Abstract
SUMMARY
Bacterial pathogens are important targets for detection and identification in medicine, food safety, public health, and security. Bacterial infection is a common cause of morbidity and mortality worldwide. In spite of the availability of antibiotics, these infections are often misdiagnosed or there is an unacceptable delay in diagnosis. Current methods of bacterial detection rely upon laboratory-based techniques such as cell culture, microscopic analysis, and biochemical assays. These procedures are time-consuming and costly and require specialist equipment and trained users. Portable stand-alone biosensors can facilitate rapid detection and diagnosis at the point of care. Biosensors will be particularly useful where a clear diagnosis informs treatment, in critical illness (e.g., meningitis) or to prevent further disease spread (e.g., in case of food-borne pathogens or sexually transmitted diseases). Detection of bacteria is also becoming increasingly important in antibioterrorism measures (e.g., anthrax detection). In this review, we discuss recent progress in the use of biosensors for the detection of whole bacterial cells for sensitive and earlier identification of bacteria without the need for sample processing. There is a particular focus on electrochemical biosensors, especially impedance-based systems, as these present key advantages in terms of ease of miniaturization, lack of reagents, sensitivity, and low cost.
INTRODUCTION
Bacterial pathogens are important targets for detection and identification in various fields, including medicine, food safety, public health, and security. Infectious diseases are among the leading causes of morbidity and mortality worldwide, causing millions of deaths and hospitalizations each year. The World Health Organization (WHO) identified infectious and parasitic diseases collectively as the second-highest cause of death worldwide in 2004, with lower respiratory tract infections (third), diarrheal diseases (fifth), and tuberculosis (seventh) being among the top 10 leading causes of death in 2011 (http://www.who.int/gho/mortality_burden_disease/causes_death/2000_2011/en/index.htmL). These types of infectious or communicable diseases are most problematic in low-income countries, such as countries in Africa, where medical facilities and methods of diagnosis and treatment are lacking. Food-borne pathogens also pose a serious health risk in higher-income countries, including the United States, where food-borne bacteria cause an estimated 76 million illnesses, 300,000 hospitalizations, and 5,000 deaths each year (1, 2). Escherichia coli O157:H7, salmonellae, Campylobacter jejuni, and Listeria monocytogenes are the leading causes of bacterial food- and waterborne illnesses.
Table 1 summarizes the burden of disease, annual cases, and mortality of the most common bacterial diseases worldwide. Despite the widespread, global availability of antibiotics, the primary cause of mortality or serious illness is delayed or inaccurate diagnosis of the bacterial infection. This underlines the urgent need for more specific and rapid analytical tests that can be employed at the point of care.
TABLE 1.
Common global diseases caused by bacterial infection and their burdens of disease and mortalitya
| Disease(s) | Causative bacterial agent(s) | Burden of disease (DALY), millions | Annual deaths, millions | Annual cases, millions | Conventional methods of diagnosis | Diagnosis time critical? | Spread prevention critical? | Reference(s) |
|---|---|---|---|---|---|---|---|---|
| Lower respiratory tract infections (e.g., pneumonia) | Streptococcus pneumoniae, Haemophilus influenzae | 94.5 | 4.2 | 430 | Physical examination, chest X-ray, sputum and blood cultures, PCR | No | No | 11 |
| Diarrheal diseases | Shigellae, Campylobacter, salmonellae, Escherichia coli O157:H7 | 72.8 | 2.1 | 4,620 | Microbiology (culture on Gram-negative-selective media), PCR, ELISA, particle agglutination assay | Can be | Yes | 12, 13 |
| TB | Mycobacterium tuberculosis | 34.2 | 1.5 | 7.8 | Chest X-ray, blood test, Mantoux TST, sputum smear and culture, staining and microscopy | No | Yes | 14 |
| Meningitis | Neisseria meningitidis, Streptococcus pneumoniae, Escherichia coli | 11.4 | 0.34 | 0.7 | Lumbar puncture, blood cultures, PCR | Yes | No | 15 |
| Sexually transmitted infections (excluding HIV) | Treponema pallidum (syphilis), Chlamydia trachomatis (chlamydia), Neisseria gonorrhoeae (gonorrhea) | 10.4 | 0.13 | 222 | Urethral/vaginal swab and culture, Gram staining and microscopy, immunoassay, particle agglutination assay | No | Yes | 16, 17 |
Abbreviations: DALY, disability-adjusted life years (i.e., number of years lost due to disease); ELISA, enzyme-linked immunosorbent assay; TB, tuberculosis; TST, tuberculin skin test; HIV, human immunodeficiency virus.
Conventional, laboratory-based methods of bacterial detection and identification typically have long processing times, can lack sensitivity and specificity, and require specialized equipment and trained users and are therefore costly and not available in all countries (3). Typically, specimens (e.g., blood, saliva, urine, or food sample) are sent for microbiological analysis using various techniques, namely, microscopy and cell culture, biochemical assays, immunological tests, or genetic analysis. Microscopy involves staining bacteria and observing their morphology and staining pattern, and it is relatively quick but not specific, whereas culturing bacteria on selective media under particular growth conditions can take up to several days. Furthermore, not all bacteria can be cultured in the laboratory. Biochemical assays include detection of particular enzymes that are bacterium specific. Immunological tests include enzyme-linked immunosorbent assays (ELISAs) and agglutination assays and are usually employed to detect particular surface epitopes. These processes are all time-consuming and costly due to the specialist technical staff and equipment required. The advent of molecular techniques such as genetic analysis has enabled more rapid identification of bacterial strains (4). PCR, an extremely sensitive technique which allows for the identification of bacteria based on their genetic material, does not require a bacterial culture step due to the small sample size required (5). PCRs need preselected genetic probes to be used to correctly pair with the target bacterial sequence. Wrong pairing may result in false-positive results, and genetically mutated strains might escape the correct probe matching. However, this is still a lengthy and expensive procedure which can take several days. Real-time PCR analysis can be completed faster, within several hours, but still requires specialist equipment and reagents (6). Critically, all of these techniques take time, require sample preparation and particular reagents and equipment, and are therefore costly. There is, therefore, an urgent demand for more rapid, cost-effective, and sensitive tests which can identify whole bacteria in the field or at the point of care, bypassing multistep processing and purification.
Particularly for clinical diagnosis and treatment, rapid identification of bacteria can be critical to the clinical outcome. For example, in the case of bacterial meningitis, there is a clear negative correlation between diagnosis time and patient survival (7) or serious and disabling sequelae such as deafness, blindness, and loss of limbs. The present diagnostic methods of lumbar puncture (which itself is hazardous) alongside neuroimaging and bacterial staining are time-consuming and delay critical administration of antibiotic therapy. A biosensor test that could detect and identify the cause of meningitis within minutes is required urgently.
For other bacterial infections, diagnostic time is less critical to clinical outcome but can be extremely important in decreasing the spread of infection, for instance, in the case of sexually transmitted infections (STIs) such as syphilis, gonorrhea, and chlamydia, which can be asymptomatic. Often, potentially infected people who attend a clinic do not return for results and treatment, particularly in low-income countries where a clinic is usually a long walk from home (8). In this instance, a point-of-care test that could provide a “while-you-wait” diagnosis would allow for immediate commencement of antibiotic therapy and the prevention of disease spread. In some clinical settings such as accident and emergency departments, screening of antibiotic-resistant “superbugs,” namely, methicillin-resistant Staphylococcus aureus (MRSA) and Clostridium difficile, may be obligatory prior to admission. Point-of-care screening would be enormously useful in providing immediate results which allow for barrier nursing and appropriate precautionary measures to be put in place to decrease the risk of infection to others.
In the case of food-borne infections arising from contaminated food or beverages, rapid and correct identification of the contaminated items, followed by their removal from sale, is desirable for the prevention of further illnesses (2). In the worst reported incident of food poisoning in the United States, consumption of soft cheese contaminated with Listeria monocytogenes resulted in 47 deaths over a period of approximately 6 months until the source was identified (9).
Following bioterrorism attacks in recent years, there is also the increasing need for field-based tests for biological warfare agents (BWAs), such as those causing anthrax (Bacillus anthracis) and plague (Yersinia pestis) (10). Two types of sensors are required here, one to provide an early-warning system for screening of potentially contaminated items and another to test potentially infected individuals for microorganisms.
BIOSENSORS FOR DETECTION OF BACTERIA
Biosensors offer a rapid and cost-effective method of bacterial detection which can be performed at the point of care without the need for a specialist user (18). This “lab-on-a-chip” method of patient diagnosis and monitoring provides a more rapid diagnosis which allows for faster and more effective therapeutic intervention, thereby preventing full-blown infection and mortality and also decreasing the spread of disease.
Biosensors essentially comprise a biorecognition element that is coupled to some form of transducer, which converts specific analyte binding to bioreceptors into a measurable or detectable readout. Biosensors can be categorized in different ways, either according to the method of signal transduction (i.e., optical, mechanical, or electrical) or by the type of bioreceptor employed (i.e., catalytic [enzyme] or affinity based [antibody, aptamer, lectin, bacteriophage, etc.]). Generally, affinity-based sensors are preferred over enzymatic biosensors for the detection of microorganisms, due to their enhanced selectivity and specificity and lack of extra reagents required. The biosensor field is expanding rapidly, with amperometric and optical techniques being the most commonly used over the last 30 years, whereas the use of more recent methods such as impedance and fiber optics is now increasing (Fig. 1A).
FIG 1.
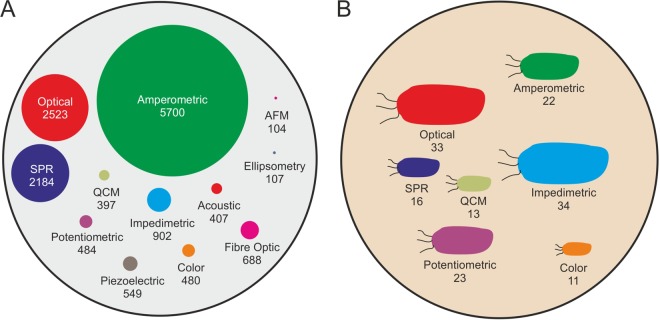
Publications on biosensors for the field in general compared with the specific detection of whole bacteria. (A) Different detection methods being used in biosensing platforms, including published literature found in ISI Web of Science using the search terms “biosensor” and “used technique” from 1983 to 2013. (B) Different techniques used for the detection of whole bacteria. The size of the circle or bacterium is proportional to the number of publications associated with that technique.
Biosensors have been developed for many different analytes, which range in size from individual ions and small molecules to nucleic acids and proteins up to whole viruses and bacteria (18). In the case of bacterial sensing, two classes of biosensors have been developed: (i) those which require sample processing to achieve bacterial disruption or lysis in order to liberate the target bacterial component and (ii) processing-free systems which target whole bacteria. In the first category, biosensors detect bacterial components such as DNA (19, 20), RNA (e.g., rRNA) (21, 22), intracellular proteins such as enzymes (23), and secreted exotoxins (24). The major disadvantage of these systems is the requirement for sample processing and extra reagents, which increases the time and cost of these tests. Therefore, biosensors for the direct, reagentless detection of whole bacteria are much more desirable for rapid, cost-effective testing at the point of care. This is particularly useful because the infectious dose of bacteria for many human pathogens is very low; for E. coli O157:H7 this has been reported to be as low as only 10 cells per gram of food or environmental sample (25).
BIOSENSORS FOR WHOLE BACTERIAL CELL DETECTION
Significant research efforts are now focused upon the detection of whole bacteria (26, 27) (Fig. 1B). It is observed that in terms of whole bacteria, impedimetric and optical methods are most commonly used. The development of biosensors for whole microorganisms is challenging because it requires detection of analytes that are much larger (micrometer scale) than typical molecular analytes such as proteins (nanometer scale), and bacteria display many surface epitopes that can lead to nonspecific interactions with the sensor surface.
Bacteria are typically between 0.5 and 5 μm in size, displaying different morphologies, including spherical cocci, rod-shaped bacilli, and spiral-shaped spirilla or spirochetes, among others. Unlike eukaryotic cells, most bacteria are encapsulated by a cell wall which is present on the outside of the cytoplasmic membrane (Fig. 2). The cell wall comprises mainly peptidoglycan, a negatively charged polymer matrix comprising of cross-linked chains of amino sugars, namely, N-acetylglucosamine and N-acetylmuramic acid. Bacteria can be classified as either Gram positive or Gram negative depending upon the architecture and thickness of the cell wall. Gram-positive bacteria retain the violet Gram stain due to their thick peptidoglycan layer on the outside of the cell membrane. In contrast, Gram-negative bacteria do not take up the stain, as their thinner peptidoglycan layer is sandwiched between two cell membranes. The outer lipid membrane of Gram-negative bacteria also contains lipopolysaccharides (LPS), which act as endotoxins and elicit a strong immune response in humans, as well as various proteins, including porins. The thick peptidoglycan wall surrounding Gram-positive bacteria contains extra components such as lipids, surface proteins, and glycoproteins. Pathogenic Gram-negative bacteria include Escherichia coli, Salmonella, Shigella, Legionella, Haemophilis influenzae, Neisseria gonorrhoeae, and Neisseria meningitides. Examples of pathogenic Gram-positive bacteria include Streptococcus, Staphylococcus, Bacillus, and Clostridium.
FIG 2.
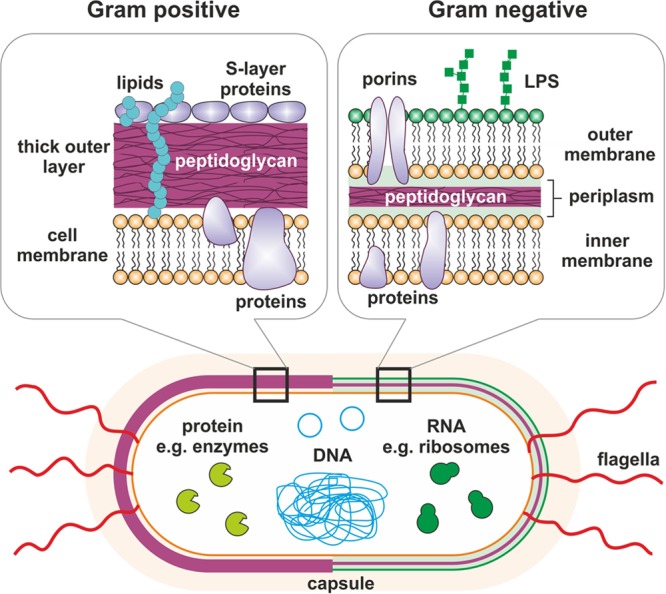
Bacterial architecture and targets for biosensing. The cell wall of Gram-positive bacteria comprises a thick layer of peptidoglycan, which also contains lipids and other protein components, surrounding a lipid membrane. In contrast, Gram-negative bacteria possess a much thinner peptidoglycan layer sandwiched in between two cell membranes. The outer membrane contains proteins, such as porins, as well as lipopolysaccharides (LPS), also known as endotoxin. The inner membranes of both types of bacteria contain various proteins. Both types of bacteria may have flagella. Intracellular targets for biosensing include proteins, DNA, and RNA.
A variety of surface antigens presented on the cell envelopes of whole bacteria, including proteins, glycoproteins, lipopolysaccharides, and peptidoglycan, can act as targets for biorecognition. Certain bioreceptors have been developed to target a specific one of these moieties; for example, lectins, a type of carbohydrate binding protein, can be employed as bioreceptors for specific cell envelope sugars (28, 29). Bacteriophages, viruses which bind to specific bacterial receptor proteins in order to infect the host cells, have also been employed for bacterial detection (30, 31). Polyclonal antibodies raised against specific bacterial strains are the most commonly used bioreceptors for whole bacterial cell detection, where the binding targets on the cell envelope are usually unknown. To increase the specificity and sensitivity of the sensor, isolated surface epitopes can be used to produce monoclonal antibodies (32, 33).
The ideal parameters for whole bacterial sensors are almost identical to the requirements for a general biosensor. Depending on the site of use, for example, stand-alone personal use at home or clinical setup, regular use in a laboratory setup, or remote regular use off site (polluted water or wastewater site), the configuration might vary, but the key properties for commercial biosensors to detect bacteria are constant. They should be inexpensive, small, easy to operate and label free, with little or no sample preparation. Important key features for an ideal bacterial biosensor are presented in Table 2.
TABLE 2.
Requirements for an ideal bacterial biosensor
| Parameter | Value or quality |
|---|---|
| Sensitivity | Less than 103 CFU/ml |
| Specificity | Can distinguish different serotypes of bacteria (e.g., can distinguish E. coli Nissle 1917 from E. coli O157:H7), minimal background, must operate in complex matrices (e.g., clinical samples such as sputum and blood, food, and beverage samples) |
| Speed | 5–10 min for a single test |
| Size | Compact, portable device that can operate at the site of interest |
| Sample processing | Label free with minimal sample processing |
| Stability | Biorecognition element must be stable at the high temperatures experienced in some countries (e.g., up to 45°C) for several months to allow for good shelf life |
| Skill of operator | No specialist training needed to use the assay, can be used by patients |
Optical Biosensors
Optical biosensors exploit analyte binding-induced changes in the optical properties of the sensor surface, which are then transduced to a detector. Optical biosensors are often divided into two categories, fluorescence based or label free (34). Examples of both are presented in Table 3. The simplest optical biosensors function by measuring a change in fluorescence or, less commonly, in absorbance or luminescence of the biosensor surface upon analyte recognition. These technologies have evolved from traditional sandwich immunoassays, where the biorecognition element comprises immobilized antibodies which allow for specific analyte detection. A secondary reagent, such as a fluorescently labeled antibody, then binds to the captured analyte on the sensor surface. This generates an optical signal, the strength of which is proportional to specific analyte binding. To convert these assays from a laboratory-based 96-well plate format to a smaller, more portable biosensor system, optical fibers have been employed for the detection of whole bacterial cells (35, 36). Fiber optic biosensors (FOB) typically comprise a source of light which passes through optical fibers containing immobilized bioreceptors to a photon detector. Analyte binding and subsequent addition of an appropriate labeling reagent give rise to a change in signal at the detector. Fluorescence-based biosensors can provide excellent sensitivity; for instance, Mouffouk and colleagues used a fluorescent dye-loaded micelle approach to detect 15 cells/ml of E. coli (37). However, the major disadvantage of using fluorescence-based optical biosensors is the requirement for sample labeling with fluorescent reagents, which adds time and cost to the procedure.
TABLE 3.
Examples of optical biosensors for detection of whole bacterial cellsa
| Target analyte(s) | Transducer signal | Sensor assembly | Bioreceptor(s) | LOD | Analyte(s) | Reference(s) |
|---|---|---|---|---|---|---|
| Various, e.g., Salmonella Typhimurium, Escherichia coli O157:H7, Shigella dysenteriae, Campylobacter jejuni | Fluorescence | NRL array sensor (fluorescence-based affinity assay) | Antibody, ganglioside receptors, oligosaccharides | 2 × 103–8 × 104 CFU/ml | Food or environmental samples | 35 |
| Salmonella enterica, Listeria monocytogenes, Escherichia coli O157:H7 | Fluorescence | Antibodies linked via biotin/avidin to optical fibers | Polyclonal antibody for capture, fluorescent monoclonal antibody or aptamer against surface protein InlA as reporter | 103 CFU/ml | Artificially contaminated meat samples | 50, 51 |
| Escherichia coli | Fluorescence | Bioconjugated magnetic beads for capture, fluorescent polymeric micelles for reporting | Polyclonal anti-E. coli antibodies | 15 cells/ml | Bacteria in buffer | 37 |
| Escherichia coli | Thin-film optical interference spectroscopy | Antibody-functionalized nanostructured oxidized porous silicon (PSiO2) | Anti-E. coli polyclonal antibody | 104 cells/ml | Bacteria in buffer | 52 |
| Salmonella Typhimurium | Light scattering | Immunoagglutination assay using anti-Salmonella-conjugated polystyrene microparticles | Anti-Salmonella polyclonal antibody | 10 CFU/ml | Liquid from processed raw chicken | 53 |
| Shewanella oneidensis | SERS | Silver nanoparticles sandwiched by analyte binding on optical fiber tip | NA | 106 cells/ml | Bacteria in buffer | 54 |
| Escherichia coli, Staphylococcus aureus, Bacillus subtilis | SPR | Lectin-functionalized anisotropic silver nanoparticles | Potato lectin | 1.5 × 104 CFU/ml | Bacteria in serum-spiked buffer | 42 |
| Escherichia coli O157:H7 | Long-range SPR | Antibodies on SAM-gold surface/antibody-functionalized magnetic nanoparticles | Anti-E. coli antibody | 50 CFU/ml | Bacteria in buffer | 39 |
| Escherichia coli | SPR | Bacteriophage covalently bound to SiO2 optical fibers | T4 bacteriophage | 103 CFU/ml | Bacteria in buffer | 41 |
Abbreviations: SERS, surface-enhanced Raman scattering; SPR, surface plasmon resonance; NA, not applicable.
Surface plasmon resonance (SPR) is a label-free method of optical sensing which has been employed for the detection of a range of analytes since the first commercially available device was launched by Biacore (GE Healthcare) in 1990 (38). SPR systems comprise a source of plane-polarized light which then passes through a glass prism, the bottom of which contacts the bioreceptor-functionalized transducer surface, which is typically a thin film of gold. Analyte binding to the transducer surface changes its refractive index, which in turn alters the angle of light exiting the prism (the SPR angle). Various SPR-based biosensors have been developed for the detection of whole bacterial cells using a variety of bioreceptors, including antibodies (39, 40), bacteriophages (31, 41), and lectins (29, 42).
The detection of whole bacteria using SPR generally yields low sensitivity compared to that using other techniques, due to factors including limited penetration of bacteria by the electromagnetic field and the similarity in refractive index between the bacterial cytoplasm and the aqueous medium (43). Localized surface plasmon resonance (LSPR), a process where noble metal nanoparticles are used to enhance the sensitivity of the system, has been used recently (44). Recent strategies to improve the sensitivity of SPR-based bacterial sensors include transducer surface modifications (45), using nanorods for multiple detection (46), sandwich-type assays including nanoparticles for analyte capture to boost the signal (42), and the use of modified SPR systems, such as long-range SPR, which are better suited to large analytes (39). For the detection of whole bacteria, LSPR is reported to be less sensitive (47) and sometimes limited by unclear sample when a biological matrix is used (48). Surface-enhanced Raman scattering (SERS) is another modification where the Raman spectrum is enhanced manyfold and has been used in combination with other techniques to detect bacterial cells even in blood medium (49) However, SPR-based systems in general still remain large, expensive pieces of equipment which have not yet been adapted for point-of-care diagnostics. Coin-size Spreeta SPR chips (Texas Instruments Inc.) have recently permitted the development of a miniaturized SPR-based biosensor, although this still required a microfluidic system and is therefore confined to the laboratory. Furthermore, interference from biological samples means that an SPR-based biosensor that operates successfully in physiological media has yet to be developed.
Mechanical Biosensors
Mechanical biosensors confer several advantages for use at the point of care; they can provide high sensitivity and quick processing times without the need for sample processing or extra reagents (55). The two main categories of mechanical biosensors are based on quartz crystal microbalance (QCM) or cantilever technology (Table 4).
TABLE 4.
Examples of mechanical biosensors for detection of whole bacterial cellsa
| Target analyte | Transducer signal | Sensor assembly | Bioreceptor | LOD | Analyte | Reference |
|---|---|---|---|---|---|---|
| E. coli O157:H7 | QCM | Antibody for capture and antibody-functionalized nanoparticles for signal enhancement | Anti-E. coli antibody | 106 cells/ml | Bacteria in buffer | 56 |
| Bacillus anthracis | QCM | Protein A/antibody-functionalized SAM on gold | Anti-B. anthracis antibody | 1 × 103 CFU or spores/ml | Vegetative cells and spores | 60 |
| Salmonella Typhimurium | QCM | Immunosensor sandwich assay using gold nanoparticles for signal amplification | Anti-Salmonella Typhimurium antibody | 10 CFU/ml | Bacteria spiked into meat samples | 58 |
| E. coli O157:H7 | PEMC | Antibody-functionalized cantilever | Anti-E. coli antibody | 1 cell/ml | Bacteria in buffer | 68 |
| Vibrio cholerae O1 | Microcantilever/DFM | Antibody-functionalized SAM on gold | Anti-V. cholerae antibody (monoclonal) | 1 × 103 CFU/ml | Bacteria in buffer | 66 |
| Listeria monocytogenes | PEMC | Protein G/antibody with postcapture antibody binding for signal amplification | Anti-L. monocytogenes antibody for capture, secondary antibody for signal amplification | 1 × 102 cells/ml | Bacteria in milk | 69 |
Abbreviations: QCM, quartz crystal microbalance; PEMC, piezoelectric-excited millimeter-size cantilever; DFM, dynamic force microscopy.
QCM sensors are label-free piezoelectric biosensors which detect the resonance frequency change that results from increased mass on the sensor surface due to analyte binding. QCM sensors have been developed for the detection of whole bacterial cells, including Escherichia coli (56, 57), Salmonella enterica serovar Typhimurium (58), Campylobacter jejuni (59) and Bacillus anthracis (60). The development of sandwich-type assays which employ nanoparticles for signal amplification has allowed for the detection of very few bacterial cells, down to 10 CFU/ml in some cases (58).
Microcantilever sensor technology is an emerging label-free technique that offers very high sensitivity, fast response times, and ease of miniaturization for the development of point-of-care sensors (61, 62). Cantilever sensors typically comprise a bioreceptor-functionalized microcantilever which oscillates at a particular resonant frequency. The resonant frequency of the cantilever changes due to induced mechanical bending upon an increase in mass on the sensor surface. Microcantilever sensors have been developed for the detection of various whole bacteria, including Escherichia coli O157:H7 (63, 64), Salmonella Typhimurium (65), Vibrio cholerae (66), and the biowarfare agent Francisella tularensis (67). The recently developed piezoelectric-excited millimeter-size cantilevers (PEMC) using antibodies as bioreceptors have been able to detect as few as one E. coli cell in buffer (68) and one hundred Listeria monocytogenes cells in milk (69). A major disadvantage of cantilever-based systems is that they are often limited by the need to operate in air as opposed to in physiological media, and there is a dearth of reports in which cantilever-based sensors have been tested in relevant matrices such as food or patient samples (70).
Electrochemical Biosensors
Electrochemical biosensors comprise potentiometric, amperometric, and impedimetric sensing techniques, with amperometric sensors the first type of biosensors to be described, in 1953 (71). Electrochemical biosensors have subsequently become the most developed group with greatest commercial success, largely due to amperometric glucose detection in diabetic monitoring (72). Their key advantages are low cost, point-of-care testing, and miniaturization capacity (73).
Potentiometric sensors.
Potentiometric biosensing uses ion-selective electrodes to measure the potential of a solution based on specific interactions with ions in the solution. This method measures the change in potential that occurs upon analyte recognition at the working electrode. Although potentiometry is widely used in the biosensor field, examples of potentiometric biosensors for the detection of whole bacterial cells are few. Compared to other methods such as impedance, potentiometry cannot provide specific and sensitive signals for large analytes such as bacteria. However, some innovative applications of potentiometry can provide reasonable limits of detection (LODs) (Table 5), as discussed briefly here.
TABLE 5.
Examples of potentiometric and amperometric electrochemical biosensors for detection of whole bacterial cellsa
| Biosensor type | Bacterium | Transducer | Technique | Bioreceptor | LOD | Comment | Reference |
|---|---|---|---|---|---|---|---|
| Potentiometric | Sulfate-reducing bacteria | Glassy carbon electrode | Potentiometric stripping analysis | None | 2.3 × 10–2.3 × 107 CFU/ml | Need bacterial processing | 74 |
| Staphylococcus aureus | Single-walled carbon nanotubes | EMF | Aptamer | 8 × 102 CFU/ml | Bacterium-spiked pig skin | 75 | |
| Amperometric | E. coli | Photolithographic gold | Immunomagnetic/amperometric in flow cells | Antibody | 55 cells/ml in PBS, 100 cells/ml in milk | No contact of biocomponent with sensor | 76 |
| E. coli K-12 | Screen-printed carbon electrodes | Phage-induced release and subsequent quantitation of bacterial intracellular enzyme | Bacteriophage | 1 CFU/100 ml | Cells not intact after analysis | 77 | |
| Heat-killed E. coli | SCE | Amperometric detection of secondary antibody with GOD | Biotinyl antibody | 3 × 101–3.2 × 106 CFU/ml, down to 15 CFU/ml | Labeling needed but tested in synthetic stool | 78 | |
| Staphylococcus aureus | DropSens screen-printed gold electrodes | HRP H2O2-mediated immunosensor | Antibody | 1 CFU/ml of raw milk | Indirect, label needed | 79 |
Abbreviations: SCE, saturated calomel electrode; GOD, glucose oxidase; EMF, electron motive force; HRP, horseradish peroxidase.
Potential stripping analysis (PSA) is a chrono-potentiometric method where the stripping time of a deposited compound can be measured at a set stripping potential. Marine pathogenic bacteria (sulfate-reducing bacteria [SRB]) have been detected using this method, where bacterial samples were preincubated with lead and nitric acid to produce sulfide (74). This sulfide can be detected by PSA, as with increasing concentration of bacterial sample, a longer time is needed for stripping. Although the detection range of PSA is good, the preincubation steps are not suitable for rapid and on-site detection methods.
Staphylococcus aureus, a common skin commensal, has been detected using label-free potentiometric detection (75). Electromotive force (EMF) was measured in a single-wall carbon nanotube-based aptamer system. The real-time EMF bacterial binding generated a linear signal with increasing concentration, with a detection limit of 8 × 102 cells/ml when the aptamer was covalently bound to the nanotubes.
Amperometric sensors.
Following the introduction of enzyme-based amperometric sensing of glucose 40 years ago (80), this technique has been applied commonly to a wide range of analytes, including whole bacteria (Table 5). Amperometric biosensors are based on direct measurement of the current generated by the oxidation or reduction of species produced in response to analyte-bioreceptor interaction. The bioreceptor component is commonly an enzyme such as glucose oxidase, which is used in all medical glucose monitors (81). The current generated is directly proportional to the analyte concentration and therefore is easily determined (72). Indeed, key advantages of amperometric biosensors are their relative simplicity and ease of miniaturization. They also generally confer excellent sensitivity. Limitations include low specificity depending on the applied potential, which if high may allow other redox-active species to interfere with the signal and lead to inaccuracies in results (82). This is of particular relevance in biological media, which may contain a wealth of potential interferents. Crucially, amperometric biosensors also require the analyte of interest to be a substrate for an enzymatic reaction, which is a fundamental limitation in attempting to broaden the use of this type of biosensor. Therefore, although in the field of biosensing amperometry is the most common detection method, in the case of whole-cell bacterial sensing this is not as widely used.
A novel method of differentiating hemolytic from nonhemolytic bacteria within a mixed population using liposome-trapped electron mediators with amperometric detection was reported (83). Hemolytic bacteria can disrupt liposomes, thus releasing electron mediators in the medium, which can be detected with the increase in current, whereas control bacteria lack this ability, with no current change in the system. However, this system yielded a low detection limit, ranging from 5 × 105 to 2 × 107 CFU/ml.
The amperometric detection of E. coli in a microfluidic system coupled with immunomagnetic capture has been reported (76). In brief, the specific antibody-conjugated magnetic particles were suspended on top of a gold electrode surface inside a flow cell by magnetic force. The bacterial sample was pumped into the cell, followed by the addition of a horseradish peroxidase (HRP)-conjugated antibody label which binds in a sandwich fashion. HRP catalyzes H2O2 in the presence of the electron mediator hydroquinone and produces measurable current. The amperometric detection limit of this sensor was 55 cells/ml of E. coli in phosphate-buffered saline (PBS) and 100 cells/ml in milk. The use of hanging bioreceptors leaves the gold electrode surface clean, limiting electrode fouling. However, the use of labeling reagents and a microfluidic system limits its point-of-care use.
Bacteriophages, or phages, are viruses with the ability to infect and lyse specific bacterial strains. Amperometric quantification of coliform E. coli K-12 was achieved by the phage-mediated release of the intracellular bacterial enzyme β-d-galactosidase from bacterial cells upon screen-printed carbon electrodes (77). Phage-mediated cell lysis increases specificity while boosting sensitivity through enzyme release to achieve a higher amperometric signal. The sensor was able to detect 1 CFU/100 ml of sample but had the disadvantage of the need for preincubation of bacterial cells with enzyme enhancer and phage.
A complex amperometric sensor was constructed to detect heat-killed E. coli strains spiked into synthetic stool samples (78). First, a biocompatible nanolayer of fullerene (C60), ferrocene (Fc), and thiolated chitosan (CHI-SH) composite was deposited on top of glassy carbon electrodes, followed by conjugation of Au-SiO2-streptavidin-biotinyl primary antibodies. Target bacteria were detected and quantified by sandwich detection using secondary antibodies tagged with Pt nanochains and glucose oxidase. Current change was measured in the presence of glucose. Although the detection limit was low (15 CFU/ml) and the system functioned in synthetic stool samples, multistep sensor construction and the use of several labels make the system complicated.
Indirect amperometric detection of Staphylococcus aureus was achieved using a competitive magnetic immunoassay with a detection limit of 1 CFU/ml (79). Commercial screen-printed gold electrodes were used to construct the immunosensor. Antibodies against protein A were immobilized on magnetic beads upon the sensor surface. S. aureus, which displays protein A on the cell surface, was captured by the antibodies and was quantitatively detected by adding HRP-protein A as a competitor. However, the system requires labels and the signal enhancer tetrathiafulvaline, again negating its point-of-care usefulness.
Impedimetric sensors.
Impedimetric biosensors are a very promising choice for the detection of whole bacteria, being label free, less costly than other systems, highly sensitive, and not affected by the presence of other analytes or colored compounds in the sample matrix. Crucially, impedimetric systems are easy to miniaturize, which facilitates their translation to point-of-care systems.
Since the late 19th century, after Oliver Heaviside coined the term “impedance,” electrochemical impedance spectroscopy (EIS) has been employed to characterize different biological systems (18). Impedimetric biosensors function by analyte-bioreceptor interaction causing a change in capacitance and electron transfer resistance across a working electrode surface (Fig. 3). As analyte binding increases with higher analyte concentration, the impedance across the electrode surface changes and is detected at a transducer. The impedance may be seen to increase or decrease depending on the analyte (84). Bioreceptors are commonly antibodies, although they may be other molecules capable of detecting a wide range of analytes from proteins up to whole bacteria and viruses (85, 86). A main advantage of impedance biosensors is the unrestricted measurement of the molecule of interest, with no requirements for the analyte to be an enzymatic substrate or for formation of electroactive species as in amperometric sensing. Currently there are no impedance biosensors that have had widespread commercial success, although this technology is increasing in use rapidly, with clear evidence of a growing number of publications within this field. Disadvantages of impedance biosensors are cited as variable reproducibility, high limits of detection, and problems with nonspecific binding (84, 85). However, with continued improvements and the advancement of miniaturization of equipment, EIS has become an increasingly attractive technique in biosensor applications. In general, impedance (Z) is complex phenomenon which can be correlated directly with analyte binding to a biosensor surface. Usually, Z is recorded over a wide range of frequency with respect to time, where two major components, i.e., resistance (R) and capacitance (C), are measured. Impedance data are often represented as Nyquist plots, where R is termed the “real component of impedance” on the x axis and C is termed the “imaginary component of impedance” on the y axis. A typical Nyquist plot is semicircular, with a 45-degree rise sometimes observed at the low-frequency end (Fig. 3B).
FIG 3.
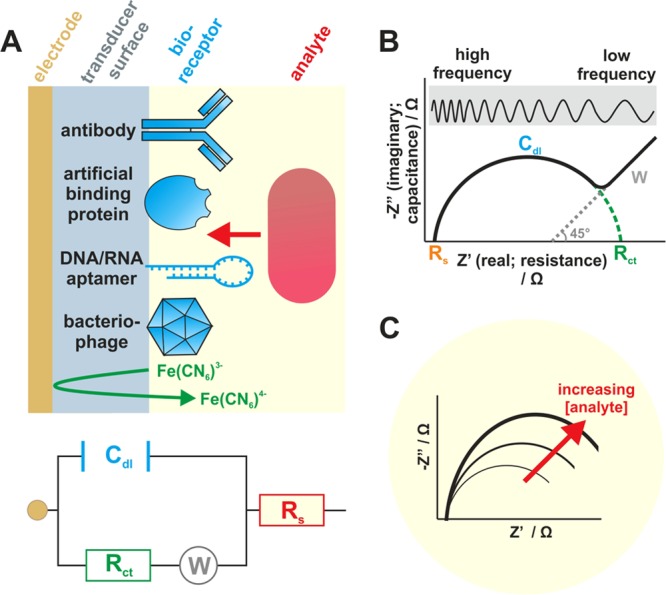
Structure and electrochemical function of impedimetric biosensors for bacterial detection. (A) Layer-by-layer sensor construction typically comprises an electrode surface functionalized (e.g., using a polymer or self-assembled monolayer) to allow for attachment of bioreceptors, including antibodies, half-antibodies, artificial binding proteins, nucleic acid aptamers, and bacteriophages. Most impedance-based systems utilize electron mediators, e.g., ferri/ferrocyanide [Fe(CN6)3−/4−] to monitor charge transfer resistance. The diagram is not to scale. The Randles circuit illustrates the components of the system: double-layer capacitance (Cdl), charge transfer resistance (Rct), solution resistance (Rs), and Warburg impedance (W) (W is observed only in some systems at low frequency). (B) Nyquist plot showing the features of the Randles circuit. (C) Impedance changes resulting from analyte-surface interactions are proportional to analyte concentration.
At high frequency, the major component of impedance derives from the resistance from solution itself (solution resistance [Rs]), whereas at lower frequency, impedance arises from the resistance to the flow of electrons or charge close to the electrode surface (charge transfer resistance [Rct]). The Nyquist plot can be translated into an equivalent circuit model proposed by Randles (18), where it is easy to isolate each individual component (Fig. 3A). Changes in impedance arising from increasing deposition on the sensor surface, upon either layer-by-layer sensor construction or analyte binding, can be plotted quantitatively (Fig. 3C).
Impedimetric detection of an analyte can be achieved in the presence or absence of an additional electron/redox mediator. In the presence of electron mediators such as Ru(NH3)63+/2+ (hexa-ammineruthenium III/II ions) and Fe(CN6)3−/4− (ferricyanide/ferrocyanide), the impedance is termed Faradaic impedance. In the absence of mediators, the observed impedance is called non-Faradaic impedance. The use of electron mediators ensures a plentiful supply of redox species to ensure that impedance does not become limited. Although impedance measurement is straightforward, the complexity depends on the choice of electrode material, base layer construction (type of self-assembled monolayer [SAM] or polymers), bioreceptor conjugation chemistry, type and size of analytes, and complexity of the sample matrix. These issues have turned the research focus toward optimizing layer-by-layer sensor construction to achieve the optimum impedance signal with minimum noise.
A plethora of reports detailing the impedimetric detection of whole bacterial cells has emerged in recent years (Fig. 1). Most of these studies have focused upon detection of the model organism E. coli (26, 87), although other bacteria have also been detected, including sulfate-reducing bacteria (88), Salmonella Typhimurium (89), Campylobacter jejuni (90), and Staphylococcus aureus (91). The reported sensor construction varies widely in the selection of base electrode materials, choice of bioreceptor, linking chemistry, and finally impedance data representation. The most common way of presenting data is the change in Rct upon analyte addition (raw Rct change or percent change); however, plotting real impedance, imaginary impedance, or absolute impedance against bacterial concentration is also employed. Chrono-impedimetric data can also be obtained by taking measurements at a fixed frequency to monitor real-time binding.
A comprehensive list of published impedimetric sensors to detect whole bacteria is presented in Table 6. Here, several recent case studies are discussed in more detail, based on their advantages and novel features, including choice of electrode material, transducer surface functionalization, choice of conjugation strategies, and readout methods.
TABLE 6.
Examples of impedimetric electrochemical biosensors for detection of whole bacterial cellsa
| Bacterium(a) | Transducer | Chemistry | Bioreceptor | LOD | Reference |
|---|---|---|---|---|---|
| E. coli O157:H7 | Gold | EDC/NHS | Antibody | 2 CFU/ml | 26 |
| E. coli O157:H7 | Nanoporous aluminum oxide membrane | Trimethoxysilane-HA-EDC/NHS | Antibody | 10 CFU/ml | 27 |
| E. coli O157:H7 | Nanoporous aluminum oxide membrane | Silane-PEG | Antibody | 10 CFU/ml | 92 |
| E. coli K-12 | Gold microelectrode, interdigitated | Physisorption | T4 bacteriophage | 104–107 CFU/ml | 87 |
| E. coli K-12 | Boron-doped UNCD microelectrode array | Physisorption | Antibody | NA | 93 |
| E. coli O157:H7 | Gold microelectrode, interdigitated | Physisorption | Antibody | 2.5 × 104 CFU/ml and 2.5 × 107 CFU/ml | 94 |
| E. coli | Gold | SAM-EDC/NHS | Antibody | 1.0 × 103 CFU/ml | 95 |
| E. coli | Gold electrode | SAM-biotin-NeutrAvidin | Biotinyl antibody | 10 CFU/ml | 96 |
| E. coli | 7% gold-tungsten plate wire | Polyethyleneamine-streptavidin | Biotinyl antibody | 103–108 CFU/ml | 97 |
| E. coli | Gold disk | mSAM | Synthetic glycan | 102–103 CFU/ml | 98 |
| E. coli | Polysilicon interdigitated electrodes | Glutaraldehyde | Antibody | 3 × 102 CFU/ml | 99 |
| E. coli O157:H7 | Gold | SAM-HA-EDC/NHS | Antibody | 7 CFU/ml | 100 |
| E. coli | Gold | SAM-PDICT cross-linker | Bacteriophage | 8 × 102 CFU/ml | 101 |
| E. coli | Graphene paper | Biotin-streptavidin | Antibody | 1.5 × 102 CFU/ml | 102 |
| E. coli | Screen-printed carbon microarrays | EDC/NHS | Bacteriophage | 104 CFU/ml for 50-μl samples | 103 |
| Sulfate-reducing bacteria | Glassy carbon | Reduced graphene sheet with chitosan plus 1% glutaraldehyde | Antibody | 1.8 × 101–1.8 × 107 CFU/ml | 88 |
| Sulfate-reducing bacteria | ITO | Chitosan-reduced grapheme sheet | Bioimprint of bacteria | 1.0 × 104–1.0 × 108 CFU/ml | 104 |
| Sulfate-reducing bacteria | Foam Ni | Nanoparticle-SAM-EDC/NHS | Antibody | 2.1 × 101–2.1 × 107 CFU/ml | 105 |
| Salmonella Typhimurium | Gold | SAM-glutaraldehyde | Antibody | NA | 89 |
| Salmonella Typhimurium | Electroplated gold on disposable printed circuit board | 16-MHDA-EDC-NHS | Monoclonal antibody | 10 CFU in 100 ml | 106 |
| Salmonella Typhimurium | Gold | Polytyramine-glutaraldehyde | Antibody | NA | 107 |
| Campylobacter jejuni | Glassy carbon | Physisorped onto O-carboxymethylchitosan surface-modified Fe3O4 nanoparticles | Monoclonal antibody | 1.0 × 103–1.0 × 107 CFU/ml | 90 |
| Listeria innocua | Gold | SAM-EDC/NHS | Endolysin (bacteriophage-encoded peptidoglycan hydrolases) | 1.1 × 104 and 105 CFU/ml | 30 |
| Staphylococcus aureus | Nanoporous alumina | Silane–1% GPMS | Antibody | 102 CFU/ml | 91 |
| Porphyromonas gingivalis, E. coli | Microfluidic cell with hydrodynamic focusing | No immobilization/impedance reading during flow of cells | None | 103 cells/ml | 108 |
Abbreviations: EDC, ethyl(dimethylaminopropyl) carbodiimide; PEG, polyethylene glycol; UNCD, ultrananocrystalline diamond; NA, not available; PDICT, 1,4-dithiocyanate; ITO, indium tin oxide; mSAM, mixed self-assembled monolayer; NHS, N-hydroxysuccinimide; SAM, self-assembled monolayer; MHDA, mecaptohexadecanoic acid; GPMS, (3-glycidoxypropyl)trimethoxysilane.
The detection of viable cells in mixed populations of live and dead cells of E. coli has been reported (99). Differentiating live cells from dead cells can be advantageous when the number of viable cells reflects the true pathogenic count. In this study, immunosensors were generated upon polycrystalline silicon interdigitated electrodes. Usually, viable cells are voluminous compared to dead cells. As live cells have a higher cell volume, their interference with the electric field is higher than that of the dead cells, which can be detected by impedance and capacitance measurement. The limit of detection for the sensor was 3 × 102 CFU/ml, and a similar signal was achieved in the presence of a large excess of dead cells, although this system has not been validated using biologically relevant samples. The more sensitive, non-Faradaic impedimetric detection of E. coli was achieved using a biotinylated whole antibody as a bioreceptor (96). Biotinyl antibodies were tethered to the biotin-presenting mixed SAM (mSAM) on a gold surface via a NeutrAvidin linkage. The sensor system gave a low detection limit of 10 CFU/ml for whole cells and was also validated by SPR. Again, however, the system was not validated in biologically relevant samples.
The use of a novel electrode material, reduced graphene oxide paper, in the construction of a nanoparticle-based immunosensor for detection of E. coli has been reported (102). Antibodies were immobilized upon electrodeposited gold nanoparticles using a biotin-streptavidin link. The sensor yielded a detection limit of 102 cells/ml with high selectivity and lower detection limits of 104 cells/ml and 103 cells/ml in contaminated ground beef and cucumber samples, respectively. This system shows promise for operation in relevant sample matrices.
Bacteriophages have high specificity toward bacteria, which makes them an attractive natural bioreceptor. In a recent study, bacteriophages were chemically tethered to SAM-functionalized gold electrodes to quantify E. coli cells (101). The sensor displayed a good detection limit of 8 × 102 CFU/ml in less than 15 min. The sensor performance was further validated by loop-mediated isothermal amplification (LAMP) of the E. coli tuf gene after cell lysis and quantitation using linear sweep voltammetry.
In a novel approach, antibody-tagged biofunctional magnetic beads were used to facilitate the migration of target bacteria to the sensor surface, (92). The immunosensor was constructed on silanized, nonporous alumina, which was separated by two compartments with fluid accessibility. Platinum wire working and reference electrodes were placed in two compartments, an unusual approach where the sensor surface was not set as the working electrode area. The antibody-coated magnetic beads with bound bacterial cells were magnetically transported on top of the alumina immunosensor surface to allow for binding. After immunoreaction, the magnetic field was removed, excess beads were washed away, and impedance readings were taken. This impedimetric method achieved a higher binding capability than the nonconcentrating method and a lower detection limit of 10 CFU/ml. Although the system is innovative, its complicated setup makes it difficult to translate into a point-of-care application.
Impedimetric detection of sulfate-reducing bacteria (SRB) was reported using nickel foam as working electrode material (105). The nickel foam has regular porous grooves; gold nanoparticles were deposited within these pores, followed by 11-mercaptoundecanoic acid (MPA) SAM-tethered antibodies. The sensor had a detection range of 2.1 × 101 to 2.1 × 107 CFU/ml with good selectivity over other strains. In another approach for SRB detection, a bioimprinting technique was used (104). In this method, biomolecules or cells can be deposited on a surface and then washed off, leaving their imprint on the surface. Briefly, multilayer reduced graphene sheets and chitosan were electrodeposited upon indium tin oxide (ITO), followed by absorption of SRB and a thin coating layer of nonconducting chitosan around the bacteria. SRB were then washed off the surface to get the bioimprint on biosensor surface. This imprint was able to capture and quantify target SRB in a range of 104 to 108 CFU/ml using EIS. It was also able to distinguish other control strains based on size and shape differences, but the authors recommended its use with other bioreceptor combinations.
Monoclonal antibodies are highly specific compared to polyclonal antibodies, offering higher sensitivity and selectivity for analyte detection. Salmonella Typhimurium has been detected by EIS using monoclonal antibodies as bioreceptors on a gold-plated disposable circuit board (106). The monoclonal antibodies were raised against Salmonella Typhimurium cell surface lipopolysaccharide (LPS), and the impedance signal at 10 Hz was able to detect the 10 bacteria in 100 ml of sample.
Although a variety of techniques are being employed to detect whole bacteria, the key challenges being faced are sensitivity, reproducibility, and miniaturization before their successful translation as a commercial product. Impedance-based biosensing shows great promise, being highly sensitive and label free. However, the present research needs to be taken forward with an emphasis on reproducible, inexpensive, and novel electrode material, stable conjugation, and strict optimization of bioreceptor configuration, orientation, and concentration. Miniaturization of impedance systems and robotic layer-by-layer construction will ultimately improve sensor performance with high reproducibility for commercialization.
CONCLUSIONS AND FUTURE PERSPECTIVES
There is a growing need for rapid and sensitive detection of bacteria, in complex samples, at the point of interest. In spite of the impressive research output in recent years, detailing specific and sensitive laboratory-based biosensor systems for the detection of bacteria, the manufacture of commercially available systems for point-of-interest application is seriously lagging behind. This is due to the issues discussed above: (i) difficulty in achieving specificity and sensitivity in complex “real-world” samples such as blood, feces, food, etc.; (ii) difficulties in reducing the size and cost of certain systems, for instance, SPR, QCM, and cantilever-based sensors; and (iii) improving the reliability of the system with novel manufacturing methods. Around 200 companies are now working in the area of biosensors and bioelectronics (109); however, the major driving force behind the commercial market (85%) is still for blood glucose monitoring.
In order to bring laboratory-based biosensor systems to market, strong collaboration between academia and industry is required to address the key issues highlighted in Fig. 4. Selection of inexpensive, reproducible, electrochemically favorable, and chemically stable base material is the initial important step toward commercial electrochemical biosensors. A wide range of base materials either alone or in combination have been explored. However, their individual suitability for particular sensor systems needs to be assessed carefully. As discussed in this paper, recent advances in transducer surface nanoengineering (e.g., increasing surface area using nanoparticles or nanofibers and the use of magnetic nanoparticles in sandwich-type assays) have shown promise in terms of boosting the sensor signal. This is important where detection of just a few bacterial cells is required. Base layers, e.g., polymers or self-assembled monolayers on which bioreceptors are immobilized, can have an influence on the electrochemical signal as well as nonspecific binding. Their thickness, surface charge, and chemical groups can be intelligently tuned for enhanced performance.
FIG 4.
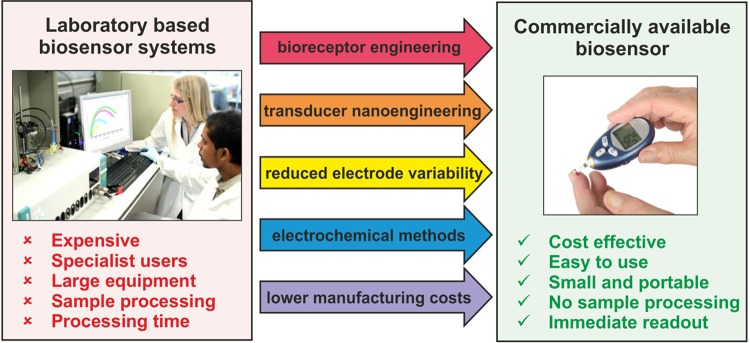
Technology translation: a summary of the current research priorities in order to bring laboratory-based biosensors for bacterial detection to market.
Equally, the development of novel bioreceptors, including bacteriophages, non-antibody binding proteins, half-antibodies, and single-chain (camelid) antibodies, offers higher specificity, which is a key advantage for detecting bacteria in complex matrices which contain many potential interferents, including human cells and commensal bacteria as well as many proteins and metabolites. Although antibodies are the most widely used bioreceptors in affinity biosensor research, their production and purification costs and stability during and after immobilization on sensor surface can be challenging. The shelf life of these antibodies on the sensor surface is not significantly long, and binding efficiency tends to decrease over time. To overcome some of these deficiencies, recent advances in engineered antibody mimetics include peptoid nanosheets (110), where antibody mimetic peptoids are self-assembled to form 3- to 5-nm-thick sheets with surface loops expressing antigen binding sites. They are chemically and biologically stable and can be produced with ease and precise control. Other remarkable engineered antibody alternatives include single-chain variable fragments (ScFv) (111), camelid-derived heavy variable-chain (VHH) antibodies (nanobodies) (112, 113), single-chain antibodies expressed via yeast surface display (114), DARPins (115), and other artificial proteins such as adhirons (116). The advantages of these alternatives are that they are comparatively small, easily customized, and conveniently mass produced in bacterial systems, avoiding traditional antibody production in mammals or birds.
Two other important aspects are regeneration of the sensor surface and multiplexing, where many bacteria can be analyzed simultaneously. Regeneration can be cost-effective, and successful regeneration can be possible with the above-mentioned stable bioreceptors, since they can often withstand harsh regeneration buffers without compromising binding capacity. Parallel multiplexing on a single chip can also reduce detection costs, providing multiple items of information from a single-shot analysis. However, all of these advancements again demand large-scale optimization, which is basically limited by funding.
Screen printing of electrodes en masse is now improving biosensor reliability. Companies making commercially available screen-printed electrodes are growing and include Metrohm USA Inc. (United States), DropSens S.L. (Spain), Gwent Sensors Ltd. (United Kingdom), Bio-Logic SAS (France), Kanichi Research Ltd. (United Kingdom), BVT Technologies Ltd. (Czech Republic), and Quasense Company Ltd. (Thailand) (18). In terms of electrochemical biosensing, specialist companies such as Uniscan Instruments Ltd. are supplying commercially available software and systems to integrate sensor chips with signal processing and readout.
However, to date, only a few commercially available biosensor systems have been employed for the detection of bacteria (117); these include SPR-based optical biosensors (Biacore), the potentiometric threshold immunoassay system (Molecular Devices Corporation), and the PCR-based universal biosensor, which employs mass spectrometry as a detection method (Ibis, San Diego, CA, USA). The immunoassay-based sensor is the only one of these that has been employed for whole bacterial cell detection (118), although this and the other sensors require sample processing. The Biacore devices and mass spectrometry-based systems are bulky and costly and require specialist users. Electrochemical methodologies offer lower manufacturing costs and ease of system miniaturization and integration, with impedance spectroscopy becoming increasingly popular due to the lack of reagents and ability to detect any analyte without the need for electroactive species. However, a commercially available impedimetric biosensor is still awaited. Unlike glucose biosensors, where sample and device size has been significantly optimized over years and a tiny blood drop can directly be tested, an impedance biosensor against bacteria might include a single dilution step before testing, depending on the detection sample. This will reduce the noise from the biological sample and produce ample volume to incubate the chip. Chip architecture and device design will also be crucial to have a user-friendly end user device.
In conclusion, the market demand and research trends presented in this review clearly demonstrate the importance of hand-held, user-friendly biosensors for whole bacterial cell detection. Electrochemical biosensors, more specifically, impedimetric sensors, can take the leading position in this area. However, the appropriate miniaturization, optimization, and clinical trials need to be done before any product is launched into market. Advancements in nanobiotechnology and biomolecular engineering and developments in particle research are moving this field quickly toward its destination. The widespread use of whole bacterial cell biosensors not only will be a milestone in the biosensor industry but will have a profound impact on food, medical, environmental, and clinical diagnostics.
ACKNOWLEDGMENTS
Asif Ahmed is funded by a University of Leeds Fully-Funded International Research Scholarship (FIRS). Natalie A. Hirst received funding from the Leeds Teaching Hospitals NHS Trust Charitable Foundation and the Bowel Disease Research Foundation.
We thank Jack Goode for photographic assistance with preparing the figures.
Biographies

Asif Ahmed received his B.Sc. (in biotechnology and genetic engineering) from Khulna University, Bangladesh, in 2002. He then completed his M.Sc. (in biomolecular sciences) at the Korea Institute of Science and Technology (KIST) in 2009 with a full scholarship. During his M.Sc. work, he designed drug candidates for the serotonin 2C receptor as antiobesity agents using molecular modeling tools. He is currently pursuing a Ph.D. in Professor Paul Millner's lab with a research focus on nanofabrication of impedimetric immunosensors against pathogenic microorganisms. This year he was awarded “best final-year Ph.D. talk” in the Annual Postgraduate Symposium in the Faculty of Biological Sciences. He has already published several papers and coauthored a book on impedimetric biosensors for medical applications. Mr. Ahmed's research interests cover molecular biotechnology and electrochemical biosensors for pathogen detection.

Jo V. Rushworth received her honors degree (B.Sc. in biochemistry with a research year studying microbiology in Paris and a National Gatsby Plant Science scholarship) and her Ph.D. (biochemistry/structural biology) from the University of Leeds, United Kingdom. During her Ph.D. work, she studied the molecular and structural biology of amyloid-beta oligomers, the causative agent of Alzheimer's disease. One of her publications arising from this study was awarded “Best in JBC Neurobiology 2013.” Subsequently, Dr. Rushworth managed Professor Paul Millner's group and developed impedimetric biosensors for specific detection of amyloid-beta oligomers. This line of research combines her interests in biomedical science and electrochemical diagnostics. Dr. Rushworth is currently a lecturer in the Faculty of Health and Life Sciences at De Montfort University, Leicester, United Kingdom, where she is setting up her own research group.

Natalie A. Hirst received a B.Sc. in experimental pathology in 2005, before receiving a medical degree in 2006 from the University of London, Barts and the London School of Medicine and Dentistry. She is a member of the Royal College of Surgeons of England, having passed the requisite examination. She is currently in her final year of study for a Ph.D. with the Bionanotechnology Group at the University of Leeds, having taken time out of full-time surgical training. Her current research is on the development of electrochemical biosensors for early detection of complications after bowel surgery.

Paul A. Millner received his B.Sc. in biochemistry and Ph.D. in plant science at the University of Leeds (United Kingdom) and then had postdoctoral fellowships at Purdue University (West Lafayette, IN, USA) and Imperial College (London, United Kingdom). He returned to Leeds in 1986 as a lecturer. After 15 years as a plant biotechnologist/protein chemist, Dr. Millner moved into the area of nano- and bionanotechnology, with a particular interest in the development of biosensors. Dr. Millner is currently the Head of the School of Biomedical Sciences at the University of Leeds and also leads the Bionanotechnology Group. Current programs in his group include work on electrochemical biosensors for diagnosis of STIs, MRSA, group A Streptococcus, and other bacteria, as well as biosensors for detecting bowel leakage after colorectal cancer resection. Dr. Millner's work is united by a deep interest in bioengineering on the nanoscale by interfacing biological reagents with surfaces to result in electrical communication or enhanced activity.
REFERENCES
- 1.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15. 10.3201/eid1701.P11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma H, Mutharasan R. 2013. Review of biosensors for foodborne pathogens and toxins. Sensors Actuat. B Chem. 183:535–549. 10.1016/j.snb.2013.03.137 [DOI] [Google Scholar]
- 3.Fournier P-E, Drancourt M, Colson P, Rolain J- M, Scola BL, Raoult D. 2013. Modern clinical microbiology: new challenges and solutions. Nat. Rev. Microbiol. 11:574–585. 10.1038/nrmicro3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. 2013. Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 26:822–880. 10.1128/CMR.00022-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burnham CA, Carroll KC. 2013. Diagnosis of Clostridium difficile infection: an ongoing conundrum for clinicians and for clinical laboratories. Clin. Microbiol. Rev. 26:604–630. 10.1128/CMR.00016-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Espy MJ, Uhl JR, Sloan LM, Buckwalter SP, Jones MF, Vetter EA, Yao JD, Wengenack NL, Rosenblatt JE, Cockerill FR, III, Smith TF. 2006. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin. Microbiol. Rev. 19:165–256. 10.1128/CMR.19.1.165-256.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van de Beek D, de Gans J, Tunkel AR, Wijdicks EFM. 2006. Community-acquired bacterial meningitis in adults. N. Engl. J. Med. 354:44–53. 10.1056/NEJMra052116 [DOI] [PubMed] [Google Scholar]
- 8.Peeling RW, Holmes KK, Mabey D, Ronald A. 2006. Rapid tests for sexually transmitted infections (STIs): the way forward. Sex. Transm. Infect. 82:V1–V6. 10.1136/sti.2006.024265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Segal M. 1988. Invisible villains; tiny microbes are biggest food hazard. FDA consumer. http://www.highbeam.com/doc/1G1-6589512.html [Google Scholar]
- 10.Gooding JJ. 2006. Biosensor technology for detecting biological warfare agents: recent progress and future trends. Anal. Chim. Acta 559:137–151. 10.1016/j.aca.2005.12.020 [DOI] [Google Scholar]
- 11.Carroll KC. 2002. Laboratory diagnosis of lower respiratory tract infections: controversy and conundrums. J. Clin. Microbiol. 40:3115–3120. 10.1128/JCM.40.9.3115-3120.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barletta F, Mercado EH, Lluque A, Ruiz J, Cleary TG, Ochoa TJ. 2013. Multiplex real-time PCR for detection of campylobacter, salmonella, and shigella. J. Clin. Microbiol. 51:2822–2829. 10.1128/JCM.01397-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfeiffer ML, DuPont HL, Ochoa TJ. 2012. The patient presenting with acute dysentery—a systematic review. J. Infect. 64:374–386. 10.1016/j.jinf.2012.01.006 [DOI] [PubMed] [Google Scholar]
- 14.Boehme CC, Saacks S, O'Brien RJ. 2013. The changing landscape of diagnostic services for tuberculosis. Semin. Respir. Crit. Care Med. 34:17–31. 10.1055/s-0032-1333468 [DOI] [PubMed] [Google Scholar]
- 15.Bamberger DM. 2010. Diagnosis, initial management, and prevention of meningitis. Am. Fam. Physician 82:1491–1498 http://www.aafp.org/afp/2010/1215/p1491.html [PubMed] [Google Scholar]
- 16.Su W-H, Tsou T-S, Chen C-S, Ho T-Y, Lee W-L, Yu Y-Y, Chen T-J, Tan C-H, Wang P-H. 2011. Are we satisfied with the tools for the diagnosis of gonococcal infection in females? J. Chin. Med. Assoc. 74:430–434. 10.1016/j.jcma.2011.08.012 [DOI] [PubMed] [Google Scholar]
- 17.Read PJ, Donovan B. 2012. Clinical aspects of adult syphilis. Intern. Med. J. 42:614–620. 10.1111/j.1445-5994.2012.02814.x [DOI] [PubMed] [Google Scholar]
- 18.Rushworth JV, Hirst NA, Goode JA, Pike DJ, Ahmed A, Millner PA. 2013. Impedimetric biosensors for medical applications: current progress and challenges. ASME, New York, NY [Google Scholar]
- 19.Paniel N, Baudart J. 2013. Colorimetric and electrochemical genosensors for the detection of Escherichia coli DNA without amplification in seawater. Talanta 115:133–142. 10.1016/j.talanta.2013.04.050 [DOI] [PubMed] [Google Scholar]
- 20.Anderson MJ, Miller HR, Alocilja EC. 2013. PCR-less DNA co-polymerization detection of Shiga like toxin 1 (stx1) in Escherichia coli O157:H7. Biosens. Bioelectron. 42:581–585. 10.1016/j.bios.2012.09.068 [DOI] [PubMed] [Google Scholar]
- 21.Gerasimova YV, Kolpashchikov DM. 2013. Detection of bacterial 16S rRNA using a molecular beacon-based X sensor. Biosens. Bioelectron. 41:386–390. 10.1016/j.bios.2012.08.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foudeh AM, Daoud JT, Faucher SP, Veres T, Tabrizian M. 2014. Sub-femtomole detection of 16s rRNA from Legionella pneumophila using surface plasmon resonance imaging. Biosens. Bioelectron. 52:129–135. 10.1016/j.bios.2013.08.032 [DOI] [PubMed] [Google Scholar]
- 23.Miranda OR, Li X, Garcia-Gonzalez L, Zhu Z-J, Yan B, Bunz UHF, Rotello VM. 2011. Colorimetric bacteria sensing using a supramolecular enzyme-nanoparticle biosensor. J. Am. Chem. Soc. 133:9650–9653. 10.1021/ja2021729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farrow B, Hong SA, Romero EC, Lai B, Coppock MB, Deyle KM, Finch AS, Stratis-Cullum DN, Agnew HD, Yang S, Heath JR. 2013. A chemically synthesized capture agent enables the selective, sensitive, and robust electrochemical detection of anthrax protective antigen. ACS Nano 7:9452–9460. 10.1021/nn404296k [DOI] [PubMed] [Google Scholar]
- 25.Johnson JL, Rose BE, Sharar AK, Ransom GM, Lattuada CP, Mcnamara AM. 1995. Methods used for detection and recovery of Escherichia coli O157h7 associated with a food-borne disease outbreak. J. Food Prot. 58:597–603 [DOI] [PubMed] [Google Scholar]
- 26.Barreiros dos Santos M, Agusil JP, Prieto-Simón B, Sporer C, Teixeira V, Samitier J. 2013. Highly sensitive detection of pathogen Escherichia coli O157:H7 by electrochemical impedance spectroscopy. Biosens. Bioelectron. 45:174–180. 10.1016/j.bios.2013.01.009 [DOI] [PubMed] [Google Scholar]
- 27.Joung C-K, Kim H-N, Lim M-C, Jeon T-J, Kim H-Y, Kim Y-R. 2013. A nanoporous membrane-based impedimetric immunosensor for label-free detection of pathogenic bacteria in whole milk. Biosens. Bioelectron. 44:210–215. 10.1016/j.bios.2013.01.024 [DOI] [PubMed] [Google Scholar]
- 28.Serra B, Gamella M, Reviejo AJ, Pingarron JM. 2008. Lectin-modified piezoelectric biosensors for bacteria recognition and quantification. Anal. Bioanal. Chem. 391:1853–1860. 10.1007/s00216-008-2141-6 [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Ye Z, Si C, Ying Y. 2013. Monitoring of Escherichia coli O157:H7 in food samples using lectin based surface plasmon resonance biosensor. Food Chem. 136:1303–1308. 10.1016/j.foodchem.2012.09.069 [DOI] [PubMed] [Google Scholar]
- 30.Tolba M, Ahmed MU, Tlili C, Eichenseher F, Loessner MJ, Zourob M. 2012. A bacteriophage endolysin-based electrochemical impedance biosensor for the rapid detection of Listeria cells. Analyst 137:5749–5756. 10.1039/c2an35988j [DOI] [PubMed] [Google Scholar]
- 31.Tawil N, Sacher E, Mandeville R, Meunier M. 2012. Surface plasmon resonance detection of E. coli and methicillin-resistant S. aureus using bacteriophages. Biosens. Bioelectron. 37:24–29. 10.1016/j.bios.2012.04.048 [DOI] [PubMed] [Google Scholar]
- 32.Ricci F, Volpe G, Micheli L, Palleschi G. 2007. A review on novel developments and applications of immunosensors in food analysis. Anal. Chim. Acta 605:111–129. 10.1016/j.aca.2007.10.046 [DOI] [PubMed] [Google Scholar]
- 33.Wang YX, Ye ZZ, Ying YB. 2012. New trends in impedimetric biosensors for the detection of foodborne pathogenic bacteria. Sensors 12:3449–3471. 10.3390/s120303449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan X, White IM, Shopova SI, Zhu H, Suter JD, Sun Y. 2008. Sensitive optical biosensors for unlabeled targets: a review. Anal. Chim. Acta 620:8–26. 10.1016/j.aca.2008.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ligler FS, Sapsford KE, Golden JP, Shriver-Lake LC, Taitt CR, Dyer MA, Barone S, Myatt CJ. 2007. The array biosensor: portable, automated systems. Anal. Sci. 23:5–10. 10.2116/analsci.23.5 [DOI] [PubMed] [Google Scholar]
- 36.Geng T, Uknalis J, Tu SI, Bhunia AK. 2006. Fiber-optic biosensor employing Alexa-Fluor conjugated antibody for detection of Escherichia coli O157:H7 from ground beef in four hours. Sensors 6:796–807. 10.3390/s6080796 [DOI] [Google Scholar]
- 37.Mouffouk F, Rosa da Costa AM, Martins J, Zourob M, Abu-Salah KM, Alrokayan SA. 2011. Development of a highly sensitive bacteria detection assay using fluorescent pH-responsive polymeric micelles. Biosens. Bioelectron. 26:3517–3523. 10.1016/j.bios.2011.01.037 [DOI] [PubMed] [Google Scholar]
- 38.Owen V. 1997. Real-time optical immunosensors—a commercial reality. Biosens. Bioelectron. 12:i–ii [Google Scholar]
- 39.Wang Y, Knoll W, Dostalek J. 2012. Bacterial pathogen surface plasmon resonance biosensor advanced by long range surface plasmons and magnetic nanoparticle assays. Anal. Chem. 84:8345–8350. 10.1021/ac301904x [DOI] [PubMed] [Google Scholar]
- 40.Baccar H, Mejri MB, Hafaiedh I, Ktari T, Aouni M, Abdelghani A. 2010. Surface plasmon resonance immunosensor for bacteria detection. Talanta 82:810–814. 10.1016/j.talanta.2010.05.060 [DOI] [PubMed] [Google Scholar]
- 41.Tripathi SM, Bock WJ, Mikulic P, Chinnappan R, Ng A, Tolba M, Zourob M. 2012. Long period grating based biosensor for the detection of Escherichia coli bacteria. Biosens. Bioelectron. 35:308–312. 10.1016/j.bios.2012.03.006 [DOI] [PubMed] [Google Scholar]
- 42.Gasparyan VK, Bazukyan IL. 2013. Lectin sensitized anisotropic silver nanoparticles for detection of some bacteria. Anal. Chim Acta 766:83–87. 10.1016/j.aca.2012.12.015 [DOI] [PubMed] [Google Scholar]
- 43.Torun O, Boyaci IH, Temur E, Tamer U. 2012. Comparison of sensing strategies in SPR biosensor for rapid and sensitive enumeration of bacteria. Biosens. Bioelectron. 37:53–60. 10.1016/j.bios.2012.04.034 [DOI] [PubMed] [Google Scholar]
- 44.Sepulveda B, Angelome PC, Lechuga LM, Liz-Marzan LM. 2009. LSPR-based nanobiosensors. Nano Today 4:244–251. 10.1016/j.nantod.2009.04.001 [DOI] [Google Scholar]
- 45.Charlermroj R, Oplatowska M, Gajanandana O, Himananto O, Grant IR, Karoonuthaisiri N, Elliott CT. 2013. Strategies to improve the surface plasmon resonance-based immmunodetection of bacterial cells. Microchim. Acta 180:643–650. 10.1007/s00604-013-0975-x [DOI] [Google Scholar]
- 46.Wang C, Irudayaraj J. 2008. Gold nanorod probes for the detection of multiple pathogens. Small 4:2204–2208. 10.1002/smll.200800309 [DOI] [PubMed] [Google Scholar]
- 47.Fu JX, Park B, Zhao YP. 2009. Limitation of a localized surface plasmon resonance sensor for Salmonella detection. Sensors Actuat. B Chem. 141:276–283. 10.1016/j.snb.2009.06.020 [DOI] [Google Scholar]
- 48.Ray PC, Khan SA, Singh AK, Senapati D, Fan Z. 2012. Nanomaterials for targeted detection and photothermal killing of bacteria. Chem. Soc. Rev. 41:3193–3209. 10.1039/c2cs15340h [DOI] [PubMed] [Google Scholar]
- 49.Cheng IF, Chang HC, Chen TY, Hu CM, Yang FL. 2013. Rapid (<5 min) identification of pathogen in human blood by electrokinetic concentration and surface-enhanced Raman spectroscopy. Sci. Rep. 3:2365. 10.1038/srep02365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohk S-H, Bhunia AK. 2013. Multiplex fiber optic biosensor for detection of Listeria monocytogenes, Escherichia coli O157:H7 and Salmonella enterica from ready-to-eat meat samples. Food Microbiol. 33:166–171 10.1016/j.fm.2012.09.013 [DOI] [PubMed] [Google Scholar]
- 51.Ohk SH, Koo OK, Sen T, Yamamoto CM, Bhunia AK. 2010. Antibody-aptamer functionalized fibre-optic biosensor for specific detection of Listeria monocytogenes from food. J. Appl. Microbiol. 109:808–817. 10.1111/j.1365-2672.2010.04709.x [DOI] [PubMed] [Google Scholar]
- 52.Massad-Ivanir N, Shtenberg G, Tzur A, Krepker MA, Segal E. 2011. Engineering nanostructured porous SiO2 surfaces for bacteria detection via “direct cell capture.” Anal. Chem. 83:3282–3289. 10.1021/ac200407w [DOI] [PubMed] [Google Scholar]
- 53.Fronczek CF, You DJ, Yoon J-Y. 2013. Single-pipetting microfluidic assay device for rapid detection of Salmonella from poultry package. Biosens. Bioelectron. 40:342–349. 10.1016/j.bios.2012.07.076 [DOI] [PubMed] [Google Scholar]
- 54.Yang X, Gu C, Qian F, Li Y, Zhang JZ. 2011. Highly sensitive detection of proteins and bacteria in aqueous solution using surface-enhanced Raman scattering and optical fibers. Anal. Chem. 83:5888–5894. 10.1021/ac200707t [DOI] [PubMed] [Google Scholar]
- 55.Arlett JL, Myers EB, Roukes ML. 2011. Comparative advantages of mechanical biosensors. Nat. Nanotechnol. 6:203–215. 10.1038/nnano.2011.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang X, Wang R, Wang Y, Su X, Ying Y, Wang J, Li Y. 2011. Evaluation of different micro/nanobeads used as amplifiers in QCM immunosensor for more sensitive detection of E. coli O157:H7. Biosens. Bioelectron. 29:23–28. 10.1016/j.bios.2011.07.059 [DOI] [PubMed] [Google Scholar]
- 57.Guo X, Lin C-S, Chen S-H, Ye R, Wu VCH. 2012. A piezoelectric immunosensor for specific capture and enrichment of viable pathogens by quartz crystal microbalance sensor, followed by detection with antibody-functionalized gold nanoparticles. Biosens. Bioelectron. 38:177–183. 10.1016/j.bios.2012.05.024 [DOI] [PubMed] [Google Scholar]
- 58.Salam F, Uludag Y, Tothill IE. 2013. Real-time and sensitive detection of Salmonella Typhimurium using an automated quartz crystal microbalance (QCM) instrument with nanoparticles amplification. Talanta 115:761–767. 10.1016/j.talanta.2013.06.034 [DOI] [PubMed] [Google Scholar]
- 59.Yakovleva ME, Moran AP, Safina GR, Wadstrom T, Danielsson B. 2011. Lectin typing of Campylobacter jejuni using a novel quartz crystal microbalance technique. Anal. Chim. Acta 694:1–5. 10.1016/j.aca.2011.03.014 [DOI] [PubMed] [Google Scholar]
- 60.Hao R, Wang D, Zhang Xe Zuo G, Wei H, Yang R, Zhang Z, Cheng Z, Guo Y, Cui Z, Zhou Y. 2009. Rapid detection of Bacillus anthracis using monoclonal antibody functionalized QCM sensor. Biosens. Bioelectron. 24:1330–1335. 10.1016/j.bios.2008.07.071 [DOI] [PubMed] [Google Scholar]
- 61.Buchapudi KR, Huang X, Yang X, Ji HF, Thundat T. 2011. Microcantilever biosensors for chemicals and bioorganisms. Analyst 136:1539–1556. 10.1039/c0an01007c [DOI] [PubMed] [Google Scholar]
- 62.Lang HP, Gerber C. 2008. Microcantilever sensors. Top. Curr. Chem. 285:1–27. 10.1007/128_2007_28 [DOI] [PubMed] [Google Scholar]
- 63.Zhang J, Ji HF. 2004. An anti E. coli O157:H7 antibody-immobilized microcantilever for the detection of Escherichia coli (E. coli). Anal. Sci. 20:585–587. 10.2116/analsci.20.585 [DOI] [PubMed] [Google Scholar]
- 64.Campbell GA, Mutharasan R. 2005. Detection of pathogen Escherichia coli O157:H7 using self-excited PZT-glass microcantilevers. Biosens. Bioelectron. 21:462–473. 10.1016/j.bios.2004.11.009 [DOI] [PubMed] [Google Scholar]
- 65.Zhu Q, Shih WY, Shih WH. 2007. In situ, in-liquid, all-electrical detection of Salmonella typhimurium using lead titanate zirconate/gold-coated glass cantilevers at any dipping depth. Biosens. Bioelectron. 22:3132–3138. 10.1016/j.bios.2007.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sungkanak U, Sappat A, Wisitsoraat A, Promptmas C, Tuantranont A. 2010. Ultrasensitive detection of Vibrio cholerae O1 using microcantilever-based biosensor with dynamic force microscopy. Biosens. Bioelectron. 26:784–789. 10.1016/j.bios.2010.06.024 [DOI] [PubMed] [Google Scholar]
- 67.Ji HF, Yan XD, Zhang J, Thundat T. 2004. Molecular recognition of biowarfare agents using micromechanical sensors. Expert Rev. Mol. Diagn. 4:859–866. 10.1586/14737159.4.6.859 [DOI] [PubMed] [Google Scholar]
- 68.Campbell GA, Mutharasan R. 2007. A method of measuring Escherichia coli O157:H7 at 1 cell/mL in 1 liter sample using antibody functionalized piezoelectric-excited millimeter-sized cantilever sensor. Environ. Sci. Technol. 41:1668–1674. 10.1021/es061947p [DOI] [PubMed] [Google Scholar]
- 69.Sharma H, Mutharasan R. 2013. Rapid and sensitive immunodetection of Listeria monocytogenes in milk using a novel piezoelectric cantilever sensor. Biosens. Bioelectron. 45:158–162. 10.1016/j.bios.2013.01.068 [DOI] [PubMed] [Google Scholar]
- 70.Longo G, Alonso-Sarduy L, Rio LM, Bizzini A, Trampuz A, Notz J, Dietler G, Kasas S. 2013. Rapid detection of bacterial resistance to antibiotics using AFM cantilevers as nanomechanical sensors. Nat. Nanotechnol. 8:522–526. 10.1038/nnano.2013.120 [DOI] [PubMed] [Google Scholar]
- 71.Clark LC, Jr, Wolf R, Granger D, Taylor Z. 1953. Continuous recording of blood oxygen tensions by polarography. J. Appl. Physiol. 6:189–193 [DOI] [PubMed] [Google Scholar]
- 72.Korotcenkov G. 2010. Chemical sensors, vol 1 General approaches. Momentum Press, New York, NY [Google Scholar]
- 73.Zelada-Guillen GA, Tweed-Kent A, Niemann M, Goeringer HU, Riu J, Xavier Rius F. 2013. Ultrasensitive and real-time detection of proteins in blood using a potentiometric carbon-nanotube aptasensor. Biosens. Bioelectron. 41:366–371. 10.1016/j.bios.2012.08.055 [DOI] [PubMed] [Google Scholar]
- 74.Wan Y, Zhang D, Hou BR. 2010. Selective and specific detection of sulfate-reducing bacteria using potentiometric stripping analysis. Talanta 82:1608–1611. 10.1016/j.talanta.2010.07.030 [DOI] [PubMed] [Google Scholar]
- 75.Zelada-Guillen GA, Sebastian-Avila JL, Blondeau P, Riu J, Rius FX. 2012. Label-free detection of Staphylococcus aureus in skin using real-time potentiometric biosensors based on carbon nanotubes and aptamers. Biosens. Bioelectron. 31:226–232. 10.1016/j.bios.2011.10.021 [DOI] [PubMed] [Google Scholar]
- 76.Laczka O, Maesa JM, Godino N, del Campo J, Fougt-Hansen M, Kutter JP, Snakenborg D, Munoz-Pascual FX, Baldrich E. 2011. Improved bacteria detection by coupling magneto-immunocapture and amperometry at flow-channel microband electrodes. Biosens. Bioelectron. 26:3633–3640. 10.1016/j.bios.2011.02.019 [DOI] [PubMed] [Google Scholar]
- 77.Neufeld T, Schwartz-Mittelmann A, Biran D, Ron EZ, Rishpon J. 2003. Combined phage typing and amperometric detection of released enzymatic activity for the specific identification and quantification of bacteria. Anal. Chem. 75:580–585. 10.1021/ac026083e [DOI] [PubMed] [Google Scholar]
- 78.Li Y, Fang LC, Cheng P, Deng J, Jiang LL, Huang H, Zheng JS. 2013. An electrochemical immunosensor for sensitive detection of Escherichia coli O157:H7 using C-60 based biocompatible platform and enzyme functionalized Pt nanochains tracing tag. Biosens. Bioelectron. 49:485–491. 10.1016/j.bios.2013.06.008 [DOI] [PubMed] [Google Scholar]
- 79.de Avila BEF, Pedrero M, Campuzano S, Escamilla-Gomez V, Pingarron JM. 2012. Sensitive and rapid amperometric magnetoimmunosensor for the determination of Staphylococcus aureus. Anal. Bioanal. Chem. 403:917–925. 10.1007/s00216-012-5738-8 [DOI] [PubMed] [Google Scholar]
- 80.Guilbault GG, Lubrano GJ. 1973. Enzyme electrode for amperometric determination of glucose. Anal. Chim. Acta 64:439–455. 10.1016/S0003-2670(01)82476-4 [DOI] [PubMed] [Google Scholar]
- 81.Wang J. 2001. Glucose biosensors: 40 years of advances and challenges. Electroanalysis 13:983–988. 10.1002/1616-8984(200201)10:13.0.CO;2-Q [DOI] [Google Scholar]
- 82.Higson SP. 2012. Biosensors for medical applications. Woodhead Publishing, Cambridge, United Kingdom [Google Scholar]
- 83.Kim HJ, Bennetto HP, Halablab MA, Choi CH, Yoon S. 2006. Performance of an electrochemical sensor with different types of liposomal mediators for the detection of hemolytic bacteria. Sensors Actuat. B Chem. 119:143–149. 10.1016/j.snb.2005.12.013 [DOI] [Google Scholar]
- 84.Daniels JS, Pourmand N. 2007. Label-free impedance biosensors: opportunities and challenges. Electroanalysis 19:1239–1257. 10.1002/elan.200603855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Berggren C, Bjarnason B, Johansson G. 2001. Capacitive biosensors. Electroanalysis 13:173–180. [DOI] [PubMed] [Google Scholar]
- 86.Katz E, Willner I. 2003. Probing biomolecular interactions at conductive and semiconductive surfaces by impedance spectroscopy: routes to impedimetric immunosensors, DNA-sensors, and enzyme biosensors. Electroanalysis 15:913–947. 10.1002/elan.200390114 [DOI] [Google Scholar]
- 87.Mejri MB, Baccar H, Baldrich E, Del Campo FJ, Helali S, Ktari T, Simonian A, Aouni M, Abdelghani A. 2010. Impedance biosensing using phages for bacteria detection: generation of dual signals as the clue for in-chip assay confirmation. Biosens. Bioelectron. 26:1261–1267. 10.1016/j.bios.2010.06.054 [DOI] [PubMed] [Google Scholar]
- 88.Wan Y, Lin Z, Zhang D, Wang Y, Hou B. 2011. Impedimetric immunosensor doped with reduced graphene sheets fabricated by controllable electrodeposition for the non-labelled detection of bacteria. Biosens. Bioelectron. 26:1959–1964. 10.1016/j.bios.2010.08.008 [DOI] [PubMed] [Google Scholar]
- 89.Mantzila AG, Maipa V, Prodromidis MI. 2008. Development of a faradic impedimetric immunosensor for the detection of Salmonella typhimurium in milk. Anal. Chem. 80:1169–1175. 10.1021/ac071570l [DOI] [PubMed] [Google Scholar]
- 90.Huang JL, Yang GJ, Meng WJ, Wu LP, Zhu AP, Jiao XA. 2010. An electrochemical impedimetric immunosensor for label-free detection of Campylobacter jejuni in diarrhea patients' stool based on O-carboxymethylchitosan surface modified Fe3O4 nanoparticles. Biosens. Bioelectron. 25:1204–1211. 10.1016/j.bios.2009.10.036 [DOI] [PubMed] [Google Scholar]
- 91.Tan F, Leung PHM, Liu ZB, Zhang Y, Xiao LD, Ye WW, Zhang X, Yi L, Yang M. 2011. A PDMS microfluidic impedance immunosensor for E. coli O157:H7 and Staphylococcus aureus detection via antibody-immobilized nanoporous membrane. Sensors Actuat. B Chem. 159:328–335. 10.1016/j.snb.2011.06.074 [DOI] [Google Scholar]
- 92.Chan KY, Ye WW, Zhang Y, Xiao LD, Leung PHM, Li Y, Yang M. 2013. Ultrasensitive detection of E. coli O157:H7 with biofunctional magnetic bead concentration via nanoporous membrane based electrochemical immunosensor. Biosens. Bioelectron. 41:532–537. 10.1016/j.bios.2012.09.016 [DOI] [PubMed] [Google Scholar]
- 93.Siddiqui S, Dai ZT, Stavis CJ, Zeng HJ, Moldovan N, Hamers RJ, Carlisle JA, Arumugam PU. 2012. A quantitative study of detection mechanism of a label-free impedance biosensor using ultrananocrystalline diamond microelectrode array. Biosens. Bioelectron. 35:284–290. 10.1016/j.bios.2012.03.001 [DOI] [PubMed] [Google Scholar]
- 94.Dweik M, Stringer RC, Dastider SG, Wu YF, Almasri M, Barizuddin S. 2012. Specific and targeted detection of viable Escherichia coli O157:H7 using a sensitive and reusable impedance biosensor with dose and time response studies. Talanta 94:84–89. 10.1016/j.talanta.2012.02.056 [DOI] [PubMed] [Google Scholar]
- 95.Geng P, Zhang X, Meng W, Wang Q, Zhang W, Jin L, Feng Z, Wu Z. 2008. Self-assembled monolayers-based immunosensor for detection of Escherichia coli using electrochemical impedance spectroscopy. Electrochim. Acta 53:4663–4668. 10.1016/j.electacta.2008.01.037 [DOI] [Google Scholar]
- 96.Maalouf R, Fournier-Wirth C, Coste J, Chebib H, Saikali Y, Vittori O, Errachid A, Cloarec JP, Martelet C, Jaffrezic-Renault N. 2007. Label-free detection of bacteria by electrochemical impedance spectroscopy: comparison to surface plasmon resonance. Anal. Chem. 79:4879–4886. 10.1021/ac070085n [DOI] [PubMed] [Google Scholar]
- 97.Lu L, Chee G, Yamada K, Jun S. 2013. Electrochemical impedance spectroscopic technique with a functionalized microwire sensor for rapid detection of foodbornepathogens. Biosens. Bioelectron. 42:492–495. 10.1016/j.bios.2012.10.060 [DOI] [PubMed] [Google Scholar]
- 98.Guo XF, Kulkarni A, Doepke A, Halsall HB, Iyer S, Heineman WR. 2012. Carbohydrate-based label-free detection of Escherichia coli ORN 178 using electrochemical impedance spectroscopy. Anal. Chem. 84:241–246. 10.1021/ac202419u [DOI] [PubMed] [Google Scholar]
- 99.de la Rica R, Baldi A, Fernandez-Sanchez C, Matsui H. 2009. Selective detection of live pathogens via surface-confined electric field perturbation on interdigitated silicon transducers. Anal. Chem. 81:3830–3835. 10.1021/ac9001854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Joung C-K, Kim H-N, Im H-C, Kim H-Y, Oh M-H, Kim Y-R. 2012. Ultra-sensitive detection of pathogenic microorganism using surface-engineered impedimetric immunosensor. Sensors Actuat. B Chem. 161:824–831. 10.1016/j.snb.2011.11.041 [DOI] [Google Scholar]
- 101.Tlili C, Sokullu E, Safavieh M, Tolba M, Ahmed MU, Zourob M. 2013. Bacteria screening, viability, and confirmation assays using bacteriophage-impedimetric/loop-mediated isothermal amplification dual-response biosensors. Anal. Chem. 85:4893–4901. 10.1021/ac302699x [DOI] [PubMed] [Google Scholar]
- 102.Wang YX, Ping JF, Ye ZZ, Wu J, Ying YB. 2013. Impedimetric immunosensor based on gold nanoparticles modified graphene paper for label-free detection of Escherichia coli O157:H7. Biosens. Bioelectron. 49:492–498. 10.1016/j.bios.2013.05.061 [DOI] [PubMed] [Google Scholar]
- 103.Shabani A, Zourob M, Allain B, Marquette CA, Lawrence MF, Mandeville R. 2008. Bacteriophage-modified microarrays for the direct impedimetric detection of bacteria. Anal. Chem. 80:9475–9482. 10.1021/ac801607w [DOI] [PubMed] [Google Scholar]
- 104.Qi P, Wan Y, Zhang D. 2013. Impedimetric biosensor based on cell-mediated bioimprinted films for bacterial detection. Biosens. Bioelectron. 39:282–288. 10.1016/j.bios.2012.07.078 [DOI] [PubMed] [Google Scholar]
- 105.Wan Y, Zhang D, Wang Y, Hou B. 2010. A 3D-impedimetric immunosensor based on foam Ni for detection of sulfate-reducing bacteria. Electrochem Commun. 12:288–291. 10.1016/j.elecom.2009.12.017 [DOI] [Google Scholar]
- 106.La Belle JT, Shah M, Reed J, Nandakumar V, Alford TL, Wilson JW, Nickerson CA, Joshi L. 2009. Label-free and ultra-low level detection of Salmonella enterica serovar Typhimurium using electrochemical impedance spectroscopy. Electroanalysis 21:2267–2271. 10.1002/elan.200904666 [DOI] [Google Scholar]
- 107.Pournaras AV, Koraki T, Prodromidis MI. 2008. Development of an impedimetric immunosensor based on electropolymerized polytyramine films for the direct detection of Salmonella typhimurium in pure cultures of type strains and inoculated real samples. Anal. Chim. Acta 624:301–307. 10.1016/j.aca.2008.06.043 [DOI] [PubMed] [Google Scholar]
- 108.Zhu T, Pei ZH, Huang JY, Xiong CY, Shi SG, Fang J. 2010. Detection of bacterial cells by impedance spectra via fluidic electrodes in a microfluidic device. Lab Chip 10:1557–1560. 10.1039/b925968f [DOI] [PubMed] [Google Scholar]
- 109.Luong JHT, Male KB, Glennon JD. 2008. Biosensor technology: technology push versus market pull. Biotechnol. Adv. 26:492–500. 10.1016/j.biotechadv.2008.05.007 [DOI] [PubMed] [Google Scholar]
- 110.Olivier GK, Cho A, Sanii B, Connolly MD, Tran H, Zuckermann RN. 2013. Antibody-mimetic peptoid nanosheets for molecular recognition. ACS Nano 7:9276–9286. 10.1021/nn403899y [DOI] [PubMed] [Google Scholar]
- 111.Ahmad ZA, Yeap SK, Ali AM, Ho WY, Alitheen NBM, Hamid M. 2012. scFv antibody: principles and clinical application. Clin. Dev. Immunol. 2012:980250. 10.1155/2012/980250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Muyldermans S. 2013. Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 82:775–797. 10.1146/annurev-biochem-063011-092449 [DOI] [PubMed] [Google Scholar]
- 113.Hassanzadeh-Ghassabeh G, Devoogdt N, De Pauw P, Vincke C, Muyldermans S. 2013. Nanobodies and their potential applications. Nanomedicine 8:1013–1026. 10.2217/nnm.13.86 [DOI] [PubMed] [Google Scholar]
- 114.Richman S, Kranz D, Stone J. 2009. Biosensor detection systems: engineering stable, high-affinity bioreceptors by yeast surface display. Methods Mol. Biol. 504:323–350. 10.1007/978-1-60327-569-9_19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Stumpp MT, Amstutz P. 2007. DARPins: a true alternative to antibodies. Curr. Opin. Drug Disc. 10:153–159 [PubMed] [Google Scholar]
- 116.Tiede C, Tang AAS, Deacon SE, Mandal U, Nettleship JE, Owen RL, George SE, Harrison DJ, Owens RJ, Tomlinson DC, McPherson MJ. 2014. Adhiron: a stable and versatile peptide display scaffold for molecular recognition applications. Protein Eng. Des. Sel. 27:145–155. 10.1093/protein/gzu007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Leonard P, Hearty S, Brennan J, Dunne L, Quinn J, Chakraborty T, O'Kennedy R. 2003. Advances in biosensors for detection of pathogens in food and water. Enzyme Microb. Technol. 32:3–13. 10.1016/S0141-0229(02)00232-6 [DOI] [Google Scholar]
- 118.Gehring AG, Patterson DL, Tu SI. 1998. Use of a light-addressable potentiometric sensor for the detection of Escherichia coli O157:H7. Anal. Biochem. 258:293–298. 10.1006/abio.1998.2597 [DOI] [PubMed] [Google Scholar]


