Abstract
Noncoding small RNAs (sRNAs) act in conjunction with the RNA chaperone Hfq to regulate gene expression in bacteria. Because Hfq is required for virulence in several bacterial pathogens, the Hfq-sRNA system is an attractive target for antibiotic development. A reporter strain in which the expression of yellow fluorescent protein (YFP) is controlled by Hfq-sRNA was engineered to identify inhibitors of this system. A reporter that is targeted by Hfq in conjunction with the RybB sRNA was used in a genetic screen to identify inhibitors from a library of cyclic peptides produced in Escherichia coli using split-intein circular ligation of peptides and proteins (SICLOPPS), an intein-based technology. One cyclic peptide identified in this screen, RI20, inhibited Hfq-mediated repression of gene expression in conjunction with both RybB and an unrelated sRNA, MicF. Gel mobility shift assays showed that RI20 inhibited binding of Hfq to RybB and MicF with similar Ki values. These data suggest that RI20 inhibits Hfq activity by blocking interactions with sRNAs and provide a paradigm for inhibiting virulence genes in Gram-negative pathogens.
INTRODUCTION
The spread of resistance to existing antibiotics represents a serious challenge to the public health sector. A recent CDC report estimated that >2 million people in the United States are infected by drug-resistant bacteria annually, causing over 20,000 fatalities (1). The limited pool of potential antibiotics in the development pipeline creates an urgent need to identify new antibiotic targets (1, 2). Targets that can be inhibited to prevent virulence and do not create a strong selective pressure to drive the spread of resistance would be especially valuable. In principle, inhibitors of bacterial pathways required for virulence but not viability can be used to treat infections. Because selective pressure for resistance to such inhibitors would be lower under some growth conditions compared to the strong selection for resistance to lethal inhibitors, the spread of resistance might be slower and the clinical lifetime of the drugs might be longer (3, 4).
One approach to targeting virulence is to inhibit regulatory pathways that control the expression of genes required for a pathogen to cause disease in a host during infection. Recent work with bacterial pathogens demonstrated that the protein Hfq, which is required for posttranscriptional regulation of gene expression by many bacterial small RNAs (sRNAs), is often required for virulence. Δhfq mutants of uropathogenic Escherichia coli, Pseudomonas aeruginosa, Salmonella enterica serovar Typhimurium, Vibrio cholerae, Yersinia pestis, Klebsiella pneumoniae, and Proteus mirabilis are all attenuated for virulence, more sensitive to an array of stresses, and often more susceptible to antibiotic treatment (5–14). Because Hfq homologues have been identified in over 50% of the sequenced bacterial genomes (15), inhibitors of this protein might be effective against a broad spectrum of pathogens.
Hfq is a member of the Sm-like family of RNA-binding proteins and acts as an RNA chaperone for regulatory sRNAs. Hfq binds with sRNAs and promotes base-pairing interactions between the sRNAs and their mRNA targets (16–18). sRNAs regulate expression of their target mRNAs in a variety of ways, often by inhibiting translation (19, 20). Hfq-sRNA activity also promotes degradation of the mRNA targets by the RNA degradosome (21). Because most sRNAs require Hfq for activity, inhibitors of Hfq are likely to disrupt a significant portion of sRNA-mediated transcriptional regulation.
To allow discovery of specific Hfq inhibitors that can be used to validate Hfq as a therapeutic target, a cell-based assay for inhibition of Hfq activity was developed and tested. The assay uses a fluorescent reporter placed under the control of the RybB sRNA in conjunction with Hfq. Libraries of cyclic peptides were generated inside bacterial cells using split-intein circular ligation of peptides and proteins (SICLOPPS), an intein-based technology (22). SICLOPPS allows the spontaneous circular ligation of peptide sequences. By randomizing codons in the SICLOPPS target sequence, libraries of cyclic peptides with large sequence diversity can be generated inside bacterial cells (23). In this work, a SICLOPPS library with five randomized codons, encoding ∼106 different cyclic peptides, was screened for potential inhibitors of Hfq-RybB. A peptide was identified that inhibited repression of target gene expression by Hfq-RybB. This peptide was also able to inhibit Hfq-dependent regulation by a second sRNA, MicF. In both cases, the peptide reduced the affinity of Hfq for the sRNA in vitro. These results indicate that Hfq is a druggable target and facilitate identification of inhibitors of Hfq from libraries of potential pharmaceuticals.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains used in this study are described in Table 1. All the reporter strains used for in vivo screening are derivatives of E. coli strain BW27786 (24). Mutant alleles lacIq and Δhfq were moved into the appropriate strains using P1 transduction, and the drug resistance markers were removed using FLP recombinase (25). E. coli strains were grown in LB at 30°C with aeration unless otherwise noted, and 100 μg/ml ampicillin, 30 μg/ml kanamycin, 30 μg/ml chloramphenicol, and 0.0002% arabinose were added where appropriate.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| BW27786 | DE(araBAD) DE(rhaBAD) DE(araFGH) ϕ(ΔaraEp PCP13-araE ΔrhaD-B) | 24 |
| SAE006 | BW27786 lacIq | This study |
| SAE008 | SAE006 pRybB pompC′-yfp | This study |
| SAE009 | SAE006 pJV300 pompC′-yfp | This study |
| SAE011 | SAE006 pMicF pompF′-yfp | This study |
| SAE012 | SAE006 pJV300 pompF′-yfp | This study |
| SAE109 | BL21(DE3) pET28a_hfq | This study |
| SAE204 | SAE011 pRI20 | This study |
| SAE221 | SAE008 pRI20_1 | This study |
| SAE222 | SAE008 pRI20_2 | This study |
| SAE223 | SAE008 pRI20_3 | This study |
| SAE224 | SAE008 pRI20_4 | This study |
| SAE225 | SAE008 pRI20_5 | This study |
| SAE229 | BW27786 Δhfq | This study |
| SAE232 | BW27786 pRI20 | This study |
| SAE250 | SAE006 pRybB pompF′-yfp | This study |
| SAE251 | SAE006 pJV300 pompF′-yfp | This study |
| SAE252 | SAE250 pRI20 | This study |
| pSB4K5 | BioBrick vector, Kanr | 46 |
| pBAD18K | pBAD vector, Kanr | 47 |
| pAS08 | R6K ori plasmid carrying tet control region with tetR, TetR-regulated promoter, and tetA, Cmr | Mark Goulian (University of Pennsylvania) |
| pRybB | Same as pFM1-1 (pBRpLac encoding rybB, Ampr) | 48 |
| pMicF | pBRpLac vector encoding micF, Ampr | 49 |
| pKMT5 | pNM12 vector encoding rybB, Ampr | 50 |
| pJV300 | sRNA control vector, Ampr | 7 |
| pARCBD | Expression plasmid for SICLOPPS, Cmr | 22 |
| pompC′-yfp | pSB4K5 with ompC′-yfp, Kanr | This study |
| pompF′-yfp | pSB4K5 with ompF′-yfp, Kanr | This study |
| pRI20 | pARCBD with RI20 (SGWKVMEG), Cmr | This study |
| pRI20_1 | pARCBD with SGWAVMEG, Cmr | This study |
| pRI20_2 | pARCBD with SGWKAMEG, Cmr | This study |
| pRI20_3 | pARCBD with SGWKVAEG, Cmr | This study |
| pRI20_4 | pARCBD with SGWKVMAG, Cmr | This study |
| pRI20_5 | pARCBD with SGWKVMEA, Cmr | This study |
| pET28a_hfq | pET28a encoding untagged hfq, Kanr | This study |
Plasmid construction.
Plasmids used in this study are described in Table 1 and oligonucleotide sequences are listed in Table 2. To make pompC′-yfp, the region of ompC that binds RybB was amplified by PCR from genomic DNA using primers ompCE and ompCB. The yfp gene was amplified by PCR using primers egfpB and egfpSA. Both PCR products were digested with BspHI and ligated using T4 DNA ligase, and the resulting ompC′-yfp fusion was amplified by PCR using primers ompCE and egfpSA, digested with EcoRI and SalI, and ligated into pBAD18K cut with the same enzymes to make the pBOY plasmid. The tet control region was amplified by PCR from pAS08 using primers pAS081 and pAS081R, and digested with EcoRI and KpnI. The ompC′-yfp fusion was amplified from pBOY by PCR using primers ompCK and egfpSp, digested with KpnI and SpeI, and ligated to the tet control region fragment. The ligated product was amplified by PCR using primers pAS081 and egfpSp, digested with EcoRI and SpeI, and ligated into pSB4K5 cut with the same enzymes to produce pompC′-yfp.
TABLE 2.
Oligonucleotides
| Name | Sequence |
|---|---|
| ompCE | TATTAGAATTCTTGCCGACTGATTAATGAGGGT |
| ompCB | ATAATTCATGACTGGGACCAGGAGGGACAGTAC |
| egfpB | TATTATCATGAGCAAGGGCGAGGAGCTGT |
| egfpSA | ATAATGTCGACTTACTTGTACAGCTCGTCCATG |
| pAS081 | ATAATGAATTCCGGAACTAGAGGATCCGCATT |
| pAS081R | ATAATGGTACCAGGGAGTGGTAAAATAACTCTAT |
| ompCK | TATTAGGTACCTTGCCGACTGATTAATGAGGGT |
| egfpSp | ATAATGTCGACTTACTTGTACAGCTCGTCCATG |
| OmpF-Brev | CGGGGTTCATGACAGGGACGATCACTGCCAG |
| OmpF-110 | CGGGGTGGTACCAGACACATAAAGACACCAAACTC |
| MicF_fwd | TAATACGACTCACTATAGGGCTATCATCATTAACTTTATTTATTACCG |
| MicF_rev | AAAAAAAACCGAATGCGAGGCATCCGGTTGAAATAGGGG |
| RybB_fwd | TAATACGACTCACTATAGGGCCACTGCTTTTCTTTGATGTCC |
| RybB_rev | AACAAAAAACCCATCAACCTTGAACCG |
| Hfqfwd_NcoI | GGCCGACCATGGCTAAGGGGCAATCTTTACAAGATCCGTT |
| Hfqrev_EcoRI | GGCCGAGAATTCTTATTCGGTTTCTTCGCTGTCCT |
| OmpC_fwd | AAAGTTAAAGTACTGTCCCTCCTGGTCC |
| OmpC_rev | GAACTGGTAAACCAGACCCAGAGCTAC |
| ECssrA11 | TGCTAAGCTTTAACGATAA |
| ECssrA13 | TTAATGGTGATGATGGTGGTGTTCGTCGTTTGCGACT |
| Ala1_top | GGGCGATCGGCCACAATTCCGGTTGGGCTGTGATGGAGGGCTGC |
| Ala1_bottom | TTAAGCAGCCCTCCATCACAGCCCAACCGGAATTGTGGCCGATCGCCCCAT |
| Ala2_top | GGGCGATCGGCCACAATTCCGGTTGGAAGGCTATGGAGGGCTGC |
| Ala2_bottom | TTAAGCAGCCCTCCATAGCCTTCCAACCGGAATTGTGGCCGATCGCCCCAT |
| Ala3_top | GGGCGATCGGCCACAATTCCGGTTGGAAGGTGGCTGAGGGCTGC |
| Ala3_bottom | TTAAGCAGCCCTCAGCCACCTTCCAACCGGAATTGTGGCCGATCGCCCCAT |
| Ala4_top | GGGCGATCGGCCACAATTCCGGTTGGAAGGTGATGGCTGGCTGC |
| Ala4_bottom | TTAAGCAGCCAGCCATCACCTTCCAACCGGAATTGTGGCCGATCGCCCCAT |
| Ala5_top | GGGCGATCGGCCACAATTCCGGTTGGAAGGTGATGGAGGCTTGC |
| Ala5_bottom | TTAAGCAAGCCTCCATCACCTTCCAACCGGAATTGTGGCCGATCGCCCCAT |
For the construction of pompF′-yfp, the region of the ompF gene targeted by sRNAs was amplified from genomic DNA by PCR using primers ompF-Brev and ompF-110, digested with BspHI and KpnI, and ligated into pompC′-yfp cut with the same enzymes.
Hfq was amplified by PCR from genomic DNA using primers Hfqfwd_NcoI and Hfqrev_EcoRI, digested with NcoI and EcoRI, and ligated into pET28a cut with the same enzymes. The SGWX5 SICLOPPS library was constructed as previously described (23).
Screen for inhibitors of Hfq-RybB.
E. coli strain SAE008 was transformed with the SICLOPPS plasmid library, and resulting colonies were scraped, grown overnight in LB with 30 μg/ml chloramphenicol, 100 μg/ml ampicillin, 30 μg/ml kanamycin, and 0.0002% arabinose at 30°C, diluted 100-fold, and grown under the same conditions to an optical density at 600 nm (OD600) of ∼0.2. Expression of RybB was induced by addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to 1 mM for 1 h, followed by induction of ompC′-yfp by addition of anhydrous tetracycline (AHT) to 100 ng/ml for 3 h. Cells were sorted by fluorescence-activated cell sorting (FACS) using a Cytopeia Influx (Becton, Dickinson) cell sorter equipped with a 200-mW 488-nm laser and a 531/40 bandpass filter for detection of yellow fluorescent protein (YFP) fluorescence intensity. Cells with the highest 0.01% YFP fluorescence intensity were selected and deposited on agar plated for clonal growth. Cells from each colony were grown in LB as described above and examined by epifluorescence microscopy, and the fluorescence intensity was measured using Simple PCI software (Compix, Inc.). Plasmid DNA was prepared from the selected clones and sequenced. Peptide sequences were determined by conceptual translation of the obtained DNA sequences.
Reporter assays.
Fluorescent reporter and control strains were grown in LB with the appropriate antibiotics and arabinose at 30°C, diluted to an OD600 of 0.02, and grown under the same conditions to an OD600 of 0.2. Expression of sRNA was induced by addition of IPTG to 1 mM for 1 h, and expression of the fluorescent reporter was induced by addition of AHT to 100 ng/ml for 3 h prior to observation and quantification by epifluorescence microscopy. All experiments were done in triplicate. To confirm expression of cyclic peptides, cells were harvested in volumes normalized to the OD600 and lysed in SDS-PAGE loading buffer. Proteins were separated on 15% SDS-polyacrylamide gels, blotted to polyvinylidene difluoride (PVDF) membranes, and the full-length SICLOPPS proteins were visualized by Western blotting using anti-chitin-binding domain (CBD) antibodies (New England BioLabs) with goat-anti-rabbit secondary antibody and ECF reagent (GE Healthcare). The full-length SICLOPPS protein contains a CBD that is removed upon cyclization. Therefore, the level of CBD is indicative of cyclic peptide expression levels. The relative amounts of proteins were determined using a Typhoon 9410 (GE Healthcare) and quantified using ImageQuant (Molecular Dynamics).
Assays with synthetic RI20 (ChinaPeptides Co., Ltd.) were performed in a similar manner, except arabinose was not included and RI20 was added to 500 μM during growth. Because RI20 was dissolved in dimethyl sulfoxide (DMSO), in control reactions cells were incubated with 1% DMSO instead of RI20.
For assays in 96-well plates, cultures were grown and induced with IPTG as described above, and 100 μl was transferred to microtiter trays. Trays were incubated at 37°C for 45 min, AHT was added, and trays were incubated at 37°C for 6 h. Fluorescence was measured with excitation at 505 nm and emission at 545 nm, and the fluorescence intensity was normalized to the cell density (OD600) of the well. Z′ values for each day were calculated as described (26), and Z′ values for 3 different days were averaged.
Northern blot analysis.
Strain SAE195 was grown with or without addition of 0.0002% arabinose to an OD600 of 1.5 in the presence of IPTG and AHT, cells were harvested by centrifugation, and RNA was prepared using the hot phenol method (27). Total RNA (5 μg) was separated by electrophoresis on a 5% urea-acrylamide gel, transferred to Hybond-N+ membrane (GE Healthcare), and incubated with 32P-labeled DNA probes at 50°C overnight. The membrane was rinsed three times with 0.5× SSPE–0.1% SDS (75 mM NaCl, 6 mM NaH2PO4, 0.6 mM EDTA, and 0.1% SDS), and bands were visualized on a Typhoon 9410 and quantified using ImageQuant. 32P-labeled DNA probes were generated from PCR products amplified from genomic DNA using primers OmpC_fwd and OmpC_rev for ompC, or primer ECssrA11 and ECssrA13 for ssrA, followed by labeling in the presence of α-[32P]dCTP using Ready-To-Go DNA-labeling beads (GE Healthcare). Labeled probes were purified using mini Quick Spin columns (Roche) to remove unincorporated nucleotides prior to hybridization.
Hfq purification.
Untagged Hfq was purified from E. coli BL21(DE3) transformed with pET28a_hfq, using Co2+-affinity chromatography as described previously (28), with modifications. Cultures were grown to an OD600 of 0.4, expression was induced by addition of IPTG to 1 mM, and cultures were grown for 4 h. Cells were harvested by centrifugation, and the cell pellet was resuspended in 50 ml lysis buffer (50 mM Tris-HCl [pH 7.5], 500 mM NH4Cl, 20 mM imidazole, and 5% wt/vol glycerol). Lysozyme was added to a final concentration of 1 mg/ml, the suspension was incubated on ice for 30 min, and cells were lysed by sonication. 1 U DNase I and 1 μg/ml RNase A were added, the lysate was incubated on ice for 1 h, and debris was removed by centrifugation. The clarified lysate was passed over a Hi-Trap metal chelation column (GE Healthcare) preloaded with CoSO4. The column was washed with five volumes of lysis buffer, followed by five volumes of wash buffer (50 mM Tris-HCl [pH 7.5], 1 M NH4Cl, and 5% wt/vol glycerol). Hfq was eluted with five column volumes of elution buffer (50 mM Tris-HCl [pH 7.5], 500 mM NH4Cl, 250 mM imidazole, and 5% wt/vol glycerol), fractions containing Hfq were combined, dialyzed against buffer A1 (50 mM Tris-HCl [pH 7.5], 100 mM NH4Cl, and 1 mM EDTA), and applied to a MonoQ HR 5/5 column (GE Healthcare) equilibrated in buffer A1. The column was washed with 5 column volumes of buffer A1, and bound protein was eluted with a linear gradient from 0 to 2 M NaCl in buffer A1. Protein was dialyzed against storage buffer (50 mM Tris-HCl [pH 7.5], 100 mM NH4Cl, 1 mM EDTA, and 10% glycerol) and stored at −20°C.
In vitro transcription of RybB and MicF RNA.
Templates for in vitro transcription were PCR products amplified from the pKMT5 plasmid using primers RybB_fwd and RybB_rev for rybB, or the pMicF plasmid using primers MicF_fwd and MicF_rev for micF. In vitro transcription was performed by mixing 500 ng template, 2 mM nucleoside triphosphates (NTPs), 1× transcription buffer (200 mM Tris-HCl [pH 8], 200 mM MgCl2, 1 M NaCl, and 100 mM spermidine), 0.5 U RNasin (Promega), 20 mM dithiothreitol (DTT), 0.1 U T7 RNA polymerase, and 0.2 μCi α-[32P]UTP, and incubating at 37°C for 2 h. Two units of RNase-free DNase was added, the reaction mixture was incubated for 30 min, and unincorporated nucleotides were removed using mini Quick Spin columns. Following extraction with phenol/chloroform/iso-amyl-alcohol (25:24:1, vol/vol/vol), the RNA was precipitated overnight at −80°C with 2 volumes ethanol and 100 μg/ml glycogen. RNA integrity was confirmed on a denaturing 6% polyacrylamide gel. Yield of purified RNA produced was estimated by determining the percentage of incorporation of α-[32P]UTP using an LS6500 scintillation counter (Beckman Coulter).
Electrophoretic mobility shift assays.
Binding assays were performed in 1× binding buffer (40 mM Tris [pH 8], 30 mM KCl, 1 mM MgCl2, 0.01% NP-40, 1 mM DTT, and 10 μg/ml bovine serum albumin [BSA]) in the presence of either DMSO or RI20 cyclic peptide. Radiolabeled RNA (0.1 nM) and 100 nM yeast RNA were incubated with Hfq in 30 μl total volume at 37°C for 10 min. Reactions were analyzed on 6% polyacrylamide gels in 0.5× Tris-borate-EDTA (TBE) and visualized using a Typhoon 9410. Binding assays were done in triplicate for every RI20 concentration using either RybB or MicF RNA. Curve fitting was performed by nonlinear regression using a ligand-binding one-site saturation equation on Sigma Plot (Systat Software, Inc.).
Stress sensitivity assays.
For hydrogen peroxide sensitivity assays, 3 ml LB top agar was inoculated with 30 μl saturated culture, plated on LB agar, and allowed to solidify. Ten microliters of 3% hydrogen peroxide was added to a 6-mm filter paper disk, and the disk was placed on the top agar. After incubation overnight, the diameter of the zone of inhibition was measured. Averages and standard deviations for >4 independent experiments were calculated.
For antibiotic sensitivity assays, broth microdilution assays were conducted as previously described (29). The MIC was determined from visual inspection, and the 50% inhibitory concentration (IC50) was determined after measuring the OD600 and plotting the percent growth inhibition versus antibiotic concentration. Experiments were performed in triplicate.
Molecular docking.
Molecular docking studies were performed using AutoDock Vina (The Scripps Research Institute) with the AutoDock Tools graphical user interface, and energy minimization was performed using the OpenBabel module. The E. coli Hfq crystal structure (PDB identification code, 1HK9) was used to calculate protein receptor grid maps using the AutoGrid module, and RI20 was docked using the Lamarckian genetic algorithm. A combination of the highest dock scores and the lowest root mean square deviation values were used as criteria to select the best binding conformation among 10 generated by the program.
RESULTS
Screen for inhibitors of Hfq-sRNA activity.
To facilitate identification of inhibitors of Hfq-sRNA activity, a cell-based assay with a positive readout for Hfq-RybB inhibition was engineered. YFP was placed under the control of Hfq-RybB by inserting the 5′ portion of ompC, including the RybB recognition sequence (30–32), upstream of the yfp coding sequence (Fig. 1A). Significant fluorescence was observed by flow cytometry and fluorescence microscopy when this ompC′-yfp gene fusion was expressed in wild-type E. coli, indicating that the ompC sequence does not interfere with YFP production or fluorescence and that the endogenous amount of RybB is not sufficient to repress ompC′-yfp expression (Fig. 1B and 2A). To determine if higher levels of Hfq-RybB repress expression of ompC′-yfp, the ompC′-yfp gene was expressed in E. coli containing the pRybB plasmid, in which rybB was overexpressed from an IPTG-inducible promoter. Fluorescence from expression of ompC′-yfp was decreased by 60% when RybB was overproduced, demonstrating that ompC′-yfp expression is repressed by Hfq-RybB (Fig. 1B and 2A). These data suggested that E. coli pRybB pompC′-yfp can be used to identify inhibitors of Hfq-RybB activity (Fig. 1B). The presence of an inhibitor of Hfq-RybB should increase expression of ompC′-yfp even when RybB is overproduced, and cells containing an inhibitor should have more YFP fluorescence (Fig. 1A).
FIG 1.
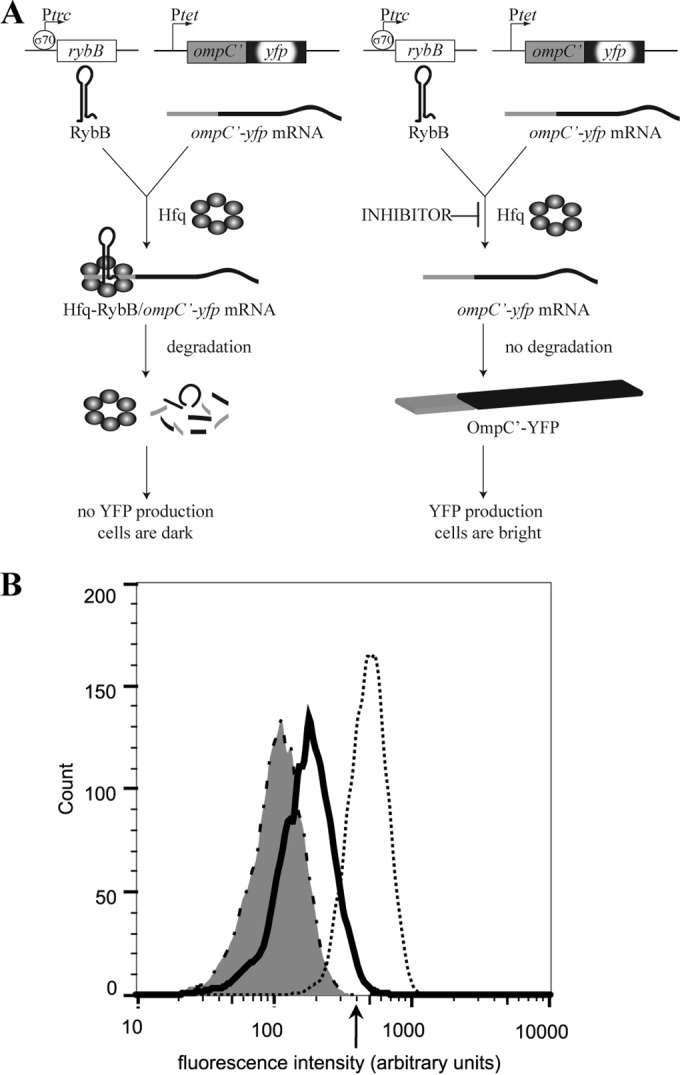
Screen for cyclic peptide inhibitors of the Hfq-RybB pathway. (A) Cartoon representation of the Hfq-RybB regulatory pathway. When the pathway is functional (left), RybB sRNA binds an Hfq hexamer. Hfq-RybB represses translation of ompC′-yfp mRNA and targets it for degradation, preventing production of OmpC′YFP. Inhibition of the pathway (right) derepresses translation, resulting in OmpC′-YFP production and fluorescent cells. (B) FACS allows isolation of cells containing an inhibitor. Histograms of fluorescence intensity from FACS showing a representative distribution (5,000 cells each) of E. coli pRybB pompC′-yfp (dashed line with gray fill), positive-control strain (pompC′-yfp [dotted line]), and pRybB pompC′-yfp transformed with the SGWX5 SICLOPPS library (black). The arrow indicates the minimum fluorescence intensity used to select cells from the SGWX5 library.
FIG 2.

RI20 inhibits Hfq-RybB repression of OmpC. (A) With no inhibitor, RybB represses OmpC′-yfp expression through base-pairing interactions (top), and E. coli pRybB pompC′-yfp cells have low YFP fluorescence intensity (RybB panel). When RI20 is produced (RybB + pRI20 panel), fluorescence intensity is similar to that in a strain with no RybB expression (E. coli pompC′-yfp [no RybB panel]). (B) Northern blotting indicates that production of RI20 increases the amount of OmpC′-yfp mRNA ∼3-fold. A representative blot from three independent experiments is shown. The normalized mRNA levels for OmpC′-yfp and SsrA (used as a loading control) are indicated. (C) Synthetic RI20 inhibits Hfq-RybB activity when added to cells in culture. Cultures of E. coli pRybB pompC′-yfp were incubated with RI20 or DMSO as a vehicle control. Fluorescence micrographs show that OmpC′-YFP fluorescence intensity is low after addition of DMSO and high after addition of RI20. Cells grown under the same conditions without pRybB (E. coli pompC′-yfp [no RybB panel]) are shown as a positive control.
To identify inhibitors, E. coli pRybB pompC′-yfp was transformed with a library of plasmids encoding randomized cyclic peptides. The encoded cyclic peptides are produced using SICLOPPS technology and have the sequence SGWX5. This library has been used to identify inhibitors of protein-protein interactions and enzymatic reactions (33–35). Production of cyclic peptides, RybB, and OmpC′-YFP was induced, and 2 × 106 cells were sorted using fluorescence-activated cell sorting (FACS) (Fig. 1B). The fluorescence histogram showed that the average cell fluorescence was ∼2-fold higher in the population expressing a cyclic peptide, suggesting that the SICLOPPS system has a small effect on fluorescence. To identify potent inhibitors, the brightest 0.01% cells were collected and propagated as clonal populations. After each strain was grown under inducing conditions, cells were examined using fluorescence microscopy. Most strains had increased levels of fluorescence, but one strain, expressing the cyclic peptide RI20, produced cells with fluorescence comparable to the E. coli pompC′-yfp positive control (Fig. 2A). The plasmid was isolated and retransformed into E. coli pRybB pompC′-yfp to verify that inhibition was due to the RI20 cyclic peptide. This strain was selected for further characterization. Sequencing of the plasmid encoding RI20 revealed that the cyclic peptide sequence was SGWKVMEG.
RI20 inhibits Hfq-mediated repression.
To quantify the efficiency of derepression by RI20, the fluorescence intensity of E. coli pRybB pompC′-yfp cells was measured with and without RI20. Because cells without RI20 serve as the negative control, a cutoff for “dark cells” that have full repression of ompC′-yfp expression was set at the average intensity plus two standard deviations from cells without RI20. Using this measure, 98% of cells producing RI20 had derepressed expression of ompC′-yfp (Fig. 2A).
Hfq-RybB promotes degradation of ompC mRNA, so inhibition of Hfq-RybB should increase the steady-state amount of ompC mRNA if synthesis rates are unchanged. With ompC′-yfp expressed from the Ptet promoter, production of RI20 increased the steady-state level of ompC′-yfp mRNA 2- to 3-fold (Fig. 2B). This result is consistent with RI20 inhibiting Hfq-RybB-mediated degradation of ompC′-yfp mRNA.
The results described above show that RI20 inhibits Hfq-RybB repression at the ompC mRNA regulatory sequence in vivo. To determine if RI20 also inhibits Hfq-RybB activity on other mRNA sequences, the ompC sequence of the ompC′-yfp fusion was replaced with the cognate RybB recognition sequence from ompF (Fig. 3A) (30, 31, 36) and the fluorescence microscopy experiment was repeated with pompF′-yfp. When RI20 was produced, 96% of cells had derepressed expression of ompF′-yfp (Fig. 3A). These results indicate that RI20 is not specific for the RybB-binding sequence in the ompC mRNA and that RI20 inhibits Hfq-RybB activity on multiple mRNA targets.
FIG 3.
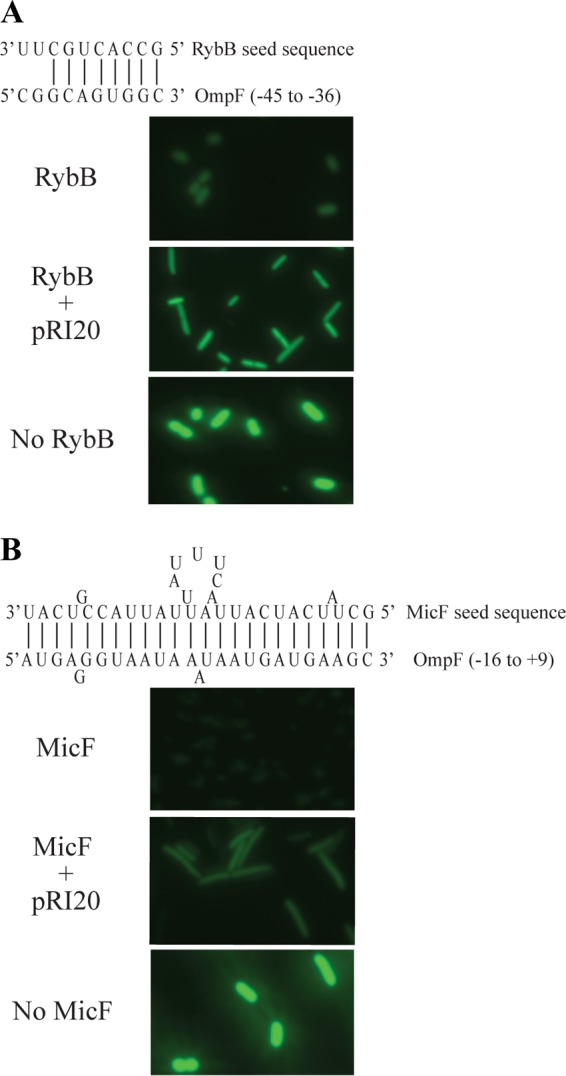
RI20 inhibits repression of OmpF by Hfq-RybB and Hfq-MicF. (A) With no inhibitor, RybB represses OmpF′-yfp expression through base-pairing interactions (top), and E. coli pRybB pompF′-yfp cells have low YPF fluorescence intensity (RybB panel). When RI20 is produced (RybB + pRI20 panel), fluorescence intensity is higher. A strain with no RybB expression (E. coli pompF′-yfp [no RybB panel]) is shown as a positive control. (B) OmpF is also repressed through base-pairing interaction with MicF (top), and E. coli pMicF pompF′-yfp cells have low YPF fluorescence intensity (MicF panel). Production of RI20 increases YFP fluorescence intensity (MicF + pRI20 panel). A strain with no MicF expression (E. coli pompF′-yfp [no MicF panel]) is shown as a positive control.
To determine if RI20 inhibits Hfq-mediated repression by other sRNAs, MicF was produced in cells expressing ompF′-yfp. Like RybB, MicF inhibits expression of ompF, but MicF makes different base contacts and represses expression more efficiently than RybB (Fig. 3B) (31, 37). In cells producing MicF, production of RI20 derepressed expression of ompF′-yfp in 40% of cells (Fig. 3B). Therefore, RI20 also inhibited Hfq-MicF activity in vivo, but not as efficiently as Hfq-RybB.
Exogenous addition of RI20 inhibits Hfq-RybB.
Most cyclic peptides produced using SICLOPPS in the SGWX5 library have no effect on Hfq-RybB activity, indicating that expression of the split intein protein and the intein products of the circularization reaction are not responsible for the inhibition (Fig. 1B). However, the reaction goes through a lariat intermediate that includes the randomized sequence and might, in principle, be responsible for the observed RI20 activity. To ensure that the in vivo activity of RI20 was due to the cyclic peptide, RI20 was chemically synthesized, purified, and added to cultures of E. coli pRybB pompC′-yfp at a final concentration of 500 μM prior to analysis by fluorescence microscopy. One hundred percent of the observed cells were derepressed, demonstrating that the cyclic peptide form of RI20 is sufficient for inhibition of Hfq-RybB in vivo (Fig. 2C).
RI20 inhibits Hfq binding to sRNAs in vitro.
Because RI20 inhibits RybB repression of multiple mRNAs and also inhibits MicF, it is likely that RI20 acts at a shared step of the repression mechanism. One candidate step is binding of Hfq to the sRNAs. Gel mobility shift assays were used to measure the affinity of Hfq for RybB and MicF and to test if RI20 inhibited binding. In the absence of peptide, the Kd (dissociation constant) for binding of Hfq and RybB was ∼90 nM, similar to previous reports (38) (Fig. 4A). RI20 inhibited binding with an apparent Ki of 111 μM (Fig. 4B to D). Cyclic peptides with sequences unrelated to RI20 (SGWEYVRP and SGWMHQVS) did not inhibit Hfq binding with RybB when added at 500 μM, suggesting that inhibition is due to the RI20 sequence and not to properties shared by all cyclic peptides. The observed Kd for Hfq binding with MicF was 2.5 nM, similar to published results (39) (Fig. 5A), and RI20 inhibited binding with an apparent Ki of 102 μM (Fig. 5B to D). These data indicate that RI20 inhibits Hfq-sRNA repression by interfering with Hfq binding to sRNAs. The similar observed Ki values for RybB and MicF suggest that RI20 inhibits Hfq binding of these sRNAs in a similar manner.
FIG 4.
RI20 inhibits Hfq binding with RybB in vitro. Gel mobility shift assays were used to measure the affinity of Hfq and RybB in the presence of different concentrations of RI20. Representative gels with (A) no RI20 and (B) with 500 μM RI20 are shown. For each gel, the fraction bound was plotted and nonlinear regression was used to determine the apparent Kd. (C) Representative curves at each RI20 concentration (0 [closed circles], 50 [squares], 100 [open triangles], 300 [closed triangles], and 500 [open circles] μM) are shown. (D) The average apparent Kd (Kd app) values for at least three repeats at each concentration were plotted versus RI20 concentration to determine the Ki. Whiskers indicate the standard deviation at each point.
FIG 5.
RI20 inhibits Hfq binding to MicF in vitro. Gel mobility shift assays were used to measure the affinity of Hfq and MicF in the presence of different concentrations of RI20. Representative gels with (A) no RI20 and (B) with 500 μM RI20 are shown. For each gel, the fraction bound was plotted and nonlinear regression was used to determine the apparent Kd. (C) Representative curves at each RI20 concentration (0 [filled circles], 50 [squares], 100 [open triangles], 300 [filled triangles], and 500 [open circles] μM) are shown. (D) The average Kd app values for at least three repeats at each concentration were plotted versus RI20 concentration to determine the Ki. Whiskers indicate the standard deviation at each point.
Sequence specificity for RI20 activity.
To identify the residues required for RI20 activity in vivo, alanine-scanning mutagenesis was performed on the KVMEG sequence. Each variant was produced using SICLOPPS in E. coli pRybB pompC′-yfp, and the expression of peptides in vivo was confirmed by Western analysis (data not shown). Quantification of fluorescence intensity revealed that substitution of alanine for the glutamate residue dramatically decreased RI20 inhibition and resulted in a fluorescence profile similar to that observed with no cyclic peptide present (Fig. 6). A smaller decrease in inhibition was observed when alanine was substituted for the methionine residue. Substitution of the remaining amino acids with alanine had little effect on RI20 inhibition. Because Western blotting showed that all mutant peptides were expressed at levels within 11% of RI20, differences in inhibition activity are unlikely to be caused by decreased expression. These results indicate that the glutamate is critical for RI20 activity in vivo, and the methionine also contributes to inhibition.
FIG 6.
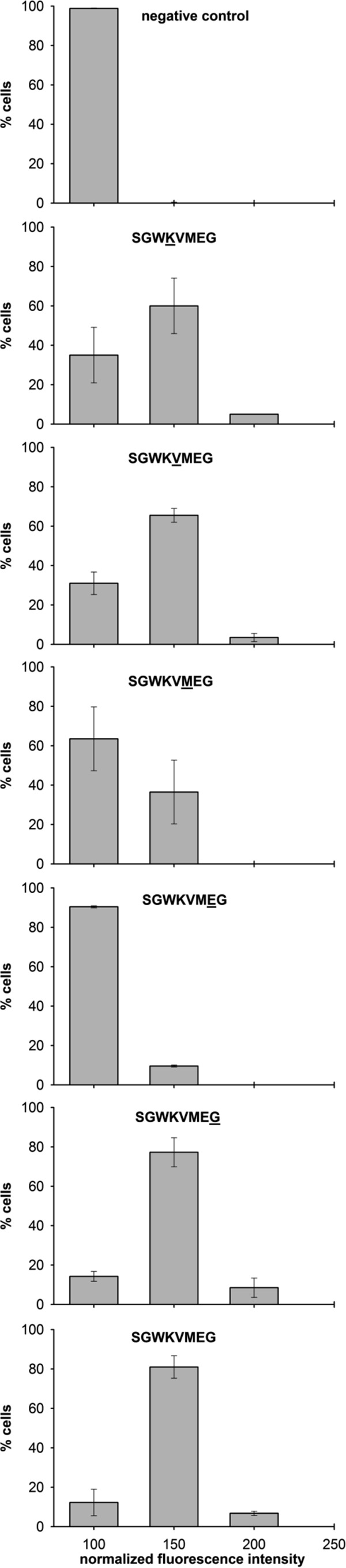
The glutamic acid residue in RI20 (SGWKVMEG) is critical for its activity. Fluorescence microscopy was used to quantify OmpF′-YPF fluorescence intensity in fields of >190 E. coli pRybB pompF′-yfp cells expressing RI20 or an alanine-substituted mutant. Intensities were normalized with the control with no cyclic peptide set to 100, binned, and the average percentage of cells in each bin over 3 trials is plotted with whiskers indicating the standard deviation. For each plot, the residue substituted with alanine is underlined.
RI20 causes defects in oxidative stress survival and antibiotic resistance.
E. coli strains deleted for hfq are more sensitive to oxidative stress and some antibiotics than wild-type cells (13, 40). To determine if RI20 production also produces these defects, strains were challenged with hydrogen peroxide in a disk diffusion assay. Wild-type cells producing RI20 and Δhfq cells both had a zone of inhibition >25% greater than wild-type cells with no RI20 plasmid, indicating a greater sensitivity to oxidative stress (Table 3). Likewise, wild-type cells producing RI20 and Δhfq cells were more sensitive to benzalkonium chloride and novobiocin than wild-type cells (Table 3). These data indicate that RI20 expression phenocopies deletion of hfq.
TABLE 3.
RI20 makes cells sensitive to hydrogen peroxide and antibiotics
| Strain | H2O2 zone of inhibition (diam [mm]) | Benzalkonium chloride |
Novobiocin |
||
|---|---|---|---|---|---|
| MIC (μg/ml) | IC50 (μg/ml) | MIC (μg/ml) | IC50 (μg/ml) | ||
| WTa | 18.5 ± 0.3 | 10 | 10 ± 3 | 125 | 19 ± 1 |
| Δhfq | 21.0 ± 0.1 | 5 | 3 ± 1 | 16 | 5 ± 1 |
| WT + pRI20 | 21.7 ± 0.7 | 5 | 2 ± 1 | 31 | 8 ± 1 |
WT, wild type.
Modeling of RI20 Hfq binding.
To gain preliminary insights into the molecular mechanism of inhibition by RI20, modeling was used to predict potential binding sites for the cyclic peptide on Hfq. RI20 was docked onto the E. coli crystallographic model of the Hfq hexamer (PDB identification code, 1HK9) using the AutoDock Vina program for modeling receptor-ligand interactions. RI20 is predicted to bind with an energy of −8.2 kcal/mol to the proximal face of Hfq, the surface responsible for interacting with sRNAs (Fig. 7A). In the predicted structure, RI20 establishes four main contact points through hydrogen bonds with three Hfq monomers (Fig. 7B). The glutamate required for RI20 function forms two hydrogen bonds with H57 of one Hfq monomer, one between the hydroxyl group of the ERI20 side chain and the backbone carbonyl group of H57Hfq, and a second between the backbone carbonyl of ERI20 and the γN of H57Hfq, which is bridged by a solvent molecule (Fig. 7B). A second Hfq monomer forms a hydrogen bond through the carbonyl backbone of H57Hfq to the indole N group of the tryptophan residue of RI20. A third Hfq monomer forms a hydrogen bond through the carbonyl backbone of H57Hfq to the backbone carbonyl of KRI20, bridged through a solvent molecule. In crystallographic studies of a hexauridine substrate bound to Hfq, a hydrogen bond is seen between the 3′-hydroxyl group of the terminal uridine and H57Hfq (41). The prediction that RI20 binds to H57Hfq in three different Hfq subunits suggests that RI20 acts as a competitive inhibitor of sRNA binding.
FIG 7.
Predicted binding of RI20 to the proximal face of Hfq using in silico analysis. (A) RI20 was docked to the crystal structure model of E. coli Hfq (PDB ID: 1HK9), and the structure of the best predicted binding conformation is shown. Each Hfq monomer is a different color. RI20 and H57Hfq residues that are contacted are shown in stick representation with nitrogen atoms in blue, oxygen atoms in red, and hydrogen atoms in white. Predicted hydrogen bonds are indicated with a yellow dashed line. (B) Closeup view of the predicted structure showing key interactions of RI20 with Hfq.
Validation of Hfq-sRNA inhibitor screen under high-HTS conditions.
Screening for high YFP fluorescence in the E. coli pRybB pompC′-yfp reporter strain successfully identified inhibitors of Hfq-sRNA activity using a FACS-based screen with a library of compounds expressed in vivo. To determine if the reporter can be used to screen compound libraries in high-throughput format, levels of YFP fluorescence from E. coli pRybB pompC′-yfp and E. coli pompC′-yfp were measured in 96-well microtiter plates. The average fluorescence in E. coli pompC′-yfp, the positive-control strain mimicking full inhibition of Hfq-RybB activity, was 4-fold higher than that in E. coli pRybB pompC′-yfp, with Z′ = 0.64 ± 0.04. These data indicate that the assay should be appropriate for high-throughput screening (HTS).
DISCUSSION
The work presented here demonstrates that regulation of gene expression by Hfq-sRNAs can be inhibited using drug-like compounds such as cyclic peptides, and that the assay system described here identifies inhibitors. Using a cell-based assay with positive readout, RI20 was identified from a library of cyclic peptides expressed inside the E. coli cell. Although the assays used the RybB sRNA, RI20 has the expected properties of an inhibitor that blocks Hfq interaction with multiple sRNAs. In vivo, it inhibited RybB-dependent repression of two mRNA targets and repression by a second sRNA, MicF, of its mRNA target. RI20 also had effects on stress resistance that were comparable to those observed with an hfq-deletion strain. In vitro assays indicated that RI20 acts by reducing the affinity of Hfq for RybB and MicF. Inhibition of RybB in vivo was more efficient than inhibition of MicF, but this difference is likely due to the higher affinity of MicF for Hfq. MicF binds Hfq with a Kd of ∼2.5 nM, whereas the Kd for Hfq binding to RybB is ∼90 nM. The Ki for RI20 in vitro was ∼100 μM for both RybB and MicF. Assuming the affinities are similar in vivo, significantly more RI20 would be required to disrupt Hfq-MicF activity than Hfq-RybB activity.
The mechanism by which Hfq binds a diverse set of cellular sRNAs is still under investigation. Current evidence suggests that Hfq has two different binding modes for sRNAs. A site on the proximal face of the Hfq hexamer binds 3′ U-rich regions, similar to those found in rho-independent transcription terminator sequences shared by many sRNAs (28, 41–43). Sites along the lateral sides of the hexamer are thought to bind to the body of the sRNAs (44). Given the similar Ki values for RybB and MicF, it is likely that RI20 prevents binding of these sRNAs to Hfq in a similar manner. RybB and MicF share little sequence similarity outside the transcriptional terminator region that binds the proximal face of Hfq, so this interaction is a strong candidate for the site of RI20 activity. Consistent with this prediction, the energy-minimized docking of RI20 to the Hfq hexamer suggests that there is a possible binding site for RI20 on the proximal face (Fig. 7). Future experimental studies are warranted to confirm these predictions.
RI20 itself is not a candidate anti-infective because the Ki is too high and high concentrations are needed to inhibit Hfq when the peptide is added to cells exogenously. Nevertheless, it has some properties that would be advantageous in an anti-infective. RI20 inhibits two sRNAs, and because the proximal face of Hfq is thought to bind all Hfq-dependent sRNAs, RI20 is likely to inhibit all Hfq-sRNA interactions. This broad specificity would prevent cells from circumventing drug activity by expressing an sRNA with redundant function and would allow a drug to be used on species that employ Hfq for virulence factor production without knowledge of the particular sRNA that is important. The predicted interaction with H57Hfq would also limit the acquisition of resistance through mutations in Hfq, because H57Hfq is required for Hfq oligomerization as well as sRNA binding (45), although mutations to other residues could potentially decrease RI20 binding and lead to resistance. A small molecule that binds to the same site as RI20 would be a candidate anti-infective. Such compounds could be rationally designed once the structure of RI20 bound to Hfq is known, or they could be identified by HTS using the assay described here.
The assay system used in this study is a cell-based assay with positive readout, which is readily adaptable to high-throughput formats and provides several advantages for high-throughput screening. Compounds added externally would have to pass through the cell membrane to inhibit Hfq-sRNA activity, minimizing hits with low permeability. The overall screening strategy is particularly powerful because the assay relies on a positive readout, gain of fluorescence, rather than loss of signal. Therefore, false-positive hits due to molecules that interfere with production of the reporter gene or cell growth will not be recovered. This assay is also easily adaptable for identification of inhibitors of other sRNAs by replacing the RybB and OmpC′ sequences with cognate sRNA and target sequences, respectively. Future identification of small-molecule inhibitors of Hfq will allow validation of Hfq as a therapeutic target in vivo.
ACKNOWLEDGMENTS
We thank Ningchun Xu and Ruth Nissly of the Microscopy and Cytometry Facility in the Huck Institutes of the Life Sciences for operation of the Influx cell sorter and flow cytometry data analysis. We thank Carol Gross, Jörg Vogel, and Susan Gottesman for sharing plasmids, and John Tomsho and Stephen Benkovic for providing the SGWX5 library.
This work was supported by NIH grant NS071542 (to S.E.A. and K.C.K.), NIH grant GM68720 (to K.C.K.), and The International Fulbright Science & Technology Award (to S.A.E).
Footnotes
Published ahead of print 7 July 2014
REFERENCES
- 1.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta, GA [Google Scholar]
- 2.Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74:417–433. 10.1128/MMBR.00016-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clatworthy AE, Pierson E, Hung DT. 2007. Targeting virulence: a new paradigm for antimicrobial therapy. Nat. Chem. Biol. 3:541–548. 10.1038/nchembio.2007.24 [DOI] [PubMed] [Google Scholar]
- 4.Allen RC, Popat R, Diggle SP, Brown SP. 2014. Targeting virulence: can we make evolution-proof drugs? Nat. Rev. Microbiol. 12:300–308. 10.1038/nrmicro3232 [DOI] [PubMed] [Google Scholar]
- 5.Kulesus RR, Diaz-Perez K, Slechta ES, Eto DS, Mulvey MA. 2008. Impact of the RNA chaperone Hfq on the fitness and virulence potential of uropathogenic Escherichia coli. Infect. Immun. 76:3019–3026. 10.1128/IAI.00022-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sonnleitner E, Hagens S, Rosenau F, Wilhelm S, Habel A, Jäger KE, Bläsi U. 2003. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb. Pathog. 35:217–228. 10.1016/S0882-4010(03)00149-9 [DOI] [PubMed] [Google Scholar]
- 7.Sittka A, Pfeiffer V, Tedin K, Vogel J. 2007. The RNA chaperone Hfq is essential for the virulence of Salmonella Typhimurium. Mol. Microbiol. 63:193–217. 10.1111/j.1365-2958.2006.05489.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding Y, Davis BM, Waldor MK. 2004. Hfq is essential for Vibrio cholerae virulence and downregulates σE expression. Mol. Microbiol. 53:345–354. 10.1111/j.1365-2958.2004.04142.x [DOI] [PubMed] [Google Scholar]
- 9.Geng J, Song Y, Yang L, Feng Y, Qiu Y, Li G, Guo J, Bi Y, Qu Y, Wang W, Wang X, Guo Z, Yang R, Han Y. 2009. Involvement of the posttranscriptional regulator Hfq in Yersinia pestis virulence. PLoS One 4:e6213. 10.1371/journal.pone.0006213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiang M-K, Lu M-C, Liu L-C, Lin C-T, Lai Y-C. 2011. Impact of Hfq on global gene expression and virulence in Klebsiella pneumoniae. PLoS One 6:e22248. 10.1371/journal.pone.0022248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang M-C, Chien H-F, Tsai Y-L, Liu M-C, Liaw S-J. 2014. The RNA chaperone Hfq is involved in stress tolerance and virulence in uropathogenic Proteus mirabilis. PLoS One 9:e85626. 10.1371/journal.pone.0085626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chao Y, Vogel J. 2010. The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 13:24–33. 10.1016/j.mib.2010.01.001 [DOI] [PubMed] [Google Scholar]
- 13.Yamada J, Yamasaki S, Hirakawa H, Hayashi-Nishino M, Yamaguchi A, Nishino K. 2010. Impact of the RNA chaperone Hfq on multidrug resistance in Escherichia coli. J. Antimicrob. Chemother. 65:853–858. 10.1093/jac/dkq067 [DOI] [PubMed] [Google Scholar]
- 14.Hayashi-Nishino M, Fukushima A, Nishino K. 2012. Impact of Hfq on the intrinsic drug resistance of Salmonella enterica serovar Typhimurium. Front. Microbiol. 3:1–5. 10.3389/fmicb.2012.00205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun X, Zhulin I, Wartell RM. 2002. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 30:3662–3671. 10.1093/nar/gkf508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogel J, Luisi BF. 2011. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 9:578–589. 10.1038/nrmicro2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagner EGH. 2013. Cycling of RNAs on Hfq. RNA Biol. 10:619–626. 10.4161/rna.24044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brennan RG, Link TM. 2007. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 10:125–133. 10.1016/j.mib.2007.03.015 [DOI] [PubMed] [Google Scholar]
- 19.Storz G, Vogel J, Wassarman KM. 2011. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell 43:880–891. 10.1016/j.molcel.2011.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Lay N, Schu DJ, Gottesman S. 2013. Bacterial small RNA-based negative regulation: Hfq and its accomplices. J. Biol. Chem. 288:7996–8003. 10.1074/jbc.R112.441386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Massé E, Escorcia FE, Gottesman S. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17:2374–2383. 10.1101/gad.1127103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott CP, Abel-Santos E, Wall M, Wahnon DC, Benkovic SJ. 1999. Production of cyclic peptides and proteins in vivo. Proc. Natl. Acad. Sci. U. S. A. 96:13638–13643. 10.1073/pnas.96.24.13638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tavassoli A, Benkovic SJ. 2007. Split-intein mediated circular ligation used in the synthesis of cyclic peptide libraries in E. coli. Nat. Protoc. 2:1126–1133. 10.1038/nprot.2007.152 [DOI] [PubMed] [Google Scholar]
- 24.Khlebnikov A, Datsenko KA, Skaug T, Wanner BL, Keasling JD. 2001. Homogenous expression of the PBAD promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology 147:3241–3247 [DOI] [PubMed] [Google Scholar]
- 25.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J-H, Chung TDY, Oldenburg KR. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4:67–73. 10.1177/108705719900400206 [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 28.Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. 2004. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat. Struct. Mol. Biol. 11:1206–1224. 10.1038/nsmb858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramadoss NS, Alumasa JN, Cheng L, Wang Y, Li S, Chambers BS, Chang H, Chatterjee AK, Brinker A, Engels IH, Keiler KC. 2013. Small molecule inhibitors of trans-translation have broad-spectrum antibiotic activity. Proc. Natl. Acad. Sci. U. S. A. 110:10282–10287. 10.1073/pnas.1302816110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Urban JH, Vogel J. 2007. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 35:1018–1037. 10.1093/nar/gkl1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gogol EB, Rhodius VA, Papenfort K, Vogel J, Gross CA. 2011. Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon. Proc. Natl. Acad. Sci. U. S. A. 108:12875–12880. 10.1073/pnas.1109379108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balbontín R, Florini F, Figueroa-Bossi N, Casadesús J, Bossi L. 2010. Recognition of heptameric seed sequence underlies multitarget regulation by RybB small RNA in Salmonella enterica. Mol. Microbiol. 78:380–394. 10.1111/j.1365-2958.2010.07342.x [DOI] [PubMed] [Google Scholar]
- 33.Horswill AR, Savinov SN, Benkovic SJ. 2004. A systematic method for identifying small-molecule modulators of protein-protein interactions. Proc. Natl. Acad. Sci. U. S. A. 101:15591–15596. 10.1073/pnas.0406999101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naumann TA, Tavassoli A, Benkovic SJ. 2008. Genetic selection of cyclic peptide Dam methyltransferase inhibitors. Chembiochem 9:194–197. 10.1002/cbic.200700561 [DOI] [PubMed] [Google Scholar]
- 35.Cheng L, Naumann TA, Horswill AR, Hong S-J, Venters BJ, Tomsho JW, Benkovic SJ, Keiler KC. 2007. Discovery of antibacterial cyclic peptides that inhibit the ClpXP protease. Protein Sci. 16:1535–1542. 10.1110/ps.072933007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papenfort K, Bouvier M, Mika F, Sharma CM, Vogel J. 2010. Evidence for an autonomous 5′ target recognition domain in an Hfq-associated small RNA. Proc. Natl. Acad. Sci. U. S. A. 107:20435–20440. 10.1073/pnas.1009784107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt M, Zheng P, Delihas N. 1995. Secondary structures of Escherichia coli antisense micF RNA, the 5′-end of the target ompF mRNA, and the RNA/RNA duplex. Biochemistry 34:3621–3631. 10.1021/bi00011a017 [DOI] [PubMed] [Google Scholar]
- 38.Papenfort K, Pfeiffer V, Mika F, Lucchini S, Hinton JCD, Vogel J. 2006. σE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 62:1674–1688. 10.1111/j.1365-2958.2006.05524.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fender A, Elf J, Hampel K, Zimmermann B, Wagner EGH. 2010. RNAs actively cycle on the Sm-like protein Hfq. Genes Dev. 24:2621–2626. 10.1101/gad.591310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wachi M, Takada A, Nagai K. 1999. Overproduction of the outer-membrane proteins FepA and FhuE responsible for iron transport in Escherichia coli hfq::cat mutant. Biochem. Biophys. Res. Commun. 264:525–529. 10.1006/bbrc.1999.1537 [DOI] [PubMed] [Google Scholar]
- 41.Sauer E, Weichenrieder O. 2011. Structural basis for RNA 3′-end recognition by Hfq. Proc. Natl. Acad. Sci. U. S. A. 108:13065–13070. 10.1073/pnas.1103420108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otaka H, Ishikawa H, Morita T, Aiba H. 2011. PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. Proc. Natl. Acad. Sci. U. S. A. 108:13059–13064. 10.1073/pnas.1107050108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishikawa H, Otaka H, Maki K, Morita T, Aiba H. 2012. The functional Hfq-binding module of bacterial sRNAs consists of a double or single hairpin preceded by a U-rich sequence and followed by a 3′ poly(U) tail. RNA 18:1062–1074. 10.1261/rna.031575.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sauer E, Schmidt S, Weichenrieder O. 2012. Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc. Natl. Acad. Sci. U. S. A. 109:9396–9401. 10.1073/pnas.1202521109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moskaleva O, Melnik B, Gabdulkhakov A, Garber M, Nikonov S, Stolboushkina E, Nikulin A. 2010. The structures of mutant forms of Hfq from Pseudomonas aeruginosa reveal the importance of the conserved His57 for the protein hexamer organization. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66:760–764. 10.1107/S1744309110017331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shetty RP, Endy D, Knight TF. 2008. Engineering BioBrick vectors from BioBrick parts. J. Biol. Eng. 2:5. 10.1186/1754-1611-2-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bouvier M, Sharma CM, Mika F, Nierhaus KH, Vogel J. 2008. Small RNA binding to 5′ mRNA coding region inhibits translational initiation. Mol. Cell 32:827–837. 10.1016/j.molcel.2008.10.027 [DOI] [PubMed] [Google Scholar]
- 49.Mandin P, Gottesman S. 2010. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 29:3094–3107. 10.1038/emboj.2010.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson KM, Rhodius VA, Gottesman S. 2007. σE regulates and is regulated by a small RNA in Escherichia coli. J. Bacteriol. 189:4243–4256. 10.1128/JB.00020-07 [DOI] [PMC free article] [PubMed] [Google Scholar]





