Abstract
SUMMARY
Human adenoviruses (HAdVs) are an important cause of infections in both immunocompetent and immunocompromised individuals, and they continue to provide clinical challenges pertaining to diagnostics and treatment. The growing number of HAdV types identified by genomic analysis, as well as the improved understanding of the sites of viral persistence and reactivation, requires continuous adaptions of diagnostic approaches to facilitate timely detection and monitoring of HAdV infections. In view of the clinical relevance of life-threatening HAdV diseases in the immunocompromised setting, there is an urgent need for highly effective treatment modalities lacking major side effects. The present review summarizes the recent progress in the understanding and management of HAdV infections.
INTRODUCTION
Since their first isolation from adenoid tissue over 60 years ago (1), human adenoviruses (HAdVs) (adénos, gland) have provided continuous challenges in a variety of clinical settings. In addition to their well-established role as infectious agents, adenoviral genomes were also shown to contain potent oncogenes, and the ability of certain types of the virus to induce tumor growth has been demonstrated in different mammalian animal models (2–4). Despite a number of studies addressing the possible role of HAdVs in human malignant disease, their putative oncogenicity in humans has remained enigmatic (5–7). Investigations of adenovirus biology have led to Nobel Prize-winning discoveries in mRNA splicing and to important progress in the understanding of antigen presentation to T cells (8). Moreover, the ability of adenoviruses to infect many cell types facilitated their exploitation as vectors for gene delivery to generate new tools for innovative treatments of important diseases, such as cancer and cardiovascular disorders (9–13). Hence, adenoviruses are highly versatile organisms with a broad spectrum of clinical roles and applications.
HAdVs were initially isolated mainly from military recruits with acute febrile respiratory disease and were subsequently associated with a number of clinical manifestations, including gastroenteritis, hepatitis, keratoconjunctivitis, meningoencephalitis, cystitis, upper and lower respiratory tract infections, and myocarditis, but also with noninflammatory conditions, such as obesity (14–21). HAdV infections are readily transmittable and, in some instances, highly contagious. Although the clinical courses are usually mild and self-limiting, infections may cause local outbreaks with severe courses, occasionally leading to a lethal outcome even in immunocompetent individuals (22–26). However, adenoviruses play a particularly important role in patients with strongly impaired immune responses, in whom viral disease is associated with high morbidity and mortality, and infections in this setting are an important focus of the present review. Progress in molecular detection methods has rendered the detection, typing, and monitoring of adenoviral infections readily applicable in the clinical routine, and the tools required for risk assessment and timely diagnosis of invasive infection are available. However, effective treatment of HAdV-related diseases in immunocompromised patients still poses great challenges.
NOMENCLATURE AND TYPING OF HUMAN ADENOVIRUSES
General Description, Structural Proteins, and Genomic Structure
HAdVs are nonenveloped viruses with a diameter of 70 to 100 nm. The external protein shell of the virus is icosahedral (eikosí, twenty; hédra, seat), with 20 triangular faces, 30 edges, and 12 vertices, and this symmetry is made up in large parts by the major virus protein, hexon. The viral capsid is composed of 252 capsomeres (capsa, box; méros, portion), including 240 hexons and 12 pentons. Five penton base proteins form individual capsomeres at the 12 vertices, where each capsomere supports a trimeric fiber protein of variable size projecting from the vertex of the capsid. The fiber protein contains three structural domains: the tail, the shaft, and the knob. The tail domain is the binding site for the penton base. The shaft domain shows various lengths between HAdV types, resulting in different flexibilities of the fiber and differences in the interaction with host cell integrins. The fiber knob domain, located at the distal, C-terminal end of the protein, binds the virus to the primary host cell receptor (27–30). Most adenoviruses bind to the classical adenovirus receptor CAR (coxsackie adenovirus receptor), but some use the membrane cofactor CD46 and/or the cell adhesion protein desmoglein 2 (DSG2) as an attachment receptor (27–29).
Minor capsid proteins IIIa, VI, VIII, and IX confer stability to the hexon shell and the entire virion and play roles in events occurring after internalization, such as endosome penetration, transcriptional activation, and nuclear reorganization (protein IX) (31, 32). Additional proteins in the core of the capsid (proteins V, VII, and X and terminal protein) interact with the viral DNA, e.g., by facilitating transportation into the nucleus of the infected cell (protein V) (30, 33).
HAdVs are double-stranded, linear DNA viruses displaying genome sizes ranging from 34 to more than 37 kb and carrying some 40 genes (33, 34). All HAdVs share a similar organization of the genome, which is divided into early, intermediate, and late regions corresponding to the infectious cycle of the virus and reflecting the transcription patterns. The early region of the genome includes four transcript families, termed E1 to E4, which are required for viral replication. The E3 transcription unit, which is highly divergent between HAdV species, also encodes proteins modulating the host immune response. The intermediate genes are represented by two transcripts, termed IX and IVa2, and the late region contains five transcript families, referred to as L1 to L5, which are involved in the production of mature virions. Moreover, the genomes display inverted terminal repeat (ITR) regions at the 3′ and 5′ ends that contain conserved sequence motifs and serve as origins of viral replication. Furthermore, depending on the HAdV type, the genomes display one or two noncoding virus-associated (VA) RNA genes involved in translational regulation, and one of them (VA RNAI) can act as a microRNA (miRNA) (30, 35).
Phylogeny and Human Adenoviral Species
Together with other mammalian adenoviruses, HAdVs are classified into the genus Mastadenovirus (mastós, breast) and are further parsed into seven species, termed A to G, with further subdivision of species B into subspecies B1 and B2 (33, 36–40). Species designation depends on several of the following characteristics: phylogenetic distance (>5 to 15%, based primarily on distance matrix analysis of the DNA polymerase amino acid sequence), genome organization (characteristically in the E3 region), nucleotide composition (G+C%), oncogenicity in rodents, host range, cross-neutralization, ability to recombine, number of VA RNA genes, and hemagglutination (33). More than 30 simian adenoviruses (SAdVs) display sequence identitites to their human counterparts to such an extent that they have also been included in the taxonomy of human adenoviruses, within species B, C, E, and G (33). Previously, HAdVs were identified, characterized, and classified by serum neutralization (SN) and hemagglutination inhibition (HI) assays, but more recently, genomic and bioinformatic analyses of the entire viral genome have superseded serological methods for the typing of novel viruses (36–42). The viruses belonging to individual HAdV species display high similarity to each other at the nucleotide level and do not commonly recombine with members of other species. The grouping into different species reflects, in part, the general cell tropism of the viruses and the resulting diseases and symptoms. Examples of common associations of individual HAdV species with infections at specific locations include gastroenteritis (HAdV-F and -G), pneumonia (HAdV-B, -C, and -E), hepatitis (HAdV-C), meningoencephalitis (HAdV-A, -B, and -D), cystitis (HAdV-B), and keratoconjunctivitis (HAdV-B and -D), but other HAdV species may also occur at the indicated sites of infection (30, 36, 43).
(Sero)typing of Human Adenoviruses
HAdV subtyping below the species level by SN and HI assays led to the identification of 51 serotypes (Table 1). The hypervariable loops (L1 and L2) of the hexon protein form the SN epitope and are the main determinants of serologic reactivity, while the fiber protein is responsible for HI typing and is a major determinant of tropism. The combination of the SN and HI tests facilitates more complete virus identification than that with either method alone (30).
TABLE 1.
Current spectrum of published human adenovirusesa
| Species | Types (serotypes/genotypes) |
|---|---|
| A | 12, 18, 31, 61 |
| B | 3, 7, 11, 14, 16, 21, 34, 35, 50, 55, 66 |
| C | 1, 2, 5, 6, 57 |
| D | 8–10, 13, 15, 17, 19, 20, 22–30, 32, 33, 36–39, 42–49, 51, 53, 54, 56, 58-60, 63-67 |
| E | 4 |
| F | 40, 41 |
| G | 52 |
The HAdV species (A to G) and types (1 to 67) belonging to individual species are indicated. While types 1 to 51 were identified by serotyping, all subsequently identified types, indicated in italics (types 52 to 67), were identified by genomic sequencing and computational analysis.
The first HAdV identified on the basis of genetic analysis was also classified as a novel species (HAdV-G) and received the chronological number 52 (36). Because all subsequently identified novel HAdVs were detected and characterized using computational analyses of genomic data (Table 1), it has been agreed to replace the term “serotype” by “type,” and criteria for the assignment of new types have been established (41, 42).
Present Controversies in HAdV Typing
There seems to be broad agreement that DNA sequence information should be exploited for HAdV typing, but differing views on the required extent of sequencing and the future role of serological methods have been presented. Some researchers take the stance that type definition should be based on the sequence of the major capsid protein, hexon, as the primary identifier, and that only strains carrying novel hexon genes should be considered “candidate new types,” because the hexon contains the major neutralizing epitope frequently targeted in molecular diagnosis (42). The designations of natural or engineered intertypic recombinant HAdVs should include the identity of the hexon gene (H) and also that of the fiber gene (F). These designations should replace those resulting from SN and HI assays, because published data support a strong correlation between identities established by sequence analyses of hexon and fiber and those determined by SN and HI assays, respectively (42). The extent of sequences required for consideration of a new candidate type is under evaluation but should not exceed the complete hexon and fiber knob or complete fiber gene sequence. The data should be integrated with the established immunotyping scheme, and serology should continue to be used in order to characterize the antigenic phenotypes. Some authors have expressed concern that the designation of new types by sequential numbers according to the order in which novel genome sequences are reported will likely lead to a large increase in their number (42).
In contrast, a proposal published by members of the adenovirus research community states that HAdVs should be identified, characterized, typed, and consecutively numbered on the basis of complete genome sequence analyses rather than by serological methods (41). Due to the power of novel genetic and bioinformatic tools, a paradigm shift in recognizing and naming HAdVs is needed. Recombination is an accepted feature of HAdV evolution, and recombinants will be classified as novel types provided that there are sufficient genomic, biological, or pathogenic differences from related types.
Serology can suggest possible recombinant viruses as intertypic strains by revealing conflicting results between SN and HI assays that are indicative of two different HAdV types. However, these methods cannot completely characterize the newly identified virus. Serologic determinants represent less than 5 to 6% of the total viral genome and thus can scarcely be regarded as adequate for full characterization of new HAdVs in the era of genomics (30). Recent data suggest that serologic and genomic analyses do not always correlate, and serotyping alone can provide misleading results (44, 45). Nevertheless, SN and HI testing should continue to be used as an additional criterion in order to comply with the current definitions of the International Committee on Taxonomy of Viruses (ICTV) (41). Recently, the full-genome sequencing of every prototype HAdV strain was completed, including a typing algorithm that includes the sequences supporting serological features (8). It is therefore conceivable that it will be possible to impute serologic test results by computing genomic sequence data, thus possibly obviating and replacing SN and HI assays.
Current Status of HAdV Types and Evolution of Human Adenoviruses
To date, 67 HAdV types have been published, and additional types are already in the pipeline. As outlined in Table 1, HAdV types 1 to 51 were characterized by serotyping, while the remaining types, identified since 2007, were detected and described by genomic and bioinformatic analyses (36, 46).
Homologous recombination (HR) and mutation are important evolutionary processes driving genetic variation within HAdV genomes (34). They are favored by the immune pressure of the host and by environmental bottlenecks. In HAdV species B, mutations seem to play a more important role, whereas among the largest HAdV species, species D, homologous recombination is the predominant mechanism contributing to genomic diversity (34). HR of tumorigenic adenoviruses in vitro was already documented in the 1970s (47, 48) and was shown to occur predominantly between HAdV types belonging to the same species, within regions of high sequence homology (30). The recent availability of whole-genome sequencing and bioinformatics has permitted the description of recombination events within genomes of HAdV species A, B, and D, particularly within the penton base, hexon, and fiber genes (37, 49–51). The requirements for recombination events appear to include coinfection of individual cells with at least two different adenoviruses displaying very similar nucleotide sequences at the recombination hot spots in the genome, as well as long-term viral persistence in the host (52, 53). The emergence of new HAdV-D types in patients with AIDS indicates a role of multiple persisting viruses under impaired immune surveillance (34, 54). HAdV-D genomes seem to recombine more frequently than other human adenoviral species, and several of the currently more than 40 HAdV-D types apparently emerged via recombination between hexon and fiber coding regions (34). The majority of novel HAdV types identified by genomic analysis belong to species D, and they were shown to include sequences derived from multiple other types from the same species. For example, HAdV-D53 resulted from recombination in the penton, hexon, and fiber regions of HAdV-D22, -D37, and -D8, respectively. Similarly, HAdV-D67 was identified as a recombinant between HAdV-D9, -D25, -D26, -D33, and -D46 (46, 55). Recent data provide evidence for the occurrence of recombination between different HAdV species, and even between HAdVs and SAdVs (56, 57). Computational analysis of HAdV-E4, the only representative of species E, indicated that this virus is of zoonotic origin and evolved through two interspecies recombination events with lateral partial gene transfer. HAdV-E4 contains 97% of a SAdV-E26-like genome chassis with a hexon containing the L1 and L2 regions from a HAdV-B16-like virus, which may provide compatibility with the new host (57). Adaptation of the virus to the new host could also be related to the acquirement of an NF-1 binding site motif, which is required for efficient viral replication, in a further recombination event.
Molecular evolution of HAdVs by homologous recombination can result in new viruses displaying different tissue tropisms and increased virulence. An improved knowledge of homologous recombination might facilitate the prediction of potential emerging HAdV types. In addition to their role in the evolution of novel HAdV types, it is important to understand the recombination mechanisms if adenoviral vectors are to be used in human patients, who might coincidentally be infected with a wild-type virus. Moreover, the occurrence of viral recombinants with lateral DNA and epitope transfers between HAdVs and SAdVs must be borne in mind when chimpanzee adenoviruses are considered as vectors for gene delivery in human patients to exploit the lack of immunoreactivity to these viruses.
PATHOGENESIS AND IMMUNITY
Prevalence of HAdV Species and Types
Most HAdV species appear to circulate globally, but predominant types differ between countries or geographic regions, and they change over time (58–60). Transmission of new strains across continents may occur and lead to replacement of hitherto dominant HAdV types (61). The adenoviruses most commonly reported to be associated with human disease worldwide are HAdV-C1, -C2, -C5, -B3, -B7, -B21, -E4, and -F41 (20, 62–66). In immunocompromised patients in the transplant setting, some of the most commonly reported adenovirus types include HAdV-C1, -C2, -C5, -A12, -A31, -B3, -B11, -B16, -B34, and -B35, with a strong predominance of species C in most instances (67–70). For example, in the transplant unit of St. Anna Children's Hospital, Vienna, Austria, HAdV species C accounts for about 80% of all adenoviral infections observed, and similar numbers were also reported from other transplant centers in different geographic regions (43, 69, 71–73). Sequential or concomitant coinfections with different adenoviruses from the same or different species are quite commonly observed in both the immunocompetent and immunocompromised patient settings (74–76) and may thus play a role in the generation of recombinant HAdV types.
Transmission
Infections in the immunocompetent host are typically caused by exposure to infected individuals via inhalation of aerosolized droplets or direct conjunctival inoculation, but transmission may also occur by fecal-oral spread, including contact with recreational freshwater or tap water, infected tissue, airflow filters, or environmental surfaces (77–81). The stability of the virus at low pH is a matter of debate, but HAdVs are resistant to gastric and biliary secretions and can therefore be detected at high levels in feces (82). Moreover, HAdVs can retain their infectious properties even after several weeks in moisture-free environments, and because they are nonenveloped viruses, they are resistant to many disinfectants. Treatment of surfaces with alcohol solutions (85 to 95%) for at least 2 min or with sodium hypochlorite for 10 min is effective at inactivating the virus (83). Efficient decontamination of surfaces is of paramount importance, particularly in transplant and intensive care units, to prevent this mode of transmission in immunosuppressed patients (84). Although exogenous infection by nosocomial or community acquisition in the inpatient setting is a rather rare cause of HAdV-related diseases, outbreaks of infections on hematology or transplant wards as well as in eye clinics, resulting in closures, have been documented (19, 85, 86, 87).
Tissue Tropism
The general affinity of HAdV species for individual tissues is outlined above, but in particular, members of the largest species, species D, show great variability in their tropisms, with growth in tissues ranging from ocular to gastrointestinal (GI) and respiratory tissues (37, 88). The basis of tissue tropism is still not well established. Adenoviral keratoconjunctivitis, which is a major cause of ocular morbidity, is most commonly caused by representatives of species D, including types 8, 19, and 37, but also by HAdV-E4, -C5, -B3, -B7, -B11, and -B14 (20). Gastrointestinal manifestations are mainly associated with HAdV-F40 and -F41, but HAdV-G52 and different members of species D, including some of the most recently identified types (types 65 and 67), have also been observed (20, 36, 43, 46, 89). Respiratory tract involvement has been associated mainly with HAdV-B3, -B7, -B16, -B21, and -E4 and various members of species C (43, 90). These examples indicate that certain adenoviruses have strong tropisms for specific tissues, but the same clinical manifestations can be caused by other HAdV types and species, thus requiring diagnostic screening methods with broad specificity.
Primary Infection and Persistence
Following HAdV transmission, the incubation period ranges from 2 days to 2 weeks, depending on the viral type and mechanism of acquisition, and the spectrum of clinical manifestations is broad (20). The majority of HAdV infections occur at a young age, and epidemics have been documented for both healthy children and adults in closed or crowded settings, including particularly military recruits (91–93). Vaccination programs for U.S. military trainees, covering the most commonly occurring HAdV types (types 4 and 7), were discontinued many years ago and were recently resumed with a newly available FDA-approved live oral vaccine against these two HAdV types. The vaccine comes as two tablets to be taken at the same time and is compatible with concomitant performance of other vaccinations. It is recommended by the Department of Defense for enlisted soldiers entering basic training but may also be encouraged for other military personnel at high risk for adenovirus infection. The vaccine is reported to prevent illness caused by these two virus types, with an efficacy of 99.3% (95% confidence interval [CI], 96.0 to 99.9%; P < 0.001), and the virus isolation rates fell dramatically after reinitiation of the vaccination program (94, 95). Updates on the vaccine are available at the website of the Centers for Disease Control and Prevention (www.cdc.gov/vaccines). Most HAdV epidemics in immunocompetent individuals are observed in winter and early spring, but infections in immunocompromised patients occur throughout the year (96, 97). Epidemiological data indicate that the majority of primary HAdV infections occur during the first 5 years of life, due to the lack of humoral immunity. In children, HAdV infections account for up to 15% of upper respiratory tract and about 5% of lower respiratory tract inflammatory diseases (98). In immunocompetent individuals, the infections are mostly mild and self-limiting, but severe and even fatal courses have been reported (26, 99, 100).
Owing to their genetic heterogeneity, HAdVs diplay broad tissue tropism and can infect several cell types. Not surprisingly, therefore, currently available evidence indicates that they can persist in a latent state in a variety of susceptible cells following primary infection. Latency is characterized by expression of viral proteins by the host cell without replication of a complete virus. A latent form of adenovirus infection was shown to persist in tonsillar lymphocytes in nearly 80% of children investigated, and the number of adenoviral genomes per lymphoid cell apparently declines with age (101–103). Moreover, latent HAdV infections were described to occur in intestinal T lymphocytes and in lung epithelial cells, where they seem to play a role in the pathogenesis of obstructive airway disease (104). Other sites of HAdV persistence have also been suggested, but the experimental evidence is limited (105–107).
Previous observations indicated that the central nervous system is apparently a sanctuary for adenoviral persistence (5), and recent findings revealed that the entire GI tract is a common location of HAdV persistence in children (unpublished data). Evasion from immune surveillance is a prerequisite for the establishment of persistent infections in permissive cells and tissues. Immune escape of adenoviruses can be mediated by different mechanisms. Specific viral proteins can block responses to anti-inflammatory and cytolytic cytokines, intrinsic cellular apoptosis, and innate and adaptive cellular immune responses (108, 109). Moreover, the viral protein E3 can downregulate major histocompatibility complex (MHC) class I molecules, thereby affecting antigen presentation and reducing T-cell attack of the infected cells (110–112).
Immune Responses to Adenoviral Infection
Similar to other viruses, HAdV is controlled by innate and adaptive immune responses (113). Rapid secretion of antiviral cytokines, such as gamma interferon (IFN-γ), tumor necrosis factor (TNF), interleukin-1 (IL-1), IL-2, and macrophage inflammatory protein, is triggered by HAdV and targets different steps in the viral life cycle, thereby limiting the amplification and spread of the virus. Moreover, innate effector cells, particularly natural killer cells, which can destroy virus-infected cells in a nonspecific fashion, are recruited and activated (104, 113). The effect of infection-induced cytokines is counteracted by viral products, such as the HAdV-encoded E1B 55-kDa protein. This protein mediates transcriptional repression of IFN-inducible genes, thereby facilitating viral replication (114).
In addition to providing the first line of defense, the innate immune system supports proliferation and differentiation of the adaptive immune response mediated by T and B cells. The generation of HAdV-specific T cells facilitates lysis of infected cells by a perforin-dependent mechanism (115). Although the large number of existing HAdV types implies that the expression of antigens that represent potential T-cell targets can be expected to be highly polymorphic, T cells raised against HAdV, including the CD4 and CD8 subsets, were shown to display cross-reactivity with different adenoviral species (116). These observations indicate that such T cells recognize conserved sequences of amino acid residues from a structural protein of HAdV (117). Indeed, one of the most important, immunodominant T-cell targets is the adenoviral hexon protein, which contains generic antigenic components common to all adenoviral species (118). Hence, exposure to adenoviruses during childhood and the ensuing generation of cross-reactive cytotoxic T cells are believed to lead to broad HAdV immunity in adults (119–121). Healthy individuals usually carry HAdV-specific T cells, which can be identified by various methods, such as gamma interferon secretion assays, cytokine flow cytometry, or detection of MHC class I multimers (118, 122, 123). The absence of HAdV-specific T cells has a negative impact on the course of HAdV infections, and conversely, reconstitution of the HAdV-specific T-cell response correlates with viral clearance (122, 124). The finding that many CD4- or CD8-restricted hexon epitopes are shared among different HAdV species and types suggests that T cells with such specificities can be protective against most, if not all, human adenonoviruses, and this fact can be exploited for vaccine-based or adoptive T-cell transfer immunotherapy for treating infections by these viruses, as outlined below.
De Novo Infection and Viral Reactivation in Transplant Recipients
In the allogeneic transplantation setting, adenoviral complications can arise from de novo infection or reactivation of persistent endogenous HAdV. Exogenous infection can occur by virus transmission from the donor via the graft or from the environment (125, 126). The occurrence of outbreaks on transplant wards demonstrates the role of environmental sources in HAdV spread (86; unpublished observations). However, endogenous reactivation of persistent HAdV appears to be the predominant cause of HAdV-associated disease in severely immunocompromised patients. This notion is supported by the absence of a seasonal pattern of infections in this setting and the finding that the HAdV strain detected prior to allogeneic hematopoietic stem cell transplantation (allo-HSCT) is generally identical to the strain isolated during the posttransplant period (126, 127). The presence of high neutralizing antibody titers against specific HAdV types before allo-HSCT permitted prediction of reactivation and viral disease caused by the same HAdV type (126). Detection of HAdV DNA in feces or nasopharyngeal aspirates of the recipient prior to HSCT has been reported as a risk factor for viral dissemination after HSCT (69, 128). Although HAdV reactivation in the immunocompromised setting could conceivably occur at different sites, observations in pediatric allo-HSCT recipients made over a period of more than 15 years suggest that viral proliferation preceding invasive infection almost invariably occurs in the GI tract (69, 71). The monitoring of viral loads in serial stool samples during the posttransplant period therefore permits timely assessment of impending disseminated disease (69), as outlined below.
RISK FACTORS, INCIDENCE, AND CLINICAL MANIFESTATIONS OF INVASIVE HAdV INFECTION IN IMMUNOCOMPROMISED PATIENTS
Risk Factors
Major factors conferring a high risk of invasive HAdV infection and disseminated disease include allogeneic stem cell (or organ) transplantation and any severe immunosuppression with a lack of cellular antiadenoviral activity. More specifically, the most prominent risk factors include allogeneic transplantations with in vivo and/or ex vivo T-cell depletion, grafts from unrelated donors or cord blood, treatment with the anti-CD52 antibody alemtuzumab (Campath) or anti-thymocyte globulin (ATG), and the presence of graft-versus-host disease (GvHD) grades III and IV associated with the use of immunosuppressive agents (129–136). Additionally, severe lymphopenia, with CD3+ cell counts of <300 per μl peripheral blood (PB), and an absence of HAdV-specific T cells play an important role in the development of viral disease (122, 124, 137–141). In contrast to the case with allo-HSCT recipients, a donor-positive and recipient-negative HAdV serostatus appears to be a risk factor for a severe course of infection in patients undergoing solid organ transplantation (43, 142, 143).
Incidence of HAdV Infections in Immunocompromised Adult and Pediatric Patients
In individuals with various congenital immunodeficiencies, particularly severe combined immunodeficiency (SCID) syndrome, severe and recurrent pulmonary HAdV infections, and even lethal disseminated disease, are not uncommon (20, 144), with reported fatality rates reaching up to 55% (43). In contrast, life-threatening disease currently appears to be relatively rare in acquired immunodeficiency associated with HIV infection (145, 146), where HAdV infections are mostly associated with acute diarrhea only (147). Other clinical manifestations in this setting have become rather exceptional (148, 149), which is attributable to the availability of highly effective antiretroviral treatment strategies (150). Before the era of effective antiretroviral therapy, a number of authors reported severe and fatal cases in patients with HIV/AIDS, associated with pneumonia, hepatitis, nephritis, meningoencephalitis, and disseminated disease (151, 152).
For patients undergoing chemotherapy for malignant diseases, respiratory infections caused by HAdV have been documented during phases of neutropenia, and lethal HAdV disease has been reported for children receiving chemotherapy for acute lymphoblastic leukemia and adults treated with alemtuzumab (153, 154). In solid organ transplant (SOT) recipients, HAdV infections can be asymptomatic, but prolonged and severe courses affecting morbidity, graft loss, and mortality may occur (155, 156). Infections can be acquired de novo or via reactivation of latent virus from the recipient or the transplanted organ (157). The occurrence of adenoviremia has been reported to be less than 10% of adult patients after kidney, heart, or liver transplantation; although the symptoms of HAdV infection are usually mild, and invasive infection does not correlate with organ rejection (157), severe and even fatal courses have also been described (158, 159). Studies in patients undergoing lung transplantation showed that pulmonary infection with HAdV can correlate with significantly elevated rates of rejection, bronchiolitis obliterans, and mortality (160–162). In line with the epidemiology of HAdV infections, detection of this virus appears to be more common in pediatric SOT recipients (155, 163), with reported rates of HAdV infection ranging from 3.5% to 38% after liver transplantation (164, 165), from 7% to 50% after lung and heart transplantation (166–168), and from 4% to 57% after intestinal or multivisceral transplantation (169, 170). In children after small bowel transplantation, biopsy specimens often revealed the presence of HAdV, but the occurrence of virus-related disease seemed to correlate primarily with the intensity of immunosuppressive treatment (170).
In the autologous HSCT setting, HAdV infections seem to be a rare event (171), while allogeneic HSCT represents a major risk factor. In allo-HSCT recipients, young age was shown to confer an elevated risk of HAdV infection (127), and life-threatening disease was invariably associated with adenoviremia (or, more precisely, HAdV DNAemia, because detection is generally based on PCR-based analysis) (71, 172). The incidences of HAdV DNAemia reported for pediatric allo-HSCT recipients range from 6% to 42% (127, 173). In the adult allo-HSCT setting, the incidences of HAdV DNAemia are apparently lower, ranging from 3% to 15% (174, 175).
Definitions of Adenoviral Infection and Disease
In the past, different groups have proposed definitions of localized and disseminated adenovirus infection as well as probable and proven/definite adenovirus disease based on various technical approaches to virus detection (69, 176–178). Owing to the fact that highly sensitive techniques, based primarily on PCR, have become the gold standard for the detection and monitoring of HAdV infections, the European Conference on Infections in Leukemia (ECIL) recently recommended the following definitions (143): (i) local infection—positive HAdV PCR, virus isolation, or antigen detection in biopsy material or fluids other than peripheral blood; (ii) systemic (invasive) infection—positive HAdV PCR (viremia/DNAemia), virus isolation, or antigen detection in peripheral blood; (iii) probable disease—HAdV infection plus corresponding symptoms and signs without histological confirmation; and (iv) proven disease—HAdV infection plus corresponding symptoms related to the infection, with histological confirmation of HAdV infection in the appropriate location.
Moreover, in previous studies, intestinal adenovirus disease was defined as reproducible detection of HAdV in stool specimens at levels detectable and quantifiable by real-time PCR, together with enteritis and in the absence of other infections or GvHD, while the mere presence of HAdV in stool was regarded as virus shedding only. Disseminated HAdV disease was defined as disease with multiple-organ involvement (e.g., hepatitis, encephalitis, and retinitis) in the presence of two or more HAdV-positive PCR assays for peripheral blood and other sites tested (e.g., cerebrospinal fluid, bronchoalveolar lavage [BAL] fluid, respiratory secretions, or urine), in the absence of other identifiable causes. HAdV-associated death was defined as multiple-organ failure in the presence of increasing or persisting adenoviral loads in peripheral blood, in association with AdV detection from multiple other sites (69, 71).
Clinical Presentations and Outcomes of HAdV Infections in Solid Organ and Allogeneic Stem Cell Transplant Recipients
The most common occurrence of HAdV disease is observed between 2 and 3 months posttransplantation, and the first symptoms include fever, enteritis, elevated liver enzymes, and secondary pancytopenia (82). In SOT recipients, the transplanted organ is often the primary site of HAdV-related disease. Clinical manifestations reported for patients receiving lung, liver, kidney, or small bowel transplants include pneumonia, hepatitis, nephritis, hemorrhagic cystitis, enteritis, and disseminated disease (97). The manifestations tend to be more severe in pediatric transplant populations, with reported mortality rates occasionally exceeding 50% (179), and surveillance of HAdV loads in peripheral blood may be instrumental for identifying patients requiring antiviral treatment to prevent fatal disease (159, 164, 180). In adult patients undergoing SOT, the incidence of viremia is lower, and the presence of HAdV is often transient and self-limited, with asymptomatic clinical courses. Routine surveillance of HAdV by PCR therefore does not seem to be indicated for adult organ transplant recipients (157, 181).
Data on HAdV infections in the hematopoietic stem cell transplantation setting are far more abundant. In allo-HSCT recipients, the spectrum of HAdV-associated diseases can range from mild gastroenteric or respiratory symptoms to severe manifestations, including hemorrhagic enteritis or cystitis, pneumonia, hepatitis, nephritis, encephalitis, myocarditis, and, occasionally, concomitant involvement of several organs, which may lead to a lethal outcome by multiorgan failure (69, 182, 183). Postmortem investigation of the affected organs, including the liver in particular, reveals massive replication of the virus, with lysis of the infected cells and release of viral particles into peripheral blood (184), underlining the diagnostic relevance of viremia. In a study performed at our institution in the pediatric allo-HSCT setting, transplant-related mortality associated with HAdV reached 6% of the entire patient cohort investigated (69, 71), but fatal disease attributable to HAdV infection has been reported in up to 50% of patients with DNAemia (69, 133). The majority of transplant-related deaths attributable to HAdV infection occur within the first 100 days posttransplantation (69).
It is important to emphasize that the occurrence and overall mortality of HAdV infections are apparently lower in adult patients undergoing allo-HSCT (<1%) but can also be very high in the presence of HAdV DNAemia (133, 175, 185). A fatal outcome is particularly frequent in cases of DNAemia associated with disseminated disease, with reported lethality rates reaching up to 60 to 80% for both children and adults (69, 71, 131, 186).
Previous observations indicate that the clinical courses of invasive HAdV infection in children undergoing allo-HSCT can be fulminant, with a fatal outcome within a few days after onset of the first clinical symptoms of viral disease (69). Timely start of treatment is important for successful control of HAdV infections in immunocompromised patients, but an immediate availability of effective therapeutic strategies is still limited. Rapid and reliable diagnosis of impending HAdV disease is therefore of paramount importance.
Predictive Value of Viremia and HAdV Proliferation in the Gastrointestinal Tract
Since spread of the virus into peripheral blood is a characteristic sign of disseminated HAdV disease and viral load values correspond to the severity of organ pathology (187), quantitative monitoring in plasma or serum has become an essential screening tool after allo-HSCT. Additionally, there is growing evidence that HAdV detection and surveillance of virus proliferation kinetics in stool provide early information on impending invasive infection and HAdV disease (69, 188, 189). Viral persistence in the GI tract and shedding of HAdV into feces are common findings which, in previous experience, occur in more than one-third of pediatric patients after allo-HSCT and may not necessarily be associated with clinical symptoms of intestinal infection (69). Detection and quantitative surveillance of HAdV in serial stool samples revealed two distinct patterns indicating the risk of invasive infection. Patients displaying very slow or absent proliferation kinetics in serial analyses, with maximum HAdV loads below 106 virus copies per gram of stool, apparently have a very low risk of adenoviremia. In contrast, patients showing rapid proliferation, exceeding the threshold of 106 virus copies per gram and sometimes revealing extremely high viral loads of >1011 copies per gram, have a very high risk of experiencing viremia and disseminated disease. In a study performed at our center, viremia occurred in more than 70% of pediatric patients with these findings, whereas none of the individuals with maximum viral loads below the indicated threshold experienced invasive HAdV infection (69). These observations were recently confirmed by other groups (188, 189) and are expected to have important implications for future diagnostic and treatment strategies.
DIAGNOSIS AND MONITORING
Diagnostic Screening
Conventional approaches to HAdV detection in affected samples, such as peripheral blood, stool, urine, BAL fluid, nasopharyngeal aspirates, or swabs, include primarily immunofluorescence staining for antigen detection and viral culture (43, 97, 190, 191). However, due to the limited sensitivity and, in case of viral culture, rather long time to readout, these methods have largely been supplanted in routine clinical diagnostics by molecular screening approaches generally relying on PCR-based techniques (71, 97, 192). Owing to the superior sensitivity and specificity of molecular tests, facilitating equally effective detection of all HAdV types in any diagnostic material, PCR assays have become a standard screening tool (143).
Despite the predominance of certain HAdV species in specific clinical settings, including immunocompromised patients, employment of broad-spectrum HAdV screening assays is necessary in order to permit reliable detection, even of rarely occurring HAdV species and types, with adequate sensitivity (Fig. 1). Several groups have established such “pan-adenoviral” assays based on PCR, exploiting the sequence information available at the respective time points (73, 192–195). HAdV screening assays target conserved regions within the HAdV genome, most commonly within the hexon gene, but the inclusion of additional target regions, e.g., within the fiber gene, may be required to ensure reliable detection of all known types with comparable sensitivities (194). Due to the fact that the spectrum of newly identified HAdV types has been expanding based on the implementation of genomic analyses, established assays need to be updated in order to facilitate reliable coverage of the entire range of human adenoviruses. Since newly identified HAdV types generally result from recombination events within the same or different human-specific species of the virus (34), the target regions of established PCR assays are preserved in most instances, thus permitting equally sensitive detection of the new recombinants. Nevertheless, this issue requires careful attention, as exemplified by the HAdV screening assay established at our center in 2005, based on the sequence information accessible at that time (194). The test was originally demonstrated to cover all 51 known HAdV (sero)types with comparable detection limits. Alignment with genomic sequences of all newly published HAdV types revealed that the current primer-probe combinations of this real-time PCR assay can be expected to reliably cover nearly all hitherto identified HAdV types, with two exceptions. The sequence of HAdV-A61 revealed a few mismatches in the target region of the downstream primer, possibly affecting the sensitivity of detection. This finding required the addition of an appropriately modified primer to the reaction mix, with subsequent confirmation of this adaption in vitro. The second exception was HAdV-G52, which displays the greatest similarity to a simian adenovirus (SAdV-1) and is not reliably covered by the assay. This example highlights the need to control and adequately adapt established diagnostic assays based on newly identified HAdV types if the test is expected to serve for “pan-adenoviral” screening. The availability of complete genomic sequences of all currently known HAdV types greatly facilitates appropriate modifications of established assays and the development of novel tests. The same requirements apply to commercial HAdV assays which are FDA approved in the United States and/or CE marked in Europe. A number of such kits have been introduced, including the Adenovirus R-gene kit (bioMérieux, Lyon, France), ELITe MGB kit (ELITech Group Molecular Diagnostics, Puteaux, France), FilmArray RP kit (BioFire Diagnostics, Inc., Salt Lake City, UT), eSensor RVP kit (GenMark Diagnostics, Carlsbad, CA), xTAG RVP Fast and xTAG RVPv1 kits (Luminex Molecular Diagnostics, Toronto, Canada), Prodesse ProAdeno+ assay (Hologic Gen-Probe, San Diego, CA), and Anyplex II RV16 kit (Seegene, South Korea), and this list may not be exhaustive. Most of the indicated kits cover multiple viruses and are only approved for qualitative analysis of respiratory specimens. Studies comparing the performances of such kits indicated a particularly high variability for HAdV detection, with inadequate identification of certain HAdV types by some of the tests (196–198). The monitoring of patients in the immunocompromised setting requires tests permitting reliable detection of all potentially relevant HAdV types in different clinical specimens, as well as accurate quantitative assessment of viral loads. Among the currently available commercial kits, this requirement appears to be met by the Adenovirus R-gene kit (199), and possibly the ELITe MGB kit, although the latter seems to be approved for whole-blood analysis only.
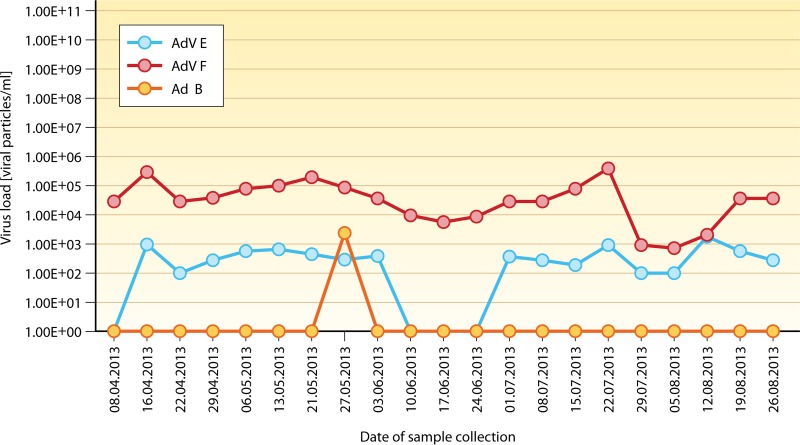
FIG 1.
Adenoviremia with multiple HAdV species. Different HAdV types or species can be detected in peripheral blood and/or other sites, both concomitantly and sequentially. The example displayed shows a rare constellation of invasive infection, associated with HAdV species E, F, and occasionally B, during the posttransplantation course (x axis) of a pediatric patient (86, 269). This observation highlights the fact that very unusual findings are possible, which must be accounted for by the implementation of appropriate diagnostic techniques for HAdV detection and monitoring. The y axis indicates the virus copy number per ml of blood determined by real-time PCR.
It is important to note that the lower detection limit for HAdV detection in clinical specimens, particularly peripheral blood, should be in the range of 102 virus copies/ml in order to prevent false-negative test results and to permit early initiation of treatment according to some published guidelines (130).
Relevance of HAdV Detection and Quantification at Specific Sites
A prospective study of pediatric patients undergoing allo-HSCT, which focused on screening of HAdV at different sites (specimen types) including the throat, stool, urine, peripheral blood, and, occasionally, other locations, revealed that detection of adenovirus at multiple (i.e., more than 2) sites reflects the presence of invasive infection (71). This observation was in line with earlier reports (200, 201), but the study revealed that peripheral blood is the only infection site indicative of a high risk of disseminated disease. A number of studies provided similar findings (73, 172, 175, 182, 186, 192), rendering peripheral blood the most important source for clinical surveillance of HAdV infections in the immunocompromised setting. The time point of HAdV DNAemia may have prognostic relevance: in studies performed at our center, patients showing DNAemia before day 100 after allo-HSCT developed life-threatening disseminated disease in more than 60% of instances, despite antiadenoviral treatment, whereas later onset of viremia did not seem to be associated with disseminated HAdV disease (69, 71).
Interestingly, other reports suggested that HAdV detection in nasopharyngeal aspirates of children prior to allo-HSCT is a strong predictor for ensuing adenoviremia and may therefore provide an indication for postponement of transplantation, if possible (128, 130). In fact, HAdV may be persistently detectable in nasopharyngeal secretions in some pediatric patients (202, 203), and these observations therefore require careful consideration.
Despite the clear diagnostic and prognostic relevance of HAdV DNAemia, only a proportion of high-risk patients with this finding develop overt disease. A number of groups have therefore attempted to identify virus load levels that could serve as a rational basis for the start of preemptive antiviral treatment. However, the thresholds suggested by different authors are highly divergent, ranging from 102 (in individuals with high risk) to >106 copies/ml (130, 135, 172, 175, 186, 204, 205), and it is difficult therefore to draw generally applicable conclusions. Moreover, the measured absolute values are, at least to some extent, dependent on the individual technique used, and in the absence of appropriate interlaboratory standardization, the values cannot readily be adopted by or exchanged between centers. Other authors have shown that rapidly rising viral loads are detectable in peripheral blood prior to the onset of clinical symptoms of HAdV disease, suggesting that the monitoring of viral titer kinetics may be a more readily applicable parameter (71, 180). Moreover, in addition to facilitating prediction of HAdV-related disease, surveillance of HAdV titer kinetics in peripheral blood is also instrumental for assessment of the response to therapy (69, 130, 206). In previous studies, a decrease in viral load of at least 1 log within 2 to 3 weeks of antiviral treatment was regarded as a minimum requirement for an adequate response (69, 71), but there are no generally accepted guidelines for diagnostic definitions of response at this time.
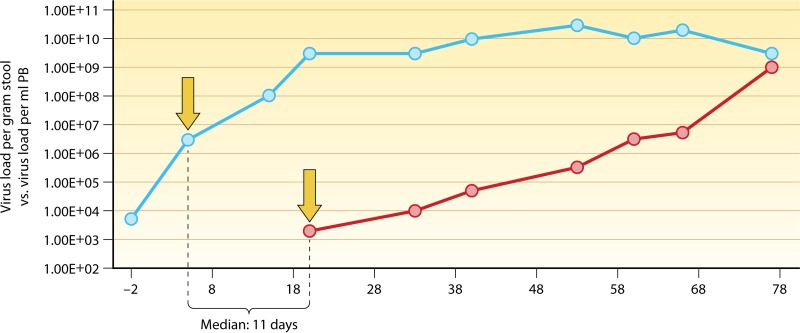
The relevance of HAdV proliferation kinetics and peak load levels in stool specimens from pediatric allo-HSCT recipients for risk assessment of invasive infection has already been indicated (see above). Since the timely initiation of antiviral therapy appears to be critical in this setting and starting treatment upon detection of HAdV viremia may be too late in a number of instances (69, 71, 207), it is essential to identify the earliest possible time points for rational initiation of therapy. In view of the high risk of invasive infection and disseminated disease in children displaying rapid HAdV proliferation kinetics in serial stool specimens, with peak levels exceeding 106 virus copies/g, systematic screening of intestinal excretions should be part of the diagnostic routine during the posttransplantation period. The median time span between detection of HAdV loads in stool exceeding the indicated threshold and the first appearance of the virus in peripheral blood was 11 days (69), thus providing a rational window of opportunity for early start of therapy, with the aim of preventing invasive infection (Fig. 2). Based on the available data and experience in the pediatric allo-HSCT setting, an algorithm for diagnosis and treatment of HAdV infections has been established (Fig. 3).
FIG 2.
Temporal correlation between intestinal HAdV infection and viremia. The median time span between the observation of rapidly rising HAdV copy numbers in serial stool specimens (blue line), exceeding the threshold of 106 virus copies per gram of stool (left arrow), and the first detection of viremia (right arrow; red line) was 11 days (69), providing a rational basis for early start of treatment.
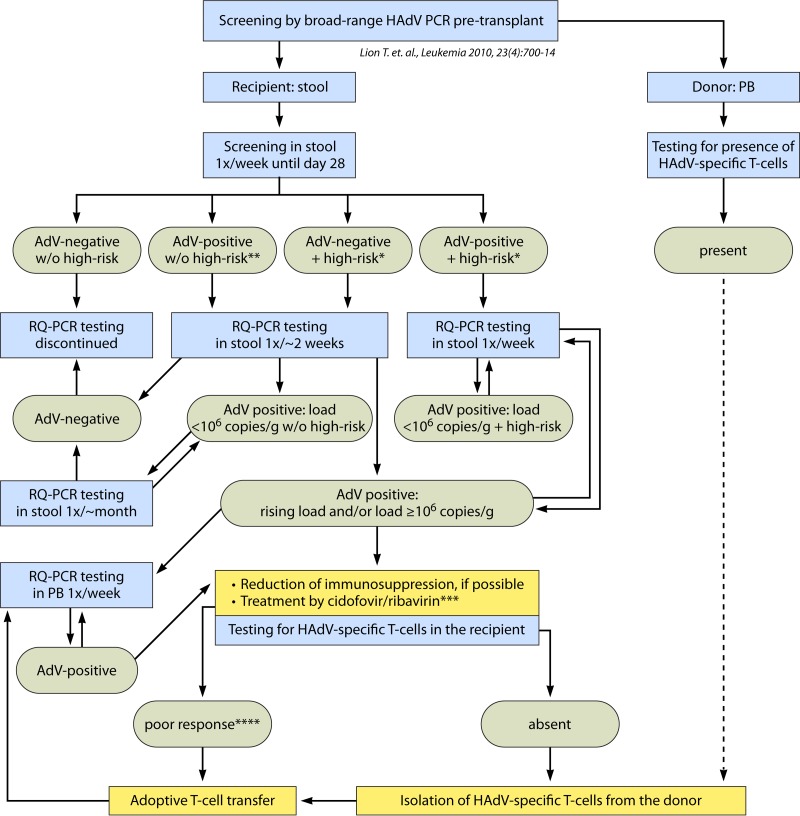
FIG 3.
Algorithm for diagnosis and treatment of HAdV infections. This algorithm was established based on the insights provided by studies performed at our center (69). It is important, however, that the indicated absolute threshold value of 106 virus copies/g of stool may be dependent on the specific real-time quantitative PCR (RQ-PCR) approach used in our study and may require adjustment when using other quantitative approaches. Initiation of antiviral therapy at the proposed preinvasive stage may inhibit or slow down proliferation of the virus until recovery of the immune system permits control of the infection. This approach may be instrumental in preventing life-threatening disseminated HAdV disease in individuals at high risk, while limiting the rate of overtreatment in patients after allogeneic HSCT. Screening of PB specimens is usually terminated after documentation of stable PCR negativity. However, the risk of relapse after successful treatment of adenovirus and resolution of HAdV DNAemia may be difficult to assess. Further molecular monitoring should therefore be based on the individual risk profile. In high-risk situations, continued monitoring of both stool and blood specimens may be warranted. *, high-risk parameters include T-cell depletion, GvHD (≥grade II), other, concomitant viral infections, and CD3+ counts of <300/μl PB; **, for patients who are HAdV positive in stool, with <103 virus copies/g after day 28, and do not display any high-risk features, the intervals of testing can be extended further; ***, ribavirin may be indicated only in the presence of HAdV species C; ****, <1-log reduction of viral load within ∼2 weeks of treatment. w/o, without. (Reprinted from reference 69 with permission.)
Adenovirus Typing
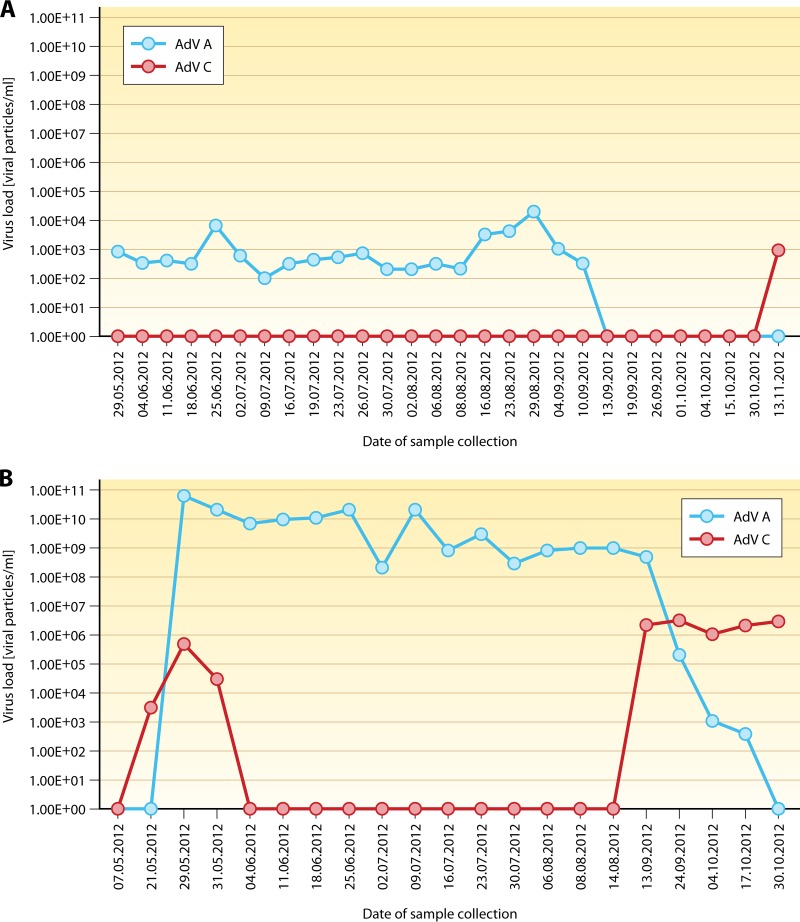
Identification of adenoviruses at the levels of species, (sero)types, and even strains is relevant for epidemiological studies and for precise documentation of nosocomial outbreaks. For the selection of optimal treatment, HAdV typing is currently of lesser importance because, with the exception of ribavirin (see below), available therapeutic strategies are independent of the HAdV species present. However, in view of the fact that different HAdV species and types may occur contemporaneously or sequentially in individual patients (52, 53, 72, 107, 208), typing may permit a better understanding of the dynamics and evolution of HAdV infection (Fig. 4).
FIG 4.
Course of adenoviremia with switch of HAdV species A to C. (Top) Kinetics of HAdV viremia during the posttransplantation course in a pediatric patient, revealing the disappearance of DNAemia caused by HAdV species A to below the detection level of real-time PCR, with a recurrence of HAdV positivity for a different HAdV species in peripheral blood after about 6 weeks of negative PCR findings. (Bottom) The switch observed in peripheral blood was preceded by corresponding kinetics of HAdV loads in serial stool specimens.
Typing was traditionally performed by serological methods (209), but these approaches have mostly been replaced by molecular techniques based on PCR amplification of specific target regions coupled with different detection formats, such as fragment length analysis, hybridization to species-specific probes, or sequencing (210–215). Molecular typing methods are more rapid and readily applicable and can provide better discriminatory capacity. Owing to the decreasing costs of next-generation sequencing (NGS) approaches, whole-genome analysis (36) may become the method of choice for detailed HAdV typing in the foreseeable future, even in the routine diagnostic setting.
Diagnostic Recommendations as a Basis for Preemptive Treatment
In adult organ transplant recipients, asymptomatic viremia is apparently common (≤22%), and the risk of progression to adenoviral disease in the presence of HAdV DNAemia with or without specific cutoff values for viral loads remains unknown (156). Routine screening for HAdV is therefore not recommended at present, although adenovirus infections in this setting can be severe and affect morbidity, mortality, and graft survival, particularly in young children (156).
According to the most recent ECIL guidelines for patients undergoing chemotherapy or autologous HSCT, monitoring is recommended only in cases of clinical suspicion of HAdV infection or disease (143). For allo-HSCT recipients, the ECIL recommends HAdV monitoring only for patients displaying at least one of the risk factors for HAdV disease (see above); in these instances, quantitative PCR monitoring of peripheral blood should be performed at weekly or shorter intervals until adequate immune reconstitution is established (143). Although the fact that molecular monitoring of viral loads in serial stool specimens can facilitate early detection of impending invasive HAdV infection is acknowledged by the ECIL, it has not been included in the current diagnostic guidelines because the level of available evidence was not deemed sufficient. Hence, detection of viremia (DNAemia) in the presence of at least one risk factor is presently the indication for initiation of preemptive antiviral treatment according to the ECIL criteria.
Somewhat older guidelines for preventing infectious complications in HSCT recipients, published on behalf of the Center for International Blood and Marrow Transplant Research (CIBMTR) and a consortium of several other societies, recommend weekly monitoring of peripheral blood for active HAdV infections by PCR during the first 6 months posttransplantation or the duration of severe immunosuppression/lymphopenia only for patients displaying the highest risk for adenoviral disease (216). This subset of patients includes individuals with refractory GvHD, recipients of T-cell-depleted stem cell grafts, haploidentical transplants, or umbilical cord transplants, and patients treated with anti-T-cell antibodies. However, these guidelines do not provide any critical value for viral loads that should trigger the initiation of therapeutic intervention.
According to another recently established guideline for preemptive treatment in the allo-HSCT setting, published on behalf of two institutions (Department of Pediatrics, University Utrecht, Utrecht, Netherlands, and the Center for Cell and Gene Therapy, Baylor College of Medicine, Houston, TX), weekly quantitative PCR monitoring of peripheral blood is also recommended, but the indication for preemptive treatment is linked to the detection of critical HAdV load levels, which vary depending on the individual patient risk for severe HAdV infection and an adverse outcome (130). The thresholds defined as critical range from >102 viral copies/ml for patients with high-risk features to >103 copies/ml for patients with intermediate risk and >104 copies/ml for individuals displaying a low-risk profile. These recommendations have raised questions pertaining to possible overtreatment, with associated side effects, when using very low thresholds of viral load as a basis for preemptive treatment (206). Conversely, other observations suggest that initiation of treatment upon detection of viremia at any level might be too late for successful control of the disease in various instances (69, 71). Diagnostic findings from serial stool specimens revealing rapid HAdV proliferation, with peak levels of >106 virus copies/g, could therefore serve as a rational basis for an earlier start of preemptive therapy, before the infection becomes invasive (69) (Fig. 3). However, more clinical data on the success of preemptive treatment based on the indicated recommendations are needed to provide unequivocal support for the currently available diagnostic guidelines.
CURRENT TREATMENT MODALITIES
Present recommendations for treatment of HAdV infections focus on immunocompromised patients, particularly allogeneic transplant recipients, who apparently carry the greatest risk of severe and life-threatening clinical courses. The approaches pursued may include prophylaxis, preemptive treatment based on virus detection prior to onset of clinical symptoms, sometimes linked to specific thresholds of viral load, or therapeutic (symptomatic) treatment in the presence of virus-related disease. At present, there is little evidence for a beneficial effect of HAdV prophylaxis, and the ECIL does not recommend prophylactic antiviral therapy with currently available virustatic drugs (143). For SOT recipients, the treatment indication for mild or asymptomatic HAdV infection is not clear, since prospective studies have shown that adenoviremia may be present without any clinical symptoms and may clear spontaneously (157). Some authors therefore recommend antiviral treatment only for symptomatic patients (20). In contrast, for patients undergoing allo-HSCT, preemptive treatment is strongly advocated by all major guidelines in order to inhibit or slow down viral replication, with the aim to prevent overt disease until immune reconstitution from the allograft permits clearance of the infection (130, 143, 216). The principal options for preemptive treatment include (i) the tapering of immunosuppressive therapy, which should be performed whenever possible; (ii) use of antiviral drugs; and (iii) immunotherapy in case of failure of the previous lines of treatment.
Antiviral Drugs
Most evidence for the in vivo efficacy of antiviral therapy against HAdV in the preemptive setting is available for cidofovir (130, 142, 177, 217–221), but the clinical effect of the drug as treatment for overt viral disease is apparently limited (134, 222). The compound is a nucleotide analog of cytosine that preferentially inhibits viral DNA polymerase and viral replication by more efficient competitive incorporation into DNA (223). Although resistant mutants have been described in vitro, cidofovir apparently displays efficacy against all HAdV species (224–226), and it is currently the primary anti-HAdV agent for preemptive therapy (69, 130, 143). It is used as induction therapy at a dose of 5 mg/kg of body weight/week for 2 weeks and at 2-week intervals thereafter (143). Alternatively, a schedule of 1 mg/kg three times a week has been suggested (130), and the required duration of therapy is linked to the clinical and molecular response, determined by a rather individually defined reduction of viral load (69, 130). The clinical results of preemptive treatment with cidofovir in the context of allo-HSCT are controversial, with some studies reporting success rates of ∼70% or more and others reporting rather poor responses (69, 130, 177, 219, 222, 227, 228). The limitations of treatment with cidofovir include its low bioavailability and poor correlation of pharmacologic effects with the prescribed dose (229). Moreover, cidofovir can display a dose-limiting nephrotoxicity, and frequent monitoring of renal and tubular function and concomitant hydration and uroprotection with probenecid are recommended (97, 130, 143).
Ribavirin is a nucleoside analog of guanosine that displays in vitro activity against DNA and RNA viruses, and the mechanisms of action may include inhibition of viral polymerases, viral RNA capping, and an increased mutation rate in newly synthesized DNA (230). Analysis of HAdV isolates revealed a consistent sensitivity of all types belonging to species C only (224), and the evidence for therapeutic efficacy of the compound in vivo is controversial (225, 230–233). Ribavirin is therefore not generally recommended for treatment of HAdV infections (143, 234). However, despite the conflicting results on the activity against HAdV in vivo, the low nephrotoxicity of ribavirin and the documented in vitro efficacy against HAdV species C may justify its use in specific clinical situations (235, 236). Oral, intravenous, and aerosol therapies with ribavirin have been used (237–239), and the compound has been applied at our center, in the pediatric allo-HSCT setting, at a dose of 20 mg/kg in combination with cidofovir as preemptive therapy in the presence of infections caused by representatives of HAdV species C (69). However, the actual clinical benefit of this treatment remains unclear.
Ganciclovir is a synthetic analog of 2′-deoxyguanosine which requires phosphorylation to ganciclovir monophosphate by a viral kinase and, subsequently, formation of ganciclovir diphosphate and triphosphate, catalyzed by cellular kinases. Ganciclovir triphosphate is a competitive inhibitor of dGTP incorporation into DNA and preferentially inhibits viral rather than cellular DNA polymerases. Moreover, it is a poor substrate for chain elongation, thereby disrupting viral DNA synthesis. A possible benefit of ganciclovir against HAdV infections in allo-HSCT recipients has been suggested (240). However, since adenoviruses (in contrast to members of the herpesvirus family) lack viral thymidine kinase, and cellular kinases are inefficient at phosphorylating the compound, the anti-HAdV efficacy of ganciclovir is predictably modest (230). Based on current data, there appears to be no justification for recommending the use of this drug for HAdV treatment (69, 130, 143).
Among other antiviral agents tested, the pyrophosphate analog foscarnet was demonstrated to display no activity against HAdV (230). A relatively recently introduced compound, brincidofovir (1-O-hexadecyloxypropyl-cidofovir; formerly known as CMX001), is an orally bioavailable lipid conjugate of cidofovir displaying substantially less nephrotoxicity than that of the parent drug. The compound has been employed successfully for treatment of viral infections in allo-HSCT recipients and other settings (241–243), but mutations conferring resistance may arise (244). Preliminary observations on the efficacy against HAdV showed promising results (245, 246), and clinical development of the drug is currently ongoing.
Immunotherapy
Measures supporting T-cell immunity play an important role in the armamentarium against invasive HAdV infections. This is attributable to the current limitations of antiviral chemotherapy and the evidence that T-cell recovery with reconstitution of HAdV-specific immune responses is essential for effective clearance of invasive infections. The initial step should therefore include reduction of immunosuppressive treatment whenever possible, as indicated above. Moreover, the transfer of HAdV-specific T cells from the original stem cell donor or third-party donors may represent the most effective currently available treatment option (143).
The proportion of HAdV-reactive T cells within the entire lymphocyte population of individuals who have been exposed to the virus is low. Infusion of unselected donor lymphocytes (DLI) can still provide antiviral immunity (247), but the potentially high frequency of alloreactive T cells and the ensuing side effects are major impediments to this approach (130). The isolation of HAdV-specific T cells from peripheral blood of the original stem cell donor has therefore become the method of choice for treatment of HAdV infections in allo-HSCT recipients not responding to antiviral chemotherapy, and different approaches to the generation of such cells have been established. Regardless of the HAdV species the donor has been exposed to, the HAdV-reactive T cells are expected to be cross-reactive with all HAdV types, because the hexon, the main constituent of the viral capsid, is the immunodominant T-cell target, containing several epitopes that are conserved among adenoviruses (116, 248). One of the early successful attempts of adoptive T-cell transfer was based on the isolation of donor-derived mononuclear cells, their stimulation ex vivo with HAdV antigen, and magnetic separation of reactive T cells secreting IFN-γ, which included both CD4+ and CD8+ T cells. The cells were infused without further in vitro expansion, and the results indicated that the efficacy of this treatment does not depend on the dose of infused cells, because even very small numbers of HAdV-specific donor-derived T cells expanded easily in vivo in the presence of the constant antigen challenge mediated by the viral infection (207). The most critical parameter for the success of treatment was appropriate timing, i.e., early T-cell transfer upon detection of viremia (207). These observations indicate that rapid availability of T cells for adoptive transfer, based on methods requiring only short or no in vitro expansion, is essential. Based on this notion, a number of different approaches to selection of HAdV-specific T cells have been introduced. The considerable variety of approaches includes, for example, different types of MHC multimers facilitating clinical-grade enrichment of HAdV-specific or multivirus-specific T cells displaying low or absent alloreactivity (249, 250). The MHC multimer technology requires knowledge of immunodominant human leukocyte antigen (HLA)-restricted peptide epitopes and facilitates the isolation of antigen-specific CD8+ T cells (MHC class I multimers) or CD4+ T cells (MHC class II multimers) of high purity (251). Short-term in vitro expansion under good manufacturing practice (GMP) conditions can render adoptive T-cell transfer available in less than 2 weeks (123, 252–254). In addition to a variety of methods based on the isolation of HAdV-specific T cells from the original stem cell donors, several approaches exploiting third-party donors are emerging (141, 255–263). Allogeneic third-party donors are a particularly important alternative option for cord blood recipients, for patients receiving allografts from HAdV-seronegative donors, and for solid organ transplantations from cadaveric donors, where donor blood is not available. Healthy seropositive individuals have been exploited to generate partially HLA-matched virus-specific T cells (VSTs) for adoptive immunotherapy (251). However, clinical implementation of this approach requires the availability of a large pool of HLA-typed healthy donors, and the use of incompletely HLA-matched T cells bears the risk of complications resulting from alloreactive side effects (264). Despite the existing concerns, this approach appears to be feasible, and current clinical results are encouraging (265). Another recently presented methodology is the generation of VST cell lines from healthy donors with common HLA polymorphisms (266). The employment of banked third-party-derived VSTs was demonstrated to represent an additional safe and readily applicable strategy for rapidly available treatment of severe viral infections in allo-HSCT recipients (266), thus further expanding the spectrum of clinical options for effective immunotherapy in immunocompromised patients. Although adoptive transfer of HAdV-specific T cells is currently one of the most promising treatment approaches for high-risk patient populations, it still needs to be regarded as experimental and should only be performed in the context of clinical trials in specialized centers.
SUMMARY AND PERSPECTIVES
Despite the considerable progress in diagnosis and treatment of adenoviral infections over the past years, a number of relevant issues remain to be solved. The steadily growing spectrum of HAdV types emanating from genomic analyses is a challenge for molecular screening assays. Although newly identified types of the virus generally result from recombination events between known HAdV representatives, and the sequences targeted by PCR tests may be preserved, it is necessary to test and occasionally adapt the assays to ensure adequate coverage of existing HAdV types. Complete HAdV genome sequences are now available in public databases, thereby permitting at least in silico control of the primers and probes used to ensure coverage of any newly identified HAdV type by the assay used. However, in view of the clinical importance of adenoviral infections, the availability of commercial, regularly updated diagnostic tests (199, 267) is highly desirable to facilitate standardized diagnostics under stringent quality control conditions. Commercial assays should permit pan-HAdV screening, quantification of viral loads, and, ideally, typing at least to the species level in order to provide a basis for more specifically targeted treatment approaches (251, 268). It is conceivable that easier access to next-generation sequencing facilities and decreasing costs of analysis will enable detailed HAdV typing, even in the routine clinical setting, in the near future.
Current recommendations for adenovirus screening and monitoring as a basis for preemptive treatment in patients at high risk for HAdV disease are still relatively diverse (69, 130, 143), and further studies are needed to provide reliable data permitting the establishment of standardized approaches. Optimized diagnostics will greatly affect the rational and timely initiation of antiviral treatment, which was shown to be a prerequisite for successful therapy. Despite current recommendations for preemptive administration of antiviral drugs, unequivocal evidence for a beneficial effect on mortality is still missing, and appropriate prospective studies are needed. The introduction of novel antiviral agents can be expected to further improve the efficacy of treatment and to reduce the toxicity of some commonly prescribed virustatic drugs. In this regard, the orally bioavailable and less nephrotoxic lipid ester of cidofovir, brincidofovir, could provide an important improvement of current antiviral therapies. Preliminary data from ongoing studies are promising, and an international therapy trial with brincidofovir in transplant recipients displaying intestinal HAdV infection, who have an elevated risk of viremia and disseminated disease (69, 188, 189), is currently in preparation. This study is expected to provide important evidence for the efficacy of early preemptive virustatic treatment. Another interesting novel antiviral compound is ganciclovir triphosphate, which does not require further phosphorylation by viral kinases and can therefore be expected to display therapeutic activity against HAdV.
In addition to the favorable properties of new antiviral drugs, advances in antiviral immunotherapy with adenovirus-specific T cells offer great potential for further improvement in the prevention or treatment of HAdV infections. Anti-HAdV T cells generated by different approaches are cross-reactive with all types of the virus and can therefore provide broad antiadenoviral immunity. It is not entirely clear, however, whether an equivalent quality of immune response to any HAdV type can be expected. It is conceivable that the generation of T cells targeted to individual HAdV types and mediating specific interactions with type-restricted epitopes (268), in addition to the broadly shared hexon epitopes, may elicit more effective immune responses (251). This notion may warrant careful attention. If the safety, efficacy, and feasibility of HAdV-specific or multivirus-specific T-cell transfer can be firmly established, it is reasonable to envision that this treatment will be employed successfully not only in the settings of preemptive and symptomatic therapy but also as prophylaxis in high-risk patients to prevent severe viral diseases. The establishment of allogeneic T-cell donor registries of HLA-typed healthy donors tested for the presence of virus-specific T cells could serve as a rapidly available source for adoptive immunotherapy in immunocompromised patients lacking a suitable T-cell donor. Such T-cell donor registries might provide readily available off-the-shelf products facilitating rapid initiation of immunotherapy in patients carrying a high risk of life-threatening viral infections. Broad availability of banked third-party virus-specific T cells and/or virus-specific T-cell lines could mark the beginning of a new era in combatting the threats of viral infections in immunocompromised patients in the foreseeable future.
ACKNOWLEDGMENT
I have no conflict of interest relevant in the context of the present paper.
Biography

Thomas Lion received his M.D. in 1984, from the University of Vienna, Vienna, Austria, and his Ph.D. in 1995, from the Charles University, Prague, Czech Republic. He was a postdoctoral research fellow at the University of Chicago and was appointed as a professor at the University of Vienna in 1997. He is a certified specialist in laboratory medicine (1993), pediatrics (with a focus on hemato-oncology; 1996), and human genetics (1997). Professor Lion is Head of the Division for Molecular Microbiology at the Children's Cancer Research Institute (CCRI), Medical Director of the diagnostic center LabDia Labordiagnostik, Vienna, Austria, and Chairman of the Scientific Board of the Austrian Society of Pediatrics. He has published numerous papers on infectious diseases in the immunocompromised host and has served as a section editor and editorial board member for various journals. He has received 16 national and international research awards and owns different patents on molecular diagnostic techniques.
REFERENCES
- 1.Rowe JP, Huebner RJ, Gilmore LK, Parrott RH, Ward TG. 1953. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc. Soc. Exp. Biol. Med. 84:570–573 [DOI] [PubMed] [Google Scholar]
- 2.Trentin JJ, Yabe Y, Taylor G. 1962. The quest for human cancer viruses. Science 137:835–841. 10.1126/science.137.3533.835 [DOI] [PubMed] [Google Scholar]
- 3.Hohlweg U, Dorn A, Hosel M, Webb D, Buettner R, Doerfler W. 2004. Tumorigenesis by adenovirus type 12 in newborn Syrian hamsters. Curr. Top. Microbiol. Immunol. 273:215–244. 10.1007/978-3-662-05599-1_7 [DOI] [PubMed] [Google Scholar]
- 4.Javier RT. 1994. Adenovirus type 9 E4 open reading frame 1 encodes a transforming protein required for the production of mammary tumors in rats. J. Virol. 68:3917–3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kosulin K, Haberler C, Hainfellner JA, Amann G, Lang S, Lion T. 2007. Investigation of adenovirus occurrence in pediatric tumor entities. J. Virol. 81:7629–7635. 10.1128/JVI.00355-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kosulin K, Hoffmann F, Clauditz TS, Wilczak W, Dobner T. 2013. Presence of adenovirus species C in infiltrating lymphocytes of human sarcoma. PLoS One 8:e63646. 10.1371/journal.pone.0063646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanchez-Prieto R, de Alava E, Palomino T, Guinea J, Fernandez V, Cebrian SLLM, Cabello P, Martin P, San Roman C, Bornstein R, Pardo J, Martinez A, Diaz-Espada F, Barrios Y, Ramon y Cajal S. 1999. An association between viral genes and human oncogenic alterations: the adenovirus E1A induces the Ewing tumor fusion transcript EWS-FLI1. Nat. Med. 5:1076–1079. 10.1038/12516 [DOI] [PubMed] [Google Scholar]
- 8.Greber UF, Arnberg N, Wadell G, Benko M, Kremer EJ. 2013. Adenoviruses—from pathogens to therapeutics: a report on the 10th International Adenovirus Meeting. Cell. Microbiol. 15:16–23. 10.1111/cmi.12031 [DOI] [PubMed] [Google Scholar]
- 9.Dicks MD, Spencer AJ, Edwards NJ, Wadell G, Bojang K, Gilbert SC, Hill AV, Cottingham MG. 2012. A novel chimpanzee adenovirus vector with low human seroprevalence: improved systems for vector derivation and comparative immunogenicity. PLoS One 7:e40385. 10.1371/journal.pone.0040385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miura Y, Yamasaki S, Davydova J, Brown E, Aoki K, Vickers S, Yamamoto M. 2013. Infectivity-selective oncolytic adenovirus developed by high-throughput screening of adenovirus-formatted library. Mol. Ther. 21:139–148. 10.1038/mt.2012.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamasaki S, Miura Y, Davydova J, Vickers SM, Yamamoto M. 2013. Intravenous genetic mesothelin vaccine based on human adenovirus 40 inhibits growth and metastasis of pancreatic cancer. Int. J. Cancer 133:88–97. 10.1002/ijc.27983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diaconu I, Cerullo V, Hirvinen ML, Escutenaire S, Ugolini M, Pesonen SK, Bramante S, Parviainen S, Kanerva A, Loskog AS, Eliopoulos AG, Pesonen S, Hemminki A. 2012. Immune response is an important aspect of the antitumor effect produced by a CD40L-encoding oncolytic adenovirus. Cancer Res. 72:2327–2338. 10.1158/0008-5472.CAN-11-2975 [DOI] [PubMed] [Google Scholar]
- 13.Shang Q, Wang H, Song Y, Wei L, Lavebratt C, Zhang F, Gu H. 2014. Serological data analyses show that adenovirus 36 infection is associated with obesity: a meta-analysis involving 5739 subjects. Obesity 22:895–900. 10.1002/oby.20533 [DOI] [PubMed] [Google Scholar]
- 14.Shauer A, Gotsman I, Keren A, Zwas DR, Hellman Y, Durst R, Admon D. 2013. Acute viral myocarditis: current concepts in diagnosis and treatment. Isr. Med. Assoc. J. 15:180–185 [PubMed] [Google Scholar]
- 15.de Ory F, Avellon A, Echevarria JE, Sanchez-Seco MP, Trallero G, Cabrerizo M, Casas I, Pozo F, Fedele G, Vicente D, Pena MJ, Moreno A, Niubo J, Rabella N, Rubio G, Perez-Ruiz M, Rodriguez-Iglesias M, Gimeno C, Eiros JM, Melon S, Blasco M, Lopez-Miragaya I, Varela E, Martinez-Sapina A, Rodriguez G, Marcos MA, Gegundez MI, Cilla G, Gabilondo I, Navarro JM, Torres J, Aznar C, Castellanos A, Guisasola ME, Negredo AI, Tenorio A, Vazquez-Moron S. 2013. Viral infections of the central nervous system in Spain: a prospective study. J. Med. Virol. 85:554–562. 10.1002/jmv.23470 [DOI] [PubMed] [Google Scholar]
- 16.Chhabra P, Payne DC, Szilagyi PG, Edwards KM, Staat MA, Shirley SH, Wikswo M, Nix WA, Lu X, Parashar UD, Vinje J. 2013. Etiology of viral gastroenteritis in children <5 years of age in the United States, 2008–2009. J. Infect. Dis. 208:790–800. 10.1093/infdis/jit254 [DOI] [PubMed] [Google Scholar]
- 17.Rocholl C, Gerber K, Daly J, Pavia AT, Byington CL. 2004. Adenoviral infections in children: the impact of rapid diagnosis. Pediatrics 113:e51–e56. 10.1542/peds.113.1.e51 [DOI] [PubMed] [Google Scholar]
- 18.Mameli C, Zuccotti GV. 2013. The impact of viral infections in children with community-acquired pneumonia. Curr. Infect. Dis. Rep. 15:197–202. 10.1007/s11908-013-0339-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Centers for Disease Control and Prevention. 2013. Adenovirus-associated epidemic keratoconjunctivitis outbreaks—four states, 2008–2010. MMWR Morb. Mortal. Wkly. Rep. 62:637–641 [PMC free article] [PubMed] [Google Scholar]
- 20.Lynch JP, 3rd, Fishbein M, Echavarria M. 2011. Adenovirus. Semin. Respir. Crit. Care Med. 32:494–511. 10.1055/s-0031-1283287 [DOI] [PubMed] [Google Scholar]
- 21.Esposito S, Preti V, Consolo S, Nazzari E, Principi N. 2012. Adenovirus 36 infection and obesity. J. Clin. Virol. 55:95–100. 10.1016/j.jcv.2012.06.009 [DOI] [PubMed] [Google Scholar]
- 22.Berciaud S, Rayne F, Kassab S, Jubert C, Faure-Della Corte M, Salin F, Wodrich H, Lafon ME, Typadeno Study Members 2012. Adenovirus infections in Bordeaux University Hospital 2008–2010: clinical and virological features. J. Clin. Virol. 54:302–307. 10.1016/j.jcv.2012.04.009 [DOI] [PubMed] [Google Scholar]
- 23.Chen SP, Huang YC, Chiu CH, Wong KS, Huang YL, Huang CG, Tsao KC, Lin TY. 2013. Clinical features of radiologically confirmed pneumonia due to adenovirus in children. J. Clin. Virol. 56:7–12. 10.1016/j.jcv.2012.08.021 [DOI] [PubMed] [Google Scholar]
- 24.Alharbi S, Van Caeseele P, Consunji-Araneta R, Zoubeidi T, Fanella S, Souid AK, Alsuwaidi AR. 2012. Epidemiology of severe pediatric adenovirus lower respiratory tract infections in Manitoba, Canada, 1991–2005. BMC Infect. Dis. 12:55. 10.1186/1471-2334-12-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis PF, Schmidt MA, Lu X, Erdman DD, Campbell M, Thomas A, Cieslak PR, Grenz LD, Tsaknardis L, Gleaves C, Kendall B, Gilbert D. 2009. A community-based outbreak of severe respiratory illness caused by human adenovirus serotype 14. J. Infect. Dis. 199:1427–1434. 10.1086/598521 [DOI] [PubMed] [Google Scholar]
- 26.Savon C, Acosta B, Valdes O, Goyenechea A, Gonzalez G, Pinon A, Mas P, Rosario D, Capo V, Kouri V, Martinez PA, Marchena JJ, Gonzalez G, Rodriguez H, Guzman MG. 2008. A myocarditis outbreak with fatal cases associated with adenovirus subgenera C among children from Havana City in 2005. J. Clin. Virol. 43:152–157. 10.1016/j.jcv.2008.05.012 [DOI] [PubMed] [Google Scholar]
- 27.Nilsson EC, Storm RJ, Bauer J, Johansson SM, Lookene A, Angstrom J, Hedenstrom M, Eriksson TL, Frangsmyr L, Rinaldi S, Willison HJ, Pedrosa Domellof F, Stehle T, Arnberg N. 2011. The GD1a glycan is a cellular receptor for adenoviruses causing epidemic keratoconjunctivitis. Nat. Med. 17:105–109. 10.1038/nm.2267 [DOI] [PubMed] [Google Scholar]
- 28.Trinh HV, Lesage G, Chennamparampil V, Vollenweider B, Burckhardt CJ, Schauer S, Havenga M, Greber UF, Hemmi S. 2012. Avidity binding of human adenovirus serotypes 3 and 7 to the membrane cofactor CD46 triggers infection. J. Virol. 86:1623–1637. 10.1128/JVI.06181-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, Li ZY, Liu Y, Persson J, Beyer I, Moller T, Koyuncu D, Drescher MR, Strauss R, Zhang XB, Wahl JK, 3rd, Urban N, Drescher C, Hemminki A, Fender P, Lieber A. 2011. Desmoglein 2 is a receptor for adenovirus serotypes 3, 7, 11 and 14. Nat. Med. 17:96–104. 10.1038/nm.2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson CM, Seto D, Jones MS, Dyer DW, Chodosh J. 2011. Molecular evolution of human species D adenoviruses. Infect. Genet. Evol. 11:1208–1217. 10.1016/j.meegid.2011.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith JG, Wiethoff CM, Stewart PL, Nemerow GR. 2010. Adenovirus. Curr. Top. Microbiol. Immunol. 343:195–224. 10.1007/82_2010_16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiethoff CM, Wodrich H, Gerace L, Nemerow GR. 2005. Adenovirus protein VI mediates membrane disruption following capsid disassembly. J. Virol. 79:1992–2000. 10.1128/JVI.79.4.1992-2000.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harrach B, Benkö M, Both GW, Brown M, Davison AJ, Echavarria M, Hess M, Jones MS, Kajon A, Lehmkuhl HD, Mautner V, Mittal SK, Wadell G. 2012. Adenoviridae—ninth report of the International Committee on Taxonomy of Viruses, p 125–141 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy. Elsevier, Oxford, United Kingdom [Google Scholar]
- 34.Robinson CM, Singh G, Lee JY, Dehghan S, Rajaiya J, Liu EB, Yousuf MA, Betensky RA, Jones MS, Dyer DW, Seto D, Chodosh J. 2013. Molecular evolution of human adenoviruses. Sci. Rep. 3:1812. 10.1038/srep01812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamel W, Segerman B, Oberg D, Punga T, Akusjarvi G. 2013. The adenovirus VA RNA-derived miRNAs are not essential for lytic virus growth in tissue culture cells. Nucleic Acids Res. 41:4802–4812. 10.1093/nar/gkt172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones MS, 2nd, Harrach B, Ganac RD, Gozum MM, Dela Cruz WP, Riedel B, Pan C, Delwart EL, Schnurr DP. 2007. New adenovirus species found in a patient presenting with gastroenteritis. J. Virol. 81:5978–5984. 10.1128/JVI.02650-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson CM, Singh G, Henquell C, Walsh MP, Peigue-Lafeuille H, Seto D, Jones MS, Dyer DW, Chodosh J. 2011. Computational analysis and identification of an emergent human adenovirus pathogen implicated in a respiratory fatality. Virology 409:141–147. 10.1016/j.virol.2010.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walsh MP, Seto J, Liu EB, Dehghan S, Hudson NR, Lukashev AN, Ivanova O, Chodosh J, Dyer DW, Jones MS, Seto D. 2011. Computational analysis of two species C human adenoviruses provides evidence of a novel virus. J. Clin. Microbiol. 49:3482–3490. 10.1128/JCM.00156-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu EB, Ferreyra L, Fischer SL, Pavan JV, Nates SV, Hudson NR, Tirado D, Dyer DW, Chodosh J, Seto D, Jones MS. 2011. Genetic analysis of a novel human adenovirus with a serologically unique hexon and a recombinant fiber gene. PLoS One 6:e24491. 10.1371/journal.pone.0024491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seto J, Walsh MP, Mahadevan P, Purkayastha A, Clark JM, Tibbetts C, Seto D. 2009. Genomic and bioinformatics analyses of HAdV-14p, reference strain of a re-emerging respiratory pathogen and analysis of B1/B2. Virus Res. 143:94–105. 10.1016/j.virusres.2009.03.011 [DOI] [PubMed] [Google Scholar]
- 41.Seto D, Chodosh J, Brister JR, Jones MS, Members of the Adenovirus Research Community 2011. Using the whole-genome sequence to characterize and name human adenoviruses. J. Virol. 85:5701–5702. 10.1128/JVI.00354-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aoki K, Benko M, Davison AJ, Echavarria M, Erdman DD, Harrach B, Kajon AE, Schnurr D, Wadell G, Members of the Adenovirus Research Community 2011. Toward an integrated human adenovirus designation system that utilizes molecular and serological data and serves both clinical and fundamental virology. J. Virol. 85:5703–5704. 10.1128/JVI.00491-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Echavarria M. 2008. Adenoviruses in immunocompromised hosts. Clin. Microbiol. Rev. 21:704–715. 10.1128/CMR.00052-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu EB, Wadford DA, Seto J, Vu M, Hudson NR, Thrasher L, Torres S, Dyer DW, Chodosh J, Seto D, Jones MS. 2012. Computational and serologic analysis of novel and known viruses in species human adenovirus D in which serology and genomics do not correlate. PLoS One 7:e33212. 10.1371/journal.pone.0033212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walsh MP, Seto J, Jones MS, Chodosh J, Xu W, Seto D. 2010. Computational analysis identifies human adenovirus type 55 as a re-emergent acute respiratory disease pathogen. J. Clin. Microbiol. 48:991–993. 10.1128/JCM.01694-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsushima Y, Shimizu H, Kano A, Nakajima E, Ishimaru Y, Dey SK, Watanabe Y, Adachi F, Mitani K, Fujimoto T, Phan TG, Ushijima H. 2013. Genome sequence of a novel virus of the species human adenovirus D associated with acute gastroenteritis. Genome Announc. 1:e00068–12. 10.1128/genomeA.00068-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takemori N. 1972. Genetic studies with tumorigenic adenoviruses. 3. Recombination in adenovirus type 12. Virology 47:157–167 [DOI] [PubMed] [Google Scholar]
- 48.Williams J, Grodzicker T, Sharp P, Sambrook J. 1975. Adenovirus recombination: physical mapping of crossover events. Cell 4:113–119. 10.1016/0092-8674(75)90117-8 [DOI] [PubMed] [Google Scholar]
- 49.Walsh MP, Chintakuntlawar A, Robinson CM, Madisch I, Harrach B, Hudson NR, Schnurr D, Heim A, Chodosh J, Seto D, Jones MS. 2009. Evidence of molecular evolution driven by recombination events influencing tropism in a novel human adenovirus that causes epidemic keratoconjunctivitis. PLoS One 4:e5635. 10.1371/journal.pone.0005635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lukashev AN, Ivanova OE, Eremeeva TP, Iggo RD. 2008. Evidence of frequent recombination among human adenoviruses. J. Gen. Virol. 89:380–388. 10.1099/vir.0.83057-0 [DOI] [PubMed] [Google Scholar]
- 51.Robinson CM, Rajaiya J, Walsh MP, Seto D, Dyer DW, Jones MS, Chodosh J. 2009. Computational analysis of human adenovirus type 22 provides evidence for recombination among species D human adenoviruses in the penton base gene. J. Virol. 83:8980–8985. 10.1128/JVI.00786-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vora GJ, Lin B, Gratwick K, Meador C, Hansen C, Tibbetts C, Stenger DA, Irvine M, Seto D, Purkayastha A, Freed NE, Gibson MG, Russell K, Metzgar D. 2006. Co-infections of adenovirus species in previously vaccinated patients. Emerg. Infect. Dis. 12:921–930. 10.3201/eid1206.050245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCarthy T, Lebeck MG, Capuano AW, Schnurr DP, Gray GC. 2009. Molecular typing of clinical adenovirus specimens by an algorithm which permits detection of adenovirus coinfections and intermediate adenovirus strains. J. Clin. Virol. 46:80–84. 10.1016/j.jcv.2009.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Curlin ME, Huang ML, Lu X, Celum CL, Sanchez J, Selke S, Baeten JM, Zuckerman RA, Erdman DD, Corey L. 2010. Frequent detection of human adenovirus from the lower gastrointestinal tract in men who have sex with men. PLoS One 5:e11321. 10.1371/journal.pone.0011321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaneko H, Aoki K, Ishida S, Ohno S, Kitaichi N, Ishiko H, Fujimoto T, Ikeda Y, Nakamura M, Gonzalez G, Koyanagi KO, Watanabe H, Suzutani T. 2011. Recombination analysis of intermediate human adenovirus type 53 in Japan by complete genome sequence. J. Gen. Virol. 92:1251–1259. 10.1099/vir.0.030361-0 [DOI] [PubMed] [Google Scholar]
- 56.Wevers D, Metzger S, Babweteera F, Bieberbach M, Boesch C, Cameron K, Couacy-Hymann E, Cranfield M, Gray M, Harris LA, Head J, Jeffery K, Knauf S, Lankester F, Leendertz SA, Lonsdorf E, Mugisha L, Nitsche A, Reed P, Robbins M, Travis DA, Zommers Z, Leendertz FH, Ehlers B. 2011. Novel adenoviruses in wild primates: a high level of genetic diversity and evidence of zoonotic transmissions. J. Virol. 85:10774–10784. 10.1128/JVI.00810-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dehghan S, Seto J, Liu EB, Walsh MP, Dyer DW, Chodosh J, Seto D. 2013. Computational analysis of four human adenovirus type 4 genomes reveals molecular evolution through two interspecies recombination events. Virology 443:197–207. 10.1016/j.virol.2013.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin KH, Lin YC, Chen HL, Ke GM, Chiang CJ, Hwang KP, Chu PY, Lin JH, Liu DP, Chen HY. 2004. A two decade survey of respiratory adenovirus in Taiwan: the reemergence of adenovirus types 7 and 4. J. Med. Virol. 73:274–279. 10.1002/jmv.20087 [DOI] [PubMed] [Google Scholar]
- 59.Ishiko H, Shimada Y, Konno T, Hayashi A, Ohguchi T, Tagawa Y, Aoki K, Ohno S, Yamazaki S. 2008. Novel human adenovirus causing nosocomial epidemic keratoconjunctivitis. J. Clin. Microbiol. 46:2002–2008. 10.1128/JCM.01835-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ampuero JS, Ocana V, Gomez J, Gamero ME, Garcia J, Halsey ES, Laguna-Torres VA. 2012. Adenovirus respiratory tract infections in Peru. PLoS One 7:e46898. 10.1371/journal.pone.0046898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kajon AE, Lu X, Erdman DD, Louie J, Schnurr D, George KS, Koopmans MP, Allibhai T, Metzgar D. 2010. Molecular epidemiology and brief history of emerging adenovirus 14-associated respiratory disease in the United States. J. Infect. Dis. 202:93–103. 10.1086/653083 [DOI] [PubMed] [Google Scholar]
- 62.Qurei L, Seto D, Salah Z, Azzeh M. 2012. A molecular epidemiology survey of respiratory adenoviruses circulating in children residing in Southern Palestine. PLoS One 7:e42732. 10.1371/journal.pone.0042732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo L, Gonzalez R, Zhou H, Wu C, Vernet G, Wang Z, Wang J. 2012. Detection of three human adenovirus species in adults with acute respiratory infection in China. Eur. J. Clin. Microbiol. Infect. Dis. 31:1051–1058. 10.1007/s10096-011-1406-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yliharsila M, Harju E, Arppe R, Hattara L, Holsa J, Saviranta P, Soukka T, Waris M. 2013. Genotyping of clinically relevant human adenoviruses by array-in-well hybridization assay. Clin. Microbiol. Infect. 19:551–557. 10.1111/j.1469-0691.2012.03926.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tabain I, Ljubin-Sternak S, Cepin-Bogovic J, Markovinovic L, Knezovic I, Mlinaric-Galinovic G. 2012. Adenovirus respiratory infections in hospitalized children: clinical findings in relation to species and serotypes. Pediatr. Infect. Dis. J. 31:680–684. 10.1097/INF.0b013e318256605e [DOI] [PubMed] [Google Scholar]
- 66.Barrero PR, Valinotto LE, Tittarelli E, Mistchenko AS. 2012. Molecular typing of adenoviruses in pediatric respiratory infections in Buenos Aires, Argentina (1999–2010). J. Clin. Virol. 53:145–150. 10.1016/j.jcv.2011.11.001 [DOI] [PubMed] [Google Scholar]
- 67.Madisch I, Wolfel R, Harste G, Pommer H, Heim A. 2006. Molecular identification of adenovirus sequences: a rapid scheme for early typing of human adenoviruses in diagnostic samples of immunocompetent and immunodeficient patients. J. Med. Virol. 78:1210–1217. 10.1002/jmv.20683 [DOI] [PubMed] [Google Scholar]
- 68.Leen AM, Rooney CM. 2005. Adenovirus as an emerging pathogen in immunocompromised patients. Br. J. Haematol. 128:135–144. 10.1111/j.1365-2141.2004.05218.x [DOI] [PubMed] [Google Scholar]
- 69.Lion T, Kosulin K, Landlinger C, Rauch M, Preuner S, Jugovic D, Potschger U, Lawitschka A, Peters C, Fritsch G, Matthes-Martin S. 2010. Monitoring of adenovirus load in stool by real-time PCR permits early detection of impending invasive infection in patients after allogeneic stem cell transplantation. Leukemia 24:706–714. 10.1038/leu.2010.4 [DOI] [PubMed] [Google Scholar]
- 70.Al Qurashi YM, Guiver M, Cooper RJ. 2011. Sequence typing of adenovirus from samples from hematological stem cell transplant recipients. J. Med. Virol. 83:1951–1958. 10.1002/jmv.22204 [DOI] [PubMed] [Google Scholar]
- 71.Lion T, Baumgartinger R, Watzinger F, Matthes-Martin S, Suda M, Preuner S, Futterknecht B, Lawitschka A, Peters C, Potschger U, Gadner H. 2003. Molecular monitoring of adenovirus in peripheral blood after allogeneic bone marrow transplantation permits early diagnosis of disseminated disease. Blood 102:1114–1120. 10.1182/blood-2002-07-2152 [DOI] [PubMed] [Google Scholar]
- 72.Kroes AC, de Klerk EP, Lankester AC, Malipaard C, de Brouwer CS, Claas EC, Jol-van der Zijde EC, van Tol MJ. 2007. Sequential emergence of multiple adenovirus serotypes after pediatric stem cell transplantation. J. Clin. Virol. 38:341–347. 10.1016/j.jcv.2007.01.001 [DOI] [PubMed] [Google Scholar]
- 73.Echavarria M, Forman M, van Tol MJ, Vossen JM, Charache P, Kroes AC. 2001. Prediction of severe disseminated adenovirus infection by serum PCR. Lancet 358:384–385. 10.1016/S0140-6736(01)05580-5 [DOI] [PubMed] [Google Scholar]
- 74.Kwon HJ, Rhie YJ, Seo WH, Jang GY, Choi BM, Lee JH, Lee CK, Kim YK. 2013. Clinical manifestations of respiratory adenoviral infection among hospitalized children in Korea. Pediatr. Int. 55:450–454. 10.1111/ped.12108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takayama R, Hatakeyama N, Suzuki N, Yamamoto M, Hayashi T, Ikeda Y, Ikeda H, Nagano H, Ishida T, Tsutsumi H. 2007. Quantification of adenovirus species B and C viremia by real-time PCR in adults and children undergoing stem cell transplantation. J. Med. Virol. 79:278–284. 10.1002/jmv.20796 [DOI] [PubMed] [Google Scholar]
- 76.Gray GC, McCarthy T, Lebeck MG, Schnurr DP, Russell KL, Kajon AE, Landry ML, Leland DS, Storch GA, Ginocchio CC, Robinson CC, Demmler GJ, Saubolle MA, Kehl SC, Selvarangan R, Miller MB, Chappell JD, Zerr DM, Kiska DL, Halstead DC, Capuano AW, Setterquist SF, Chorazy ML, Dawson JD, Erdman DD. 2007. Genotype prevalence and risk factors for severe clinical adenovirus infection, United States 2004–2006. Clin. Infect. Dis. 45:1120–1131. 10.1086/522188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lessa FC, Gould PL, Pascoe N, Erdman DD, Lu X, Bunning ML, Marconi VC, Lott L, Widdowson MA, Anderson LJ, Srinivasan A. 2009. Health care transmission of a newly emergent adenovirus serotype in health care personnel at a military hospital in Texas, 2007. J. Infect. Dis. 200:1759–1765. 10.1086/647987 [DOI] [PubMed] [Google Scholar]
- 78.Russell KL, Broderick MP, Franklin SE, Blyn LB, Freed NE, Moradi E, Ecker DJ, Kammerer PE, Osuna MA, Kajon AE, Morn CB, Ryan MA. 2006. Transmission dynamics and prospective environmental sampling of adenovirus in a military recruit setting. J. Infect. Dis. 194:877–885. 10.1086/507426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu Z, Zhang Y, Xu S, Yu P, Tian X, Wang L, Liu Z, Tang L, Mao N, Ji Y, Li C, Yang Z, Wang S, Wang J, Li D, Xu W. 2009. Outbreak of acute respiratory disease in China caused by B2 species of adenovirus type 11. J. Clin. Microbiol. 47:697–703. 10.1128/JCM.01769-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wan GH, Huang CG, Huang YC, Huang JP, Yang SL, Lin TY, Tsao KC. 2012. Surveillance of airborne adenovirus and Mycoplasma pneumoniae in a hospital pediatric department. PLoS One 7:e33974. 10.1371/journal.pone.0033974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Soller JA, Bartrand T, Ashbolt NJ, Ravenscroft J, Wade TJ. 2010. Estimating the primary etiologic agents in recreational freshwaters impacted by human sources of faecal contamination. Water Res. 44:4736–4747. 10.1016/j.watres.2010.07.064 [DOI] [PubMed] [Google Scholar]
- 82.Matthes-Martin S, Boztug H, Lion T. 2013. Diagnosis and treatment of adenovirus infection in immunocompromised patients. Expert Rev. Anti Infect. Ther. 11:1017–1028. 10.1586/14787210.2013.836964 [DOI] [PubMed] [Google Scholar]
- 83.Rutala WA, Peacock JE, Gergen MF, Sobsey MD, Weber DJ. 2006. Efficacy of hospital germicides against adenovirus 8, a common cause of epidemic keratoconjunctivitis in health care facilities. Antimicrob. Agents Chemother. 50:1419–1424. 10.1128/AAC.50.4.1419-1424.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Calkavur S, Olukman O, Ozturk AT, Kilic FK, Gulfidan G, Devrim I, Malatyali R, Oruc Y, Atlihan F. 2012. Epidemic adenoviral keratoconjunctivitis possibly related to ophthalmological procedures in a neonatal intensive care unit: lessons from an outbreak. Ophthalmic Epidemiol. 19:371–379. 10.3109/09286586.2012.718402 [DOI] [PubMed] [Google Scholar]
- 85.Leruez-Ville M, Chardin-Ouachee M, Neven B, Picard C, Le Guinche I, Fischer A, Rouzioux C, Blanche S. 2006. Description of an adenovirus A31 outbreak in a paediatric haematology unit. Bone Marrow Transplant. 38:23–28. 10.1038/sj.bmt.1705389 [DOI] [PubMed] [Google Scholar]
- 86.Mattner F, Sykora KW, Meissner B, Heim A. 2008. An adenovirus type F41 outbreak in a pediatric bone marrow transplant unit: analysis of clinical impact and preventive strategies. Pediatr. Infect. Dis. J. 27:419–424. 10.1097/INF.0b013e3181658c46 [DOI] [PubMed] [Google Scholar]
- 87.Jalal H, Bibby DF, Tang JW, Bennett J, Kyriakou C, Peggs K, Cubitt D, Brink NS, Ward KN, Tedder RS. 2005. First reported outbreak of diarrhea due to adenovirus infection in a hematology unit for adults. J. Clin. Microbiol. 43:2575–2580. 10.1128/JCM.43.6.2575-2580.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Henquell C, Boeuf B, Mirand A, Bacher C, Traore O, Dechelotte P, Labbe A, Bailly JL, Peigue-Lafeuille H. 2009. Fatal adenovirus infection in a neonate and transmission to health-care workers. J. Clin. Virol. 45:345–348. 10.1016/j.jcv.2009.04.019 [DOI] [PubMed] [Google Scholar]
- 89.Matsushima Y, Shimizu H, Kano A, Nakajima E, Ishimaru Y, Dey SK, Watanabe Y, Adachi F, Suzuki K, Mitani K, Fujimoto T, Phan TG, Ushijima H. 2012. Novel human adenovirus strain, Bangladesh. Emerg. Infect. Dis. 18:846–848. 10.3201/eid1805.111584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Metzgar D, Gibbins C, Hudson NR, Jones MS. 2010. Evaluation of multiplex type-specific real-time PCR assays using the LightCycler and joint biological agent identification and diagnostic system platforms for detection and quantitation of adult human respiratory adenoviruses. J. Clin. Microbiol. 48:1397–1403. 10.1128/JCM.01600-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moura PO, Roberto AF, Hein N, Baldacci E, Vieira SE, Ejzenberg B, Perrini P, Stewien KE, Durigon EL, Mehnert DU, Harsi CM. 2007. Molecular epidemiology of human adenovirus isolated from children hospitalized with acute respiratory infection in Sao Paulo, Brazil. J. Med. Virol. 79:174–181. 10.1002/jmv.20778 [DOI] [PubMed] [Google Scholar]
- 92.Kajon AE, Moseley JM, Metzgar D, Huong HS, Wadleigh A, Ryan MA, Russell KL. 2007. Molecular epidemiology of adenovirus type 4 infections in US military recruits in the postvaccination era (1997–2003). J. Infect. Dis. 196:67–75. 10.1086/518442 [DOI] [PubMed] [Google Scholar]
- 93.Chang SY, Lee CN, Lin PH, Huang HH, Chang LY, Ko W, Chang SF, Lee PI, Huang LM, Kao CL. 2008. A community-derived outbreak of adenovirus type 3 in children in Taiwan between 2004 and 2005. J. Med. Virol. 80:102–112. 10.1002/jmv.21045 [DOI] [PubMed] [Google Scholar]
- 94.Hoke CH, Jr, Snyder CE., Jr 2013. History of the restoration of adenovirus type 4 and type 7 vaccine, live oral (adenovirus vaccine) in the context of the Department of Defense acquisition system. Vaccine 31:1623–1632. 10.1016/j.vaccine.2012.12.029 [DOI] [PubMed] [Google Scholar]
- 95.Kuschner RA, Russell KL, Abuja M, Bauer KM, Faix DJ, Hait H, Henrick J, Jacobs M, Liss A, Lynch JA, Liu Q, Lyons AG, Malik M, Moon JE, Stubbs J, Sun W, Tang D, Towle AC, Walsh DS, Wilkerson D, Adenovirus Vaccine Efficacy Trial Consortium 2013. A phase 3, randomized, double-blind, placebo-controlled study of the safety and efficacy of the live, oral adenovirus type 4 and type 7 vaccine, in U.S. military recruits. Vaccine 31:2963–2971. 10.1016/j.vaccine.2013.04.035 [DOI] [PubMed] [Google Scholar]
- 96.Mitchell LS, Taylor B, Reimels W, Barrett FF, Devincenzo JP. 2000. Adenovirus 7a: a community-acquired outbreak in a children's hospital. Pediatr. Infect. Dis. J. 19:996–1000. 10.1097/00006454-200010000-00011 [DOI] [PubMed] [Google Scholar]
- 97.Ison MG. 2006. Adenovirus infections in transplant recipients. Clin. Infect. Dis. 43:331–339. 10.1086/505498 [DOI] [PubMed] [Google Scholar]
- 98.Hong JY, Lee HJ, Piedra PA, Choi EH, Park KH, Koh YY, Kim WS. 2001. Lower respiratory tract infections due to adenovirus in hospitalized Korean children: epidemiology, clinical features, and prognosis. Clin. Infect. Dis. 32:1423–1429. 10.1086/320146 [DOI] [PubMed] [Google Scholar]
- 99.Carr MJ, Kajon AE, Lu X, Dunford L, O'Reilly P, Holder P, De Gascun CF, Coughlan S, Connell J, Erdman DD, Hall WW. 2011. Deaths associated with human adenovirus-14p1 infections, Europe, 2009–2010. Emerg. Infect. Dis. 17:1402–1408. 10.3201/1708.101760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Siminovich M, Murtagh P. 2011. Acute lower respiratory tract infections by adenovirus in children: histopathologic findings in 18 fatal cases. Pediatr. Dev. Pathol. 14:214–217. 10.2350/10-05-0838-OA.1 [DOI] [PubMed] [Google Scholar]
- 101.Garnett CT, Erdman D, Xu W, Gooding LR. 2002. Prevalence and quantitation of species C adenovirus DNA in human mucosal lymphocytes. J. Virol. 76:10608–10616. 10.1128/JVI.76.21.10608-10616.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Roy S, Calcedo R, Medina-Jaszek A, Keough M, Peng H, Wilson JM. 2011. Adenoviruses in lymphocytes of the human gastro-intestinal tract. PLoS One 6:e24859. 10.1371/journal.pone.0024859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Garnett CT, Talekar G, Mahr JA, Huang W, Zhang Y, Ornelles DA, Gooding LR. 2009. Latent species C adenoviruses in human tonsil tissues. J. Virol. 83:2417–2428. 10.1128/JVI.02392-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hogg JC. 2001. Role of latent viral infections in chronic obstructive pulmonary disease and asthma. Am. J. Respir. Crit. Care Med. 164:S71–S75. 10.1164/ajrccm.164.supplement_2.2106063 [DOI] [PubMed] [Google Scholar]
- 105.Guarner J, de Leon-Bojorge B, Lopez-Corella E, Ferebee-Harris T, Gooding L, Garnett CT, Shieh WJ, Dawson J, Erdman D, Zaki SR. 2003. Intestinal intussusception associated with adenovirus infection in Mexican children. Am. J. Clin. Pathol. 120:845–850. 10.1309/LBRNGF9MJW2MHT97 [DOI] [PubMed] [Google Scholar]
- 106.Leung AY, Chan M, Cheng VC, Yuen KY, Kwong YL. 2005. Quantification of adenovirus in the lower respiratory tract of patients without clinical adenovirus-related respiratory disease. Clin. Infect. Dis. 40:1541–1544. 10.1086/429627 [DOI] [PubMed] [Google Scholar]
- 107.Zheng X, Lu X, Erdman DD, Anderson EJ, Guzman-Cottrill JA, Kletzel M, Katz BZ. 2008. Identification of adenoviruses in specimens from high-risk pediatric stem cell transplant recipients and controls. J. Clin. Microbiol. 46:317–320. 10.1128/JCM.01585-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schneider-Brachert W, Tchikov V, Merkel O, Jakob M, Hallas C, Kruse ML, Groitl P, Lehn A, Hildt E, Held-Feindt J, Dobner T, Kabelitz D, Kronke M, Schutze S. 2006. Inhibition of TNF receptor 1 internalization by adenovirus 14.7K as a novel immune escape mechanism. J. Clin. Invest. 116:2901–2913. 10.1172/JCI23771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wold WS, Doronin K, Toth K, Kuppuswamy M, Lichtenstein DL, Tollefson AE. 1999. Immune responses to adenoviruses: viral evasion mechanisms and their implications for the clinic. Curr. Opin. Immunol. 11:380–386. 10.1016/S0952-7915(99)80064-8 [DOI] [PubMed] [Google Scholar]
- 110.Lichtenstein DL, Toth K, Doronin K, Tollefson AE, Wold WS. 2004. Functions and mechanisms of action of the adenovirus E3 proteins. Int. Rev. Immunol. 23:75–111. 10.1080/08830180490265556 [DOI] [PubMed] [Google Scholar]
- 111.Sester M, Koebernick K, Owen D, Ao M, Bromberg Y, May E, Stock E, Andrews L, Groh V, Spies T, Steinle A, Menz B, Burgert HG. 2010. Conserved amino acids within the adenovirus 2 E3/19K protein differentially affect downregulation of MHC class I and MICA/B proteins. J. Immunol. 184:255–267. 10.4049/jimmunol.0902343 [DOI] [PubMed] [Google Scholar]
- 112.Hansen TH, Bouvier M. 2009. MHC class I antigen presentation: learning from viral evasion strategies. Nat. Rev. Immunol. 9:503–513. 10.1038/nri2575 [DOI] [PubMed] [Google Scholar]
- 113.Guidotti LG, Chisari FV. 2001. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu. Rev. Immunol. 19:65–91. 10.1146/annurev.immunol.19.1.65 [DOI] [PubMed] [Google Scholar]
- 114.Chahal JS, Gallagher C, DeHart CJ, Flint SJ. 2013. The repression domain of the E1B 55-kilodalton protein participates in countering interferon-induced inhibition of adenovirus replication. J. Virol. 87:4432–4444. 10.1128/JVI.03387-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Heemskerk B, van Vreeswijk T, Veltrop-Duits LA, Sombroek CC, Franken K, Verhoosel RM, Hiemstra PS, van Leeuwen D, Ressing ME, Toes RE, van Tol MJ, Schilham MW. 2006. Adenovirus-specific CD4+ T cell clones recognizing endogenous antigen inhibit viral replication in vitro through cognate interaction. J. Immunol. 177:8851–8859. 10.4049/jimmunol.177.12.8851 [DOI] [PubMed] [Google Scholar]
- 116.Leen AM, Christin A, Khalil M, Weiss H, Gee AP, Brenner MK, Heslop HE, Rooney CM, Bollard CM. 2008. Identification of hexon-specific CD4 and CD8 T-cell epitopes for vaccine and immunotherapy. J. Virol. 82:546–554. 10.1128/JVI.01689-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Veltrop-Duits LA, Heemskerk B, Sombroek CC, van Vreeswijk T, Gubbels S, Toes RE, Melief CJ, Franken KL, Havenga M, van Tol MJ, Schilham MW. 2006. Human CD4+ T cells stimulated by conserved adenovirus 5 hexon peptides recognize cells infected with different species of human adenovirus. Eur. J. Immunol. 36:2410–2423. 10.1002/eji.200535786 [DOI] [PubMed] [Google Scholar]
- 118.Serangeli C, Bicanic O, Scheible MH, Wernet D, Lang P, Rammensee HG, Stevanovic S, Handgretinger R, Feuchtinger T. 2010. Ex vivo detection of adenovirus specific CD4+ T-cell responses to HLA-DR-epitopes of the hexon protein show a contracted specificity of T(HELPER) cells following stem cell transplantation. Virology 397:277–284. 10.1016/j.virol.2009.10.049 [DOI] [PubMed] [Google Scholar]
- 119.Hutnick NA, Carnathan D, Demers K, Makedonas G, Ertl HC, Betts MR. 2010. Adenovirus-specific human T cells are pervasive, polyfunctional, and cross-reactive. Vaccine 28:1932–1941. 10.1016/j.vaccine.2009.10.091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Leen AM, Sili U, Vanin EF, Jewell AM, Xie W, Vignali D, Piedra PA, Brenner MK, Rooney CM. 2004. Conserved CTL epitopes on the adenovirus hexon protein expand subgroup cross-reactive and subgroup-specific CD8+ T cells. Blood 104:2432–2440. 10.1182/blood-2004-02-0646 [DOI] [PubMed] [Google Scholar]
- 121.Heemskerk B, Veltrop-Duits LA, van Vreeswijk T, ten Dam MM, Heidt S, Toes RE, van Tol MJ, Schilham MW. 2003. Extensive cross-reactivity of CD4+ adenovirus-specific T cells: implications for immunotherapy and gene therapy. J. Virol. 77:6562–6566. 10.1128/JVI.77.11.6562-6566.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Feuchtinger T, Lucke J, Hamprecht K, Richard C, Handgretinger R, Schumm M, Greil J, Bock T, Niethammer D, Lang P. 2005. Detection of adenovirus-specific T cells in children with adenovirus infection after allogeneic stem cell transplantation. Br. J. Haematol. 128:503–509. 10.1111/j.1365-2141.2004.05331.x [DOI] [PubMed] [Google Scholar]
- 123.Geyeregger R, Freimuller C, Stevanovic S, Stemberger J, Mester G, Dmytrus J, Lion T, Rammensee HG, Fischer G, Eiz-Vesper B, Lawitschka A, Matthes S, Fritsch G. 2013. Short-term in-vitro expansion improves monitoring and allows affordable generation of virus-specific T-cells against several viruses for a broad clinical application. PLoS One 8:e59592. 10.1371/journal.pone.0059592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zandvliet ML, Falkenburg JH, van Liempt E, Veltrop-Duits LA, Lankester AC, Kalpoe JS, Kester MG, van der Steen DM, van Tol MJ, Willemze R, Guchelaar HJ, Schilham MW, Meij P. 2010. Combined CD8+ and CD4+ adenovirus hexon-specific T cells associated with viral clearance after stem cell transplantation as treatment for adenovirus infection. Haematologicaae 95:1943–1951. 10.3324/haematol.2010.022947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Runde V, Ross S, Trenschel R, Lagemann E, Basu O, Renzing-Kohler K, Schaefer UW, Roggendorf M, Holler E. 2001. Adenoviral infection after allogeneic stem cell transplantation (SCT): report on 130 patients from a single SCT unit involved in a prospective multi center surveillance study. Bone Marrow Transplant. 28:51–57. 10.1038/sj.bmt.1703083 [DOI] [PubMed] [Google Scholar]
- 126.Veltrop-Duits LA, van Vreeswijk T, Heemskerk B, Thijssen JC, El Seady R, Jol-van der Zijde EM, Claas EC, Lankester AC, van Tol MJ, Schilham MW. 2011. High titers of pre-existing adenovirus serotype-specific neutralizing antibodies in the host predict viral reactivation after allogeneic stem cell transplantation in children. Clin. Infect. Dis. 52:1405–1413. 10.1093/cid/cir231 [DOI] [PubMed] [Google Scholar]
- 127.van Tol MJ, Kroes AC, Schinkel J, Dinkelaar W, Claas EC, Jol-van der Zijde CM, Vossen JM. 2005. Adenovirus infection in paediatric stem cell transplant recipients: increased risk in young children with a delayed immune recovery. Bone Marrow Transplant. 36:39–50. 10.1038/sj.bmt.1705003 [DOI] [PubMed] [Google Scholar]
- 128.de Pagter AP, Haveman LM, Schuurman R, Schutten M, Bierings M, Boelens JJ. 2009. Adenovirus DNA positivity in nasopharyngeal aspirate preceding hematopoietic stem cell transplantation: a very strong risk factor for adenovirus DNAemia in pediatric patients. Clin. Infect. Dis. 49:1536–1539. 10.1086/644739 [DOI] [PubMed] [Google Scholar]
- 129.Kampmann B, Cubitt D, Walls T, Naik P, Depala M, Samarasinghe S, Robson D, Hassan A, Rao K, Gaspar H, Davies G, Jones A, Cale C, Gilmour K, Real M, Foo M, Bennett-Rees N, Hewitt A, Amrolia P, Veys P. 2005. Improved outcome for children with disseminated adenoviral infection following allogeneic stem cell transplantation. Br. J. Haematol. 130:595–603. 10.1111/j.1365-2141.2005.05649.x [DOI] [PubMed] [Google Scholar]
- 130.Lindemans CA, Leen AM, Boelens JJ. 2010. How I treat adenovirus in hematopoietic stem cell transplant recipients. Blood 116:5476–5485. 10.1182/blood-2010-04-259291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.La Rosa AM, Champlin RE, Mirza N, Gajewski J, Giralt S, Rolston KV, Raad I, Jacobson K, Kontoyiannis D, Elting L, Whimbey E. 2001. Adenovirus infections in adult recipients of blood and marrow transplants. Clin. Infect. Dis. 32:871–876. 10.1086/319352 [DOI] [PubMed] [Google Scholar]
- 132.Myers GD, Krance RA, Weiss H, Kuehnle I, Demmler G, Heslop HE, Bollard CM. 2005. Adenovirus infection rates in pediatric recipients of alternate donor allogeneic bone marrow transplants receiving either antithymocyte globulin (ATG) or alemtuzumab (Campath). Bone Marrow Transplant. 36:1001–1008. 10.1038/sj.bmt.1705164 [DOI] [PubMed] [Google Scholar]
- 133.Taniguchi K, Yoshihara S, Tamaki H, Fujimoto T, Ikegame K, Kaida K, Nakata J, Inoue T, Kato R, Fujioka T, Okada M, Soma T, Ogawa H. 2012. Incidence and treatment strategy for disseminated adenovirus disease after haploidentical stem cell transplantation. Ann. Hematol. 91:1305–1312. 10.1007/s00277-012-1440-3 [DOI] [PubMed] [Google Scholar]
- 134.Robin M, Marque-Juillet S, Scieux C, Peffault de Latour R, Ferry C, Rocha V, Molina JM, Bergeron A, Devergie A, Gluckman E, Ribaud P, Socie G. 2007. Disseminated adenovirus infections after allogeneic hematopoietic stem cell transplantation: incidence, risk factors and outcome. Haematologicaae 92:1254–1257. 10.3324/haematol.11279 [DOI] [PubMed] [Google Scholar]
- 135.Lee YJ, Chung D, Xiao K, Papadopoulos EB, Barker JN, Small TN, Giralt SA, Zheng J, Jakubowski AA, Papanicolaou GA. 2013. Adenovirus viremia and disease: comparison of T cell-depleted and conventional hematopoietic stem cell transplantation recipients from a single institution. Biol. Blood Marrow Transplant. 19:387–392. 10.1016/j.bbmt.2012.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hiwarkar P, Gaspar HB, Gilmour K, Jagani M, Chiesa R, Bennett-Rees N, Breuer J, Rao K, Cale C, Goulden N, Davies G, Amrolia P, Veys P, Qasim W. 2013. Impact of viral reactivations in the era of pre-emptive antiviral drug therapy following allogeneic haematopoietic SCT in paediatric recipients. Bone Marrow Transplant. 48:803–808. 10.1038/bmt.2012.221 [DOI] [PubMed] [Google Scholar]
- 137.Feuchtinger T, Lang P, Handgretinger R. 2007. Adenovirus infection after allogeneic stem cell transplantation. Leuk. Lymphoma 48:244–255. 10.1080/10428190600881157 [DOI] [PubMed] [Google Scholar]
- 138.Myers GD, Bollard CM, Wu MF, Weiss H, Rooney CM, Heslop HE, Leen AM. 2007. Reconstitution of adenovirus-specific cell-mediated immunity in pediatric patients after hematopoietic stem cell transplantation. Bone Marrow Transplant. 39:677–686. 10.1038/sj.bmt.1705645 [DOI] [PubMed] [Google Scholar]
- 139.Heemskerk B, Lankester AC, van Vreeswijk T, Beersma MF, Claas EC, Veltrop-Duits LA, Kroes AC, Vossen JM, Schilham MW, van Tol MJ. 2005. Immune reconstitution and clearance of human adenovirus viremia in pediatric stem-cell recipients. J. Infect. Dis. 191:520–530. 10.1086/427513 [DOI] [PubMed] [Google Scholar]
- 140.Guerin-El Khourouj V, Dalle JH, Pedron B, Yakouben K, Bensoussan D, Cordeiro DJ, Peltier L, Ouachee-Chardin M, Baruchel A, Sterkers G. 2011. Quantitative and qualitative CD4 T cell immune responses related to adenovirus DNAemia in hematopoietic stem cell transplantation. Biol. Blood Marrow Transplant. 17:476–485. 10.1016/j.bbmt.2010.09.010 [DOI] [PubMed] [Google Scholar]
- 141.Zandvliet ML, van Liempt E, Jedema I, Kruithof S, Kester MG, Guchelaar HJ, Falkenburg JH, Meij P. 2011. Simultaneous isolation of CD8(+) and CD4(+) T cells specific for multiple viruses for broad antiviral immune reconstitution after allogeneic stem cell transplantation. J. Immunother. 34:307–319. 10.1097/CJI.0b013e318213cb90 [DOI] [PubMed] [Google Scholar]
- 142.Anderson EJ, Guzman-Cottrill JA, Kletzel M, Thormann K, Sullivan C, Zheng X, Katz BZ. 2008. High-risk adenovirus-infected pediatric allogeneic hematopoietic progenitor cell transplant recipients and preemptive cidofovir therapy. Pediatr. Transplant. 12:219–227. 10.1111/j.1399-3046.2007.00851.x [DOI] [PubMed] [Google Scholar]
- 143.Matthes-Martin S, Feuchtinger T, Shaw PJ, Engelhard D, Hirsch HH, Cordonnier C, Ljungman P, Fourth European Conference on Infections in Leukemia 2012. European guidelines for diagnosis and treatment of adenovirus infection in leukemia and stem cell transplantation: summary of ECIL-4 (2011). Transpl. Infect. Dis. 14:555–563. 10.1111/tid.12022 [DOI] [PubMed] [Google Scholar]
- 144.Al-Herz W, Moussa MA. 2012. Survival and predictors of death among primary immunodeficient patients: a registry-based study. J. Clin. Immunol. 32:467–473. 10.1007/s10875-011-9636-1 [DOI] [PubMed] [Google Scholar]
- 145.Nebbia G, Chawla A, Schutten M, Atkinson C, Raza M, Johnson M, Geretti A. 2005. Adenovirus viraemia and dissemination unresponsive to antiviral therapy in advanced HIV-1 infection. AIDS 19:1339–1340. 10.1097/01.aids.0000180115.26561.27 [DOI] [PubMed] [Google Scholar]
- 146.Adeyemi OA, Yeldandi AV, Ison MG. 2008. Fatal adenovirus pneumonia in a person with AIDS and Burkitt lymphoma: a case report and review of the literature. AIDS Read. 18:196–198, 201–202, 206–207 [PubMed] [Google Scholar]
- 147.Thomas PD, Pollok RC, Gazzard BG. 1999. Enteric viral infections as a cause of diarrhoea in the acquired immunodeficiency syndrome. HIV Med. 1:19–24. 10.1046/j.1468-1293.1999.00004.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Dikov D, Chatelet FP, Dimitrakov J. 2005. Pathologic features of necrotizing adenoviral prostatitis in an AIDS patient. Int. J. Surg. Pathol. 13:227–231. 10.1177/106689690501300218 [DOI] [PubMed] [Google Scholar]
- 149.Vincentelli C, Schniederjan MJ, Brat DJ. 2010. 35-year-old HIV-positive woman with basal forebrain mass. Brain Pathol. 20:265–268. 10.1111/j.1750-3639.2009.00347.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Huppmann AR, Orenstein JM. 2010. Opportunistic disorders of the gastrointestinal tract in the age of highly active antiretroviral therapy. Hum. Pathol. 41:1777–1787. 10.1016/j.humpath.2010.06.007 [DOI] [PubMed] [Google Scholar]
- 151.Khoo SH, Bailey AS, de Jong JC, Mandal BK. 1995. Adenovirus infections in human immunodeficiency virus-positive patients: clinical features and molecular epidemiology. J. Infect. Dis. 172:629–637. 10.1093/infdis/172.3.629 [DOI] [PubMed] [Google Scholar]
- 152.Schnurr D, Bollen A, Crawford-Miksza L, Dondero ME, Yagi S. 1995. Adenovirus mixture isolated from the brain of an AIDS patient with encephalitis. J. Med. Virol. 47:168–171. 10.1002/jmv.1890470210 [DOI] [PubMed] [Google Scholar]
- 153.Steiner I, Aebi C, Ridolfi Luthy A, Wagner B, Leibundgut K. 2008. Fatal adenovirus hepatitis during maintenance therapy for childhood acute lymphoblastic leukemia. Pediatr. Blood Cancer 50:647–649. 10.1002/pbc.21120 [DOI] [PubMed] [Google Scholar]
- 154.Roch N, Salameire D, Gressin R, Morand P, Epaulard O, Pavese P, Brion JP, Stahl JP. 2008. Fatal adenoviral and enteroviral infections and an Epstein-Barr virus positive large B-cell lymphoma after alemtuzumab treatment in a patient with refractory Sezary syndrome. Scand. J. Infect. Dis. 40:343–346. 10.1080/00365540701684817 [DOI] [PubMed] [Google Scholar]
- 155.Hoffman JA. 2009. Adenovirus infections in solid organ transplant recipients. Curr. Opin. Organ Transplant. 14:625–633. 10.1097/MOT.0b013e3283324e1b [DOI] [PubMed] [Google Scholar]
- 156.Florescu DF, Hoffman JA, AST Infectious Diseases Community of Practice 2013. Adenovirus in solid organ transplantation. Am. J. Transplant. 13(Suppl 4):206–211. 10.1111/ajt.12112 [DOI] [PubMed] [Google Scholar]
- 157.Humar A, Kumar D, Mazzulli T, Razonable RR, Moussa G, Paya CV, Covington E, Alecock E, Pescovitz MD, PV16000 Study Group 2005. A surveillance study of adenovirus infection in adult solid organ transplant recipients. Am. J. Transplant. 5:2555–2559. 10.1111/j.1600-6143.2005.01033.x [DOI] [PubMed] [Google Scholar]
- 158.Florescu MC, Miles CD, Florescu DF. 2013. What do we know about adenovirus in renal transplantation? Nephrol. Dial. Transplant. 28:2003–2010. 10.1093/ndt/gft036 [DOI] [PubMed] [Google Scholar]
- 159.Watcharananan SP, Avery R, Ingsathit A, Malathum K, Chantratita W, Mavichak V, Chalermsanyakorn P, Jirasiritham S, Sumethkul V. 2011. Adenovirus disease after kidney transplantation: course of infection and outcome in relation to blood viral load and immune recovery. Am. J. Transplant. 11:1308–1314. 10.1111/j.1600-6143.2011.03479.x [DOI] [PubMed] [Google Scholar]
- 160.Clark NM, Lynch JP, 3rd, Sayah D, Belperio JA, Fishbein MC, Weigt SS. 2013. DNA viral infections complicating lung transplantation. Semin. Respir. Crit. Care Med. 34:380–404. 10.1055/s-0033-1348473 [DOI] [PubMed] [Google Scholar]
- 161.Vu DL, Bridevaux PO, Aubert JD, Soccal PM, Kaiser L. 2011. Respiratory viruses in lung transplant recipients: a critical review and pooled analysis of clinical studies. Am. J. Transplant. 11:1071–1078. 10.1111/j.1600-6143.2011.03490.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bridges ND, Spray TL, Collins MH, Bowles NE, Towbin JA. 1998. Adenovirus infection in the lung results in graft failure after lung transplantation. J. Thorac. Cardiovasc. Surg. 116:617–623. 10.1016/S0022-5223(98)70168-0 [DOI] [PubMed] [Google Scholar]
- 163.Moulik M, Breinholt JP, Dreyer WJ, Kearney DL, Price JF, Clunie SK, Moffett BS, Kim JJ, Rossano JW, Jefferies JL, Bowles KR, O'Brian Smith E, Bowles NE, Denfield SW, Towbin JA. 2010. Viral endomyocardial infection is an independent predictor and potentially treatable risk factor for graft loss and coronary vasculopathy in pediatric cardiac transplant recipients. J. Am. Coll. Cardiol. 56:582–592. 10.1016/j.jacc.2010.02.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Engelmann G, Heim A, Greil J, Schmitt CP, Flechtenmacher C, Daum E, Kusters U, Schmidt J, Meyburg J, Schnitzler P. 2009. Adenovirus infection and treatment with cidofovir in children after liver transplantation. Pediatr. Transplant. 13:421–428. 10.1111/j.1399-3046.2008.01014.x [DOI] [PubMed] [Google Scholar]
- 165.de Mezerville MH, Tellier R, Richardson S, Hebert D, Doyle J, Allen U. 2006. Adenoviral infections in pediatric transplant recipients: a hospital-based study. Pediatr. Infect. Dis. J. 25:815–818. 10.1097/01.inf.0000233542.48267.fd [DOI] [PubMed] [Google Scholar]
- 166.Liu M, Worley S, Arrigain S, Aurora P, Ballmann M, Boyer D, Conrad C, Eichler I, Elidemir O, Goldfarb S, Mallory GB, Mogayzel PJ, Parakininkas D, Visner G, Sweet S, Faro A, Michaels M, Danziger-Isakov LA. 2009. Respiratory viral infections within one year after pediatric lung transplant. Transpl. Infect. Dis. 11:304–312. 10.1111/j.1399-3062.2009.00397.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Liu M, Mallory GB, Schecter MG, Worley S, Arrigain S, Robertson J, Elidemir O, Danziger-Isakov LA. 2010. Long-term impact of respiratory viral infection after pediatric lung transplantation. Pediatr. Transplant. 14:431–436. 10.1111/j.1399-3046.2010.01296.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Shirali GS, Ni J, Chinnock RE, Johnston JK, Rosenthal GL, Bowles NE, Towbin JA. 2001. Association of viral genome with graft loss in children after cardiac transplantation. N. Engl. J. Med. 344:1498–1503. 10.1056/NEJM200105173442002 [DOI] [PubMed] [Google Scholar]
- 169.Pinchoff RJ, Kaufman SS, Magid MS, Erdman DD, Gondolesi GE, Mendelson MH, Tane K, Jenkins SG, Fishbein TM, Herold BC. 2003. Adenovirus infection in pediatric small bowel transplantation recipients. Transplantation 76:183–189. 10.1097/01.TP.0000072808.93060.0F [DOI] [PubMed] [Google Scholar]
- 170.Florescu DF, Islam MK, Mercer DF, Grant W, Langnas AN, Freifeld AG, Sudan D, Basappa R, Dimaio D, Kalil AC. 2010. Adenovirus infections in pediatric small bowel transplant recipients. Transplantation 90:198–204. 10.1097/00007890-201007272-00380, [DOI] [PubMed] [Google Scholar]
- 171.Abinun M, Flood TJ, Cant AJ, Veys P, Gennery AR, Foster HE, Friswell M, Baildam E, Davidson J, Southwood TR, Livermore P, Wedderburn LR. 2009. Autologous T cell depleted haematopoietic stem cell transplantation in children with severe juvenile idiopathic arthritis in the UK (2000–2007). Mol. Immunol. 47:46–51. 10.1016/j.molimm.2008.12.029 [DOI] [PubMed] [Google Scholar]
- 172.Erard V, Huang ML, Ferrenberg J, Nguy L, Stevens-Ayers TL, Hackman RC, Corey L, Boeckh M. 2007. Quantitative real-time polymerase chain reaction for detection of adenovirus after T cell-replete hematopoietic cell transplantation: viral load as a marker for invasive disease. Clin. Infect. Dis. 45:958–965. 10.1086/521851 [DOI] [PubMed] [Google Scholar]
- 173.Walls T, Hawrami K, Ushiro-Lumb I, Shingadia D, Saha V, Shankar AG. 2005. Adenovirus infection after pediatric bone marrow transplantation: is treatment always necessary? Clin. Infect. Dis. 40:1244–1249. 10.1086/429235 [DOI] [PubMed] [Google Scholar]
- 174.Chakrabarti S, Mautner V, Osman H, Collingham KE, Fegan CD, Klapper PE, Moss PA, Milligan DW. 2002. Adenovirus infections following allogeneic stem cell transplantation: incidence and outcome in relation to graft manipulation, immunosuppression, and immune recovery. Blood 100:1619–1627. 10.1182/blood-2002-02-0377 [DOI] [PubMed] [Google Scholar]
- 175.Gustafson I, Lindblom A, Yun Z, Omar H, Engstrom L, Lewensohn-Fuchs I, Ljungman P, Broliden K. 2008. Quantification of adenovirus DNA in unrelated donor hematopoietic stem cell transplant recipients. J. Clin. Virol. 43:79–85. 10.1016/j.jcv.2008.04.014 [DOI] [PubMed] [Google Scholar]
- 176.Flomenberg P, Babbitt J, Drobyski WR, Ash RC, Carrigan DR, Sedmak GV, McAuliffe T, Camitta B, Horowitz MM, Bunin N. 1994. Increasing incidence of adenovirus disease in bone marrow transplant recipients. J. Infect. Dis. 169:775–781. 10.1016/j.jacc.2010.02.060 [DOI] [PubMed] [Google Scholar]
- 177.Ljungman P, Ribaud P, Eyrich M, Matthes-Martin S, Einsele H, Bleakley M, Machaczka M, Bierings M, Bosi A, Gratecos N, Cordonnier C, Infectious Diseases Working Party of the European Group for Blood and Marrow Transplantation 2003. Cidofovir for adenovirus infections after allogeneic hematopoietic stem cell transplantation: a survey by the Infectious Diseases Working Party of the European Group for Blood and Marrow Transplantation. Bone Marrow Transplant. 31:481–486. 10.1038/sj.bmt.1703798 [DOI] [PubMed] [Google Scholar]
- 178.Suparno C, Milligan DW, Moss PA, Mautner V. 2004. Adenovirus infections in stem cell transplant recipients: recent developments in understanding of pathogenesis, diagnosis and management. Leuk. Lymphoma 45:873–885. 10.1080/10428190310001628176 [DOI] [PubMed] [Google Scholar]
- 179.Kojaoghlanian T, Flomenberg P, Horwitz MS. 2003. The impact of adenovirus infection on the immunocompromised host. Rev. Med. Virol. 13:155–171. 10.1002/rmv.386 [DOI] [PubMed] [Google Scholar]
- 180.Seidemann K, Heim A, Pfister ED, Koditz H, Beilken A, Sander A, Melter M, Sykora KW, Sasse M, Wessel A. 2004. Monitoring of adenovirus infection in pediatric transplant recipients by quantitative PCR: report of six cases and review of the literature. Am. J. Transplant. 4:2102–2108. 10.1111/j.1600-6143.2004.00631.x [DOI] [PubMed] [Google Scholar]
- 181.Humar A, Doucette K, Kumar D, Pang XL, Lien D, Jackson K, Preiksaitis J. 2006. Assessment of adenovirus infection in adult lung transplant recipients using molecular surveillance. J. Heart Lung Transplant. 25:1441–1446. 10.1016/j.healun.2006.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kalpoe JS, van der Heiden PL, Barge RM, Houtzager S, Lankester AC, van Tol MJ, Kroes AC. 2007. Assessment of disseminated adenovirus infections using quantitative plasma PCR in adult allogeneic stem cell transplant recipients receiving reduced intensity or myeloablative conditioning. Eur. J. Haematol. 78:314–321. 10.1111/j.1600-0609.2007.00821.x [DOI] [PubMed] [Google Scholar]
- 183.Omar H, Yun Z, Lewensohn-Fuchs I, Perez-Bercoff L, Orvell C, Engstrom L, Vuong GK, Ljungman P. 2010. Poor outcome of adenovirus infections in adult hematopoietic stem cell transplant patients with sustained adenovirus viremia. Transpl. Infect. Dis. 12:465–469. 10.1111/j.1399-3062.2010.00528.x [DOI] [PubMed] [Google Scholar]
- 184.Forstmeyer D, Henke-Gendo C, Brocker V, Wildner O, Heim A. 2008. Quantitative temporal and spatial distribution of adenovirus type 2 correlates with disease manifestations and organ failure during disseminated infection. J. Med. Virol. 80:294–297. 10.1002/jmv.21071 [DOI] [PubMed] [Google Scholar]
- 185.Avivi I, Chakrabarti S, Milligan DW, Waldmann H, Hale G, Osman H, Ward KN, Fegan CD, Yong K, Goldstone AH, Linch DC, Mackinnon S. 2004. Incidence and outcome of adenovirus disease in transplant recipients after reduced-intensity conditioning with alemtuzumab. Biol. Blood Marrow Transplant. 10:186–194. 10.1016/j.bbmt.2003.11.001 [DOI] [PubMed] [Google Scholar]
- 186.Ganzenmueller T, Buchholz S, Harste G, Dammann E, Trenschel R, Heim A. 2011. High lethality of human adenovirus disease in adult allogeneic stem cell transplant recipients with high adenoviral blood load. J. Clin. Virol. 52:55–59. 10.1016/j.jcv.2011.06.005 [DOI] [PubMed] [Google Scholar]
- 187.Heim A. 2011. Advances in the management of disseminated adenovirus disease in stem cell transplant recipients: impact of adenovirus load (DNAemia) testing. Expert Rev. Anti Infect. Ther. 9:943–945. 10.1586/eri.11.113 [DOI] [PubMed] [Google Scholar]
- 188.Jeulin H, Salmon A, Bordigoni P, Venard V. 2011. Diagnostic value of quantitative PCR for adenovirus detection in stool samples as compared with antigen detection and cell culture in haematopoietic stem cell transplant recipients. Clin. Microbiol. Infect. 17:1674–1680. 10.1111/j.1469-0691.2011.03488.x [DOI] [PubMed] [Google Scholar]
- 189.Legoff J, Feghoul L, Mercier-Delarue S, Dalle JH, Scieux C, Cherot J, de Fontbrune FS, Baruchel A, Socie G, Simon F. 2013. Broad-range PCR-electrospray ionization mass spectrometry for detection and typing of adenovirus and other opportunistic viruses in stem cell transplant patients. J. Clin. Microbiol. 51:4186–4192. 10.1128/JCM.01978-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kim YJ, Boeckh M, Englund JA. 2007. Community respiratory virus infections in immunocompromised patients: hematopoietic stem cell and solid organ transplant recipients, and individuals with human immunodeficiency virus infection. Semin. Respir. Crit. Care Med. 28:222–242. 10.1055/s-2007-976494 [DOI] [PubMed] [Google Scholar]
- 191.Lee J, Choi EH, Lee HJ. 2010. Comprehensive serotyping and epidemiology of human adenovirus isolated from the respiratory tract of Korean children over 17 consecutive years (1991–2007). J. Med. Virol. 82:624–631. 10.1002/jmv.21701 [DOI] [PubMed] [Google Scholar]
- 192.Lankester AC, van Tol MJ, Claas EC, Vossen JM, Kroes AC. 2002. Quantification of adenovirus DNA in plasma for management of infection in stem cell graft recipients. Clin. Infect. Dis. 34:864–867. 10.1086/339073 [DOI] [PubMed] [Google Scholar]
- 193.Huang ML, Nguy L, Ferrenberg J, Boeckh M, Cent A, Corey L. 2008. Development of multiplexed real-time quantitative polymerase chain reaction assay for detecting human adenoviruses. Diagn. Microbiol. Infect. Dis. 62:263–271. 10.1016/j.diagmicrobio.2008.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ebner K, Suda M, Watzinger F, Lion T. 2005. Molecular detection and quantitative analysis of the entire spectrum of human adenoviruses by a two-reaction real-time PCR assay. J. Clin. Microbiol. 43:3049–3053. 10.1128/JCM.43.7.3049-3053.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Heim A, Ebnet C, Harste G, Pring-Akerblom P. 2003. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J. Med. Virol. 70:228–239. 10.1002/jmv.10382 [DOI] [PubMed] [Google Scholar]
- 196.Popowitch EB, O'Neill SS, Miller MB. 2013. Comparison of the Biofire FilmArray RP, Genmark eSensor RVP, Luminex xTAG RVPv1, and Luminex xTAG RVP Fast multiplex assays for detection of respiratory viruses. J. Clin. Microbiol. 51:1528–1533. 10.1128/JCM.03368-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kim HK, Oh SH, Yun KA, Sung H, Kim MN. 2013. Comparison of Anyplex II RV16 with the xTAG respiratory viral panel and Seeplex RV15 for detection of respiratory viruses. J. Clin. Microbiol. 51:1137–1141. 10.1128/JCM.02958-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Couturier MR, Barney T, Alger G, Hymas WC, Stevenson JB, Hillyard D, Daly JA. 2013. Evaluation of the FilmArray(R) respiratory panel for clinical use in a large children's hospital. J. Clin. Lab. Anal. 27:148–154. 10.1002/jcla.21576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Jeulin H, Salmon A, Bordigoni P, Venard V. 2010. Comparison of in-house real-time quantitative PCR to the Adenovirus R-Gene kit for determination of adenovirus load in clinical samples. J. Clin. Microbiol. 48:3132–3137. 10.1128/JCM.00976-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Legrand F, Berrebi D, Houhou N, Freymuth F, Faye A, Duval M, Mougenot JF, Peuchmaur M, Vilmer E. 2001. Early diagnosis of adenovirus infection and treatment with cidofovir after bone marrow transplantation in children. Bone Marrow Transplant. 27:621–626. 10.1038/sj.bmt.1702820 [DOI] [PubMed] [Google Scholar]
- 201.Baldwin A, Kingman H, Darville M, Foot AB, Grier D, Cornish JM, Goulden N, Oakhill A, Pamphilon DH, Steward CG, Marks DI. 2000. Outcome and clinical course of 100 patients with adenovirus infection following bone marrow transplantation. Bone Marrow Transplant. 26:1333–1338. 10.1038/sj.bmt.1702716 [DOI] [PubMed] [Google Scholar]
- 202.Kalu SU, Loeffelholz M, Beck E, Patel JA, Revai K, Fan J, Henrickson KJ, Chonmaitree T. 2010. Persistence of adenovirus nucleic acids in nasopharyngeal secretions: a diagnostic conundrum. Pediatr. Infect. Dis. J. 29:746–750. 10.1097/INF.0b013e3181d743c8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Jaggi P, Kajon AE, Mejias A, Ramilo O, Leber A. 2013. Human adenovirus infection in Kawasaki disease: a confounding bystander? Clin. Infect. Dis. 56:58–64. 10.1093/cid/cis807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Teramura T, Naya M, Yoshihara T, Kanoh G, Morimoto A, Imashuku S. 2004. Adenoviral infection in hematopoietic stem cell transplantation: early diagnosis with quantitative detection of the viral genome in serum and urine. Bone Marrow Transplant. 33:87–92. 10.1038/sj.bmt.1704320 [DOI] [PubMed] [Google Scholar]
- 205.Claas EC, Schilham MW, de Brouwer CS, Hubacek P, Echavarria M, Lankester AC, van Tol MJ, Kroes AC. 2005. Internally controlled real-time PCR monitoring of adenovirus DNA load in serum or plasma of transplant recipients. J. Clin. Microbiol. 43:1738–1744. 10.1128/JCM.43.4.1738-1744.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Ganzenmueller T, Heim A. 2012. Adenoviral load diagnostics by quantitative polymerase chain reaction: techniques and application. Rev. Med. Virol. 22:194–208. 10.1002/rmv.724 [DOI] [PubMed] [Google Scholar]
- 207.Feuchtinger T, Matthes-Martin S, Richard C, Lion T, Fuhrer M, Hamprecht K, Handgretinger R, Peters C, Schuster FR, Beck R, Schumm M, Lotfi R, Jahn G, Lang P. 2006. Safe adoptive transfer of virus-specific T-cell immunity for the treatment of systemic adenovirus infection after allogeneic stem cell transplantation. Br. J. Haematol. 134:64–76. 10.1111/j.1365-2141.2006.06108.x [DOI] [PubMed] [Google Scholar]
- 208.Echavarria M, Maldonado D, Elbert G, Videla C, Rappaport R, Carballal G. 2006. Use of PCR to demonstrate presence of adenovirus species B, C, or F as well as coinfection with two adenovirus species in children with flu-like symptoms. J. Clin. Microbiol. 44:625–627. 10.1128/JCM.44.2.625-627.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.De Jong JC, Wermenbol AG, Verweij-Uijterwaal MW, Slaterus KW, Wertheim-Van Dillen P, Van Doornum GJ, Khoo SH, Hierholzer JC. 1999. Adenoviruses from human immunodeficiency virus-infected individuals, including two strains that represent new candidate serotypes Ad50 and Ad51 of species B1 and D, respectively. J. Clin. Microbiol. 37:3940–3945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Xu W, McDonough MC, Erdman DD. 2000. Species-specific identification of human adenoviruses by a multiplex PCR assay. J. Clin. Microbiol. 38:4114–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Vabret A, Gouarin S, Joannes M, Barranger C, Petitjean J, Corbet S, Brouard J, Lafay F, Duhamel JF, Guillois B, Freymuth F. 2004. Development of a PCR- and hybridization-based assay (PCR Adenovirus Consensus) for the detection and the species identification of adenoviruses in respiratory specimens. J. Clin. Virol. 31:116–122. 10.1016/j.jcv.2004.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Casas I, Avellon A, Mosquera M, Jabado O, Echevarria JE, Campos RH, Rewers M, Perez-Brena P, Lipkin WI, Palacios G. 2005. Molecular identification of adenoviruses in clinical samples by analyzing a partial hexon genomic region. J. Clin. Microbiol. 43:6176–6182. 10.1128/JCM.43.12.6176-6182.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Sarantis H, Johnson G, Brown M, Petric M, Tellier R. 2004. Comprehensive detection and serotyping of human adenoviruses by PCR and sequencing. J. Clin. Microbiol. 42:3963–3969. 10.1128/JCM.42.9.3963-3969.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Ebner K, Rauch M, Preuner S, Lion T. 2006. Typing of human adenoviruses in specimens from immunosuppressed patients by PCR-fragment length analysis and real-time quantitative PCR. J. Clin. Microbiol. 44:2808–2815. 10.1128/JCM.00048-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Lu X, Erdman DD. 2006. Molecular typing of human adenoviruses by PCR and sequencing of a partial region of the hexon gene. Arch. Virol. 151:1587–1602. 10.1007/s00705-005-0722-7 [DOI] [PubMed] [Google Scholar]
- 216.Tomblyn M, Chiller T, Einsele H, Gress R, Sepkowitz K, Storek J, Wingard JR, Young JA, Boeckh MJ. 2009. Guidelines for preventing infectious complications among hematopoietic cell transplant recipients: a global perspective. Preface. Bone Marrow Transplant. 44:453–455. 10.1038/bmt.2009.254 [DOI] [PubMed] [Google Scholar]
- 217.Hoffman JA, Shah AJ, Ross LA, Kapoor N. 2001. Adenoviral infections and a prospective trial of cidofovir in pediatric hematopoietic stem cell transplantation. Biol. Blood Marrow Transplant. 7:388–394. 10.1053/bbmt.2001.v7.pm11529489 [DOI] [PubMed] [Google Scholar]
- 218.Leruez-Ville M, Minard V, Lacaille F, Buzyn A, Abachin E, Blanche S, Freymuth F, Rouzioux C. 2004. Real-time blood plasma polymerase chain reaction for management of disseminated adenovirus infection. Clin. Infect. Dis. 38:45–52. 10.1086/380450 [DOI] [PubMed] [Google Scholar]
- 219.Yusuf U, Hale GA, Carr J, Gu Z, Benaim E, Woodard P, Kasow KA, Horwitz EM, Leung W, Srivastava DK, Handgretinger R, Hayden RT. 2006. Cidofovir for the treatment of adenoviral infection in pediatric hematopoietic stem cell transplant patients. Transplantation 81:1398–1404. 10.1097/01.tp.0000209195.95115.8e [DOI] [PubMed] [Google Scholar]
- 220.Neofytos D, Ojha A, Mookerjee B, Wagner J, Filicko J, Ferber A, Dessain S, Grosso D, Brunner J, Flomenberg N, Flomenberg P. 2007. Treatment of adenovirus disease in stem cell transplant recipients with cidofovir. Biol. Blood Marrow Transplant. 13:74–81. 10.1016/j.bbmt.2006.08.040 [DOI] [PubMed] [Google Scholar]
- 221.Doan ML, Mallory GB, Kaplan SL, Dishop MK, Schecter MG, McKenzie ED, Heinle JS, Elidemir O. 2007. Treatment of adenovirus pneumonia with cidofovir in pediatric lung transplant recipients. J. Heart Lung Transplant. 26:883–889. 10.1016/j.healun.2007.06.009 [DOI] [PubMed] [Google Scholar]
- 222.Symeonidis N, Jakubowski A, Pierre-Louis S, Jaffe D, Pamer E, Sepkowitz K, O'Reilly RJ, Papanicolaou GA. 2007. Invasive adenoviral infections in T-cell-depleted allogeneic hematopoietic stem cell transplantation: high mortality in the era of cidofovir. Transpl. Infect. Dis. 9:108–113. 10.1111/j.1399-3062.2006.00184.x [DOI] [PubMed] [Google Scholar]
- 223.Lenaerts L, Naesens L. 2006. Antiviral therapy for adenovirus infections. Antiviral Res. 71:172–180. 10.1016/j.antiviral.2006.04.007 [DOI] [PubMed] [Google Scholar]
- 224.Morfin F, Dupuis-Girod S, Mundweiler S, Falcon D, Carrington D, Sedlacek P, Bierings M, Cetkovsky P, Kroes AC, van Tol MJ, Thouvenot D. 2005. In vitro susceptibility of adenovirus to antiviral drugs is species-dependent. Antivir. Ther. 10:225–229 [PubMed] [Google Scholar]
- 225.Morfin F, Dupuis-Girod S, Frobert E, Mundweiler S, Carrington D, Sedlacek P, Bierings M, Cetkovsky P, Kroes AC, van Tol MJ, Thouvenot D. 2009. Differential susceptibility of adenovirus clinical isolates to cidofovir and ribavirin is not related to species alone. Antivir. Ther. 14:55–61 [PubMed] [Google Scholar]
- 226.Kinchington PR, Araullo-Cruz T, Vergnes JP, Yates K, Gordon YJ. 2002. Sequence changes in the human adenovirus type 5 DNA polymerase associated with resistance to the broad spectrum antiviral cidofovir. Antiviral Res. 56:73–84. 10.1016/S0166-3542(02)00098-0 [DOI] [PubMed] [Google Scholar]
- 227.Nagafuji K, Aoki K, Henzan H, Kato K, Miyamoto T, Eto T, Nagatoshi Y, Ohba T, Obama K, Gondo H, Harada M. 2004. Cidofovir for treating adenoviral hemorrhagic cystitis in hematopoietic stem cell transplant recipients. Bone Marrow Transplant. 34:909–914. 10.1038/sj.bmt.1704682 [DOI] [PubMed] [Google Scholar]
- 228.Bhadri VA, Lee-Horn L, Shaw PJ. 2009. Safety and tolerability of cidofovir in high-risk pediatric patients. Transpl. Infect. Dis. 11:373–379. 10.1111/j.1399-3062.2009.00391.x [DOI] [PubMed] [Google Scholar]
- 229.Cundy KC. 1999. Clinical pharmacokinetics of the antiviral nucleotide analogues cidofovir and adefovir. Clin. Pharmacokinet. 36:127–143. 10.2165/00003088-199936020-00004 [DOI] [PubMed] [Google Scholar]
- 230.Naesens L, Lenaerts L, Andrei G, Snoeck R, Van Beers D, Holy A, Balzarini J, De Clercq E. 2005. Antiadenovirus activities of several classes of nucleoside and nucleotide analogues. Antimicrob. Agents Chemother. 49:1010–1016. 10.1128/AAC.49.3.1010-1016.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Abe S, Miyamura K, Oba T, Terakura S, Kasai M, Kitaori K, Sasaki T, Kodera Y. 2003. Oral ribavirin for severe adenovirus infection after allogeneic marrow transplantation. Bone Marrow Transplant. 32:1107–1108. 10.1038/sj.bmt.1704276 [DOI] [PubMed] [Google Scholar]
- 232.Lankester AC, Heemskerk B, Claas EC, Schilham MW, Beersma MF, Bredius RG, van Tol MJ, Kroes AC. 2004. Effect of ribavirin on the plasma viral DNA load in patients with disseminating adenovirus infection. Clin. Infect. Dis. 38:1521–1525. 10.1086/420817 [DOI] [PubMed] [Google Scholar]
- 233.Homma M, Inoue Y, Hasegawa Y, Kojima H, Kohda Y. 2008. Blood ribavirin concentration in high-dose ribavirin for adenovirus-induced haemorrhagic cystitis—a case report. J. Clin. Pharm. Ther. 33:75–78. 10.1111/j.1365-2710.2008.00874.x [DOI] [PubMed] [Google Scholar]
- 234.Kinchington PR, Romanowski EG, Jerold Gordon Y. 2005. Prospects for adenovirus antivirals. J. Antimicrob. Chemother. 55:424–429. 10.1093/jac/dki057 [DOI] [PubMed] [Google Scholar]
- 235.Riner A, Chan-Tack KM, Murray JS. 2009. Intravenous ribavirin—review of the FDA's Emergency Investigational New Drug Database (1997–2008) and literature review. Postgrad. Med. 121:139–146. 10.3810/pgm.2009.05.2014, [DOI] [PubMed] [Google Scholar]
- 236.Breuer S, Rauch M, Matthes-Martin S, Lion T. 2012. Molecular diagnosis and management of viral infections in hematopoietic stem cell transplant recipients. Mol. Diagn. Ther. 16:63–77. 10.1007/BF03256431 [DOI] [PubMed] [Google Scholar]
- 237.Gavin PJ, Katz BZ. 2002. Intravenous ribavirin treatment for severe adenovirus disease in immunocompromised children. Pediatrics 110:e9. 10.1542/peds.110.1.e9 [DOI] [PubMed] [Google Scholar]
- 238.Darr S, Madisch I, Heim A. 2008. Antiviral activity of cidofovir and ribavirin against the new human adenovirus subtype 14a that is associated with severe pneumonia. Clin. Infect. Dis. 47:731–732. 10.1086/590970 [DOI] [PubMed] [Google Scholar]
- 239.Shah DP, Ghantoji SS, Mulanovich VE, Ariza-Heredia EJ, Chemaly RF. 2012. Management of respiratory viral infections in hematopoietic cell transplant recipients. Am. J. Blood Res. 2:203–218 [PMC free article] [PubMed] [Google Scholar]
- 240.Bruno B, Gooley T, Hackman RC, Davis C, Corey L, Boeckh M. 2003. Adenovirus infection in hematopoietic stem cell transplantation: effect of ganciclovir and impact on survival. Biol. Blood Marrow Transplant. 9:341–352. 10.1016/S1083-8791(03)00102-2 [DOI] [PubMed] [Google Scholar]
- 241.Marty FM, Winston DJ, Rowley SD, Vance E, Papanicolaou GA, Mullane KM, Brundage TM, Robertson AT, Godkin S, Mommeja-Marin H, Boeckh M, CMX001-201 Clinical Study Group 2013. CMX001 to prevent cytomegalovirus disease in hematopoietic-cell transplantation. N. Engl. J. Med. 369:1227–1236. 10.1056/NEJMoa1303688 [DOI] [PubMed] [Google Scholar]
- 242.Bonnafous P, Bogaert S, Godet AN, Agut H. 2013. HDP-CDV as an alternative for treatment of human herpesvirus-6 infections. J. Clin. Virol. 56:175–176. 10.1016/j.jcv.2012.11.006 [DOI] [PubMed] [Google Scholar]
- 243.De Clercq E. 11 March 2013. A cutting-edge view on the current state of antiviral drug development. Med. Res. Rev. 10.1002/med.21281 [DOI] [PubMed] [Google Scholar]
- 244.James SH, Price NB, Hartline CB, Lanier ER, Prichard MN. 2013. Selection and recombinant phenotyping of a novel CMX001 and cidofovir resistance mutation in human cytomegalovirus. Antimicrob. Agents Chemother. 57:3321–3325. 10.1128/AAC.00062-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Paolino K, Sande J, Perez E, Loechelt B, Jantausch B, Painter W, Anderson M, Tippin T, Lanier ER, Fry T, DeBiasi RL. 2011. Eradication of disseminated adenovirus infection in a pediatric hematopoietic stem cell transplantation recipient using the novel antiviral agent CMX001. J. Clin. Virol. 50:167–170. 10.1016/j.jcv.2010.10.016 [DOI] [PubMed] [Google Scholar]
- 246.Florescu DF, Pergam SA, Neely MN, Qiu F, Johnston C, Way S, Sande J, Lewinsohn DA, Guzman-Cottrill JA, Graham ML, Papanicolaou G, Kurtzberg J, Rigdon J, Painter W, Mommeja-Marin H, Lanier R, Anderson M, van der Horst C. 2012. Safety and efficacy of CMX001 as salvage therapy for severe adenovirus infections in immunocompromised patients. Biol. Blood Marrow Transplant. 18:731–738. 10.1016/j.bbmt.2011.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Chakrabarti S, Collingham KE, Fegan CD, Pillay D, Milligan DW. 2000. Adenovirus infections following haematopoietic cell transplantation: is there a role for adoptive immunotherapy? Bone Marrow Transplant. 26:305–307. 10.1038/sj.bmt.1702508 [DOI] [PubMed] [Google Scholar]
- 248.Tang J, Olive M, Pulmanausahakul R, Schnell M, Flomenberg N, Eisenlohr L, Flomenberg P. 2006. Human CD8+ cytotoxic T cell responses to adenovirus capsid proteins. Virology 350:312–322. 10.1016/j.virol.2006.01.024 [DOI] [PubMed] [Google Scholar]
- 249.Tischer S, Kaireit T, Figueiredo C, Hiller O, Maecker-Kolhoff B, Geyeregger R, Immenschuh S, Blasczyk R, Eiz-Vesper B. 2012. Establishment of the reversible peptide-major histocompatibility complex (pMHC) class I histamer technology: tool for visualization and selection of functionally active antigen-specific CD8(+) T lymphocytes. Int. Immunol. 24:561–572. 10.1093/intimm/dxs059 [DOI] [PubMed] [Google Scholar]
- 250.Schmitt A, Tonn T, Busch DH, Grigoleit GU, Einsele H, Odendahl M, Germeroth L, Ringhoffer M, Ringhoffer S, Wiesneth M, Greiner J, Michel D, Mertens T, Rojewski M, Marx M, von Harsdorf S, Dohner H, Seifried E, Bunjes D, Schmitt M. 2011. Adoptive transfer and selective reconstitution of streptamer-selected cytomegalovirus-specific CD8+ T cells leads to virus clearance in patients after allogeneic peripheral blood stem cell transplantation. Transfusion 51:591–599. 10.1111/j.1537-2995.2010.02940.x [DOI] [PubMed] [Google Scholar]
- 251.Eiz-Vesper B, Maecker-Kolhoff B, Blasczyk R. 2012. Adoptive T-cell immunotherapy from third-party donors: characterization of donors and set up of a T-cell donor registry. Front. Immunol. 3:410. 10.3389/fimmu.2012.00410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Chakupurakal G, Onion D, Bonney S, Cobbold M, Mautner V, Moss P. 2013. HLA-peptide multimer selection of adenovirus-specific T cells for adoptive T-cell therapy. J. Immunother. 36:423–431. 10.1097/CJI.0b013e3182a8029e [DOI] [PubMed] [Google Scholar]
- 253.Uhlin M, Gertow J, Uzunel M, Okas M, Berglund S, Watz E, Brune M, Ljungman P, Maeurer M, Mattsson J. 2012. Rapid salvage treatment with virus-specific T cells for therapy-resistant disease. Clin. Infect. Dis. 55:1064–1073. 10.1093/cid/cis625 [DOI] [PubMed] [Google Scholar]
- 254.Gerdemann U, Vera JF, Rooney CM, Leen AM. 2011. Generation of multivirus-specific T cells to prevent/treat viral infections after allogeneic hematopoietic stem cell transplant. J. Vis. Exp. 2011:2736. 10.3791/2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Comoli P, Schilham MW, Basso S, van Vreeswijk T, Bernardo ME, Maccario R, van Tol MJ, Locatelli F, Veltrop-Duits LA. 2008. T-cell lines specific for peptides of adenovirus hexon protein and devoid of alloreactivity against recipient cells can be obtained from HLA-haploidentical donors. J. Immunother. 31:529–536. 10.1097/CJI.0b013e31817b9c6b [DOI] [PubMed] [Google Scholar]
- 256.Feuchtinger T, Richard C, Joachim S, Scheible MH, Schumm M, Hamprecht K, Martin D, Jahn G, Handgretinger R, Lang P. 2008. Clinical grade generation of hexon-specific T cells for adoptive T-cell transfer as a treatment of adenovirus infection after allogeneic stem cell transplantation. J. Immunother. 31:199–206. 10.1097/CJI.0b013e31815ef862 [DOI] [PubMed] [Google Scholar]
- 257.Leen AM, Christin A, Myers GD, Liu H, Cruz CR, Hanley PJ, Kennedy-Nasser AA, Leung KS, Gee AP, Krance RA, Brenner MK, Heslop HE, Rooney CM, Bollard CM. 2009. Cytotoxic T lymphocyte therapy with donor T cells prevents and treats adenovirus and Epstein-Barr virus infections after haploidentical and matched unrelated stem cell transplantation. Blood 114:4283–4292. 10.1182/blood-2009-07-232454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Khanna N, Stuehler C, Conrad B, Lurati S, Krappmann S, Einsele H, Berges C, Topp MS. 2011. Generation of a multipathogen-specific T-cell product for adoptive immunotherapy based on activation-dependent expression of CD154. Blood 118:1121–1131. 10.1182/blood-2010-12-322610 [DOI] [PubMed] [Google Scholar]
- 259.Sili U, Leen AM, Vera JF, Gee AP, Huls H, Heslop HE, Bollard CM, Rooney CM. 2012. Production of good manufacturing practice-grade cytotoxic T lymphocytes specific for Epstein-Barr virus, cytomegalovirus and adenovirus to prevent or treat viral infections post-allogeneic hematopoietic stem cell transplant. Cytotherapy 14:7–11. 10.3109/14653249.2011.636963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Gerdemann U, Keirnan JM, Katari UL, Yanagisawa R, Christin AS, Huye LE, Perna SK, Ennamuri S, Gottschalk S, Brenner MK, Heslop HE, Rooney CM, Leen AM. 2012. Rapidly generated multivirus-specific cytotoxic T lymphocytes for the prophylaxis and treatment of viral infections. Mol. Ther. 20:1622–1632. 10.1038/mt.2012.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Lugthart G, Albon SJ, Ricciardelli I, Kester MG, Meij P, Lankester AC, Amrolia PJ. 2012. Simultaneous generation of multivirus-specific and regulatory T cells for adoptive immunotherapy. J. Immunother. 35:42–53. 10.1097/CJI.0b013e31823569e2 [DOI] [PubMed] [Google Scholar]
- 262.Haveman LM, Bierings M, Klein MR, Beekman JM, de Jager W, Kuis W, Albani S, Prakken BJ. 2013. Selection of perforin expressing CD4+ adenovirus-specific T-cells with artificial antigen presenting cells. Clin. Immunol. 146:228–239. 10.1016/j.clim.2013.01.002 [DOI] [PubMed] [Google Scholar]
- 263.Qasim W, Gilmour K, Zhan H, Derniame S, McNicol AM, Ip W, Hiwarkar P, Veys P, Gaspar HB. 2013. Interferon-gamma capture T cell therapy for persistent adenoviraemia following allogeneic haematopoietic stem cell transplantation. Br. J. Haematol. 161:449–452. 10.1111/bjh.12251 [DOI] [PubMed] [Google Scholar]
- 264.Qasim W, Derniame S, Gilmour K, Chiesa R, Weber M, Adams S, Rao K, Amrolia P, Goulden N, Veys P, Gaspar H. 2011. Third-party virus-specific T cells eradicate adenoviraemia but trigger bystander graft-versus-host disease. Br. J. Haematol. 154:150–153. 10.1111/j.1365-2141.2011.08579.x [DOI] [PubMed] [Google Scholar]
- 265.Sukdolak C, Tischer S, Dieks D, Figueiredo C, Goudeva L, Heuft HG, Verboom M, Immenschuh S, Heim A, Borchers S, Mischak-Weissinger E, Blasczyk R, Maecker-Kolhoff B, Eiz-Vesper B. 2013. CMV-, EBV- and ADV-specific T cell immunity: screening and monitoring of potential third-party donors to improve post-transplantation outcome. Biol. Blood Marrow Transplant. 19:1480–1492. 10.1016/j.bbmt.2013.07.015 [DOI] [PubMed] [Google Scholar]
- 266.Leen AM, Bollard CM, Mendizabal AM, Shpall EJ, Szabolcs P, Antin JH, Kapoor N, Pai SY, Rowley SD, Kebriaei P, Dey BR, Grilley BJ, Gee AP, Brenner MK, Rooney CM, Heslop HE. 2013. Multicenter study of banked third-party virus-specific T cells to treat severe viral infections after hematopoietic stem cell transplantation. Blood 121:5113–5123. 10.1182/blood-2013-02-486324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Pillet S, Lardeux M, Dina J, Grattard F, Verhoeven P, Le Goff J, Vabret A, Pozzetto B. 2013. Comparative evaluation of six commercialized multiplex PCR kits for the diagnosis of respiratory infections. PLoS One 8:e72174. 10.1371/journal.pone.0072174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Imahashi N, Nishida T, Ito Y, Kawada J, Nakazawa Y, Toji S, Suzuki S, Terakura S, Kato T, Murata M, Naoe T. 2013. Identification of a novel HLA-A*24:02-restricted adenovirus serotype 11-specific CD8(+) T-cell epitope for adoptive immunotherapy. Mol. Immunol. 56:399–405. 10.1016/j.molimm.2013.05.232 [DOI] [PubMed] [Google Scholar]
- 269.Slatter MA, Read S, Taylor CE, Crooks BN, Abinun M, Flood TJ, Cant AJ, Wright C, Gennery AR. 2005. Adenovirus type F subtype 41 causing disseminated disease following bone marrow transplantation for immunodeficiency. J. Clin. Microbiol. 43:1462–1464. 10.1128/JCM.43.3.1462-1464.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]