dual combination study of VX-222 and telaprevir (17).
MATERIALS AND METHODS
Patient population and study design.
In the phase 1b study (ClinicalTrials.gov identifier NCT00911963), VX-222 was administered as a monotherapy for 3 days in treatment-naive patients who had chronic genotype 1 HCV infection. This study contained 4 treatment cohorts, where 32 patients were randomized to receive VX-222 at doses of 250, 500, and 750 mg twice daily or 1,500 mg once daily, or else placebo, for 3 consecutive days with a treatment: placebo ratio of 6:2 in each dose group (8 patients/cohort). At the end of 3 days, patients were offered peginterferon and ribavirin for 48 weeks.
The phase 2a study (ClinicalTrials.gov identifier NCT01080222) investigated the safety, tolerability, and antiviral activity of VX-222 at 100 mg or 400 mg twice daily, combined with various telaprevir-containing regimens in chronically genotype 1 HCV-infected patients. The treatment-naive patients received VX-222 and telaprevir only (dual regimen; n = 47) or with ribavirin (triple regimen; n = 46) or peginterferon and ribavirin (quadruple regimen; n = 59) for 12 weeks. Patients with detectable HCV RNA at weeks 2 and/or 8 received peginterferon and ribavirin for an additional 24 (dual and triple regimens) or 12 (quadruple regimen) weeks (17).
The studies were conducted in full compliance with the guidelines of Good Clinical Practice and of the World Medical Assembly Declaration of Helsinki. The study protocols and informed consent forms were approved by the institutional review boards at each site. All patients provided written informed consent before participating in any study-related activities.
Measurement of viral load.
Plasma HCV RNA levels in phase 1b and 2a studies were assessed using Roche COBAS AmpliPrep/COBAS TaqMan HCV real-time PCR (v2.1; LLOQ = 43 IU/ml; limit of detection [LOD] = 15 IU/ml) and Roche COBAS TaqMan HCV/HPS assay (v2.0; LLOQ = 25 IU/ml; LOD = 15 IU/ml), respectively.
Amplification and sequencing of HCV from plasma.
For VX-222 3-day monotherapy, amplification and population sequencing of the NS5B polymerase were performed on samples that had been collected at baseline (predose), during dosing (day 3 and day 4, which was the end of dosing), and at follow-up (days 5, 10, and 20, and long-term up to 6 months). For combination therapy with VX-222 and telaprevir, amplification and population sequencing of the NS3 protease and NS5B polymerase domains were performed at baseline, viral breakthrough, and follow-up time points (17).
A 3-ml blood sample was drawn, and approximately 1.5 ml of plasma was collected for viral sequencing. Viral RNA was extracted from plasma and amplified through the nested reverse transcription-PCR (RT-PCR) described previously (24). Purified PCR products were sequenced using Big-Dye terminator sequencing methods (Beckman-Coulter, Danvers, MA). The lower limit of detection (LLOD) of RT-PCR amplification is 1,000 IU/ml HCV RNA (24).
In a subset of patients (n = 7) in VX-222 3-day monotherapy, clonal sequence analysis was performed to determine the linkage of amino acid substitutions. RT-PCR products of plasma HCV RNA were cloned into pCR-XL-TOPO vector (Invitrogen, Carlsbad, CA) and transformed into Top-10 electrocompetent One Shot Escherichia coli cells (Invitrogen, Carlsbad, CA). Plasmid DNA was prepared and sequenced (Sanger sequencing at Beckman-Coulter, Danvers, MA) from 96 E. coli colonies per patient per time point.
Sequence alignment and analysis.
Sequence traces encompassing the first 1,665 nucleotides (555 amino acids) of the NS5B polymerase (for VX-222 3-day monotherapy and dual therapy with VX-222 and telaprevir) and 2,085 nucleotides (695 amino acids) of the NS3 protease (for dual therapy) were analyzed using the software Mutation Surveyor (SoftGenetics, State College, PA). Genotype-specific references for genotype 1a (H77; GenBank accession no. AF009606) and genotype 1b (Con1; GenBank accession no. AJ238799) were used to align traces for the baseline of each patient. Amino acid substitutions from postbaseline samples were detected by comparing sequences to the patient-specific baseline sequence.
Due to the small number of patients in VX-222 3-day monotherapy, the use of a statistical analysis to identify variants that were enriched from VX-222 treatment was not possible. Therefore, the frequency of NS5B substitutions at positions 419, 422, 423, 482, 486, and 494 that were previously identified as being associated with resistance to VX-222 (our unpublished in vitro selection data) or other thumb-binding NNIs (18–21, 23) was assessed. Additionally, an independent exploratory genotypic analysis was conducted to identify other variants that were observed in more than 1 patient.
Variants with substitutions in both NS3 and NS5B regions were assessed in the dual combination therapy of VX-222 and telaprevir as described previously (17).
Compounds.
The nonnucleoside NS5B polymerase inhibitors VX-222, filibuvir (20, 25), HCV-796 (26), and CMPD 55 (27), the NS3-4A protease inhibitor telaprevir (28, 29), and the NS5A inhibitor daclatasvir (30) were synthesized at Vertex Pharmaceuticals Incorporated (Boston, MA, USA). The nucleoside polymerase inhibitor mericitabine (31) was obtained from SAI Advantium (Hyderabad, India). All compounds were dissolved in dimethyl sulfoxide (DMSO) to a final concentration of 10 or 20 mM and stored at −20°C. Human recombinant interferon-alfa (IFN-α) (Calbiochem, La Jolla, CA) and ribavirin (Sigma-Aldrich, St. Louis, MO) were reconstituted in water and stored at −80°C and −20°C, respectively.
Construction of variant replicons.
Mutations were introduced into the genotype 1b subgenomic replicon plasmid pFK I341PiLuc/NS3–3′/ET, which was kindly provided by R. Bartenschlager (adapted from the work of Lohmann et al. [32]) or the genotype 1a subgenomic replicon plasmid 1a-PiLuci-1a6* (33), using PCR-based site-directed mutagenesis and standard recombinant DNA technologies. The integrity of all variant constructs was confirmed by DNA sequencing.
EC50 determination for variant replicons.
The EC50s of variant replicons were determined in Huh-7-ET-cured cells that were established by curing Huh-7-ET cells containing the subgenomic replicon pFK I389luc-ubi-neo/NS3–3′/5.1 (34) as described previously (33). Briefly, full-length HCV subgenomic replicon RNA was generated from linearized DNA templates using the Megascript T7 kit (Life Technologies, Grand Island, NY). The resulting RNA was transfected into 4 × 106 Huh-7-ET-cured cells resuspended in Ingenio (Mirus Bio LLC, Madison, WI) using a Bio-Rad Gene Pulser II electroporator. Transfected cells were resuspended in cell culture medium and plated in 96-well, tissue-culture-treated plates at 1 × 104 cells in 100 μl. After incubation at 37°C for 24 h, the cultured cells were mixed with 100 μl of medium containing serially diluted compounds and allowed to grow for 3 days. The cells were lysed with lysis buffer (Promega, Madison, WI), and luciferase activity was measured using a luciferase assay system (Promega, Madison, WI). The EC50s were calculated from dose-response curves using a 4-parameter curve-fitting method in the Softmax Pro program (Molecular Devices Corp., Sunnyvale, CA). Multiple independent assays were conducted for each viral replicon, and the means and standard deviations (SD) of the replicon EC50s were calculated. The change (fold) in sensitivity to HCV inhibitors was calculated by dividing the mean EC50 for each variant by the mean EC50 for the wild-type replicon. Variants were considered to have reduced susceptibility to the inhibitors if the increase in the EC50 compared with that of the wild type was >3-fold, because the variation of the EC50s in the assay was ≤3-fold.
Replication capacity of variant replicons.
The replication capacity of variant replicons was determined as previously described (33). Briefly, replicon RNA was in vitro transcribed and transfected into Huh-7-ET-cured cells. Transfected cells were plated into duplicated 96-well plates and cultured at 37°C for 2.5 h for the first set of plates and 96 h for the second set of plates. The cells were lysed and kept frozen at −80°C until assayed for luciferase activity. For any given variant replicon, a normalized luciferase signal was calculated by dividing the luciferase signal at 96 h postelectroporation with that of the same variant replicon at 2.5 h postelectroporation. The relative replication capacity of the variant is expressed as the percentage of the normalized luciferase signal of the variant replicon compared with that of the wild-type replicon (as 100%) and that of a HCV polymerase-defective replicon containing AAG or GND for genotype 1a or 1b, respectively, at the GDD active site (as 0%).
RESULTS
Genotypic analysis of HCV NS5B polymerase was performed at baseline and during and after dosing for all patients receiving VX-222 3-day monotherapy in the phase 1b study. Considering the small number of patients in the study and the limited time points during the critical time window through day 4 of VX-222 dosing, population sequencing was conducted for all samples collected, including those with HCV RNA below the LLOD of RT-PCR amplification. NS5B variants with substitutions at positions 419, 422, 423, 482, 486, or 494 that were previously identified as being associated with resistance to VX-222 (our unpublished in vitro selection data) or other thumb-binding NNIs (18–21, 23) were assessed. Variants with substitutions identified at these positions are shown in Table 1. The profile of resistance against VX-222 and other HCV inhibitors as well as the replication capacity of these variants was evaluated in transient replicon systems (Tables 1 and 2). Variants identified from individual patients on VX-222 treatment are summarized in Table 3. Clonal sequence analysis was performed for a subset of patients to determine the genomic linkage of multiple variants observed at different resistance positions by population sequence analysis (Table 4). To identify other potential NS5B variants that may be associated with resistance to VX-222, besides those with substitutions at L419, R422, M423, I482, A486, or V494, an additional independent exploratory genotypic analysis for substitutions occurring in more than 1 patient was performed. Four additional variants (T130A, Q309R, E440G, and K510R variants) were identified from this analysis.
TABLE 1.
Phenotypic characterization of NS5B variants observed in VX-222 3-day monotherapya
| Variant | VX-222 |
Filibuvir |
Replication capacity (%) | ||
|---|---|---|---|---|---|
| EC50 (μM) | FC | EC50 (μM) | FC | ||
| WT | 0.0037 ± 0.0008 | 1.0 ± 0.2 | 0.029 ± 0.009 | 1.0 ± 0.3 | 100 ± 0 |
| L419C | 0.87 ± 0.03 | 234 ± 9 | 0.016 ± 0.003 | 0.6 ± 0.1 | 42 ± 3 |
| L419I | 0.048 ± 0.023 | 13 ± 6 | 0.020 ± 0.002 | 0.7 ± 0.1 | 65 ± 9 |
| L419M | 0.19 ± 0.03 | 50 ± 7 | 0.045 ± 0.018 | 1.6 ± 0.6 | 98 ± 7 |
| L419S | 2.5 ± 0.4 | 683 ± 102 | 0.045 ± 0.001 | 1.6 ± 0.03 | 5 ± 1 |
| L419V | 0.13 ± 0.02 | 35 ± 4 | 0.022 ± 0.007 | 0.8 ± 0.2 | 64 ± 5 |
| R422K | 1.8 ± 0.5 | 472 ± 131 | 6.9 ± 2.2 | 239 ± 76 | 47 ± 14 |
| M423I | 0.0071 ± 0.0018 | 1.9 ± 0.5 | 12 ± 2 | 425 ± 81 | 68 ± 14 |
| M423T | 0.045 ± 0.011 | 12 ± 3 | >25 | >867 | 56 ± 7 |
| M423V | 0.025 ± 0.006 | 6.8 ± 1.5 | >25 | >867 | 22 ± 6 |
| I482L | 0.27 ± 0.04 | 72 ± 12 | 0.12 ± 0.03 | 4.2 ± 0.9 | 106 ± 25 |
| I482N | 0.28 ± 0.05 | 76 ± 13 | 1.1 ± 0.2 | 40 ± 7 | 44 ± 4 |
| I482T | 0.028 ± 0.010 | 7.4 ± 2.7 | 0.19 ± 0.02 | 6.7 ± 0.6 | 38 ± 1 |
| A486S | 0.0029 ± 0.0001 | 0.8 ± 0.02 | 0.012 ± 0.003 | 0.4 ± 0.1 | 74 ± 12 |
| A486T | 0.090 ± 0.022 | 24 ± 6 | 0.051 ± 0.006 | 1.8 ± 0.2 | 77 ± 3 |
| A486V | 0.25 ± 0.09 | 67 ± 24 | 0.073 ± 0.013 | 2.5 ± 0.4 | 77 ± 4 |
| V494A | 0.029 ± 0.010 | 7.9 ± 2.7 | 0.15 ± 0.02 | 5.1 ± 0.8 | 68 ± 9 |
Data were obtained from the genotype 1b replicon. All values are means ± SD. The EC50s were derived from 12 to 14 and 3 to 6 individual experiments for the wild-type (WT) and variant replicons, respectively. The mean fold change (FC) values were determined by dividing the mean EC50 of a given variant replicon by that of the WT replicon. The SD of the fold change were determined by dividing the SD of the EC50 of a given variant replicon by the mean EC50 of the WT replicon. The mean replication capacity and SD was derived from 3 individual experiments. The “>” symbol denotes that the value is greater than the value presented; the actual value could not be determined, since no significant reduction of HCV RNA level was observed at the maximum concentration of VX-222 or filibuvir used (25 μM). Data for the L419P variant were not available due to the replicon's inability to replicate.
TABLE 2.
Cross-resistance profile of VX-222-resistant variants to IFN-α, ribavirin, and other HCV inhibitorsa
| Variant | IFN-α |
Ribavirin |
Telaprevir |
Daclatasvir |
HCV-796 |
CMPD 55 |
Mericitabine |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EC50 (U/ml) | FC | EC50 (μM) | FC | EC50 (μM) | FC | EC50 (pM) | FC | EC50 (μM) | FC | EC50 (μM) | FC | EC50 (μM) | FC | |
| WT | 1.4 | 1.0 | 24 | 1.0 | 0.27 | 1.0 | 4.5 | 1.0 | 0.0072 | 1.0 | 0.086 | 1.0 | 0.87 | 1.0 |
| L419M | 1.2 | 0.9 | 25 | 1.0 | 0.15 | 0.6 | 4.2 | 0.9 | 0.0086 | 1.2 | 0.089 | 1.0 | 0.44 | 0.5 |
| R422K | 1.2 | 0.8 | 22 | 0.9 | 0.15 | 0.5 | 3.6 | 0.8 | 0.0049 | 0.7 | 0.068 | 0.8 | 0.45 | 0.5 |
| M423T | 1.5 | 1.1 | 29 | 1.2 | 0.25 | 0.9 | 3.8 | 0.8 | 0.0043 | 0.6 | 0.060 | 0.7 | 0.89 | 1.0 |
| I482L | 1.5 | 1.0 | 23 | 0.9 | 0.22 | 0.8 | 6.0 | 1.3 | 0.0072 | 1.0 | 0.19 | 2.3 | 0.44 | 0.5 |
| A486V | 1.2 | 0.8 | 27 | 1.1 | 0.36 | 1.3 | 3.1 | 0.7 | 0.0031 | 0.4 | 0.10 | 1.2 | 0.75 | 0.9 |
| V494A | 1.3 | 0.9 | 24 | 1.0 | 0.16 | 0.6 | 3.9 | 0.9 | 0.0028 | 0.4 | 0.29 | 3.3 | 0.58 | 0.7 |
The EC50s are the means of the results of 3 individual experiments for the wild-type (WT) and variant replicons. The fold change (FC) was determined by dividing the mean EC50 of a given variant replicon by that of the WT replicon.
TABLE 3.
Population sequence analysis at baseline and during and after VX-222 dosing in VX-222 3-day monotherapy
| Treatment groupa | Patientb | Genotype | HCV RNA reduction at day 4 (log10 IU/ml) | VX-222-resistant variantc |
||||
|---|---|---|---|---|---|---|---|---|
| At baseline | During treatment (by day 4) | At follow-upd |
||||||
| Day 5 | Day 10 | Day 20 | ||||||
| 250 mg bid | 1 | 1b | −2.8 | A486Se | R422R/K, A486S | M423M/T, A486S, T130A | A486S | A486S |
| 2 | 1a | −3.4 | − | L419L/S, A486A/V | A486A/V | − | − | |
| 3 | 1a | −3.7f | − | R422R/K | M423M/T, A486A/V | M423M/Ig | − | |
| 4 | 1a | −2.0 | − | L419M | R422R/K, A486A/V | − | NA | |
| 5 | 1a | −2.9 | − | R422R/K, M423M/V | − | − | − | |
| 6 | 1a | −3.9 | − | L419C | − | − | − | |
| 500 mg bid | 7 | 1b | −3.2 | − | I482I/N, A486A/V, T130A, E440G | I482I/T, A486A/T | − | − |
| 8 | 1a | −3.2 | − | A486A/V, V494V/A, Q309R | Q309R | Q309R | − | |
| 9 | 1a | −3.7 | − | R422R/K, I482L | A486A/V | − | ND | |
| 10 | 1b | −3.6 | − | A486V | NA | K510R | NA | |
| 11 | 1a | −3.2 | − | NA | R422R/K | − | NA | |
| 12 | 1a | −3.6 | − | R422R/K, A486A/V | − | − | − | |
| 750 mg bid | 13 | 1a | −3.8 | − | L419L/P, R422R/K | L419L/P, R422R/K | − | − |
| 14 | 1a | −3.5 | − | L419L/S, R422R/K | L419L/V, A486A/V | − | NA | |
| 15 | 1a | −3.5 | − | L419L/S, R422R/K | L419L/S, A486A/V | M423M/T, K510R | NA | |
| 16 | 1b | −3.3 | − | R422R/K, A486A/T | M423M/T, V494V/A | A486A/V | A486A/V | |
| 17 | 1b | −2.6 | − | L419L/I, M423M/V, V494V/A | − | − | − | |
| 18 | 1a | −2.3 | − | R422R/K, M423M/T, I482I/L, A486A/V | I482I/N, A486A/V | − | − | |
| 1,500 mg qd | 19 | 1a | −3.5 | − | NA | NA | NA | NA |
| 20 | 1b | −3.1 | − | NA | R422R/K, I482I/L | L419M | Q309R | |
| 21 | 1a | −3.9 | − | R422K | NA | − | NA | |
| 22 | 1a | −3.2 | − | R422R/K, A486A/V | A486A/V | − | − | |
| 23 | 1b | −3.2 | − | NA | NA | NA | NA | |
| 24 | 1b | −3.6 | − | R422K, M423T | R422R/K, A486A/V | − | E440G | |
bid, twice daily; qd, once daily.
Data for patients on VX-222 treatment (n = 24) are presented. Patients on placebo (n = 8) are not included; no VX-222 resistance-associated variants were observed in these patients throughout the study.
All variants with the substitutions at the VX-222 resistance-associated positions 419, 422, 423, 482, 486, or 494 and variants observed in exploratory analysis (in italics) are shown. −, no substitutions at these positions were observed, and no variants were observed in exploratory analysis. NA, not available because the HCV RNA level was below the LLOD of RT-PCR amplification (1,000 IU/ml); ND, not determined due to the lack of sample.
Data for follow-up beyond day 20 are not shown.
The A486S variant, which is not a resistant variant, was present in a significant proportion of the viral population at baseline and throughout the duration of the study in patient 1.
The day 3 value is shown.
The M423I variant is not a resistant variant.
TABLE 4.
Clonal sequence analysis for potential linkage of VX-222-resistant variants observed in VX-222 3-day monotherapy
| Patient | Time point | No. of clones | Most prevalent substitution |
2nd most prevalent substitution |
Combination of substitutions |
|||
|---|---|---|---|---|---|---|---|---|
| Substitution | % of clones | Substitution | % of clones | Combination | % of clones | |||
| 2 | Day 4 | 75 | L419S | 30.7 | A486V | 21.3 | L419S+A486V | 1.3 |
| 3 | Day 5 | 58 | A486V | 27.6 | M423T | 24.1 | None | NAa |
| 4 | Day 5 | 75 | R422K | 53.3 | A486V | 46.7 | R422K+A486V | 1.3 |
| 5 | Day 4 | 76 | M423V | 79.0 | R422K | 15.8 | None | NA |
| 14 | Day 3 | 78 | R422K | 41.0 | L419S | 35.9 | None | NA |
| 15 | Day 3 | 79 | L419S | 38.0 | R422K | 38.0 | None | NA |
| 18 | Day 3 | 65 | A486V | 49.2 | M423T | 32.3 | None | NA |
NA, not available.
Variants with substitutions in both the NS3 and NS5B regions were observed in patients receiving the combination of VX-222 and telaprevir in a phase 2a study (17). The susceptibility to telaprevir and VX-222 as well as the replication capacity of these variants was evaluated in transient replicon systems (Table 5).
TABLE 5.
Resistance profile of NS3 and NS5B variants observed in the dual combination therapy of VX-222 and telaprevira
| Replicon | Variant |
EC50 (μM) |
Fold change |
Replication capacity (%) | |||
|---|---|---|---|---|---|---|---|
| NS3 | NS5B | Telaprevir | VX-222 | Telaprevir | VX-222 | ||
| Genotype 1b | WT | WT | 0.261 ± 0.078 | 0.0035 ± 0.0009 | 1.0 ± 0.3 | 1.0 ± 0.2 | 100 ± 0 |
| V36A | R422K | 1.73 ± 0.28 | 2.8 ± 0.7 | 6.7 ± 1.1 | 822 ± 210 | 18 ± 7 | |
| V36A | — | 1.12 ± 0.54 | 0.0020 ± 0.0009 | 4.3 ± 2.1 | 0.6 ± 0.2 | 20 ± 6 | |
| — | R422K | 0.147 ± 0.031 | 1.8 ± 0.5 | 0.6 ± 0.1 | 506 ± 143 | 47 ± 14 | |
| V36A+R155K | L419S | 10.5 ± 2.7 | 4.1 ± 0.5 | 40 ± 10 | 1,179 ± 133 | 1 ± 0.2 | |
| V36A+R155K | — | 6.61 ± 2.48 | 0.0026 ± 0.0005 | 25 ± 9 | 0.7 ± 0.1 | 14 ± 2 | |
| — | L419S | 0.164 ± 0.030 | 2.5 ± 0.3 | 0.6 ± 0.1 | 737 ± 89 | 4 ± 1 | |
| A156T | R422K | >25 | 2.8 ± 0.6 | >96 | 810 ± 166 | 6 ± 1 | |
| A156T | — | >25 | 0.0018 ± 0.0005 | >96 | 0.5 ± 0.2 | 6 ± 1 | |
| — | R422K | 0.147 ± 0.031 | 1.8 ± 0.5 | 0.6 ± 0.1 | 506 ± 143 | 47 ± 14 | |
| Genotype 1a | WT | WT | 0.805 ± 0.210 | 0.0028 ± 0.0013 | 1.0 ± 0.3 | 1.0 ± 0.5 | 100 ± 0 |
| V36M+R155K | R422K | >25 | 0.93 ± 0.34 | >31 | 329 ± 121 | 0.6 ± 0.1 | |
| V36M+R155K | — | >25 | 0.0042 ± 0.0010 | >31 | 1.5 ± 0.3 | 20 ± 2 | |
| — | R422K | 1.15 ± 0.19 | 0.94 ± 0.10 | 1.4 ± 0.2 | 335 ± 35 | 1 ± 0.6 | |
| V36L+A156S | R422K | >25 | 0.97 ± 0.26 | >31 | 345 ± 94 | 0.6 ± 0.2 | |
| V36L+A156S | — | >25 | 0.0039 ± 0.0016 | >31 | 1.4 ± 0.6 | 11 ± 1 | |
| — | R422K | 1.15 ± 0.19 | 0.94 ± 0.10 | 1.4 ± 0.2 | 335 ± 35 | 1 ± 0.6 | |
All values are means ± SD. The EC50 was derived from 10 to 25 and 3 to 4 individual experiments for the wild-type (WT) and variant replicons, respectively. The fold change was determined by dividing the mean EC50 of a given variant replicon by that of the WT replicon. The SD of the fold change was determined by dividing the SD of the EC50 of a given variant replicon by the mean EC50 of the WT replicon. The mean replication capacity and SD were derived from 3 to 6 individual experiments. The “>” symbol denotes that the value is greater than the value presented; the actual value could not be determined, since no significant reduction of HCV RNA level was observed at the maximum concentration of telaprevir used (25 μM). —, WT sequence.
Phenotypic susceptibility of VX-222-resistant variants observed in VX-222 3-day monotherapy.
The EC50s of the NS5B variants that were observed in VX-222 3-day monotherapy (L419C/I/M/P/S/V, R422K, M423I/T/V, I482L/N/T, A486S/T/V, and V494A variants) were determined in genotype 1b replicons (Table 1). Of these 17 variants, 14 showed reduced susceptibility to VX-222 compared with the wild type in HCV replicon cells, with the L419C/S and R422K variants showing higher levels of resistance (>200-fold) than the rest of the variants (6.8- to 76-fold). The M423I and A486S variants remained susceptible to VX-222, with changes in EC50 of 1.9- and 0.8-fold compared with wild-type replicon, respectively. The EC50 for the L419P variant could not be obtained due to the poor replication of the replicon.
The susceptibility to VX-222 was also determined for the T130A variant, one of the 4 NS5B variants that were identified in the exploratory genotypic analysis; this variant was fully sensitive to VX-222, with a change in EC50 of 0.5-fold compared with the wild-type replicon (data not shown in Table 1).
Susceptibility of VX-222-resistant variants to filibuvir and other classes of HCV inhibitors.
Filibuvir (PF-00868554) is a dihydropyranone derivative nonnucleoside polymerase inhibitor that, like VX-222, also binds to the thumb domain of NS5B (20, 25). Most of the NS5B variants identified in patients receiving VX-222 showed a different profile of resistance to filibuvir in the replicon system (Table 1). The L419C/I/M/S/V and A486T/V variants showed reduced susceptibility to VX-222 but remained sensitive to filibuvir. The I482L variant showed a lower level of resistance to filibuvir than to VX-222, whereas the M423I/T/V variants showed much higher levels of resistance to filibuvir than to VX-222. The R422K, I482N/T, and V494A variants showed similar levels of resistance to both filibuvir and VX-222.
One variant for each of the VX-222 resistance-associated positions 419, 422, 423, 482, 486, and 494 was evaluated for cross-resistance to IFN-α, ribavirin, and other classes of HCV inhibitors. The L419M, R422K, M423T, I482L, and A486V variants that exhibited reduced susceptibility to VX-222 remained sensitive to IFN-α, ribavirin, an NS3 protease inhibitor (telaprevir [28, 29]), an NS5A inhibitor (daclatasvir [30]), NS5B polymerase NNIs binding to the palm domain (HCV-796 [26]) or to the finger loop domain (CMPD 55 [27]), and an NS5B polymerase nucleoside inhibitor (mericitabine [31]) (Table 2). The V494A variant showed a 3.3-fold increase in EC50 against CMPD 55 but remained susceptible to IFN-α, ribavirin, and other DAAs (Table 2).
In vitro replication capacity of VX-222-resistant variants.
Of the 17 NS5B variants identified in VX-222 3-day monotherapy, 15 had a lower replication capacity than that of the wild type (Table 1). A reduced replication capacity ranging from 5 to 77% of that of the wild type (100%) was observed for 14 variants. The L419C/S, R422K, M423V, and I482N/T variants showed the greatest reduction in replication capacity (<50%) compared with the wild type. The replication capacity of the L419P variant was extremely low; therefore, no accurate data could be obtained. The L419M and I482L variants had replication capacities of 98% and 106%, respectively, similar to that of wild-type replicon.
Genotypic analysis of baseline samples in VX-222 3-day monotherapy.
No amino acid substitutions were observed at L419, R422, M423, I482, A486, and V494 for any patients (n = 32) at baseline from population sequence analysis of the NS5B polymerase (GenBank accession numbers KC123434 to KC127656) (Table 3). This result is consistent with the high conservation of the consensus amino acid at these positions (>98.7% of patients) and the low prevalence of the variants (<1% of patients) in a large DAA-naïve sequence data set of genotype 1 HCV (35). A variant with the A486S substitution, which had not been previously identified as a drug-resistant variant, was observed in patient 1 (Table 3) at baseline and throughout the duration of the study. This variant was susceptible to VX-222 in the replicon system (0.8-fold change in EC50) (Table 1). Additionally, the presence of this variant prior to treatment did not significantly alter the response of the patient to VX-222, with a 2.8-log10 HCV RNA decline being achieved by the end of dosing on day 4 (Table 3).
Genotypic analysis during dosing in VX-222 3-day monotherapy.
Population sequence of the NS5B region was obtained for 20 of 24 patients during (day 3) and at the end of dosing (day 4); the remaining 4 patients (patients 11, 19, 20, and 23) had HCV RNA levels below the LLOD of RT-PCR amplification, and the sequence could not be obtained (Table 3). Variants with substitutions at one or more of positions 419, 422, 423, 482, 486, and 494 were detected in all 20 patients for whom sequence data were available, with substitutions at each of these 6 positions being observed: L419, 35% (7/20); R422, 65% (13/20); M423, 20% (4/20); I482, 15% (3/20); A486, 40% (8/20; A486S excluded); and V494, 10% (2/20). The most commonly observed variants were R422K (65%), A486V (35%), and L419S (15%). All other variants (L419C/I/M/P, M423T/V, I482L/N, A486T, and V494A variants) were seen in 10% of patients or less. A similar resistance pattern was observed for the 4 treatment cohorts with different doses of VX-222: the R422K variant, a higher-level resistant variant (472-fold increase in EC50), and the A486V variant, a variant with a relatively high level of resistance (67-fold increase in EC50), were observed in all treatment cohorts, while the other variants with a range of sensitivity to VX-222 (6.8- to 683-fold increase in EC50) were observed sporadically. In addition, no obvious difference in variant pattern was observed between genotypes 1a and 1b.
Some patients had multiple variants observed as a minor species at different resistance positions. For instance, minority populations of L419S and A486V variants were observed for patient 2 at day 4, and minority populations of M423T and A486V variants were observed for patient 3 at day 5 (Table 3). To determine if these multiple variants were linked on a single viral genome, clonal sequence analysis was performed on a representative subset of 7 patients (Table 4). In 2 patients (patients 2 and 4), 1 of 75 clones analyzed indicated linkage of these variants, suggesting that a viral species with double substitutions may exist but was observed in only a small proportion (1%) of the viral populations. No linkage between the variants was observed in the remaining 5 patients.
Genotypic analysis after dosing in VX-222 3-day monotherapy.
NS5B variants with substitutions at positions 419, 422, 423, 482, 486, and 494 were analyzed at days 5, 10, and 20 and 6 months, after the end of VX-222 dosing (Table 3). Six of 24 patients had HCV RNA below the LLOD of RT-PCR amplification during treatment (by day 4) or at day 5, and the change of viral populations could not be assessed. There were 18 patients that had sequence data available during treatment and at day 5 postdosing. Of these, only 1 patient (patient 13) had the same viral population (L419L/P and R422R/K) at day 5 that was present during treatment. Seventeen patients had different viral populations after treatment at day 5 compared to the end of treatment. The majority of the 17 patients had variants replaced by those with lower levels of resistance (n = 10) or had no variants with substitutions at L419, R422, M423, I482, A486, or V494 (n = 5). By day 10, 22 of 24 patients had sequence data available, and among these 22, the wild-type NS5B sequence was observed for 19, including 1 patient (patient 3) with the M423I variant, which was shown to be susceptible to VX-222 in vitro (Table 1). Three of the 22 patients (patients 15, 16, and 20) had one of the L419M, M423T, or A486V variants. Of these 3 patients, 2 had sequence data available at day 20: patient 20 had the wild-type NS5B sequence, and for patient 16, the A486V variant was still the predominant variant. The A486V variant observed in patient 16 was the only variant that was detected from the 15 patients for whom sequence data were available at day 20; a wild-type sequence was observed for the remaining 14 patients. No variants were observed beyond day 20, and all variants returned to wild type by the last follow-up time point (data not shown in Table 3).
Exploratory genotypic analysis of NS5B variants observed in VX-222 3-day monotherapy.
To identify other potential NS5B variants that may be associated with resistance to VX-222, an additional independent analysis for substitutions occurring in more than 1 patient in the 3-day monotherapy was performed. Four additional variants, the T130A, Q309R, E440G, and K510R variants, were identified from this analysis. Each of these variants was observed in 2 patients by day 10 or later, and none of the variants were observed in multiple patients during the dosing period where the highest selective pressure for potential VX-222-resistant variants occurs. The natural prevalence of these variants was examined in a large data set of DAA treatment-naive HCV sequences (35). The T130A variant had the lowest prevalence and was observed in only <1% of the population. Similarly, the E440G variant was also not commonly observed (2%). On the other hand, the Q309R and K510R variants were more predominant, being observed in 25% and 40% of the population, respectively.
Phenotypic characteristics of NS3 and NS5B variants observed in combination therapy with VX-222 and telaprevir.
In the study of the combination of telaprevir and VX-222, variants resistant to both telaprevir and VX-222 were identified by population sequence analysis. These variants had the V36A, R155I/T, A156T/V, V36A/M+R155K, or V36L+A156S substitutions in the NS3 protease domain, which were associated with resistance to telaprevir, and also had the L419S or R422K substitution in the NS5B polymerase domain, which was associated with resistance to VX-222 (17). The majority of the mutations in NS3 and NS5B regions were predominant in the population sequence analysis, indicating that these mutations are linked and present in a single viral genome. This was further confirmed in clonal sequence analysis of a subset of the samples (data not shown).
To phenotypically characterize the NS3 and NS5B variants, double-mutant replicons were generated (Table 5). A genotype 1b replicon was constructed for variant NS3–V36A+NS5B-R422K, since this variant was observed in a patient infected with genotype 1b HCV. Although all other NS3 and NS5B variants were observed in patients infected with genotype 1a HCV, only 2 genotype 1a variant replicons (NS3–V36M+R155K+NS5B-R422K and NS3–V36L+A156S+NS5B-R422K) could be obtained with replicative capability. For the rest of the variants, only genotype 1b replicons could be generated (NS3–V36A+R155K+NS5B-L419S and NS3–A156T+NS5B-R422K), or no replicons with measurable replicative capability could be obtained (variants containing R155I/T or A156V).
The NS3 and NS5B variants showed reduced susceptibility to both telaprevir and VX-222 in HCV replicons (Table 5). The resistance to telaprevir conferred by these NS3 and NS5B variants was comparable to that of the single NS3 variants, of which the V36A variant had lower-level resistance, with a 4.3-fold increase in EC50 compared with that of the wild-type replicon, and the A156T, V36A/M+R155K, and V36L+A156S variants had higher-level resistance, with EC50s increased 25-fold or higher. Similarly, the reduced susceptibility to VX-222 conferred by the NS3 and NS5B variants was comparable to that of the NS5B variants with the single substitutions L419S and R422K, which conferred 335- to 737-fold resistance in genotype 1a or 1b replicons.
The NS3 and NS5B variants had a lower replication capacity than the wild type but were able to replicate at a level comparable to that of the single variant with the lower replication capacity (Table 5). For instance, the variant NS3–V36A+NS5B-R422K had a replication capacity of 18%, which was closer to the 20% for the V36A variant than the 47% for the R422K variant. Similarly, the replication capacity of the NS3–V36A+R155K+NS5B-L419S variant (1%) was comparable to that of the L419S variant (4%) rather than that of the V36A+R155K variant, which had a higher replication capacity (14%). Therefore, no apparent replication benefits or disadvantages were observed for variants with resistance-associated substitutions in both regions. In general, variants replicated less efficiently in genotype 1a than genotype 1b replicons. NS5B variant R422K had replication capacities of 1% and 47% in genotype 1a and genotype 1b replicons, respectively. Similarly, the NS3 variant V36M+R155K had a lower replication capacity in the genotype 1a replicon (20%) than that previously obtained in the genotype 1b replicon (42%) (33). Furthermore, the replication capacity of the NS3 V36A, A156T, and V36A+R155K variants obtained in genotype 1b (6% to 20%) was lower than that previously determined in a different genotype 1b replicon (16% to 104%) (33).
DISCUSSION
An understanding of drug resistance is important in optimizing DAA treatment regimens to increase success rate and minimize the clinical impact of resistance. The HCV genome exhibits significant genetic heterogeneity, with high sequence diversity both between and within the various genotypes and subtypes (36, 37). The low fidelity of the HCV polymerase, high viral replication rate, and strong selective pressure on the virus result in a unique and diverse viral quasispecies in each patient (38). New HCV populations with every potential substitution are likely generated many times each day, some of which convey various degrees of resistance to DAAs (37, 39, 40). Thus, it is likely that all patients have some DAA-resistant variants prior to treatment. Along with the availability of replication space, the prevalence of a resistant variant in a patient's viral quasispecies is generally determined by its replicative fitness and selective advantage compared with the rest of the viral population in the presence of drug-selective pressure (39). Minor populations of preexisting resistant variants are usually present at levels below the detection limits of current sequencing techniques, as they are less fit than wild-type virus but have a fitness advantage over wild-type virus in the presence of a drug and become the dominant viral species (39, 40).
Substitutions at L419, R422, M423, I482, A486, and V494 in NS5B polymerase were previously identified as being associated with resistance to thumb-binding NNIs. Our cocrystal structure analysis showed that VX-222 binds to NS5B polymerase at the predominantly hydrophobic pocket in the thumb domain that has been described as a common binding site for thiophene-based NNIs (13). Among the residues that line the binding pocket (L419, R422, M423, L474, H475, S476, Y477, I482, A486, L489, L497, R501, and W528 [13]), L419, R422, M423, I482, and A486 are in direct contact with VX-222 (Fig. 1B), and substitutions of these residues can affect the hydrophobic or hydrogen bond interactions between NS5B polymerase and VX-222 to various extents, resulting in decrease in binding affinity. V494 is part of loop 2, which connects the T helix with the surface helix; it has no direct contact with VX-222 (Fig. 1B). However, substitutions of V494 can influence the orientation of the T helix and impact the inhibitor binding in an indirect manner. In addition, the carboxylate group of VX-222 makes hydrogen bonds to the main chain of S476 (2.8 Å) and Y477 (2.7 Å) of loop 1 on the flexible side of the pocket (Fig. 1B).
In a phase 1b study of VX-222, no amino acid substitutions were detected at positions 419, 422, 423, 482, 486, and 494 at baseline. However, a variety of substitutions were observed in patients during dosing: L419C/I/M/P/S, R422K, M423T/V, I482L/N, A486T/V, and V494A, with R422K, A486V, and L419S being the most commonly observed variants. Both the R422K and L419S variants showed higher levels of resistance (472- to 683-fold increase in EC50), and the A486V variant showed a relatively high level of resistance (67-fold increase in EC50, but with a higher replication capacity than the R422K and L419S variants) to VX-222 in vitro. Thus, these variants were most likely to be selected during VX-222 dosing. In addition, the R422K and A486V variants were observed in all treatment cohorts, while the L419S variant and other variants were observed sporadically in some treatment cohorts, indicating a similar resistance profile for all dose cohorts and a lack of correlation between the level of resistance and the dose of VX-222. The majority of variants observed required a single nucleotide change from the consensus codon, with minimal differences between genotypes 1a and 1b; therefore, no obvious difference in variant selection pattern was observed between these two subtypes. The postdosing time points allowed evaluation of the evolution and fitness of the viral population containing VX-222-resistant variants in patients. At day 5 postdosing, VX-222-resistant variants were replaced by wild-type virus (n = 5) or variants with lower levels of resistance (n = 10) in most of the patients (83% [15/18]). By day 10 and day 20 after the end of dosing, the majority of patients (86% [19/22] for day 10 and 93% [14/15] for day 20) had wild-type NS5B sequences. All patients (10/10) showed wild-type virus at the last follow-up at 6 months. Clearly, the levels of VX-222-resistant variants that were selected during dosing declined over time after treatment. The replication capacity of the variants was evaluated with site-directed mutants, which may have limited relevance for the fitness for some of the viruses harboring the same mutations in the native context. However, most of the VX-222-resistant variants showed reduced replication capacity compared with the wild type, providing a mechanism by which variants are replaced by wild-type virus over time in the absence of drug-selective pressure in patients.
VX-222-resistant variants were susceptible to IFN-α, ribavirin, an NS3 protease inhibitor (telaprevir), an NS5A inhibitor (daclatasvir), NS5B polymerase NNIs that bind to the palm domain (HCV-796) or the finger loop domain (CMPD 55), and an NS5B polymerase nucleoside inhibitor (mericitabine), demonstrating that VX-222 has the potential for combination treatment consisting of DAAs with different mechanisms of actions. However, it has been reported that residue V494 is involved in the interaction of NS5B polymerase with finger loop-binding inhibitors, including CMPD 55 (41). In this study, the V494A variant showed a 3.3-fold increase in EC50 against CMPD 55, slightly higher than the cutoff set for reduced susceptibility (3-fold). In addition, most of the VX-222-resistant variants showed a different profile of resistance to filibuvir, a dihydropyranone derivative nonnucleoside polymerase inhibitor that also binds to the thumb domain. Substitutions at M423 were reported as the predominant amino acid changes for filibuvir, arising in 46% of filibuvir-treated patients in monotherapy studies (42). Previously published results showed that these variants conferred 706 to >2,202-fold resistance to filibuvir in replicon cells (20), which is consistent with the data reported here. In a different study, it was shown that M423T was at least 100-fold more resistant to filibuvir than to VX-222, and this resistance was the result of a 250-fold loss in the binding affinity of the mutated enzyme to filibuvir (43).
In addition to the analysis of NS5B variants with the substitutions at the positions that were previously identified as being associated with resistance to VX-222 and other thumb-binding NNIs, 4 novel variants (T130A, Q309R, E440G, and K510R variants) were observed in VX-222 3-day monotherapy. Each of these 4 variants was observed in only 2 patients, and none was observed in multiple patients during the dosing period. Thus, it is unlikely that these variants confer resistance to VX-222, considering that the highest selective pressure for potential VX-222-resistant variants occurs during the dosing period. Additionally, the Q309R and K510R variants are commonly observed at these positions, and therefore, the observation of these variants after the end of dosing is likely unrelated to VX-222. Although the T130A variant had a low prevalence before treatment (<1%), it did not confer a decrease in susceptibility to VX-222 in the replicon system. Furthermore, the structural analysis shows that all 4 variants are located >17 Å from the VX-222 binding region (Fig. 1C) and are not likely to have direct interactions with VX-222. Substitutions of these residues are not predicted to have any impact on VX-222 binding. While the data do not support a role for any of the exploratory variants in VX-222 resistance, larger data sets in future clinical studies will allow a better understanding of the clinical relevance of these variants.
Variant populations resistant to both telaprevir and VX-222 emerged under the selection of these two drugs in the dual-combination study. The resistance level to telaprevir or VX-222 conferred by NS3 and NS5B variants was comparable to that conferred by the single NS3 or NS5B variants, indicating that the linkage of the substitutions in NS3 and NS5B did not increase the resistance of the single variants to either telaprevir or VX-222. The NS3 and NS5B variants had a lower replication capacity than the wild type but were able to replicate at a level comparable to that of the single variant with the lower replication capacity. Variants replicated less efficiently in genotype 1a than genotype 1b replicons, probably due to the impact of the genetic background of the replicons. Likely for the same reason, the replication capacity of some of the variants obtained from assays with different genotype 1b replicons also showed a difference.
In summary, NS5B variants with a wide range of decreased sensitivity to VX-222 were selected during 3 days of dosing, but their levels declined over time after treatment due to the lower fitness compared with the wild type. NS3 and NS5B variants selected as a result of the combination of VX-222 and telaprevir showed reduced susceptibility to both drugs and had lower replication capacity than the wild type.
ACKNOWLEDGMENTS
This study was sponsored by Vertex Pharmaceuticals Incorporated.
M. Jiang, E. Zhang, A. Ardzinski, J. Sullivan, O. Nicolas, D. Bartels, R. Rijnbrand, and T. Kieffer are employees of Vertex Pharmaceuticals Incorporated and may own stock or options in that company. A. Tigges, A. Davis, M. Nelson, J. Spanks, J. Dorrian, and B. G. Rao were employed at Vertex Pharmaceuticals Incorporated at the time this research was conducted and may own or have owned stock or options in that company.
Footnotes
Published ahead of print 30 June 2014
REFERENCES
- 1.WHO. 2013. Hepatitis C: WHO fact sheet no. 164. World Health Organization, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs164/en/index.html [Google Scholar]
- 2.Pol S, Vallet-Pichard A, Corouge M, Mallet VO. 2012. Hepatitis C: epidemiology, diagnosis, natural history and therapy. Contrib. Nephrol. 176:1–9. 10.1159/000332374 [DOI] [PubMed] [Google Scholar]
- 3.Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Goncales FL, Jr, Haussinger D, Diago M, Carosi G, Dhumeaux D, Craxi A, Lin A, Hoffman J, Yu J. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975–982. 10.1056/NEJMoa020047 [DOI] [PubMed] [Google Scholar]
- 4.Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Koury K, Ling M, Albrecht JK. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958–965. 10.1016/S0140-6736(01)06102-5 [DOI] [PubMed] [Google Scholar]
- 5.Schaefer EA, Chung RT. 2012. Anti-hepatitis C virus drugs in development. Gastroenterology 142:1340–1350 e1341. 10.1053/j.gastro.2012.02.015 [DOI] [PubMed] [Google Scholar]
- 6.Casey LC, Lee WM. 2013. Hepatitis C virus therapy update 2013. Curr. Opin. Gastrenterol. 29:243–249. 10.1097/MOG.0b013e32835ff972 [DOI] [PubMed] [Google Scholar]
- 7.Pawlotsky JM. 2011. The results of phase III clinical trials with telaprevir and boceprevir presented at the Liver Meeting 2010: a new standard of care for hepatitis C virus genotype 1 infection, but with issues still pending. Gastroenterology 140:746–754. 10.1053/j.gastro.2011.01.028 [DOI] [PubMed] [Google Scholar]
- 8.Sarrazin C, Zeuzem S. 2010. Resistance to direct antiviral agents in patients with hepatitis C virus infection. Gastroenterology 138:447–462. 10.1053/j.gastro.2009.11.055 [DOI] [PubMed] [Google Scholar]
- 9.The Medical Letter on Drugs and Therapeutics. 2014. Simeprevir (Olysio) for chronic hepatitis C. Med. Lett. Drugs Ther. 56:1–3 [PubMed] [Google Scholar]
- 10.Keating GM, Vaidya A. 2014. Sofosbuvir: first global approval. Drugs 74:273–282. 10.1007/s40265-014-0179-7 [DOI] [PubMed] [Google Scholar]
- 11.Dabbouseh NM, Jensen DM. 2013. Future therapies for chronic hepatitis C. Nat. Rev. Gastroenterol. Hepatol. 10:268–276. 10.1038/nrgastro.2013.17 [DOI] [PubMed] [Google Scholar]
- 12.Biswal BK, Cherney MM, Wang M, Chan L, Yannopoulos CG, Bilimoria D, Nicolas O, Bedard J, James MN. 2005. Crystal structures of the RNA-dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors. J. Biol. Chem. 280:18202–18210. 10.1074/jbc.M413410200 [DOI] [PubMed] [Google Scholar]
- 13.Biswal BK, Wang M, Cherney MM, Chan L, Yannopoulos CG, Bilimoria D, Bedard J, James MN. 2006. Non-nucleoside inhibitors binding to hepatitis C virus NS5B polymerase reveal a novel mechanism of inhibition. J. Mol. Biol. 361:33–45. 10.1016/j.jmb.2006.05.074 [DOI] [PubMed] [Google Scholar]
- 14.Wang M, Ng KK, Cherney MM, Chan L, Yannopoulos CG, Bedard J, Morin N, Nguyen-Ba N, Alaoui-Ismaili MH, Bethell RC, James MN. 2003. Non-nucleoside analogue inhibitors bind to an allosteric site on HCV NS5B polymerase. Crystal structures and mechanism of inhibition. J. Biol. Chem. 278:9489–9495. 10.1074/jbc.M209397200 [DOI] [PubMed] [Google Scholar]
- 15.Bedard J, Nicolas O, Bilimoria D, L'Heureux L, Fex P, David M, Chan L. 2009. Identification and characterization of VCH-222, a novel potent and selective non-nucleoside HCV polymerase inhibitor. J. Hepatology 50(Suppl. 1):S340. 10.1016/S0168-8278(09)60937-5 [DOI] [Google Scholar]
- 16.Rodriguez-Torres M, Lawitz E, Conway B, Kaita K, Sheikh AM, Ghalib R, Adrover R, Cooper C, Silva M, Rosario M, Bourgault B, Proulx L, McHutchison JG. 2010. Safety and antiviral activity of the HCV non-nucleoside polymerase inhibitor VX-222 in treatment-naive genotype 1 HCV-infected patients. J. Hepatology 52(Suppl. 1):S14. 10.1016/S0168-8278(10)60033-5 [DOI] [Google Scholar]
- 17.Di Bisceglie AM, Sulkowski M, Gane E, Jacobson IM, Nelson D, DeSouza C, Alves K, George S, Kieffer T, Zhang EZ, Kauffman R, Asmal M, Koziel MJ. 2014. VX-222, a non-nucleoside NS5B polymerase inhibitor, in telaprevir-based regimens for genotype 1 hepatitis C virus infection. Eur. J. Gastroenterol. Hepatol. 26:761–773. 10.1097/MEG.0000000000000084 [DOI] [PubMed] [Google Scholar]
- 18.Cooper C, Lawitz EJ, Ghali P, Rodriguez-Torres M, Anderson FH, Lee SS, Bedard J, Chauret N, Thibert R, Boivin I, Nicolas O, Proulx L. 2009. Evaluation of VCH-759 monotherapy in hepatitis C infection. J. Hepatol. 51:39–46. 10.1016/j.jhep.2009.03.015 [DOI] [PubMed] [Google Scholar]
- 19.Shi ST, Herlihy KJ, Graham JP, Fuhrman SA, Doan C, Parge H, Hickey M, Gao J, Yu X, Chau F, Gonzalez J, Li H, Lewis C, Patick AK, Duggal R. 2008. In vitro resistance study of AG-021541, a novel nonnucleoside inhibitor of the hepatitis C virus RNA-dependent RNA polymerase. Antimicrob. Agents Chemother. 52:675–683. 10.1128/AAC.00834-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi ST, Herlihy KJ, Graham JP, Nonomiya J, Rahavendran SV, Skor H, Irvine R, Binford S, Tatlock J, Li H, Gonzalez J, Linton A, Patick AK, Lewis C. 2009. Preclinical characterization of PF-00868554, a potent nonnucleoside inhibitor of the hepatitis C virus RNA-dependent RNA polymerase. Antimicrob. Agents Chemother. 53:2544–2552. 10.1128/AAC.01599-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Troke PJ, Lewis M, Simpson P, Gore K, Hammond J, Craig C, Westby M. 2012. Characterization of resistance to the nonnucleoside NS5B inhibitor filibuvir in hepatitis C virus-infected patients. Antimicrob. Agents Chemother. 56:1331–1341. 10.1128/AAC.05611-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gane EJ, Stedman CA, Hyland RH, Ding X, Svarovskaia E, Subramanian GM, Symonds WT, McHutchison JG, Pang PS. 2014. Efficacy of nucleotide polymerase inhibitor sofosbuvir plus the NS5A inhibitor ledipasvir or the NS5B non-nucleoside inhibitor GS-9669 against HCV genotype 1 infection. Gastroenterology 146:736–743. 10.1053/j.gastro.2013.11.007 [DOI] [PubMed] [Google Scholar]
- 23.Fenaux M, Eng Leavitt SSA, Lee YJ, Mabery EM, Tian Y, Byun D, Canales E, Clarke MO, Doerffler E, Lazerwith SE, Lew W, Liu Q, Mertzman M, Morganelli P, Xu L, Ye H, Zhang J, Matles M, Murray BP, Mwangi J, Zhang J, Hashash A, Krawczyk SH, Bidgood AM, Appleby TC, Watkins WJ. 2013. Preclinical characterization of GS-9669, a thumb site II inhibitor of the hepatitis C virus NS5B polymerase. Antimicrob. Agents Chemother. 57:804–810. 10.1128/AAC.02052-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang EZ, Bartels DJ, Frantz JD, Seepersaud S, Lippke JA, Shames B, Zhou Y, Lin C, Kwong A, Kieffer TL. 2013. Development of a sensitive RT-PCR method for amplifying and sequencing near full-length HCV genotype 1 RNA from patient samples. Virol. J. 10:53. 10.1186/1743-422X-10-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Tatlock J, Linton A, Gonzalez J, Jewell T, Patel L, Ludlum S, Drowns M, Rahavendran SV, Skor H, Hunter R, Shi ST, Herlihy KJ, Parge H, Hickey M, Yu X, Chau F, Nonomiya J, Lewis C. 2009. Discovery of (R)-6-cyclopentyl-6-(2-(2,6-diethylpyridin-4-yl)ethyl)-3-((5,7-dimethyl-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)methyl)-4-hydroxy-5,6-dihydropyran-2-one (PF-00868554) as a potent and orally available hepatitis C virus polymerase inhibitor. J. Med. Chem. 52:1255–1258. 10.1021/jm8014537 [DOI] [PubMed] [Google Scholar]
- 26.Kneteman NM, Howe AY, Gao T, Lewis J, Pevear D, Lund G, Douglas D, Mercer DF, Tyrrell DL, Immermann F, Chaudhary I, Speth J, Villano SA, O'Connell J, Collett M. 2009. HCV796: A selective nonstructural protein 5B polymerase inhibitor with potent anti-hepatitis C virus activity in vitro, in mice with chimeric human livers, and in humans infected with hepatitis C virus. Hepatology 49:745–752. 10.1002/hep.22717 [DOI] [PubMed] [Google Scholar]
- 27.Harper S, Avolio S, Pacini B, Di Filippo M, Altamura S, Tomei L, Paonessa G, Di Marco S, Carfi A, Giuliano C, Padron J, Bonelli F, Migliaccio G, De Francesco R, Laufer R, Rowley M, Narjes F. 2005. Potent inhibitors of subgenomic hepatitis C virus RNA replication through optimization of indole-N-acetamide allosteric inhibitors of the viral NS5B polymerase. J. Med. Chem. 48:4547–4557. 10.1021/jm050056+ [DOI] [PubMed] [Google Scholar]
- 28.Lin K, Perni RB, Kwong AD, Lin C. 2006. VX-950, a novel hepatitis C virus (HCV) NS3-4A protease inhibitor, exhibits potent antiviral activities in HCV replicon cells. Antimicrob. Agents Chemother. 50:1813–1822. 10.1128/AAC.50.5.1813-1822.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin C, Lin K, Luong YP, Rao BG, Wei YY, Brennan DL, Fulghum JR, Hsiao HM, Ma S, Maxwell JP, Cottrell KM, Perni RB, Gates CA, Kwong AD. 2004. In vitro resistance studies of hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061: structural analysis indicates different resistance mechanisms. J. Biol. Chem. 279:17508–17514. 10.1074/jbc.M313020200 [DOI] [PubMed] [Google Scholar]
- 30.Gao M, Nettles RE, Belema M, Snyder LB, Nguyen VN, Fridell RA, Serrano-Wu MH, Langley DR, Sun JH, O'Boyle DR, II, Lemm JA, Wang C, Knipe JO, Chien C, Colonno RJ, Grasela DM, Meanwell NA, Hamann LG. 2010. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature 465:96–100. 10.1038/nature08960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ali S, Leveque V, Le Pogam S, Ma H, Philipp F, Inocencio N, Smith M, Alker A, Kang H, Najera I, Klumpp K, Symons J, Cammack N, Jiang WR. 2008. Selected replicon variants with low-level in vitro resistance to the hepatitis C virus NS5B polymerase inhibitor PSI-6130 lack cross-resistance with R1479. Antimicrob. Agents Chemother. 52:4356–4369. 10.1128/AAC.00444-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113. 10.1126/science.285.5424.110 [DOI] [PubMed] [Google Scholar]
- 33.Jiang M, Mani N, Lin C, Ardzinski A, Nelson M, Reagan D, Bartels D, Zhou Y, Nicolas O, Rao BG, Muh U, Hanzelka B, Tigges A, Rijnbrand R, Kieffer TL. 2013. In vitro phenotypic characterization of hepatitis C virus NS3 protease variants observed in clinical studies of telaprevir. Antimicrob. Agents Chemother. 57:6236–6245. 10.1128/AAC.01578-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vrolijk JM, Kaul A, Hansen BE, Lohmann V, Haagmans BL, Schalm SW, Bartenschlager R. 2003. A replicon-based bioassay for the measurement of interferons in patients with chronic hepatitis C. J. Virol. Methods 110:201–209. 10.1016/S0166-0934(03)00134-4 [DOI] [PubMed] [Google Scholar]
- 35.Bartels DJ, Sullivan JC, Zhang EZ, Tigges AM, Dorrian JL, De Meyer S, Takemoto D, Dondero E, Kwong AD, Picchio G, Kieffer TL. 2013. Hepatitis C virus variants with decreased sensitivity to direct-acting antivirals (DAAs) were rarely observed in DAA-naive patients prior to treatment. J. Virol. 87:1544–1553. 10.1128/JVI.02294-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ray SC TD. 2009. Hepatitis C, p 2157–2187 In Mandell GL, Bennett JE, Dolan R. (ed), Mandell. Douglas, and Bennett's principles and practice of infectious diseases. Churchill Livingstone-Elsevier, Philadelphia, PA [Google Scholar]
- 37.Zeuzem S. 1999. Clinical implications of hepatitis C viral kinetics. J. Hepatol. 31(Suppl 1):S61–S64 [DOI] [PubMed] [Google Scholar]
- 38.Bukh J, Miller RH, Purcell RH. 1995. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin. Liver Dis. 15:41–63. 10.1055/s-2007-1007262 [DOI] [PubMed] [Google Scholar]
- 39.Adiwijaya BS, Herrmann E, Hare B, Kieffer T, Lin C, Kwong AD, Garg V, Randle JC, Sarrazin C, Zeuzem S, Caron PR. 2010. A multi-variant, viral dynamic model of genotype 1 HCV to assess the in vivo evolution of protease-inhibitor resistant variants. PLoS Comput. Biol. 6:e1000745. 10.1371/journal.pcbi.1000745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rong L, Dahari H, Ribeiro RM, Perelson AS. 2010. Rapid emergence of protease inhibitor resistance in hepatitis C virus. Sci. Transl. Med. 2:30ra32. 10.1126/scitranslmed.3000544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Shi ST. 2010. Non-nucleoside inhibitors of hepatitis C virus polymerase: current progress and future challenges. Future Med. Chem. 2:121–141. 10.4155/fmc.09.148 [DOI] [PubMed] [Google Scholar]
- 42.Troke P, Lewis M, Simpson P, van der Ryst E, Hammond J, Graig C, Perros M, Westby M. 2009. Genotypic characterization of HCV NS5B following 8-day monotherapy with the polymerase inhibitor PF-00868554 in HCV-infected subjects. J. Hepatol. 50(Suppl 1):S351. 10.1016/S0168-8278(09)60970-3 [DOI] [Google Scholar]
- 43.Yi G, Deval J, Fan B, Cai H, Soulard C, Ranjith-Kumar CT, Smith DB, Blatt L, Beigelman L, Kao CC. 2012. Biochemical study of the comparative inhibition of hepatitis C virus RNA polymerase by VX-222 and filibuvir. Antimicrob. Agents Chemother. 56:830–837. 10.1128/AAC.05438-11 [DOI] [PMC free article] [PubMed] [Google Scholar]