Abstract
Valley fever (VF) is difficult to diagnose, partly because the symptoms of VF are confounded with those of other community-acquired pneumonias. Confirmatory diagnostics detect IgM and IgG antibodies against coccidioidal antigens via immunodiffusion (ID). The false-negative rate can be as high as 50% to 70%, with 5% of symptomatic patients never showing detectable antibody levels. In this study, we tested whether the immunosignature diagnostic can resolve VF false negatives. An immunosignature is the pattern of antibody binding to random-sequence peptides on a peptide microarray. A 10,000-peptide microarray was first used to determine whether valley fever patients can be distinguished from 3 other cohorts with similar infections. After determining the VF-specific peptides, a small 96-peptide diagnostic array was created and tested. The performances of the 10,000-peptide array and the 96-peptide diagnostic array were compared to that of the ID diagnostic standard. The 10,000-peptide microarray classified the VF samples from the other 3 infections with 98% accuracy. It also classified VF false-negative patients with 100% sensitivity in a blinded test set versus 28% sensitivity for ID. The immunosignature microarray has potential for simultaneously distinguishing valley fever patients from those with other fungal or bacterial infections. The same 10,000-peptide array can diagnose VF false-negative patients with 100% sensitivity. The smaller 96-peptide diagnostic array was less specific for diagnosing false negatives. We conclude that the performance of the immunosignature diagnostic exceeds that of the existing standard, and the immunosignature can distinguish related infections and might be used in lieu of existing diagnostics.
INTRODUCTION
Coccidioidomycosis, commonly known as valley fever (VF), is caused by the fungi Coccidioides immitis (California strain) or Coccidioides posadasii and is found in the arid soil of the southwestern desert regions of United States and South America. Human disease is caused by inhalation of the arthroconidia (spores) of the fungus and presents primarily with flu-like symptoms or, progressively, pneumonia. VF affects an estimated 150,000 (1) people in the United States every year, primarily in the states of Arizona (2), California (3), Nevada, New Mexico, and Utah. A major problem in the management of the disease is the failure to detect (sensitivity) 30% of the infected individuals. We have tested whether a new diagnostic technology, immunosignatures, can address this problem.
Sixty percent (4) of VF-exposed individuals are either asymptomatic or have mild symptoms, with the infection usually being self-limiting. The remaining 40% (5) of exposed individuals demonstrate symptoms, such as skin rashes and respiratory ailment, lasting from months to years. In 5 to 10% (4, 6) of these, infection disseminates, affecting other organs, the skin, bones, and nervous system. Individuals from non-Caucasian ethnicities (1), such as African Americans, Filipinos, and Asians, as well as those who are ≥65 years, pregnant women, and patients with immunocompromised immune systems, are more susceptible to VF, particularly the disseminated form of the disease. As per the Arizona Department of Health Services (ADHS), VF patients visit physicians three times on average before they are tested for VF, and more so if patients visiting AZ from regions nonendemic for the disease are diagnosed by physicians unacquainted with diseases of the American Southwest (7). VF alone is known to account for $86 million in hospital charges in Arizona in the year 2007 (7), but the burden is difficult to estimate outside AZ and CA.
The confirmatory diagnostic test for VF is an immunodiffusion (ID) assay, which detects antibodies against antigens within fungal coccidioidin causing complement fixation (CF) and tube precipitation (TP). Coccidioidin is a culture filtrate of the mycelial form of C. immitis, the heat-treated portion of which is used to detect IgM antibodies, and the untreated portion of which is used to detect IgG antibodies (8). The sensitivity of IDCF is 77%, and the sensitivity of IDTP is between 75 and 91% (9). An alternative is to culture the organism from body fluids or tissue, but a concern is infection risk for technicians (10). Although culture is a preliminary diagnostic for pneumonia, the sensitivity of this approach for VF ranges from a low of 23% to a high of 100%, depending on clinical status (11). The recovery rate of this pathogen through culture ranges from 0.4% from blood to 8.3% from respiratory tract specimens (12). As noted, the most clinically pressing issue is the low sensitivities of these diagnostics as primary tests.
We propose utilizing the immunosignature diagnostic technology (13) to address some or all of the limitations of current diagnostics for VF, particularly as a diagnosis for patients misclassified at the first test. The immunosignature technology utilizes a high-density array of non-life-space peptides to provide mimics of epitopes, even those that are discontinuous or nonproteins. We present data using microarrays of 10,000 unique random-sequence peptides. The peptides are 20 amino acids long, with 17 variable positions and a 3-residue linker at the attachment end. As opposed to single-antigen enzyme-linked immunosorbent assay (ELISA)-based methods, the disease-specific signature signal in an immunosignature comes from multiple peptides that form a distinct disease-specific pattern of antibody binding. Most antibody-based immunological tests examine the presence of new antibodies in infected individuals. An immunosignature can display both the presence of new antibodies relative to infection or chronic disease and any suppression of preexisting antibodies (measured as the loss of signal) that were commonly present in healthy controls. These signals may reflect memory responses to vaccinations and common pathogen exposures, but with standard immunologic tests, this loss would be difficult to detect. An immunosignature, unlike many genetic or immunological tests, is both sample sparing and robust to sample handling (14). Because the sensitivity of an immunosignature is higher than that of ELISA-based serological tests (13, 15), and discrimination across multiple diseases is possible, we asked whether the platform was suited to be used as a valley fever diagnostic method both for diagnosing VF and differentiating between other common respiratory infections. Current VF diagnostic methods may produce false-negative results, resulting in late recognition of the disease (16–18), which adversely affects patient outcomes. We proposed a series of tests to determine whether an immunosignature assay performs better than existing VF diagnostics. We postulated that this assay would detect VF earlier, with a greater sensitivity and at a lower cost than conventional methods. The ability of immunosignatures to distinguish VF from 3 other infections was tested. The same array was used to test whether VF-negative patients could be correctly classified as having VF. The effect of rescaling the platform by reducing the total number of peptides from 10,000 to <100 disease-specific ones was also evaluated.
MATERIALS AND METHODS
Serum samples used in this study.
All patient serum samples used in this study are listed in Table 1. Patient serology was determined by the tests shown in Table S1 in the supplemental material.
TABLE 1.
Patient sample cohorts utilized in this study
| Source and cohort no. | Infectiona | No. of patients | No. of samples |
|---|---|---|---|
| Confounding infection pilot studyb | |||
| 1 | Aspergillus | 20 | 20 |
| 2 | Chlamydia | 20 | 20 |
| 3 | Mycoplasma | 19 | 19 |
| 4 | Normal | 31 | 31 |
| VF training set | 18 | 18 | |
| Valley fever patient sera with non-VF controlsc | |||
| 1 | VF training set (U of A) | 35 | 55 |
| 2 | VF test set (U of A) | 25 | 67 |
| 3 | Normal individuals (ASU) | 41 | 41 |
| 4 | Influenza vaccinees (2006-2007 seasonal vaccine) (ASU) | 7 | 7 |
U of A, University of Arizona; ASU, Arizona State University.
The samples for the confounding infection pilot study were obtained from SeraCare Life Sciences (Milford, MA).
The samples from the VF training set were obtained from John Galgiani, MD, Valley Fever Center for Excellence (Tucson, AZ).
Confounding infection samples.
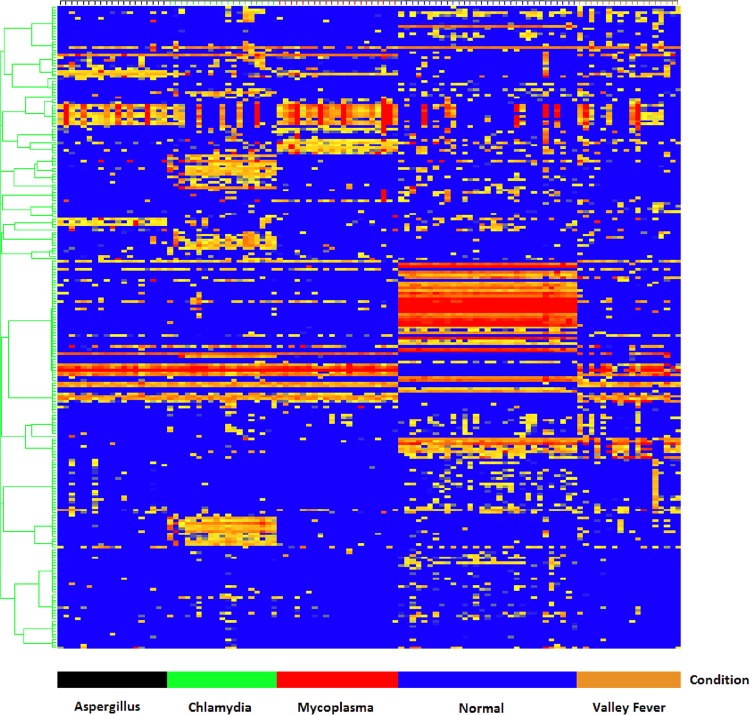
In order to test whether different infections were discernible, patient samples representing 19 Aspergillus fumigatus, 19 Mycoplasma pneumoniae, and 19 Chlamydia pneumoniae isolates were processed alongside 18 VF and 31 healthy sera on the 10,000-peptide microarray. The A. fumigatus, M. pneumoniae, and C. pneumoniae samples were acquired from SeraCare Life Sciences (Milford, MD) and were tested by commercial ELISAs for the presence of antibodies to the respective infections by SeraCare (See Table S1 in the supplemental material). The valley fever samples were obtained from John Galgiani (University of Arizona, Tucson, AZ; institutional review board [IRB] no. FWA00004218), and the healthy controls were obtained locally (IRB no. 0905004024). The results are shown in Fig. 1.
FIG 1.
Hierarchical clustering of informative peptides across five diseases. Peptides (y axis) are colored by intensity, with blue corresponding to low intensity and red to high intensity. Patients (x axis) are grouped by their corresponding peptide values with Aspergillus (black), Mycoplasma (red), Chlamydia (green), normal (blue), and valley fever (brown) grouping by cohort, as computed by GeneSpring 7.3.1 (Agilent, Santa Clara, CA). The peptides were selected by Fisher's exact test.
Valley fever and normal donor serum samples used in this study.
A training cohort of 55 VF samples and a blinded test set of 67 samples were obtained as deidentified human patient sera from John Galgiani. The nondisease serum samples included 7 influenza vaccine (2006 to 2007) recipient samples prevaccine and postvaccine plus 41 locally obtained healthy donor samples. Immunosignatures were obtained on the 100-peptide diagnostic subarray. Following the submission of our classification results to John Galgiani, the test set was unblinded and revealed to contain 25 patients with two or more serum samples collected longitudinally per patient during subsequent clinic visits. For each patient in the test cohort, the initial sample had an IDCF titer of zero but seroconverted at a later date as the infection progressed. All samples were serologically characterized by John Galgiani's laboratory for IDCF and IDTP titers. Tables 2 and 3 describe the CF titer distribution for the patients in the training and test cohorts, respectively.
TABLE 2.
Diagnosis (via IDCF) of 55 unique patient samples from the VF training cohort
| CF titer | No. of samples |
|---|---|
| 0 | 6 |
| 1 | 4 |
| 2 | 8 |
| 4 | 5 |
| 8 | 3 |
| 16 | 8 |
| 32 | 11 |
| 64 | 5 |
| 128 | 3 |
| 256 | 2 |
TABLE 3.
Diagnosis (via IDCF) of 67 blinded samples from the VF test cohort
| CF titer | No. of samples (no. of patients) |
|---|---|
| 0 | 48 (25) |
| 1 | 5 (4) |
| 2 | 7 (7) |
| 4 | 3 (3) |
| 8 | 1 (1) |
| 16 | 2 (2) |
| 32 | 1 (1) |
Blinded test patient sample set.
The test sample set included 25 patients with two or more serum samples per individual, for a total of 67 samples. Twenty-four of these symptomatic patients had an IDCF titer of zero and were given a negative diagnosis for VF after their first clinic visit. All 24 patients returned to the clinic for a follow-up appointment between 7 and 27 days after the first visit; blood samples were drawn for the second time, at which point 12 were still seronegative by the IDCF test. Of the 12 IDCF-negative patients, only 6 returned for the third follow-up visit due to continued symptoms, and 6 others returned either for monitoring of increasing CF titers or retesting due to a positive IDTP result. The time interval for the third visit ranged between 4 and 159 days after the second visit. Four of these patient samples were drawn again between 96 and 147 days, at which time a verified IDCF titer was observed in 2 patients who were given a positive VF diagnosis. One symptomatic patient returned for a fifth visit and remained seronegative on both the IDCF and IDTP tests 113 days later, despite being symptomatic for valley fever and IDTP positive in the fourth visit.
Microarray production and processing.
The 10,000-nonnatural sequence peptide immunosignature array and the 96-peptide VF diagnostic arrays were produced and processed as described in Legutki et al. (13, 19). Briefly, the peptides were spotted onto standard slides using a noncontact Piezo printer. The average spot diameter was ∼140 μm; 10,000 peptides were printed in 2-up format, enabling 2 separate arrays per slide. The slides were exposed to sera diluted 500-fold in sample buffer for 1 h and were washed in sample buffer, and the primary antibodies were detected with a fluorescent anti-human secondary antibody. The 16-bit 10-μm tagged image file format (TIFF) images from the Agilent C scanner were aligned using the GenePix 6.0 software (Axon Instruments, Union City, CA), and the data files were imported into GeneSpring 7.3.1 (Agilent, Santa Clara, CA) and R (20) for further analysis. Each training patient sample was processed in triplicate on the 10,000-peptide array. The 10,000-peptide array data were median normalized per chip and per feature. Any array with a Pearson's correlation coefficient of <0.80 across technical replicates was reprocessed. Patient samples were excluded from further analyses if they consistently produced extremely high background and/or consistently failed to provide reproducible results across technical replicates.
Statistical classification of disease groups.
The statistical classification of disease groups was done using naive Bayes from the R klaR package (21) combined with the leave-one-out cross-validation (LOOCV) and hold-out algorithm as implemented in the R package DMwR (22). Testing the classifier was done using a data-holdout experiment in which the training and test data sets were combined, and 70% of the randomly chosen data were used to train and predict the remaining 30% of the data set. This procedure was repeated 20 times to ensure every sample was predicted more than once by training on multiple combinations of other samples. The evaluation of immunosignaturing was compared to the IDCF test using sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy parameters.
Statistical classification of confounding infections.
To assess whether the random peptide microarray could specifically distinguish multiple confounding infections, serum samples were processed on the 10,000 random-peptide microarray. The slides were printed in batches of 136 slides each over a period of 2 days. Six different slide print batches were mixed together and used to process these serum samples. ComBat normalization was applied to the median-normalized data to eliminate differences between the samples due to batch effects (23, 24). A total of 243 random peptides capable of distinguishing between the five classes (VF, Aspergillus, Mycoplasma, Chlamydia, and normal) were selected using Fisher's exact association test, as implemented in GeneSpring GX 7.3.1. Since a physically separate training and test data set were missing for this analysis, the stringent holdout cross-validation as implemented in R package DMwR (22) was used on this data set to assess classification performance.
RESULTS
Valley fever immunosignature is distinct from those of other infections.
Our initial question was whether VF infection would produce an immunosignature that was distinguishable from those of other infections. The concern was that a general inflammatory response to infection may dominate the signature. To test this, we used serum samples from individuals infected with A. fumigatus, M. pneumoniae, and C. pneumoniae. Figure 1 and Table 4 show the results from an experiment in which the disease cohorts were classified and cross-validated using a 70% train/30% hold-out method, described in Materials and Methods. Figure 1 shows the relative intensities of 243 peptides found by Fisher's exact test, with the grouping of the individual cohorts shown on the x axis, and with hierarchical clustering applied to the peptides and the persons with Pearson's correlation used as the similarity measure. Each disease cohort groups together in the heat map. A quantitative assessment of the classification using the naive Bayes algorithm is presented in Table 4. The accuracy of simultaneous classification was 97% for Aspergillus and Mycoplasma and 98% for Chlamydia and VF. These results support the conclusion that a VF-specific signature can be distinguished from those of other potentially confounding infections.
TABLE 4.
| Infection type | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) |
|---|---|---|---|---|---|
| Aspergillus | 92 | 98 | 93 | 98 | 97 |
| Chlamydia | 95 | 99 | 97 | 99 | 98 |
| Mycoplasma | 98 | 98 | 87 | 100 | 97 |
| VF | 88 | 99 | 97 | 98 | 98 |
Naive Bayes was used to simultaneously classify the 108 patients into their respective groups using hold-out (70% train, 30% test, 20 iterations) cross-validation to estimate error.
Valley fever immunosignature is distinct from that of uninfected individuals.
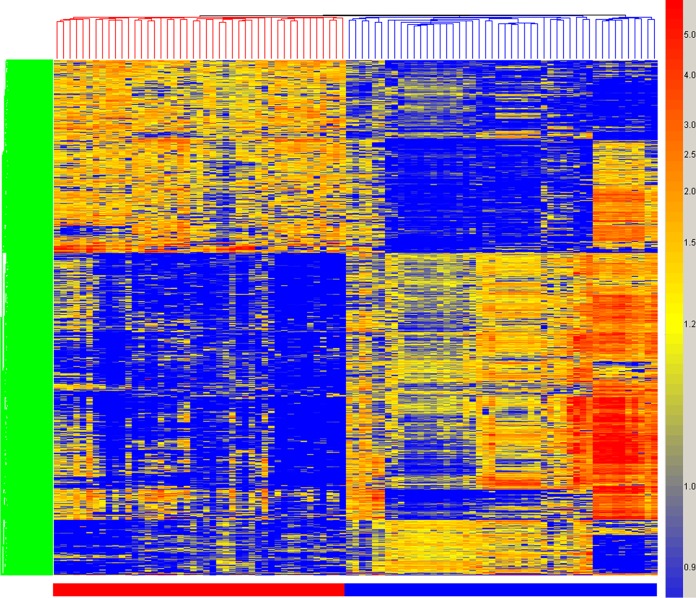
We then examined the immunosignatures of 45 VF clinical samples, shown in Table 2, on the 10,000-peptide microarray and identified 1,586 peptides from a 1-way analysis of variance (ANOVA) (5% family-wise error rate [FWER] correction), with a threshold of a P value of <1 × 10−14 indicating significance between the 45 VF training samples and 34 nondisease controls and 7 influenza vaccine recipients both prevaccine and 21 days postvaccine. The influenza virus signature was included to exclude a common potentially confounding signal. This signature is presented as a heat map in Fig. 2. Note that the differences between the non-VF and VF samples include reactivity that is higher in disease, as expected, but also signals that are lower in disease. Seventy percent of the samples were used to define a signature, and the remaining 30% were tested using the classifier. Repeating this 20 times yielded a perfect separation of infected from noninfected samples each time.
FIG 2.
Hierarchical clustering of valley fever immunosignature. A total of 1,586 peptides from a 1-way ANOVA between VF-infected and uninfected individuals are plotted on the y axis. The coloring is based on the signal intensities obtained from relative binding on the 10,000-peptide array, with blue representing low relative intensity and red representing high signal intensity. Each column represents the immunosignature of one individual, with VF patients (red), uninfected individuals, and pre- and post-influenza vaccine sera (blue).
Creating a 96-peptide VF diagnostic microarray.
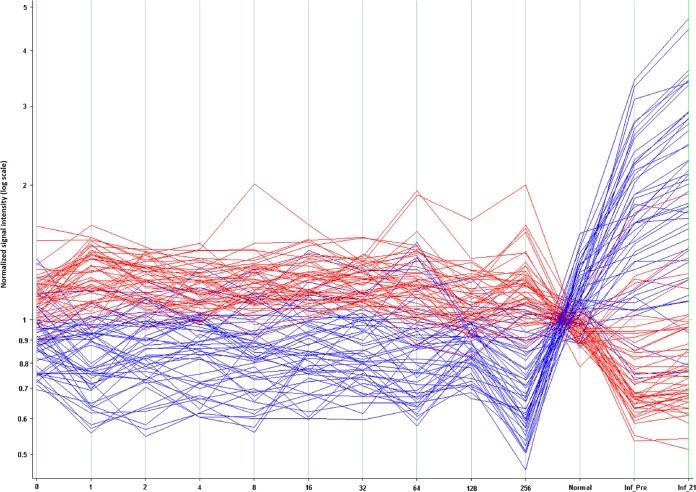
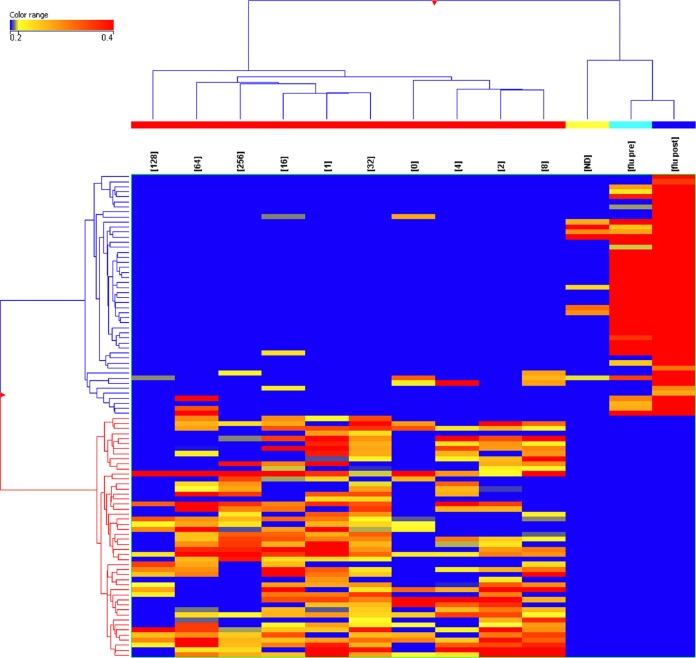
Under some circumstances, it may be useful to use the 10,000-peptide array as a discovery platform for informative peptides and then create a smaller diagnostic array from a subset of those peptides. To test this idea, we selected 96 peptides from the 1,586-peptide signature described above using pattern-matching algorithms within GeneSpring GX. We selected 96 peptides, as this number is easily handled on standard microtiter plates. Forty-eight peptides were chosen based on the criteria of capturing consistently high antibody signal across the VF samples and low antibody signal in the influenza vaccine samples. The other 48 were chosen based on the criteria of having a consistently low antibody signal in the VF samples but a high signal in the influenza vaccine samples. The log10 median-normalized signals for each of the 96 peptides are depicted in Fig. 3 in a line plot across patients whose signals were averaged by their CF titer. To test the robustness of these signature peptides, we performed a permuted t test by randomly reassigning the patient identifiers to the samples. The best P value obtainable after confusing the patient identifiers was <2.8 × 10−3, 9 orders of magnitude larger than when patient data were correctly labeled. It is therefore unlikely that the selected peptides were obtained by random chance. Figure 4 shows a heat map representation of these same 96 peptides, averaged per CF titer or influenza vaccine status. Hierarchical clustering was used to cluster the patient groups and peptides, with the colors within the cells representing high (red) to low (blue) intensities. Table 5 lists the performance of pairwise comparisons using 70% training/30% test averaged over 20 reiterations. The best performance was in distinguishing VF infection from noninfection (100% sensitivity, 97% specificity), and the worst was for VF infection versus influenza vaccines (100% sensitivity, 82% specificity). Based on this performance in the context of the 10,000-peptide array, these 96 predictor peptides were resynthesized (Sigma-Genosys, St. Louis, MO) and printed on a smaller array to test the performance of the VF focused array.
FIG 3.
Signal intensity (y axis) for 96 peptides from the 10,000 peptide microarray that distinguish VF and influenza vaccine recipients. The x axis indicates signal response averaged across patients for each CF titer. On the far right are signals averaged for the influenza vaccine recipients and normal donors. These data originated from the full 10,000-peptide array. Forty-eight peptides that captured high antibody binding in VF patients and low signals in normal/influenza vaccine recipients are colored in red. Forty-eight peptides showing higher signals in normal/influenza vaccine recipients and low signals for VF patients are colored in blue. Consistency was seen across the valley fever patients, and a reversal in signal was seen for non-VF patients. Inf_Pre, influenza vaccine recipients; Inf_21, patients 21 days postvaccine.
FIG 4.
Heat map showing normalized average signals from the 96 predictor peptides as in Fig. 2 but displaying the cohort separation. The data were averaged per CF titer and for 45 VF patients (red bars), 34 healthy controls (yellow bar), 7 pre-2006 influenza vaccine recipients (cyan bar) (flu pre), and 21-day postvaccine patients (dark blue bar) (flu post) (x axis). A t test identified 96 peptides (y axis) as being highly significant for distinguishing VF and healthy donors (ND).
TABLE 5.
Naive Bayes classification results from the VF training cohort on the 10,000-peptide microarray using the 96 predictor peptidesa
| Data set used, training (hold-out expt) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) |
|---|---|---|---|---|---|
| VF, normal | 100 | 97 | 97 | 100 | 98 |
| VF, influenza vaccine | 100 | 91 | 99 | 100 | 99 |
| VF, normal, influenza vaccine | 100 | 96 | 96 | 100 | 98 |
| 0 (CF titer, influenza vaccine | 100 | 82 | 76 | 100 | 88 |
| LOOCV, no-hold-out (all data) | 100 | 92 | 92 | 100 | 96 |
| For comparison | |||||
| CF titer (IDCF results) | 87 | 100 | 100 | 50 | 88 |
Hold-out splits all data randomly into 70% train/30% predict. The results are from 20 iterations of random hold-outs.
Performance of a 96-peptide VF diagnostic microarray.
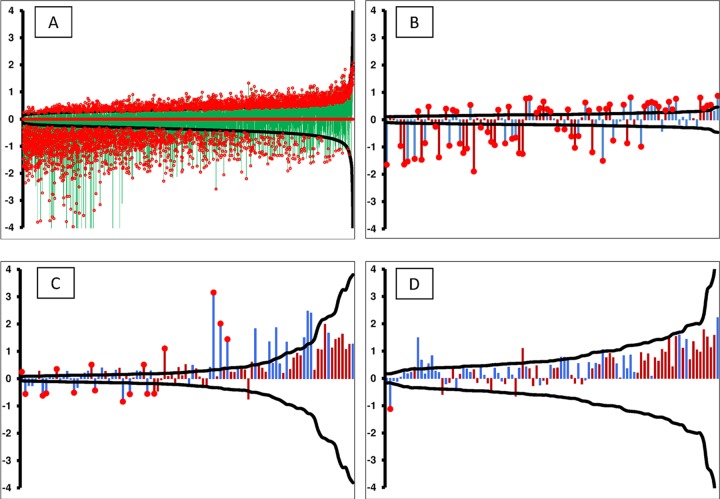
The performance of the VF diagnostic subarray was tested using a smaller set of training and non-VF control samples. Upon verification, the complete training and blinded test samples (67 blinded samples and 13 non-VF controls) were processed on the VF diagnostic 96-peptide subarray under similar conditions as those for the 10,000-peptide array. Table 6 shows the resulting classification performance. Of note is the performance against the samples with a CF titer of 0. While the VF diagnostic peptide set clearly had higher sensitivity than that of the ID assay, there was a substantial drop in the specificity compared to that of the ID assay and the performance on the 10,000-peptide array. Because there was a measurable difference in the performances of the different microarrays, we examined the detection limits for each peptide in the context of the actual fold change values between the healthy and VF patients. Figure 5 is a graph combining actual fold change values for every peptide plotted against the detection limit and the P value obtained from a t test between the VF versus normal cohorts. The smaller the delta, the more sensitive the peptide was to a signal and, consequently, the smaller the fold change needed to exceed this limit. Of note are Fig. 5B and C, which compare the performance differences between 96 VF diagnostic peptides within the context of the 10,000-peptide arrays and the same peptides that were resynthesized and independently printed on the VF diagnostic arrays. This comparison demonstrates the higher performance of the 96 peptides in the context of the 10,000-peptide array.
TABLE 6.
Naive Bayes classification resultsa from 96-peptide VF diagnostic subarray
| Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) | |
|---|---|---|---|---|---|
| CF titer by data set used | |||||
| CF titer (IDCF results) | 28 | 100 | 100 | 13 | 35 |
| CF titer = 0 | 100 | 43 | 92 | 100 | 93 |
| All data (0 and other titers) | 99 | 43 | 94 | 75 | 93 |
| Training and test hold-out, 20 iterationsb | |||||
| CF titer (IDCF results) | 52 | 100 | 100 | 19 | 57 |
| CF titer = 0 | 91 | 85 | 96 | 70 | 90 |
| All data (0 and other titers) | 82 | 92 | 99 | 37 | 83 |
| Training LOOCVc | |||||
| CF titer (IDCF results) | 87 | 100 | 100 | 50 | 88 |
| CF titer = 0 | 100 | 67 | 75 | 100 | 83 |
| All data (0 and other titers) | 100 | 67 | 96 | 100 | 96 |
Ninety-six-peptide diagnostic array data were tested for performance on a blinded cohort of false-negative VF patients.
Performance using all possible patient samples, including test and training samples.
Performance using the training data set only.
FIG 5.
Limits of detection graphed from a post hoc power calculation. The black curve in each figure represents the ±delta (minimum detectable fold change) calculated from the statistical precision of each peptide independently. The probes along the x axis are sorted by the calculated power, thus forming a smooth curve. Delta was calculated using α as 1/number of peptides/microarray, β of 0.20, and n of number of patients per group. The vertical bars (y axis) represent the log2 ratio between the healthy and VF-infected patients, with red bars indicating a peptide selected to predict VF and blue bars representing peptides selected for detection of non-VF conditions. The red circles on top of certain bars specify statistically significant fold changes at a P value of <0.01. The peptides used were 10,440 random peptides (training data set) using VF and healthy controls (A), 96 VF predictor peptides (training data set) within the 10,000 microarray (B), 96 resynthesized VF predictor peptides (training data set) for the VF-diagnostic assay (C), and 96 resynthesized VF predictor peptides (test data set) for the VF-diagnostic assay (D).
DISCUSSION
Our objectives in this study were to determine if the immunosignature diagnostic method had the potential to address the clinical problem of detecting infection in patients with an IDCF titer of 0, and if so, what was the best array format. We first demonstrated that VF infection as assayed on the 10,000-peptide array has a distinct immunosignature relative to those of 2 bacterial and one other fungal infection. We then showed using a 70% training/30% test format that the 10,000-peptide array accurately discriminated samples from patients with VF from non-VF-infected patients and influenza vaccinees. A total of 1,586 peptides were statistically significantly different between the classes. A portion of the signature was from peptides that had less reactivity in the samples from patients with VF than in those from the noninfected controls. Ninety-six peptides from the 1,586 that had good signature performances in the context of the 10,000-peptide array were resynthesized and used to create a smaller VF diagnostic subarray. When tested against the VF infection and control samples, this array demonstrated increased sensitivity (100%) but poor specificity compared to that of the conventional IDCF assay. The individual statistical analysis of the 96 peptides demonstrated that all performed better in the context of the 10,000-peptide array than in the subarray format.
We previously published studies demonstrating that influenza virus infection in mice (25) and the influenza vaccine in humans (13) can be distinguished from normal controls by immunosignatures. Here, we extend this list, showing that the immunosignatures of two different species of bacteria and two fungi are distinct. Only 283 peptides of the 10,000-peptide array were required to simultaneously distinguish the 4 infections with >97% accuracy. The development of the immunosignature diagnostic method for clinical application will require further validation testing against other common agents of community-acquired pneumonias and infections causing flu-like symptoms.
As noted, for VF, a clinically important issue is the people who report symptoms caused by infection with VF and yet do not test as seropositive by the standard immunological tests, the patients with a CF titer of 0. Using the 10,000-peptide immunosignature array, we demonstrated that there were 1,586 peptides that were reproducibly different between the samples from patients with VF and those from patients without VF. The non-VF samples included ones from patients who had received the influenza vaccine as part of an effort to exclude influenza infection signatures. Noteworthy is that a large portion of this signature was composed of peptides that had lower signal in the VF infection samples than the noninfection samples. We have noted this phenomenon before (13, 19, 26). This type of reactivity is not easily detected in standard ELISA-like methods. We pose that this may be due to the infection causing suppression or elimination of B cell-producing antibodies that are normally present in most healthy persons.
A reasonable strategy for developing immunosignatures is to use the large 10,000-peptide array as a discovery format and then produce subarrays with smaller numbers of peptides for the clinical diagnostic. The possible advantages of the smaller arrays are that they may be less expensive to manufacture, since fewer peptides are required, the peptides might be of higher quality, and the peptides may be simpler to read. To test this approach, we chose 96 peptides from the 1,586 peptides in the 10,000-peptide signature and pattern-matching analysis between the disease and nondisease groups. Forty-eight peptides were chosen that were consistently high in VF infection samples but low in influenza vaccine samples, and 48 were chosen with the converse signature. From a practical perspective, 96 is convenient, as it is the basic unit used in peptide synthesis and is prevalent in fluidics chambers, gaskets, and robotics. The peptides were selected based on their consistent signal over all titers, even in the samples with a CF titer of 0 (false negatives). We did not determine whether signatures that distinguish the titers can be selected to monitor VF progression, since there were too few matched longitudinal patient samples. The implication, however, is that the antibody reactivity that these peptides measure is independent of that measured in the ID assay.
This VF diagnostic subarray was tested in a blinded test against the VF infection and noninfection samples. The infection samples included the samples with a CF titer of 0. While this subarray was significantly more sensitive than the IDCF assay, it was less specific. This increase in sensitivity but loss of specificity was evident in the samples with a CF titer of 0. The implication is that this subarray at least needs to be used in combination with the standard ID assay to obtain maximum specificity and sensitivity.
Interestingly, the performance of the subarray was poorer than that of the 10,000-peptide array. This may in part be due to the selection criteria for the 96 peptides, which were positive for influenza vaccine recipients only. The peptides selected against a wider assortment of non-VF infection samples might perform better. It may also be that the additional peptides on the 10,000-peptide array distribute the antibody response to infection with a finer resolution, enabling high sensitivity and specificity. The 10,000-peptide format may have the advantage of being used to discriminate multiple infections on the same platform, a true test of specificity for any diagnostic method.
We have demonstrated that the immunosignature platform has clinical diagnostic potential relative to VF infection. It can address the clinical problem of the infections producing a CF titer of 0, either in the 10,000-peptide format or the subarray format in combination with the standard ID assay. There are ∼50 million people in the regions endemic for VF, with an estimated 30% being exposed over time to the infectious agent. However, since most people have little, if any, symptoms, it is unlikely that a diagnostic would be used generally to screen for VF infection. There is an existing standard antifungal treatment (fluconazole) and a new one in development (Nikkomycin Z). An improved diagnostic can at least identify symptomatic patients more accurately as having VF and may allow more effective use of treatments rapidly following the onset of symptoms and diagnosis.
Supplementary Material
ACKNOWLEDGMENTS
We thank John Lainson for contact printing of experimental peptide microarrays. We also thank Elizabeth Lambert, Mara Gardner, John Lainson, and Bart Legutki for processing patient serum samples on the 10,000 (version 1) random peptide arrays as per the stipulated protocols and experimental layout. We also thank Neal Woodbury for critical suggestions for the manuscript.
The arrays described here are available through the Peptide Array Core at Arizona State University.
This work was partially supported by startup funds from the Arizona State Technology Research Infrastructure Fund to S.A.J. This work was also supported by the Chemical Biological Technologies Directorate contracts HDTRA-11-1-0010 from the Department of Defense Chemical and Biological Defense program through the Defense Threat Reduction Agency (DTRA) to S.A.J.
Footnotes
Published ahead of print 25 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/CVI.00228-14.
REFERENCES
- 1.Galgiani JN, Ampel NM, Blair JE, Catanzaro A, Johnson RH, Stevens DA, Williams PL. 2005. Coccidioidomycosis. Clin. Infect. Dis. 41:1217. 10.1086/496991 [DOI] [PubMed] [Google Scholar]
- 2.Kaitlin B, Tsang C, Tabnak F, Chiller T. 2013. Increase in reported coccidioidomycosis–United States, 1998–2011. MMWR Morb. Mortal. Wkly. Rep. 62:217–221 [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2009. Increase in coccidioidomycosis–California, 2000–2007. MMWR Morb. Mortal. Wkly. Rep. 58:105–109 [PubMed] [Google Scholar]
- 4.DiCaudo DJ. 2006. Coccidioidomycosis: a review and update. J. Am. Acad. Dermatol. 55:929. 10.1016/j.jaad.2006.04.039 [DOI] [PubMed] [Google Scholar]
- 5.Tamerius JD, Comrie AC. 2011. Coccidioidomycosis incidence in Arizona predicted by seasonal precipitation. PLoS One 6:e21009. 10.1371/journal.pone.0021009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galgiani JN. 1999. Coccidioidomycosis: a regional disease of national importance: rethinking approaches for control. Ann. Intern. Med. 130:293. 10.7326/0003-4819-130-4-199902160-00015 [DOI] [PubMed] [Google Scholar]
- 7.Becker-Merok A, Nikolaisen C, Nossent HC. 2006. B-lymphocyte activating factor in systemic lupus erythematosus and rheumatoid arthritis in relation to autoantibody levels, disease measures and time. Lupus 15:570–576. 10.1177/0961203306071871 [DOI] [PubMed] [Google Scholar]
- 8.Lindsley MD, Warnock DW, Morrison CJ. 2006. Serological and molecular diagnosis of fungal infections, p 569–605 In Detrick BH, Robert Folds G, James D. (ed), Manual of molecular and clinical laboratory immunology, 7th ed. ASM Press, Washington, DC [Google Scholar]
- 9.Pappagianis D. 2001. Serologic studies in Coccidioidomycosis. Semin. Respir. Infect. 16:242–250. 10.1053/srin.2001.29315 [DOI] [PubMed] [Google Scholar]
- 10.Stevens DA, Clemons KV, Levine HB, Pappagianis D, Baron EJ, Hamilton JR, Deresinski SC, Johnson N. 2009. Expert opinion: what to do when there is Coccidioides exposure in a laboratory. Clin. Infect. Dis. 49:919–923. 10.1086/605441 [DOI] [PubMed] [Google Scholar]
- 11.DiTomasso JP, Ampel NM, Sobonya RE, Bloom JW. 1994. Bronchoscopic diagnosis of pulmonary coccidioidomycosis. Comparison of cytology, culture, and transbronchial biopsy. Diagn. Microbiol. Infect. Dis. 18:83–87. 10.1016/0732-8893(94)90070-1 [DOI] [PubMed] [Google Scholar]
- 12.Saubolle MA. 2007. Laboratory aspects in the diagnosis of coccidioidomycosis. Ann. N. Y. Acad. Sci. 1111:301–314. 10.1196/annals.1406.049 [DOI] [PubMed] [Google Scholar]
- 13.Legutki JB, Magee DM, Stafford P, Johnston SA. 2010. A general method for characterization of humoral immunity induced by a vaccine or infection. Vaccine 28:4529–4537. 10.1016/j.vaccine.2010.04.061 [DOI] [PubMed] [Google Scholar]
- 14.Chase BA, Johnston SA, Legutki JB. 2012. Evaluation of biological sample preparation for immunosignature-based diagnostics. Clin. Vaccine Immunol. 19:352–358. 10.1128/CVI.05667-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sykes KF, Legutki JB, Stafford P. 2013. Immunosignaturing: a critical review. Trends Biotechnol. 31:45–51. 10.1016/j.tibtech.2012.10.012 [DOI] [PubMed] [Google Scholar]
- 16.Mathisen G, Shelub A, Truong J, Wigen C. 2010. Coccidioidal meningitis: clinical presentation and management in the fluconazole era. Medicine (Baltimore) 89:251–284. 10.1097/MD.0b013e3181f378a8 [DOI] [PubMed] [Google Scholar]
- 17.Drake KW, Adam RD. 2009. Coccidioidal meningitis and brain abscesses: analysis of 71 cases at a referral center. Neurology 73:1780–1786. 10.1212/WNL.0b013e3181c34b69 [DOI] [PubMed] [Google Scholar]
- 18.Bouza E, Dreyer JS, Hewitt WL, Meyer RD. 1981. Coccidioidal meningitis. An analysis of thirty-one cases and review of the literature. Medicine (Baltimore) 60:139–172 [PubMed] [Google Scholar]
- 19.Stafford P, Halperin R, Legutki JB, Magee DM, Galgiani J, Johnston SA. 2012. Physical characterization of the “immunosignaturing effect.” Mol. Cell. Proteomics 11:M111.011593. 10.1074/mcp.M111.011593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ihaka R, Gentleman R. 1996. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 5:299–314 [Google Scholar]
- 21.Weihs C, Ligges U, Luebke K, Raabe N. 2005. klaR analyzing German business cycles. Springer-Verlag, Berlin, Germany [Google Scholar]
- 22.Torgo L. 2010. Data Mining with R: learning with case studies, 1st ed. Chapman and Hall/CRC, Boca Raton, FL [Google Scholar]
- 23.Johnson WE, Li C, Rabinovic A. 2007. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8:118–127. 10.1093/biostatistics/kxj037 [DOI] [PubMed] [Google Scholar]
- 24.Chen C, Grennan K, Badner J, Zhang D, Gershon E, Jin L, Liu C. 2011. Removing batch effects in analysis of expression microarray data: an evaluation of six batch adjustment methods. PLoS One 6:e17238. 10.1371/journal.pone.0017238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Legutki JB, Johnston SA. 2013. Immunosignatures can predict vaccine efficacy. Proc. Natl. Acad. Sci. U. S. A. 110:18614–18619. 10.1073/pnas.1309390110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kukreja M, Johnston S, Stafford P. 2012. Immunosignaturing microarrays distinguish antibody profiles of related pancreatic diseases. J. Proteomics Bioinformatics S. 6:001. 10.4172/jpb.S6-001 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.