ABSTRACT
Influenza virus neuraminidase (NA) is an interesting target of small-molecule antiviral drugs. We isolated a set of H5N1 NA-specific single-domain antibodies (N1-VHHm) and evaluated their in vitro and in vivo antiviral potential. Two of them inhibited the NA activity and in vitro replication of clade 1 and 2 H5N1 viruses. We then generated bivalent derivatives of N1-VHHm by two methods. First, we made N1-VHHb by genetically joining two N1-VHHm moieties with a flexible linker. Second, bivalent N1-VHH-Fc proteins were obtained by genetic fusion of the N1-VHHm moiety with the crystallizable region of mouse IgG2a (Fc). The in vitro antiviral potency against H5N1 of both bivalent N1-VHHb formats was 30- to 240-fold higher than that of their monovalent counterparts, with 50% inhibitory concentrations in the low nanomolar range. Moreover, single-dose prophylactic treatment with bivalent N1-VHHb or N1-VHH-Fc protected BALB/c mice against a lethal challenge with H5N1 virus, including an oseltamivir-resistant H5N1 variant. Surprisingly, an N1-VHH-Fc fusion without in vitro NA-inhibitory or antiviral activity also protected mice against an H5N1 challenge. Virus escape selection experiments indicated that one amino acid residue close to the catalytic site is required for N1-VHHm binding. We conclude that single-domain antibodies directed against influenza virus NA protect against H5N1 virus infection, and when engineered with a conventional Fc domain, they can do so in the absence of detectable NA-inhibitory activity.
IMPORTANCE Highly pathogenic H5N1 viruses are a zoonotic threat. Outbreaks of avian influenza caused by these viruses occur in many parts of the world and are associated with tremendous economic loss, and these viruses can cause very severe disease in humans. In such cases, small-molecule inhibitors of the viral NA are among the few treatment options for patients. However, treatment with such drugs often results in the emergence of resistant viruses. Here we show that single-domain antibody fragments that are specific for NA can bind and inhibit H5N1 viruses in vitro and can protect laboratory mice against a challenge with an H5N1 virus, including an oseltamivir-resistant virus. In addition, plant-produced VHH fused to a conventional Fc domain can protect in vivo even in the absence of NA-inhibitory activity. Thus, NA of influenza virus can be effectively targeted by single-domain antibody fragments, which are amenable to further engineering.
INTRODUCTION
Zoonotic influenza A virus infections are a persistent threat because of their pandemic potential. In particular, highly pathogenic avian influenza viruses (HPAIV) of the H5N1, H7N1, and H7N7 subtypes occasionally cross the species barrier between domesticated birds and humans. These viruses could become transmissible between humans through reassortment with circulating swine or human influenza viruses or by gradually accumulating mutations (1, 2). In the last decade, zoonotic outbreaks have had a major effect on public health. HPAIV H5N1 (3), the swine influenza (H1N1) outbreak in 2009 (4), and more recently, human infections with H7N9 in southern Asia (5) illustrate our poor preparedness for pandemic influenza (6). HPAIV H5N1 infection in humans has a confirmed case fatality rate of approximately 60%. The high pathogenicity of HPAIV H5N1 in humans can be attributed to a high replication rate and a broad cellular tropism that can lead to systemic virus spread. In addition, deregulated induction of proinflammatory cytokines and chemokines (“cytokine storm”) is associated with severe HPAIV H5N1 infections and can result in a disproportionate immunological response (7).
Influenza virus neuraminidase (NA) is a homotetrameric type II membrane glycoprotein with sialidase activity. The NA catalytic site is located at the top of each monomer, opposite the tetramer interface. NA plays an essential role in the spread of influenza viruses by cleaving sialic acids from the host cell receptors and from virions. NA activity also contributes to virus entry by cleaving decoy receptors present in mucins that line the layer of respiratory epithelial cells (8). Immunologically, NA is the second major humoral antigenic determinant (after hemagglutinin [HA]) and is subject to antigenic drift and occasional shift. In addition, experimental influenza vaccines supplemented with NA have improved efficacy (9–11). NA is also a codeterminant of influenza A virus (IAV) pathogenicity (12–14) and is involved in limiting IAV superinfections and reassortment (15). Decreased NA activity has been correlated with H5N1 adaptation to the human airway epithelium (16), and antibodies (Abs) against NA contribute to protection against an H5N1 virus challenge in a mouse model (17).
HA, the other major antigen, and NA cooperate in a tightly controlled way. For example, the fitness of mutant IAV lacking NA activity can be rescued by the selection of HA mutants with a decreased affinity for receptors containing sialic acid (18–20). These data demonstrate the importance of NA during IAV infection, so targeting of NA is a rational strategy. Indeed, three licensed influenza antivirals, oseltamivir, zanamivir, and peramivir, target NA. Influenza viruses that are resistant to oseltamivir frequently emerge in humans. In addition, NA-specific Abs protect mice and serum anti-NA Abs are associated with resistance to an IAV challenge in humans (21).
Effective prevention and treatment strategies are needed to control H5N1 infections, and antivirals based on single-domain Ab-based technology have been described as promising (22). Naturally occurring single-heavy-chain Abs have been found in sharks and camelids (23, 24). Unlike conventional Abs that are typically composed of light and heavy polypeptide chains that together determine epitope specificity, these natural single-chain Abs are composed of heavy chains only, and hence, their epitope specificity is confined to a single domain (so-called VHH, variable domain of heavy chain). Single-domain Ab fragments derived from camelids are highly specific, robust, easy to produce, and flexible. Hence, they are often used as novel therapeutics for many diseases (25, 26). VHHs usually have extended Ab loop structures and can interact with cognate antigens with affinities matching the best affinities of conventional Abs (27–29). In addition, VHHs that potently inhibit enzymes have been described (30, 31), and they have been used to stabilize membrane proteins in order to determine crystal structure (32). The VHH tends to target concave-shaped epitopes, in contrast to the planar epitopes that are typically recognized by “classical” light- and heavy-chain Abs (33). In addition, several examples of VHHs with potent antiviral activity have been reported (34–36). We have previously described an HA-specific H5N1-neutralizing VHH (35), and more recently, an H5N2-neutralizing VHH was reported (37). Moreover, a VHH directed against the influenza virus M2 protein inhibited the replication of amantadine-sensitive and -resistant viruses and protected mice against a lethal influenza virus challenge (38). Recently, Harmsen et al. described a panel of NA-binding VHHs, some of which displayed cross-subtype NA binding, but the antiviral potential of these VHHs was not evaluated (39).
Here, we report the isolation and characterization of a set of H5N1 NA-binding VHHs (N1-VHHm) some of which have potent NA-inhibitory activity and in vitro and in vivo antiviral activities. We also describe the production of bivalent, in-tandem N1-VHHm formats in Escherichia coli and N1-VHHm proteins fused to a mouse IgG2a Fc domain in seeds of transgenic Arabidopsis plants. Further, we compared their in vitro antiviral activities against H5N1 clade 1 (oseltamivir-sensitive and -resistant strains) with those against clade 2 viruses, and we evaluated the N1-VHHm-based prophylactic protection of mice against a potentially lethal infection with H5N1 virus.
MATERIALS AND METHODS
Influenza viruses.
H5N1 IAV strains NIBRG-14 and NIBRG-23 were obtained from the UK National Institute for Biological Standards and Control, a center of the Health Protection Agency. NIBRG-14 and NIBRG-23 are 2:6 reverse genetics-derived reassortants with NA and HA segments derived from A/Vietnam/1194/2004 (H5N1) and A⃥turkey⃥Turkey⃥2005 (H5N1), respectively, and the other six segments from A/PR/8/34 (H1N1) viruses. The H5N1 H274Y virus described here is a 1:1:6 reverse genetics-derived reassortant, with NA derived from A/crested eagle/Belgium/01/2004 (40) carrying the H274Y mutation introduced by site-specific mutagenesis, HA from NIBRG-14, and the remaining six genome segments from A/PR/8/34 (41). This virus was rescued by transfection of cocultured HEK-293T and MDCK cells. The supernatant from these cells was used for endpoint dilution to obtain a clonal H5N1 H274Y virus sample that was subsequently amplified on MDCK cells, pelleted from the cell supernatant, and mouse adapted by serial passage in BALB/c mice. All of the HA segments of these H5N1 viruses lack the coding information for the polybasic cleavage site. Following adaptation to BALB/c mice, the HA and NA coding regions of mouse-adapted NIBRG-14 (NIBRG-14ma) and H5N1 H274Y (H5N1 H274Yma) were sequenced and found to be identical to those of the parental viruses. Pandemic H1N1 (pH1N1) (kindly provided by Bernard Brochier, Scientific Institute of Public Health, Brussels, Belgium) is derived from a clinical isolate of the pH1N1 virus of 2009 and was adapted to mice by serial passage (42). A/New Caledonia/20/99 was kindly provided by Alan Hay (Medical Research Council [MRC] National Institute for Medical Research, London, United Kingdom). The median tissue culture infective dose (TCID50) and median lethal dose (LD50) of the NIBRG-14ma and H5N1 H274Yma viruses were calculated by the method of Reed and Muench (43). All of the H5N1 and pH1N1 experiments described above were performed in biosafety level 2+ rooms.
Baculovirus-based production of recombinant soluble tetrameric NA.
Soluble and enzymatically active tetrameric NA (N1rec) derived from A/crested eagle/Belgium/01/2004 (H5N1) was produced by proprietary technology (WO 2002/074795). Briefly, the N1rec expression cassette consisted of the secretion signal of IAV HA (ssHA, 16 residues), a tetramerizing variant of the leucine zipper domain of the Saccharomyces cerevisiae GCN4 transcription factor (tGCN4, 32 residues) (44, 45), and the extracellular part of NA derived from A/crested eagle/Belgium/01/2004 (H5N1; amino acid residues 53 to 449) (40) (Fig. 1A). This N1rec expression cassette was cloned into the pAcMP2 baculovirus transfer vector to produce pAcMP2n1rec. Recombinant baculovirus was generated by cotransfection of pAcMP2n1rec and linearized DNA of Autographa californica nuclear polyhedrosis virus (AcNPV) (BaculoGold; BD Biosciences) into Sf9 insect cells (derived from Spodoptera frugiperda) with the ESCORT IV Transfection Reagent (Sigma-Aldrich) to generate N1rec encoding recombinant baculovirus. Sf9 cells infected with AcNPVN1rec were incubated at 28°C in rolling bottles in TC-100 medium (Invitrogen) supplemented with 10% fetal calf serum and penicillin plus streptomycin. Four days after infection, the supernatant was harvested and clarified by centrifugation (1 h at 50,000 × g).
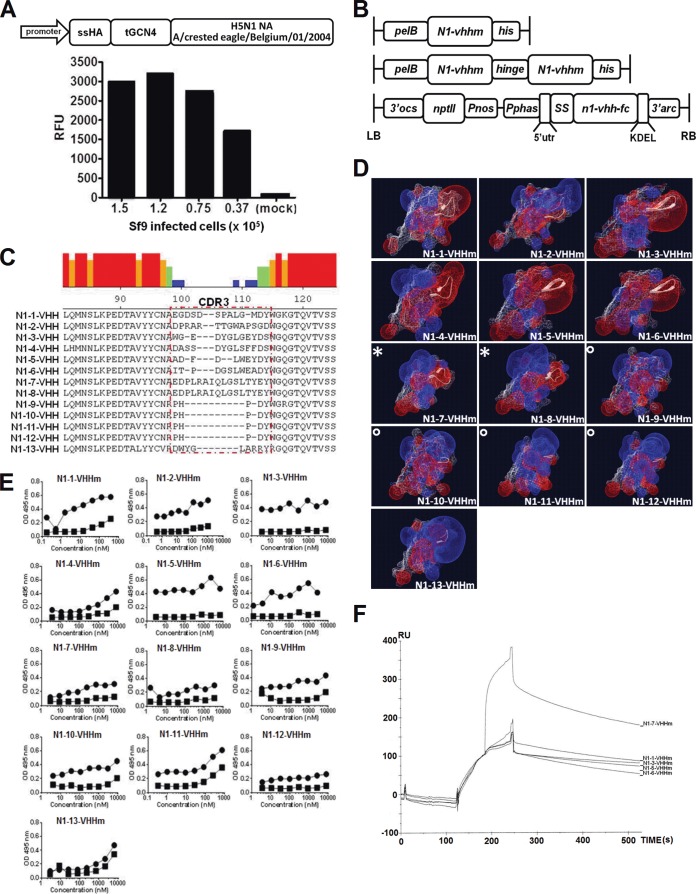
FIG 1.
Characterization of monovalent N1-VHH candidates that bind recombinant H5N1-derived NA. (A) Soluble N1rec derived from A/crested eagle/Belgium/01/2004 (H5N1) expressed in Sf9 cells. Diagram of the n1rec baculovirus expression cassette at the top: promoter, baculovirus basic protein promoter; ssHA, secretion signal of HA; tGCN4, tetramerizing GCN4-derived leucine zipper; H5N1 NA, extracellular domain of NA (amino acid residues 53 to 449). The graph at the bottom shows NA activity in the culture supernatants of different amounts of AcNPVN1rec-infected cells or mock-infected cells, with 4-MUNANA as the substrate. RFU, relative fluorescence units. (B) Schematic representation of the expression cassettes of N1rec-binding VHH domains and derived bivalent formats. Bacterium-produced monovalent N1-VHHm (N1-vhhm), with the amino-terminal periplasmic signal sequence (pelB), the n1-vhhm coding information, and a carboxy-terminal hexahistidine tag (his) (top). Bacterium-produced bivalent N1-VHHb with two n1-vhhm moieties fused by a flexible linker (llama IgG2c hinge) and a hexahistidine tag (his) (middle). For expression in transgenic A. thaliana seeds, the N1-VHH-Fc insert was cloned into the seed-specific expression cassette of the PphasGW vector. LB, left border of T-DNA; 3′ocs, 3′ end of the octopine synthase gene; nptII, neomycin phosphotransferase II open reading frame; Pnos, nopaline synthase gene promoter; Pphas, β-phaseolin gene promoter; 5′utr, 5′ untranslated region of the arc5-I gene; SS, signal sequence of the A. thaliana 2S2 seed storage protein gene; n1-vhh-fc, coding sequence of the N1-VHH gene fused to mouse CH2 and CH3 IgG2a; KDEL, endoplasmic reticulum retention signal; 3′arc, 3′-flanking regulatory sequences of the arc5-I gene; RB, T-DNA right border (bottom). (C) CDR3 sequences of 13 N1rec-binding VHH candidates. VHH CDR3 is highlighted by a red dotted box. The colored histogram denotes the homology between the sequences as follows: red, 100%; orange, 60 to 80%; green, 40 to 60%; light blue, 20 to 40%; dark blue, <20%. (D) In silico prediction of the three-dimensional structure of monomeric N1rec-binding VHHs (N1-VHHm). Structures were predicted with ESyPred3D, and diagrams were generated with DeepView software. Positive (blue) and negative (red) electrical potentials of the N1-VHHm surface are shown. The VHH CDR3 regions are represented in a white ribbon structure. The symbols * and ° indicate N1-VHHm from the same clonal families. (E) ELISA of the binding of the 13 N1-VHHm candidates to N1rec protein. N1rec (N1rec-tGCN4, filled circles) or recombinant M2e-tGCN4 (filled squares) at 10 μg/ml was used for coating. Dilution series of the 13 N1-VHHm candidates (His tagged) were incubated in the wells of coated plates, and mouse anti-His tag-HRP was used to reveal binding. OD, optical density. (F) SPR sensorgram of N1-VHHm binding to N1rec. The x axis shows time (seconds), and the y axis shows resonance units (RU), reflecting the increase in mass. N1rec was immobilized on a CM5 chip by chemical linkage. At 120 s, N1-3-VHHm was injected into the chip channel. At 180 s, N1-1-VHHm, N1-3-VHHm, N1-5-VHHm, N1-6-VHHm, or N1-7-VHHm was injected into the N1rec/N1-3VHHm-loaded channels.
Recombinant NA purification.
All purification steps were carried out at 4°C. Two parts (vol/vol) of n-butanol were added to three parts of cleared, AcNPVN1rec-infected Sf9 cell supernatant, and the mixture was shaken vigorously for 5 min. The aqueous phase, containing soluble N1rec, was separated from the organic phase, diluted 2.5 times in 5 mM KH2PO4 (pH 6.6), and passed through a 0.22-μm filter. The diluted aqueous phase was subsequently applied to an XK26⃥70 column (GE Healthcare) packed with HA Ultrogel hydroxyapatite chromatography sorbent (Pall) and eluted with a gradient of 5 to 400 mM KH2PO4 (pH 6.6) in 4% butanol. Eluted fractions with high NA activity were pooled and loaded onto a Blue Sepharose (Sigma-Aldrich) column equilibrated with 50 mM 2-(N-morpholino)ethanesulfonic acid (MES; pH 6.6) containing 5% glycerol and 8 mM CaCl2. Bound proteins were eluted with 50 mM MES (pH 6.6) containing 5% glycerol, 8 mM CaCl2, and 1.5 M NaCl. Fractions with NA activity were loaded onto a HiLoad 16/60 Superdex 200 (GE Healthcare) gel filtration column preequilibrated with 50 mM MES (pH 6.6) with 5% glycerol, 8 mM CaCl2, and 150 mM NaCl. Peak fractions containing NA activity were pooled, aliquoted, and stored at −80°C until used. All chromatography steps were performed with an Akta purification station (GE Healthcare).
NA activity assays.
NA activity was quantified by measuring the rate of cleavage of the fluorogenic substrate 4-MUNANA (2′-4-methylumbelliferyl-α-d-N-acetylneuraminic acid, sodium salt hydrate; Sigma-Aldrich) into 4-methylumbelliferone. The NA activity reaction was performed with 200 mM sodium acetate (NaAc)–2 mM CaCl2 with 1% butanol and 1 mM 4-MUNANA and measured in a kinetic mode with an excitation wavelength of 365 nm and an emission wavelength of 450 nm in a FLUOstar Optima. A standard curve of increasing concentrations of soluble 4-methylumbelliferone was included to correlate the fluorescence intensity with the molar amount of 4-methylumbelliferone. One unit of NA activity is defined as the activity needed to generate 1 nmol of 4-methylumbelliferone/min.
Fetuin (5 μg/ml; Sigma-Aldrich) was used to coat Nunc 96-well plates overnight at 4°C. Excess fetuin was washed away with phosphate-buffered saline (PBS), and IAV dilutions (in PBS with 1 mM CaCl2 and 0.5 mM MgCl2), with or without added single-domain Abs, were added and incubated for 1 h at 37°C. The amount of desialylated fetuin was measured by colorimetry to determine the binding of horseradish peroxidase (HRP)-coupled peanut agglutinin (PNA; Sigma-Aldrich). The plates were washed three times with PBS–0.1% Tween 20 and then incubated with 50 μl PNA-HRP (2.5 μg/ml in PBS–0.05% Tween 20) at room temperature. The plates were then washed three times with PBS, after which 50 μl of 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (Pharmingen BD) was added, and absorbance at 450 nm was measured with a reference at 655 nm. In the accelerated viral inhibition assay (AVINA) (46), IAV dilutions containing the N1-VHHm concentrations indicated were transferred to a black 96-well plate and 75 μl of 20 μM 4-MUNANA was added and incubated 1 h at 37°C. A 100-μl volume of stop solution (0.1 M glycine [pH 10.7], 25% ethanol) was then added to each well, and fluorescence was determined. For the AVINA and fetuin substrate assays, the amounts of IAV used were 7 × 105 PFU of NIBRG-14ma, 1 × 104 PFU of H5N1 H274Yma, and 3 × 104 PFU of pH1N1.
Camelid immunization, phage library construction, and panning.
An alpaca (Vicugna pacos) was injected subcutaneously with 125 μg of N1rec in the presence of incomplete Freund's adjuvant. Injections were administered once a week for 5 weeks. On day 39, anticoagulated blood was collected, lymphocytes were isolated with a UNI-SEP density gradient separation kit (NOVAmed), and total RNA was extracted. cDNA was prepared with oligo(dT) primers, and the VH and VHH genes were amplified with primers CALL001 (GTCCTGGCTGCTCTTCTACAAGG) and CALL002 (GGTACGTGCTGTTGAACTGTTCC). PstI and NotI restriction sites were inserted into the amplified sequences with primers A6E (GATGTGCAGCTGCAGGAGTCTGGRGGAGG) and 38 (GGACTAGTGCGGCCGCTGGAGACGGTGACCTGGGT). The resulting PCR product (550 bp) and the pHEN4 vector were digested with PstI and NotI, ligated (47), and transformed into electrocompetent E. coli TG1 cells. Transformants were grown in 2×TY medium (supplemented with 100 μg/ml ampicillin and 1% glucose) and infected with helper phage M13K07. The resulting VHH phage display library was panned four times with solid-phase coated N1rec.
Production and purification of monovalent NA-binding VHH (N1-VHHm).
The VHH coding sequence of the selected N1rec-binding phages (n = 13) was amplified by PCR with primers A6E and 38. The N1rec-binding VHH genes obtained by PCR and the pHEN6c vector were digested with PstI and BstEII, ligated, and transformed into E. coli WK6 cells. In the pHEN6c vector, the coding sequences of the N1rec-binding VHH candidates were cloned downstream of an N-terminal PelB secretion signal sequence and a carboxy-terminal hexahistidine tag was introduced (Fig. 1B) (48). For production and purification of soluble VHH, pHEN6c-transformed WK6 cells were grown in TB medium supplemented with 100 μg/ml ampicillin, 2 mM CaCl2, and 0.1% glucose. After 16 h, VHH production was induced with 1 mM isopropyl-β-d-thiogalactopyranoside, and the periplasmic fraction was extracted by osmotic shock with 0.2 M Tris (pH 8.0)–0.5 mM EDTA–0.5 M sucrose. Periplasmic extracts were centrifuged at 8,000 rpm and 4°C, and the supernatant was applied to a His Select Nickel Affinity gel (Sigma-Aldrich). After loading, the column was washed with PBS and VHHs were eluted with 0.5 M imidazole and dialyzed against PBS at 4°C. Monovalent N1rec-binding VHH (N1-VHHm) was concentrated with a Vivaspin 5,000-kDa cutoff filter (Vivascience). The total protein concentration was determined with the bicinchoninic acid protein assay kit (Thermo Scientific), and endotoxin levels was determined with a commercial Limulus amebocyte lysate-based assay (Toxinsensor Chromogenic LAL Endotoxin Assay kit; GenScript Corp.). We obtained 13 recombinant N1 NA-specific VHH proteins, which were named N1-1-VHHm to N1-13-VHHm. An irrelevant monovalent VHH against Arabidopsis thaliana albumin (SA-VHHm) was used as a negative control (49). The N1-VHHm sequences were aligned with the MegAlign software (DNAStar), and the VHH complementarity-determining region 3 (CDR3) electronegativity surface potential was predicted with the open source software ESyPred3D and visualized with DeepView (50) (Fig. 1C and D).
Enzyme-linked immunosorbent assays (ELISAs).
N1rec or M2e-tGCN4 (44) at 10 μg/ml was used to coat 96-well MaxiSorp (Nunc) plates overnight at 100 μl/well. Blocking was performed overnight with 5% skim milk in PBS. Dilution series of the 13 N1-VHHm candidates (from 10 μM to 0.1 nM) were incubated for 2 h at 37°C, and excess VHH was removed by three washes with PBS containing 0.05% Tween 20. Binding of the N1-VHHmto N1rec tGCN4 or NA globular head moieties was assessed by a mouse anti-His tag Ab fused with HRP (1:5,000). After three washes with PBS, 100 μl of TMB substrate (Pharmingen BD) was added, and absorbance at 450 nm was measured.
N1-VHHm SPR analysis.
The affinity of monovalent N1-3-VHHm, N1-5-VHHm, and N1-7-VHHm for N1rec was determined by surface plasmon resonance (SPR) on a Biacore 3000 platform (GE Healthcare). N1rec antigen was first immobilized (2,000 resonance units) in Series S Sensor Chip CM5 (GE Healthcare) by chemical NH2 coupling in 10 mM NaAc (pH 4.5). N1rec-loaded channels were regenerated with 0.02% SDS. Each monovalent N1-VHHm was diluted in HBS buffer (0.01 M HEPES, 0.15 M NaCl, 0.005% Tween 20, pH 6.4) to obtain a gradient ranging from 1.95 to 1,000 nM. The different N1-VHHs were injected over the N1rec-coated chip to record their binding kinetics. Biacore T00 evaluation software was used to calculate association rate constants (Kon), dissociation rate constants (Koff), and equilibrium dissociation constants (KD = Koff/Kon).
Construction of bacterium-produced bivalent N1-VHH (N1-VHHb).
The coding information for N1-3-VHHm and N1-5-VHHm was amplified with primers MH (CATGCCATGGGAGCTTTGGGAGCTTTGGAGCTGGGGGTCTTCGCTGTGGTGCGCTGAGGAGACGGTGACCTGGGT) and A4short (CATGCCATGATCCGCGGCCCAGCCGGCCATGGCTGATGTGCAGCTGGTGGAGTCT) to introduce an NcoI restriction enzyme site at each end of the amplified fragment. The MH primer also introduced the hinge sequence of llama γ2c (AHHSEDPSSKAPKAPMA) (51) to separate the two N1-VHHm domains. PCR amplicons of N1-3-VHHm and N1-5-VHHm were cloned into plasmid pHEN6c containing the n1-3-vhh or n1-5-vhh gene in order to generate homobivalent N1-3-VHHb and N1-5-VHHb expression constructs, which also contained a carboxy-terminal hexahistidine tag (Fig. 1B). These bivalent constructs were expressed and purified from the periplasmic fraction of E. coli as outlined above. We used an irrelevant, bivalent, in-tandem-fused VHH specific for bacterial β-lactamase (BL-VHHb) as a negative control (52) (kindly provided by Trong Nguyen-Duc, VUB, Brussels, Belgium). The endotoxin levels of the bivalent VHH preparations were determined as described above.
Production of bivalent N1-VHHb fused to the mouse IgG2 Fc fragment (N1-VHH-Fc) in seeds of stably transformed A. thaliana lines.
The three different n1-vhh-fc sequences coding for N1-3-VHHm, N1-5-VHHm, and N1-7-VHHm fused at the amino-terminal end with the 2S2 signal peptide and at the carboxy-terminal end with the Fc region of mouse IgG2a and the KDEL retention signal were synthesized commercially (GenScript) and cloned into a PphasGW binary transfer DNA (T-DNA) vector, resulting in the PphasGWn1-vhh-fc vector (Fig. 1B). The coronavirus GP4 subunit protein fused to the same mouse IgG2a Fc moiety (GP4-Fc; kindly provided by Robin Piron, VIB and Ghent University) was used as an irrelevant control in the antiviral assays. Agrobacterium C58C1Rifr(pMP90) was transformed with the PphasGWn1-3-vhh-fc, PphasGWn1-5-vhh-fc, PphasGWn1-7-vhh-fc, or PphasGWGP4-fc construct.
Agrobacterium C58C1Rifr(pMP90) was transformed with the PphasGWn1-3-vhh-fc, PphasGWn1-5-vhh-fc, PphasGWn1-7-vhh-fc, and PphasGWGP4-fc binary T-DNA vectors, and the resulting strains were used for A. thaliana floral-dip transformation (53, 54). Generations T1 to T3 were produced and screened for the best N1-VHH-Fc expressers. For protein purification, 1 g of transformed A. thaliana seeds was crushed in 25 ml of 50 mM Tris-HCl (pH 8.0)–200 mM NaCl–5 mM EDTA–0.1% (vol/vol) Tween 20–Complete protease inhibitor tablets (Roche). The seed extracts were clarified by centrifugation and applied to a protein G Sepharose column (GE Healthcare). N1-VHH-Fc proteins were eluted in 0.1 M glycine (pH 3.0) and immediately neutralized with 1 M Tris-HCl (pH 9.0). Between 1 and 10 mg of N1-VHH-Fc was recovered. The endotoxin levels of the VHH-Fc preparations were determined as described above.
Plaque assays.
Monolayers of MDCK cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum, 1% penicillin–streptomycin, and 1% glutamine and nonessential amino acids at 37°C in 5% CO2. TMPRSS2-expressing MDCK cells (kindly provided by Wolfgang Garten, Philipps-Universität Marburg, Marburg, Germany) were cultured in DMEM supplemented with the ingredients described above, as well as Geneticin (0.3 mg/ml) and puromycin (2 μg/ml). Expression of TMPRSS2 was induced with doxycycline (0.5 μg/ml) (55). At 70% confluence, the MDCK cells were infected at a multiplicity of infection of 10 with NIBRG-14ma, NIBRG-23, or H5N1 H274Yma. Samples containing monovalent N1-3/5/7-VHHm, bivalent N1-3/5-VHHb, bivalent N1-3/5/7/GP4-VHH-Fc, oseltamivir, or control medium were mixed with 0.8% Avicel RC-591 as an overlay (56). After 2 to 4 days of incubation, the cells were fixed with 4% paraformaldehyde in PBS for 30 min. The cells were permeabilized with 20 mM glycine containing 0.5% (vol/vol) Triton X-100. After blocking with bovine serum albumin (BSA), the cells were incubated for 2 h with polyclonal anti-NIBRG-14 (1:1,000) or anti-M2e monoclonal Ab (MAb; 1:5,000). After washing, anti-mouse whole IgG-HRP was used to visualize plaques with the TrueBlue peroxidase substrate (KPL).
Mouse experiments.
All mouse experiments were conducted according to national (Belgian Law 14/08/1986 and 22/12/2003 and Belgian Royal Decree 06/04/2010) and European legislation (EU Directives 2010/63/EU and 86/609/EEG) on animal regulations. All experiments on mice were approved by the ethics committee of Ghent University (permit number LA1400091), and efforts were made to avoid or diminish animal suffering. Specific-pathogen-free female BALB/c mice 7 to 9 weeks old were purchased from Charles River (Germany) and used in all experiments. Mice were housed in ventilated cages with high-efficiency particulate air filters in temperature-controlled, air-conditioned facilities and had food and water ad libitum. Mice were anesthetized by intraperitoneal injection of xylazine (10 μg/g) and ketamine (100 μg/g) before the intranasal administration of N1-VHHm, N1-VHHb, or N1-VHH-Fc. The N1-VHHm in any format were diluted in endotoxin-free PBS with 1% (wt/vol) BSA and applied intranasally in doses ranging from 0.25 to 5 mg/kg, as indicated in the figure legends. Mice were challenged 4 or 24 h later, as indicated, with 6 LD50s (H5N1 H274Yma) or 4 LD50s (NIBRG-14ma) of virus (50 μl, divided equally between the nostrils). A 30% loss of body weight was the endpoint at which sick mice were euthanized. On day 4 after the challenge, lung homogenates were prepared in PBS, cleared by centrifugation at 4°C, and used for virus titration. Monolayers of MDCK cells were infected with 50 μl of serial 1:10 dilutions of the lung homogenates in the wells of a 96-well plate in serum-free Dulbecco's modified Eagle medium (Invitrogen) supplemented with l-glutamine, nonessential amino acids, penicillin, and streptomycin. After 1 h, the inoculum was replaced with medium containing 2 μg/ml of l-1-tosylamide-2-phenylethyl chloromethyl ketone (TPCK)-treated trypsin (Sigma-Aldrich). Endpoint virus titers were determined by hemagglutination of chicken red blood cells and expressed as TCID50s per milliliter. IAV RNA levels were determined by quantitative reverse transcription-PCR (RT-qPCR). RNA was isolated from 150 μl of cleared lung homogenate with the NucleoSpin RNA virus kit (Macherey-Nagel). The relative amount of NIBRG-14ma genomic RNA was determined by preparing viral cDNA and performing quantitative PCR with M-genomic segment primers CGAAAGGAACAGCAGAGT and CCAGCTCTATGCTGACAAAATG and probe GGATGCTG (probe no. 89; Universal Probe Library, Roche) and the LightCycler 480 real-time PCR system (Roche).
Selection and characterization of H5N1 N1-VHHm escape mutant viruses.
NIBRG-14ma virus was serially diluted and used to infect MDCK cells in the presence of N1-3-VHHm (4.2 μM) or N1-5-VHHm (3.7 μM), a dose corresponding to 10 times the 50% inhibitory concentration (IC50) (see Table 2). Escape mutants were screened by monitoring cytopathic effects. After nine passages in the presence of N1-3-VHHm or N1-5-VHHm, escape viruses were plaque purified by growth on MDCK cells overlaid with 0.6% low-melting-point agarose in the presence of N1-3-VHHm or N1-5-VHHm. Escape virus isolates were amplified on MDCK cells, still in the presence of 10 times the IC50 of N1-3-VHHm or N1-5-VHHm. Total RNA was extracted from the supernatant and used to clone the cDNA corresponding to the NA viral RNA segment (57). Deduced amino acid substitutions in the NA sequence of escape viruses were modeled with PyMol (Delano Scientific; http://www.pymol.org) by using the H5N1 NA structure derived from A/Vietnam/1194/2004 (Protein Data Bank [PDB] code 2HTY). N1-VHH binding assays were performed as described previously (35). Briefly, 4 to 8 HA units of wild-type or plaque-purified candidate escape virus was used to coat the wells of ELISA plates. After blocking, the plates were incubated with different concentrations of N1-3-VHHm, N1-3-VHHb, N1-5-VHHm, or N1-5-VHHb. Binding of N1-VHHs was revealed by incubation with an anti-His tag MAb, followed by a secondary sheep anti-mouse IgG Ab conjugated to HRP and visualized by the addition of TMB substrate, and absorbance at 450 and 655 nm was measured.
TABLE 2.
N1-3-VHH and N1-5-VHH in a mono- or bivalent format inhibit N1rec activity and reduce the plaque size of H5N1 virus-infected cells
| N1-VHH format | N1rec inhibition (IC50, nM)a | Foldc | NIBRG-14 (IC50, nM)b | Foldc | H5N1 H274Yb | Foldc | NIBRG-23b | Foldc |
|---|---|---|---|---|---|---|---|---|
| N1-3-VHHm | 425.2 ± 149.4 | 1,844 ± 344.3 | 6,440 ± 1,247 | 4,295 ± 2,835.4 | ||||
| N1-5-VHHm | 374.9 ± 118.5 | 848.5 ± 380.2 | >26,500 | 3,492 ± 2,967.6 | ||||
| N1-7-VHHm | >15,448 | >32,896 | >32,896 | >32,896 | ||||
| SA-VHHmd | >15,448 | >32,896 | >32,896 | >32,896 | ||||
| N1-3-VHHb | 0.157 ± 0.206 | 2,708 | 7.6 ± 3.0 | 240 | 52.2 ± 36.8 | 123 | 24.6 ± 31.9 | 174 |
| N1-5-VHHb | 0.69 ± 0.231 | 543 | 14.5 ± 11.1 | 58 | >707.5 | 23.6 ± 3.8 | 148 | |
| BL-VHHbe | >15,448 | >32,896 | >32,896 | >32,896 | ||||
| N1-3-VHH-Fc | 1.31 ± 0.605 | 324 | 23.08 | 79 | 89.4 ± 7.8 | 71 | 24.2 ± 3.7 | 177 |
| N1-5-VHH-Fc | 1.49 ± 0.746 | 251 | 28.14 | 30 | >17,083 | 27.03 | 129 | |
| N1-7-VHH-Fc | >7,900 | >5,260 | >5,260 | >5,260 | ||||
| GP4-Fcf | >7,900 | >5,260 | >5,260 | >5,260 | ||||
| Oseltamivir | 586.1 ± 120.5 | 15,030 ± 10,039 | >243,000 | 73,000 |
Mean IC50 from three independent NA inhibition assays with 4-MUNANA and 160 ng of N1rec.
Mean plaque IC50 from two independent plaque assays performed in duplicate in which the VHH concentration reduced the plaque size by 50% compared with mock VHH.
Fold increase in the inhibitory potency of the N1-VHH bivalent format compared with its monovalent format.
Monomeric VHH against seed storage albumin.
Bivalent VHH against bacterial β-lactamase.
Coronavirus GP4 protein fused to mouse IgG2a Fc.
Statistical analysis.
GraphPad software (GraphPad Prism, version 6) was used for statistical analysis. To compare two groups in the fetuin and AVINAs, a t test was used. Differences between mouse groups were tested by two-way analysis of variance (ANOVA). When this test demonstrated a significant difference between groups (P < 0.05), t tests were used to compare two groups on each day. Kaplan-Meier survival curves were plotted, and differences in survival were analyzed by the Mantel-Cox log rank test.
RESULTS
Production of recombinant tetrameric NA.
A baculovirus expression vector system was used to produce recombinant soluble tetrameric and enzymatically active H5N1 NA derived from A/crested eagle/Belgium/01/2004 (N1rec). The N1rec coding sequence was cloned into a pAcMP2 expression vector (pAcMP2n1rec) under the control of the AcNPV basic protein promoter, which is active in the late phase of the baculovirus infection cycle (Fig. 1A). Infection of S. frugiperda (Sf9) cells with recombinant baculovirus AcNPVN1rec resulted in sialidase activity in the cell supernatant, suggesting that the N1rec product was soluble and enzymatically active NA (Fig. 1A). N1rec protein was purified from the supernatant of infected Sf9 cells by three successive chromatography steps, including size exclusion chromatography to collect tetrameric N1rec (see Materials and Methods). Using 4-MUNANA as a substrate, we calculated that purified N1rec had a specific activity of 56,851 NA U/mg.
Immunization and VHH phage library construction.
N1rec was used as an antigen for the generation and selection of NA-specific VHH. From an immunized alpaca, we obtained a VHH phage library of 2 × 108 independent transformants, 57% of which had a cDNA insert of the correct size (data not shown). Seventy-eight positive clones were selected, 13 of which had unique VHH sequences. CDR3 had the most variable amino acid sequence of the 13 N1rec-binding VHHs (N1-VHHm) (Fig. 1C). In silico structure prediction of the monomeric N1-VHHm candidates showed that the CDR3 of N1-1-VHHm, N1-3-VHHm, N1-4-VHHm, N1-5-VHHm, and N1-6-VHHm had an electronegative protruding paratope (Fig. 1D).
Production and characterization of soluble monomeric N1-VHHm.
The binding of the N1-VHHm proteins to N1rec was assessed by ELISA and compared with binding to the recombinant M2e-tGCN4 protein, which contains the same tetramerizing leucine zipper as N1rec (Table 1 and Fig. 1E). Except for N1-11-VHHm and N1-13-VHHm, none of the N1-VHHm proteins bound substantially to the recombinant M2e-tGCN4 protein. We next used the 4-MUNANA substrate to assess the ability of the N1-VHHm candidates to inhibit the enzymatic activity of N1rec. Four candidates, N1-1-VHHm, N1-3-VHHm, N1-5-VHHm, and N1-6-VHHm, inhibited the catalytic activity of N1rec (Table 1). N1-3-VHHm and N1-5-VHHm inhibited N1rec in the submicromolar range (Table 2) and, together with N1-7-VHHm, was analyzed by SPR to determine its affinity for immobilized N1rec. Both N1-3-VHHm and N1-5-VHHm had a high affinity for N1rec, with a KD in the low nanomolar (N1-5-VHHm) to high picomolar (N1-3-VHHm) range. N1-7-VHHm showed approximately 10- to 100-fold lower affinity for N1rec than N1-5-VHHm and N1-3-VHHm, respectively (Table 1). These binding affinities of N1-3-VHHm and N1-5-VHHm for N1rec resemble the affinities reported for a monomeric VHH that inhibits the activity of lysozyme (33). Competitive SPR analysis also showed that prior binding of N1-3-VHHm to N1rec abolished the subsequent binding of N1-1-VHHm, N1-5-VHHm, and N1-6-VHHm. Conversely, N1-7-VHHm binding to N1rec was not affected by prior incubation of N1rec with N1-3-VHHm or N1-5-VHHm (Fig. 1F). This suggests that N1-3-VHHm and N1-5-VHHm bind an overlapping epitope within N1rec or that binding of N1-3-VHHm sterically hinders the binding of N1-5-VHHm while N1-7-VHHm binds a different epitope.
TABLE 1.
Characteristics of N1-VHHm binding to purified recombinant N1rec as determined by ELISA and SPR
| N1-VHHm | N1rec bindinga | N1rec inhibition | Kon (M−1 s−1) | Koff (s−1) | KD (M) |
|---|---|---|---|---|---|
| 1 | + | + | NDb | ND | ND |
| 2 | + | − | ND | ND | ND |
| 3 | + | + | 3.6 E+6 | 1.3 E−3 | 3.7 E−10 |
| 4 | + | − | ND | ND | ND |
| 5 | + | + | 1.2 E−5 | 5.8 E−4 | 4.7 E−9 |
| 6 | + | + | ND | ND | ND |
| 7 | + | − | 4.4 E+4 | 2.8 E−3 | 6.3 E−8 |
| 8 | + | − | ND | ND | ND |
| 9 | + | − | ND | ND | ND |
| 10 | + | − | ND | ND | ND |
| 11 | − | − | ND | ND | ND |
| 12 | + | − | ND | ND | ND |
| 13 | − | − | ND | ND | ND |
Binding of N1rec (recombinant tetrameric N1 NA) was determined by ELISA. A minus sign indicates binding to the tetramerizing zipper and to N1rec.
ND, not done.
We next analyzed the antiviral potential of N1-3-VHHm and N1-5-VHHm by using H5N1 strain NIBRG-14ma. The amino acid sequences of the NA of NIBRG-14ma (derived from A/Vietnam/1194/2004) and N1rec (derived from A/crested eagle/Belgium/01/2004) are highly homologous, with only 13 amino acid differences, none of which map in or close to the NA catalytic site. In the presence of N1-3-VHHm or N1-5-VHHm, the size of NIBRG-14ma plaques was reduced in a concentration-dependent manner, with IC50s in the low nanomolar range (Table 2). In contrast, N1-7-VHHm did not affect the size or number of NIBRG-14ma plaques (Table 2). These results indicate that the in vitro antiviral potential of NA-specific VHH proteins depends on their NA-inhibitory activity.
Bivalent N1-VHHb formats have enhanced NA-inhibitory and antiviral potential.
It has been reported that the generation of multivalent formats of VHHs increases their functional affinity by introducing avidity (34). For example, the introduction of avidity drastically decreased the Koff of the VHH molecules directed against lysozyme and significantly improved their enzyme-inhibitory activity over that of their monovalent format (51). To increase the avidity of N1-VHHm, we compared two different bivalent formats. In one approach, we used the llama IgG2c hinge (17 amino acid residues) as a flexible linker to fuse two identical N1-VHHm gene moieties in tandem, resulting in the bivalent n1-3-vhhb and n1-5-vhhb expression elements (Fig. 1B and 2A). Despite repeated attempts, we could not generate an N1-7-VHHb expression plasmid. The corresponding proteins were expressed in E. coli and purified, and their in vitro N1-VHHb NA-inhibitory and antiviral activities were compared with those of their monovalent counterparts. N1rec inhibition by N1-3-VHHb and N1-5-VHHb was 2,000- and 500-fold stronger than that by the corresponding N1-3-VHHm and N1-5-VHHm formats, respectively (Table 2). Surprisingly, in a plaque assay using MDCK cells infected with NIBRG-14ma virus, both N1-3-VHHb and N1-5-VHHb seemingly had antiviral activities that were comparable to the activities of their monovalent counterparts (data not shown). In this assay, exogenous trypsin is used to facilitate the maturation of HA in newly produced virions to allow multicycle replication of the recombinant NIBRG-14ma virus. We noticed that the llama IgG2c hinge linker used to generate bivalent VHH formats was sensitive to trypsin cleavage (Fig. 2A). To circumvent the use of exogenously added trypsin, we took advantage of TMPRSS2-expressing MDCK cells, which are transformed with the doxycycline-inducible serine protease TMPRSS2 and allow multicycle replication of IAV in the absence of exogenous trypsin (55). By using monolayers of TMPRSS2-expressing MDCK cells for infection with NIBRG-14ma, the antiviral effect of N1-3-VHHb and N1-5-VHHb was 240- and 58-fold greater, respectively, than that of their monovalent formats (Table 2). These results obtained with N1-VHHb indicate that the NA-inhibitory and antiviral activities of both bivalent N1-VHHb formats are significantly greater than those of the monovalent N1-VHHm formats.
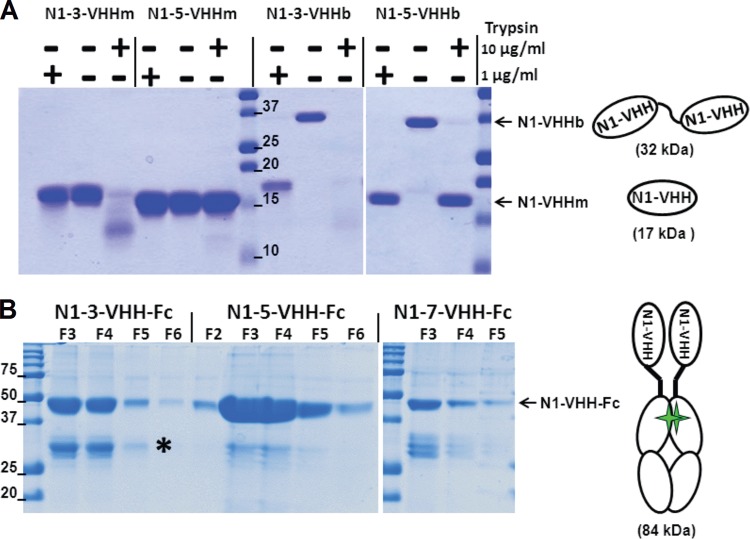
FIG 2.
N1-VHH monovalent and bivalent formats. (A) The llama IgG2c-derived hinge is sensitive to trypsin. Shown is Coomassie-stained reducing SDS-PAGE of purified N1-3-VHHm, N1-5-VHHm, N1-3-VHHb, and N1-5-VHHb incubated for 60 min at 37°C in the presence or absence of trypsin at 1 or 10 μg/ml. The N1-VHHb molecules migrate at about 32 kDa, and N1-VHHm and the cleavage products of N1-VHHb migrate as bands of about 17 kDa. The diagrams on the right depict N1-VHHm and N1-VHHb (consisting of two N1-VHHm moieties in tandem, linked by a llama IgG2c hinge of 17 amino acid residues). (B) Coomassie-stained reducing SDS-PAGE of the fractions eluted from a protein G column purification step with seed extracts of transgenic A. thaliana T3 plants expressing the N1-VHH-Fc proteins indicated. The N1-VHH-Fc constructs migrate at about 42 kDa. A degradation product (*) of about 28 kDa, corresponding to the Fc moiety only, is visible. The diagram on the right shows an N1-VHH-Fc molecule consisting of two N1-VHHm moieties fused to mouse IgG2a Fc, dimerized through a disulfide bond (green stars). The values beside the lanes are molecular sizes in kilodaltons.
We next engineered N1-VHH variants fused to a mouse IgG2a-derived Fc domain as an alternative bivalent format (N1-Vhh-Fc). To produce these more complex proteins, we used a robust plant seed-based expression system with a reported high yield of recombinant Abs (58, 59). For this, the n1-3-vhh, n1-5-vhh, and n1-7-vhh genes were fused to the sequence encoding the hinge and Fc tail of mouse IgG2a (n1-vhh-fc). These n1-vhh-fc constructs were cloned into the binary vector Pphas as a T-DNA expression cassette (Fig. 1B and 2B). Subsequently, transgenic A. thaliana plants were generated by Agrobacterium-mediated floral-dip transformation. Seed extracts of segregating T3 Arabidopsis transformants were screened by ELISA with Abs against the Fc part for the best expressers of the recombinant dimeric N1-VHH-Fc proteins. N1-3-VHH-Fc and N1-5-VHH-Fc accumulated to high levels in several transformants, and total soluble seed protein levels of 16% were found. On the contrary, N1-7-VHH-Fc did not accumulate well and the best expressors contained VHH-Fc at about 1% of their total soluble seed protein (data not shown). A good correlation was observed between the ELISA result and NA-inhibitory activity in crude seed extracts from different T3 transformants (data not shown). N1-3-VHH-Fc, N1-5-VHH-Fc, and N1-7-VHH-Fc were purified from seed extracts by protein G affinity chromatography. Reducing SDS-PAGE and Coomassie staining showed that the N1-VHH-Fc monomers migrated at approximately 42 kDa (Fig. 2B). N1-3-VHH-Fc and N1-5-VHH-Fc had stronger N1rec inhibition capacity and in vitro antiviral activity than their monovalent N1-3-VHHm and N1-5-VHHm counterparts (Table 2). Bivalent N1-7-VHH-Fc bound to N1rec in ELISAs (data not shown), but it failed to inhibit N1rec and the H5N1 viruses in vitro. Taken together, these results indicate that in vitro NA inhibition and antiviral activity against NIBRG-14ma are enhanced at least 30-fold by using bivalent N1-VHHb and N1-VHH-Fc formats, compared with those of the monovalent N1-VHHm domain.
In vitro antiviral activity of N1-VHH against clade 2.2 H5N1, oseltamivir-resistant H5N1, and 2009 pandemic H1N1 viruses.
Like NA from NIBRG-14, N1rec NA (derived from A/crested eagle/Belgium/01/2004) belongs to clade 1 of the H5N1 NAs. To assess the potential cross-reactivity of the NA activity inhibition (NAI) of the isolated VHHs in the three available formats (N1-VHHm, N1-VHHb, and N1-VHH-Fc), we focused first on NIBRG-23 NA (derived from A/turkey/Turkey/01/2005), a clade 2.2 H5N1 virus (60). Although there are only a few laboratory-confirmed human infections with H5N1, it appears that clade 2 H5N1 viruses are responsible for most of these zoonotic infections with HPAIV (61). N1-3-VHHm and N1-5-VHHm reduced the in vitro growth of the NIBRG-23 virus with an IC50 in the low micromolar range (Table 2). N1-3-VHHb, N1-5-VHHb, N1-3-VHH-Fc, and N1-5-VHH-Fc displayed about 150-fold higher in vitro antiviral activity than the corresponding monovalent N1-VHHm form against this clade 2 virus in a plaque size reduction assay. On the basis of these findings, we conclude that the two NA-inhibitory VHHs inhibit the in vitro replication of representative H5N1 IAV from clades 1 and 2 with comparable efficiency, suggesting that they target an epitope that the NAs of these viruses have in common.
Oseltamivir-resistant IAV frequently emerges and spreads in the human population. Several mutations have been reported to contribute to oseltamivir resistance, but among them, the H274Y mutation (N2 numbering) is the one most commonly found in oseltamivir-resistant viruses (62). We used reverse genetics to generate a clade 1 H5N1 virus harboring the H274Y mutation in NA (derived from A/crested eagle/Belgium/01/2004), while the HA segment was derived from NIBRG-14 and the remaining six segments were from PR/8, resulting in an H274Y mutant H5N1 virus. Next, we determined if N1-3-VHHm, N1-5-VHHm, N1-3-VHHb, N1-5-VHHHb, N1-3-VHH-Fc, and N1-5-VHH-Fc could inhibit this virus. As all of the N1-3-VHH formats tested so far performed like N1-5-VHH formats in vitro, we were surprised that only N1-3-VHH (monovalent and both bivalent formats), but not N1-5-VHH in any format, reduced the growth of H5N1 H274Y (Table 2). Compared to NIBRG-14ma, the H5N1 H274Y IC50s were 3- to 7-fold higher but still in the low nanomolar range. Even though the competitive SPR analysis suggested that the N1-3-VHHm and N1-5-VHHm epitopes in NA overlap (Fig. 1F), the contact residues necessary for their binding are probably not identical. We conclude that the three formats of N1-3-VHH evaluated can inhibit the growth of the H5N1 H274Y virus in vitro and that both bivalent formats have enhanced in vitro antiviral activity. In addition, the H274Y mutation seems to be sufficient to abolish the in vitro antiviral effect of the N1-5-VHH formats.
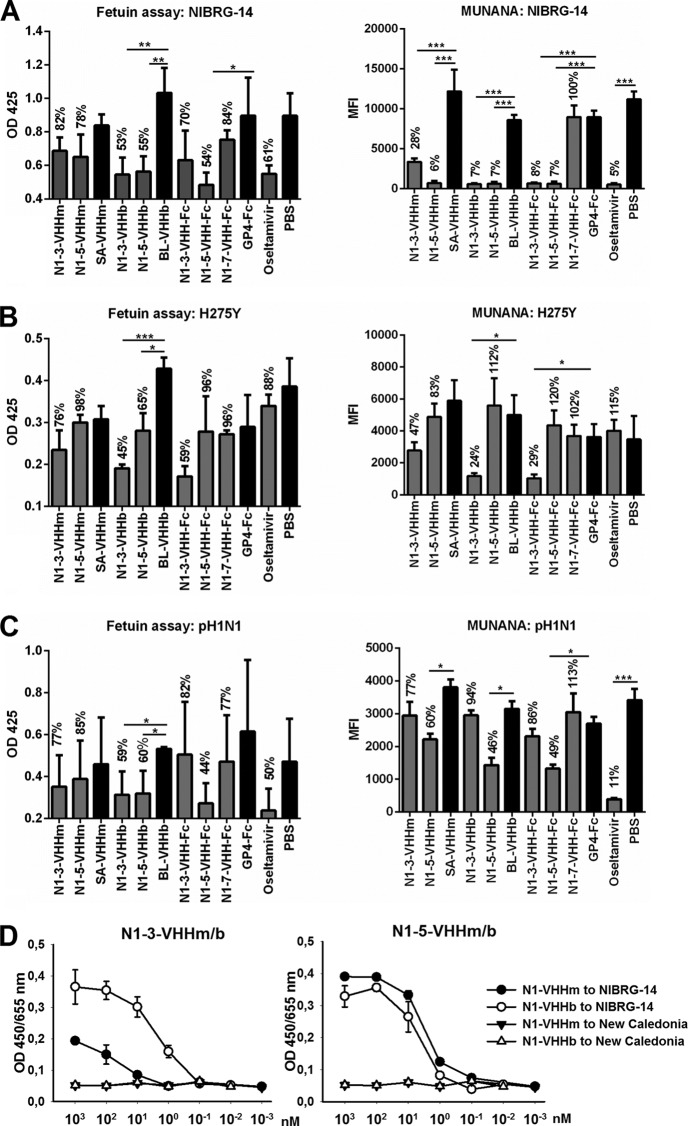
Next, we tested the antiviral potential of all of the N1-3-VHH and N1-5-VHH formats against a pandemic H1N1 2009 virus isolate (pH1N1). We used fetuin and 4-MUNANA (AVINA) as two alternative substrates for virion-associated NA activity. With NIBRG-14ma, and on the basis of 4-MUNANA hydrolysis, monovalent N1-3-VHHm and N1-5-VHHm had significant inhibitory activity, compared with the negative controls SA-VHHm (P < 0.05) and PBS (P < 0.01), although this tendency was not significant in the fetuin assay (Fig. 3A). We attribute these differences between the two assays to the types of substrate used, the small molecule 4-MUNANA versus fetuin, which is a large protein carrying multiple sialylated glycans. In the case of fetuin, HA plays an important role in recruiting NA to the substrate, which may alter the accessibility of the NA target for the N1-VHHs. We found that the bivalent molecules N1-3-VHHb, N1-5-VHHb, N1-3-VHH-Fc, and N1-5-VHH-Fc significantly inhibited NA activity on both substrates (P < 0.01, Fig. 3A).
FIG 3.
N1-VHH inhibits NA activity from NIBRG-14ma, H5N1 H274Y, and pH1N1 virions. Fetuin (left) and 4-MUNANA (right) were used as substrates. (A) NIBRG-14ma NA inhibition. (B) H5N1 H274Y oseltamivir-resistant NAI. (C) Pandemic H1N1 2009 NAI. SA-VHHm, monovalent VHH directed against seed storage albumin; BL-VHHb, bivalent VHH directed against bacterial β-lactamase; GP4-Fc, coronavirus GP4 protein fused to mouse IgG2a Fc. To determine the significance of differences between groups, a t test was used (*, P < 0.05; **, P < 0.01). Results are representative of two independent experiments. The bars represent the mean optical density (OD) at 425 nm (n = 3) in the fetuin assay and the mean fluorescence intensity (n = 2) after 1 h of incubation in the 4-MUNANA assay. Error bars represent the standard deviation of the mean. The values above the bars are normalized to the respective monovalent (SA-VHHm), bivalent (BL-VHHb), and Fc chimera (GP4-Fc) controls and PBS. The significance of differences between groups and their respective controls was determined by a t test (*, P < 0.05; **, P < 0.01; ***, P < 0.005). (D) Mono- and bivalent NA-specific VHHs bind to H5N1 virus but not to seasonal A/New Caledonia/20/99 virus. ELISA plates were coated with 8 HA units of NIBRG-14 or A/New Caledonia/20/99 (New Caledonia) virus. After blocking, wells were incubated with different concentrations of N1-3-VHHm, N1-3-VHHb, N1-5-VHHm, or N1-5-VHHb as indicated (nanomolar concentration of VHH). Binding of N1-VHHs was revealed by incubation with an anti-His tag MAb, followed by a secondary sheep anti-mouse IgG Ab conjugated to HRP and visualized by the addition of TMB. The y axis shows the absorbance at 450 nm subtracted from the absorbance at 655 nm.
In line with our previous results with H5N1 H274Y IAV, only N1-3-VHHb and N1-3-VHH-Fc showed NAI activity on both substrates (P < 0.05 or P < 0.01) (Fig. 3B). N1-5-VHHb and N1-5-VHH-Fc showed inhibitory potential against pH1N1 in both assays (P < 0.05) (Fig. 3C). We could not detect binding of the N1-VHHs to A/NewCaledonia/20/99 or inhibition of NA activity of this virus (Fig. 3D and data not shown). These AVINA and fetuin results for NIBRG-14ma and H5N1 H274Y NA are in accordance with the N1rec inhibition and the plaque size reduction assays mentioned before (Table 2). In addition, both bivalent N1-5-VHHb formats inhibited pH1N1 virion-associated NA activity, suggesting a degree of intrasubtype inhibitory effect of the N1-VHHs.
N1-3-VHHm and N1-5-VHHm treatments reduce early morbidity but not viral lung titers in H5N1-challenged mice.
We next evaluated the in vivo antiviral effect of N1-3-VHHm and N1-5-VHHm. The endotoxin levels of the VHH preparations that were used in vivo were low and comparable for the NA-specific VHH proteins and their irrelevant control counterparts (Table 3). In the first experiment we administered 100 μg of N1-3-VHHm, N1-5-VHHm, or N1-7-VHHm intranasally to BALB/c mice 4 h before a challenge with 4 LD50s of NIBRG-14ma. Two groups were included as positive controls, (i) a group of mice that received 30 μg of H5-VHHb, a bivalent NIBRG-14ma HA-neutralizing VHH (36), and (ii) a group of mice that received a large daily oral dose of oseltamivir (45 mg/kg/day). Mice treated with N1-3-VHHm or N1-5-VHHm showed significantly reduced morbidity at 72 and 96 h after infection compared with the groups treated with N1-7-VHHm and PBS (P < 0.001) (Fig. 4A). Four days after infection, the mice were sacrificed to determine lung virus titers by endpoint dilution in a TCID50 assay. All of the groups of treated mice (N1-3-VHHm, N1-5-VHHm, N1-7-VHHm, SA-VHHm, PBS, and oseltamivir), except the H5-VHHb group, had large lung virus loads ranging from 104.75 to 106.48 TCID50s/ml (data not shown). We also measured the amounts of viral RNA in the lung homogenates by a genome strand-specific RT-qPCR method. Except for the samples derived from the H5-VHHb-treated mice (P < 0.001), all of the N1-VHH-treated groups showed high viral RNA levels 96 h after infection, comparable to those of the PBS-treated group (Fig. 4B). NA activity in the lung homogenates of the treated groups, measured by AVINA, indicated that there was no difference between the N1-VHH- and PBS-treated groups, whereas NA activity was strongly reduced in the H5-VHHb and oseltamivir groups (Fig. 4C). These results are in line with the RT-qPCR results and confirmed the presence of a high viral titer in the lung homogenates of the N1-VHH groups, despite low morbidity. We speculate that the N1-VHH treatment did not have a major impact on virus entry in this in vivo challenge model but rather reduced virus spread within the lung compartment. A recent systems analysis of IAV-associated pathology in the mouse revealed a positive correlation between virus spread in the lung tissue and high pathogenicity irrespective of the number of infectious virus particles that were produced (63). In addition, shearing forces that are generated during the preparation of lung homogenates may have released virions from the cell membranes, which release was hindered in the intact lung by NA-inhibitory Nanobodies. We conclude that intranasal administration of N1-3-VHHm and N1-5-VHHm prevents body weight loss after a challenge with NIBRG-14ma virus during the early stage of H5N1 IAV infection, without an apparent decrease in viral titer in the lung homogenates.
TABLE 3.
Endotoxin levels in purified VHH preparations
| VHH | Mean LPS content (EU/ml)a ± SEM |
|---|---|
| N1-3-VHHm | 0.094 ± 0.007 |
| N1-5-VHHm | 0.096 ± 0.017 |
| N1-7-VHHm | 0.095 ± 0.012 |
| SA-VHHm | 0.094 ± 0.008 |
| N1-3-VHHb | 0.092 ± 0.005 |
| N1-5-VHHb | 0.093 ± 0.007 |
| BL-VHHb | 0.107 ± 0.024 |
| N1-3-VHH-Fc | 0.100 ± 0.020 |
| N1-5-VHH-Fc | 0.103 ± 0.019 |
| N1-7-VHH-Fc | 0.076 ± 0.011 |
| GP4-Fc | 0.100 ± 0.016 |
| PBS | 0.07 ± 0.015 |
The endotoxin (lipopolysaccharide [LPS]) levels (endotoxin units [EU]) in purified, E. coli-derived (VHHm/b) and A. thaliana-derived (VHH-Fc and GP4-Fc) proteins were determined by a Limulus amebocyte lysate-based chromogenic test and calculated relative to an endotoxin standard.
FIG 4.
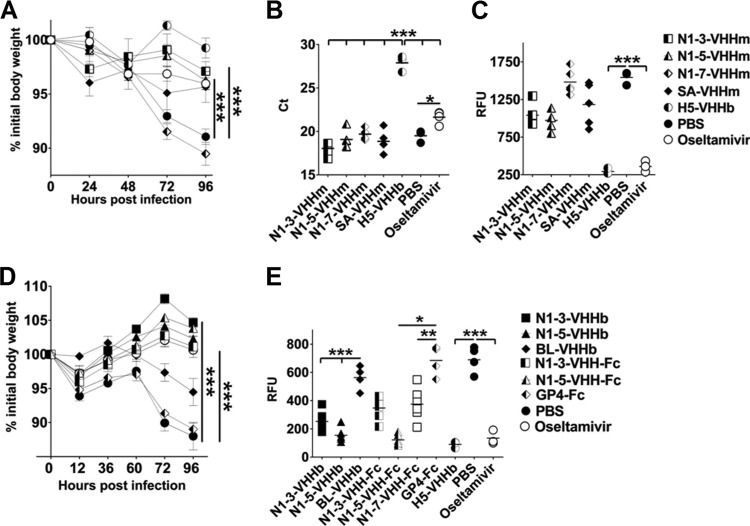
Prophylactic effect of treatment with inhibitory N1-VHHm, N1-VHHb, and N1-VHH-Fc on H5N1-challenged mice. (A) Groups of 6- to 8-week-old BALB/c mice (n = 5 per group, except oseltamivir and SA-VHHm [n = 4] and H5-VHHb and PBS [n = 3]) were given 100 μg of the N1-VHHm indicated intranasally 4 h before a challenge with 4 LD50s of NIBRG-14ma. Intranasal administration of 30 μg of H5-VHHb and daily oral administration of oseltamivir (45 mg/kg/day, starting 4 h before a challenge) were included as positive controls. Body weight was monitored daily after a challenge and is expressed as a percentage of the initial body weight. (B) Mice were sacrificed on day 4 after a challenge, and lung homogenates were prepared. Viral genomic RNA-specific RT-qPCR was used as a readout of the lung viral load. The CT values of the individual mice are plotted; a horizontal line indicates the mean. (C) AVINA of lung homogenates sampled on day 4 after infection. (D) Bivalent formats of N1-3-VHH and N1-5-VHH administered 4 h before a challenge with 4 LD50s of NIBRG-14ma reduced morbidity. Four hours before a challenge with 4 LD50s of NIBRG-14ma, groups of BALB/c mice (n = 6 per group, except BL-VHHb and PBS [n = 4] and oseltamivir and PBS [n = 3]) were treated intranasally with 60 μg of VHHb (N1-3-VHHb, N1-5-VHHb, or BL-VHHb) or 84 μg of VHH-Fc (N1-3-VHH-Fc, N1-5-VHH-Fc, or GP4-Fc). Mice treated with neutralizing H5-VHHb (30 μg) or oseltamivir (45 mg/kg/day, daily by gavage) were included as positive controls. (E) NA activity in lung homogenates of mice treated with bivalent N1-VHH and challenged with H5N1, as determined by AVINA. In panels A and D, each datum point represents the average body weight of all of the mice alive, and error bars represent standard deviations. Differences between mouse groups were determined by two-way ANOVA. Kaplan-Meier curves were used for survival analysis (*, P < 0.05; **, P < 0.01; ***, P < 0.001). RFU, relative fluorescence units.
Protection by bivalent N1-VHHb against an H5N1 challenge.
The in vitro results indicated that the potency of the bivalent formats of the inhibitory N1-VHHs against the tested H5N1 viruses was 30- to 240-fold higher than that of their monovalent counterparts (Table 2). We therefore assessed if this heightened antiviral effect would also be reflected in an in vivo challenge experiment. We first determined the protective potential during the early stages of H5N1 infection. Four hours before a challenge with 4 LD50s of NIBRG-14ma, groups of BALB/c mice were intranasally given 60 μg of N1-3-VHHb, N1-5-VHHb, or BL-VHHb (a bivalent VHH directed against the irrelevant bacterial target β-lactamase) and 84 μg of N1-3-VHH-Fc, N1-5-VHH-Fc, or GP4-Fc (a plant-produced coronavirus GP4-Fc fusion protein). Treatment with N1-3-VHHb or N1-3-VHH-Fc significantly improved morbidity at 72 and 96 h postinfection (hpi) compared with that of the GP4-Fc and PBS negative-control groups (P < 0.001) (Fig. 4D). The protection against weight loss was comparable to that observed in the positive controls (H5-VHHb and oseltamivir [the latter was given daily at 45 mg/kg/day by gavage]). Treatment with the bivalent formats of N1-5-VHH resulted in a reduction of morbidity that was significant at 60, 72, and 96 hpi compared with that of the negative controls (P < 0.001) (Fig. 4D). Determination of the lung virus load on day 4 after a challenge revealed that all of the challenged groups, except H5-VHHb-treated mice (no virus detectable), had large and comparable lung virus loads (data not shown). In addition, the NA activity in the lung homogenates of the groups treated with N1-VHH bivalent formats was assessed by an AVINA. The lung homogenates treated with bivalent N1-VHHs (N1-3-VHHb, N1-5-VHHb, N1-3-VHH-Fc, and N1-5-VHH-Fc) showed 2- to 5-fold decreases in NA activity compared to that of the BL-VHHb (P < 0.001)-, GP4-Fc (P < 0.05 or P < 0.01)-, and PBS (P < 0.01)-treated groups (Fig. 4E). This could be attributable to residual inhibitory bivalent N1-VHHb in the lung preparation, possibly because of an increased half-life of the bivalent N1-VHHs, relative to that of N1-VHHm, for which no significant reduction in lung-associated NA activity was demonstrated (Fig. 4C). Taken together, these results show that treatments with bivalent formats of N1-3-VHH and N1-5-VHH improve protection during the first 4 days following an NIBRG-14ma challenge and reduce lung-associated NA activity, a surrogate for the viral load, in H5N1-infected mouse lungs.
Dose-dependent protection against an H5N1 challenge.
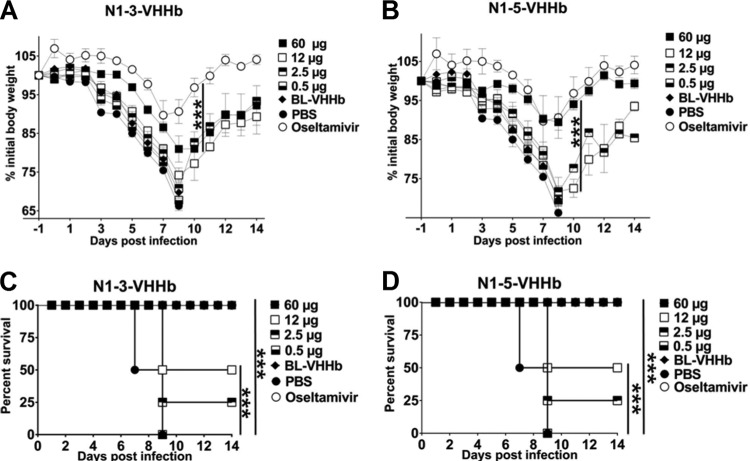
To test if prophylactic administration of N1-3-VHHb, N1-3-VHH-Fc, N1-5-VHHb, or N1-5-VHH-Fc can protect mice against a lethal challenge with NIBRG-14ma (4 LD50s), we repeated the protection studies and monitored the mice for 2 weeks. We focused on the most potent N1-VHHb bivalent formats and assessed their protective efficacy in a dose-response experiment. Groups of four BALB/c mice were treated intranasally with 60, 12, 2.5, or 0.5 μg of N1-3-VHHb or N1-5-VHHb. In parallel, another group was treated daily by oral administration of oseltamivir (45 mg/kg/day). Negative-control groups were treated with 60 μg of BL-VHHb or with PBS before a challenge. Mice that had received 60 μg of N1-3-VHHb, N1-5-VHHb, or BL-VHHb before the challenge also received a second intranasal dose of 60 μg of the same bivalent VHH on day 6 after the challenge. All of the mice in the PBS and BL-VHHb treatment groups died 9 to 10 days after infection (Fig. 5C and D). In contrast, treatment with oseltamivir and a large intranasal dose of N1-3-VHHb or N1-5-VHHb (60 μg) protected the mice against death. In agreement with this, oseltamivir and N1-5-VHHb (60 μg) reduced postchallenge body weight loss compared to that of control-treated mice (Fig. 5A and B). Although administration of the largest amount of N1-3-VHHb prevented death, it did not affect body weight loss. A single intranasal dose of 12 or 2.5 μg of N1-3-VHHb or N1-5-VHHb provided partial protection against death but failed to reduce morbidity (Fig. 5A to D).
FIG 5.
The bacterium-produced N1-VHHb formats protect H5N1-challenged mice against morbidity and death in a dose-dependent way. Groups of BALB/c mice (n = 4 per group, except the BL-VHHb, oseltamivir, and PBS groups [n = 3 per group]) were given the indicated amount of N1-VHHb intranasally 24 h before a challenge with 4 LD50s of NIBRG-14ma virus. Oseltamivir (45 mg/day/kg) was given by daily gavage starting 24 h before the challenge until day 14 after the challenge. A single additional intranasal dose of N1-3-VHHb, N1-5VHHb, or BL-VHHb was given on day 6 after infection for the highest dose (60 μg) of these VHHs only. (A) Mice treated with N1-3-VHHb. (B) Treatment with N1-5-VHHb. (C) Survival of the N1-3-VHHb group. (D) Survival of the N1-5-VHHb group. All mice were treated in parallel, and data for the PBS and oseltamivir groups in panels B and D are identical. In panels A and B, each point represents the average body weight of all of the mice alive and error bars represent standard deviations. Differences in body weight between mice were determined by two-way ANOVA. Kaplan-Meier curves were used for survival analysis (***, P < 0.001).
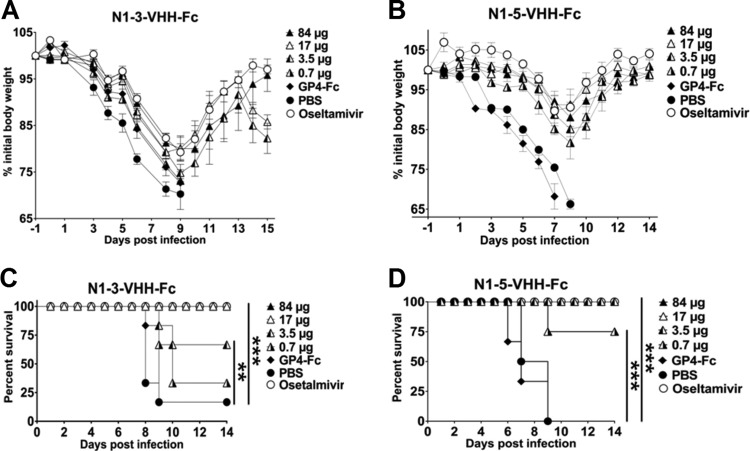
We next evaluated protection against a potentially lethal challenge with NIBRG-14ma by prior single-dose intranasal administration of the plant-produced N1-3-VHH-Fc and N1-5-VHH-Fc formats. N1-3-VHH-Fc (80 and 17 μg), as well as oseltamivir, prevented death following a challenge with NIBRG-14ma (Fig. 6C), whereas treatment with 3.5 μg of N1-3-VHH-Fc prevented the deaths of some animals. However, the challenge was associated with significant body weight loss for all of the doses and treatments (Fig. 6A and B).
FIG 6.
Plant-produced N1-VHH-Fc protects against morbidity and death after an H5N1 challenge. Groups of BALB/c mice were treated intranasally with the amounts of N1-VHH-Fc indicated 24 h before a challenge with 4 LD50s of NIBRG-14ma. Oseltamivir (45 mg/day/kg) was given to a group of three mice by daily gavage starting 24 h before the challenge until day 14 after the challenge. An additional intranasal dose of 84 μg of N1-3-VHH-Fc, N1-5-VHH-Fc, or GP4-Fc was given on day 6 after infection. (A) Relative body weights after treatment with N1-3-VHH-Fc. (B) Relative body weights after treatment with N1-5-VHH-Fc. (C, D) The survival of N1-3-VHH-Fc-treated groups is dose dependent. The experiments shown in panels B and D were performed in parallel with those in Fig. 4, and the PBS and oseltamivir datum points depicted are identical to those shown in Fig. 4. In panels A and B, each point represents the average body weight of all of the mice alive and error bars represent standard deviations. Differences between mouse groups were calculated by two-way ANOVA. Kaplan-Meier curves were used for survival analysis (**, P < 0.01; ***, P < 0.001).
When the protective efficacies of the two bivalent formats of N1-3-VHH and N1-5-VHH were compared, it appeared that the Fc moiety in the N1-VHH-Fc formats provides an extra protective effect against morbidity (compare 60 μg N1-3-VHHb with 84 μg N1-3-VHH-Fc in Fig. 5A and 6B) and mortality, following a challenge with NIBRG-14ma (Fig. 5 and 6). We conclude that a single dose of both bivalent N1-3/5-VHH formats can protect mice against a potentially lethal challenge with an H5N1 virus and that Fc-fused VHHs provide better protection, suggesting that Fc receptor-binding molecules contribute to protection.
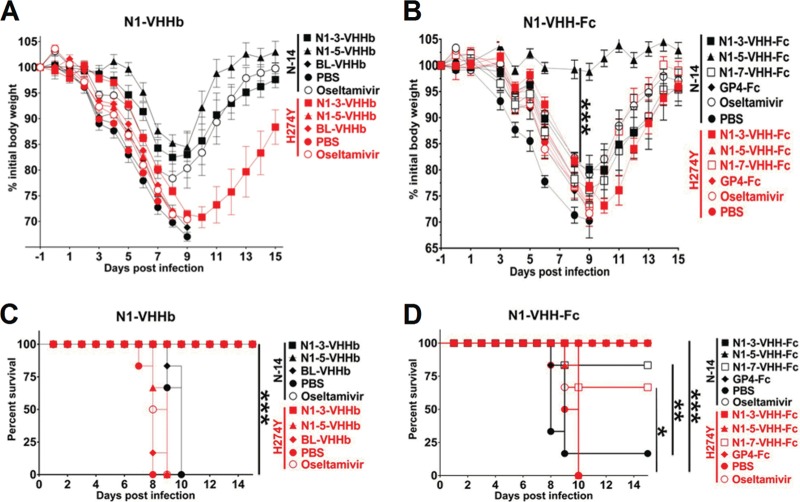
N1-3-VHHb and N1-3-VHH-Fc protect against a challenge with oseltamivir-resistant H5N1.
Our in vitro analysis demonstrated that monovalent and bivalent formats of N1-3-VHH, but not N1-5-VHH, reduced the growth of oseltamivir-resistant H5N1 H274Y virus (Table 2). To evaluate these findings in vivo, groups of six BALB/c mice received 30 μg of N1-3-VHHb, N1-5-VHHb, or BL-VHHb by intranasal administration 24 h before a challenge with 4 LD50s of NIBRG-14ma or 6 LD50s of H5N1 H274Yma. In parallel, a group was treated by daily oral administration of oseltamivir (1 mg/kg/day), a dose that has been reported to fully protect laboratory mice against a challenge with highly pathogenic H5N1 (64). In line with our previous results, mice treated with N1-3-VHHb, N1-5-VHHb, or oseltamivir suffered from substantial but transient weight loss, but they survived a challenge with the NIBRG-14ma virus, whereas mice receiving PBS or BL-VHHb died before day 10 after the challenge (Fig. 7). This result demonstrates that a single intranasal administration of N1-3-VHHb or N1-5-VHHb is sufficient to protect against a subsequent lethal challenge (24 h later) with an H5N1 virus. Following a challenge with the H5N1 H274Y mutant virus, N1-3-VHHb-treated mice were severely sick but all of them survived (Fig. 7A and C). All of the other groups, including the oseltamivir-treated mice, died after a challenge with the H5N1 H274Y virus by day 10 after infection (Fig. 7A and C). Taken together, these results show that treatment of mice with bivalent N1-3-VHHb before a challenge with an oseltamivir-resistant H5N1 virus does not control morbidity adequately but results in full recovery and survival. Furthermore, bivalent N1-5-VHHb does not protect mice against this challenge, suggesting that the N1-VHHb formats (i.e., tandem repeats of VHH separated by a short linker) require NA-inhibitory activity for in vivo protection.
FIG 7.
N1-3-VHHb and N1-3-VHH-Fc, but not N1-5-VHHb and N1-5-VHH-Fc, protect against a challenge with oseltamivir-resistant H5N1 H274Y virus. Twenty-four hours before a challenge with 4 LD50s of NIBRG-14 (N-14)ma or 6 LD50s of H274Y H5N1 virus (H274Y), groups of six BALB/c mice were given 60 μg of N1-3-VHHb, 60 μg of N1-5-VHHb, 60 μg of BL-VHHb, 84 μg of N1-3-VHH-Fc, 84 μg of N1-5-VHH-Fc, 84 μg of N1-7-VHH-Fc, or 84 μg of GP4-Fc. An additional group of six mice was treated with oseltamivir (1 mg/day/kg) starting 24 h before the challenge. (A) Relative body weights. (C) Survival over time after infection. NIBRG-14ma-infected mice treated with N1-3-VHHb, N1-5-VHHb, or oseltamivir survived the challenge. Treatment with N1-3-VHHb rescued all of the H5N1 H274Y mutant-infected mice (***, P < 0.001). (B) Relative body weights. (D) Survival over time after infection. In panels A and B, each datum point represents the average body weight of all of the mice alive and error bars represent standard deviations. Differences between groups were evaluated by two-way ANOVA for the body weight graphs. Kaplan-Meier curves were used for survival analysis (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Finally, we compared the protective potentials of intranasally instilled N1-3-VHH-Fc, N1-5-VHH-Fc, and N1-7-VHH-Fc in our lethal-challenge model. Following a challenge with 4 LD50s of NIBRG-14ma, all of the mice in the N1-3-VHH-Fc and oseltamivir groups lost body weight until day 7 after the challenge (Fig. 7B) and then started to recover, and all of them survived the challenge (Fig. 7D). In contrast, all of the PBS- or GP4-Fc-treated mice, except one, became morbid and died by 10 days postinfection (Fig. 7B and D). In the N1-7-VHH-Fc group, only one out of six mice died after the challenge. This is very surprising because N1-7-VHHm and N1-7-VHH-Fc have no detectable antiviral activity in vitro. Remarkably, in this experiment, mice that had received N1-5-VHH-Fc did not lose weight after a challenge with NIBRG-14ma (Fig. 7B). A challenge with 6 LD50s of H5N1 H274Y mutant virus was lethal to all of the mice except those that had been pretreated with N1-3-VHH-Fc (all of the mice survived) or N1-7-VHH-Fc (four out of six mice survived), though all of the animals lost substantial body weight (Fig. 7B and D). We conclude that protection against a potentially lethal challenge with oseltamivir-resistant H5N1 virus can be obtained with a single dose of N1-3-VHH-Fc or with N1-7-VHH-Fc, although the latter construct is less potent. This indicates that the IgG2a Fc moiety contributes to protection, even in the absence of NA-inhibitory activity.
N1-5-VHHm escape mutants.
To identify the amino acid residues involved in the binding of N1-3-VHHm and N1-5-VHHm to H5N1 NA, escape viruses were selected and plaque purified after serial passage of NIBRG-14ma virus in MDCK cells in the presence of a dose corresponding to 10 times the IC50 of N1-3-VHHm or N1-5-VHHm, essentially as described before (35). After nine passages, candidate escape mutants were plaque purified and the NA sequences of four (N1-5-VHHm selection) and three (N1-3-VHHm selection) isolated escape viruses were determined. One of the escape viruses that were selected with N1-5-VHHm contained a mutation in the NA coding region that resulted in a change from isoleucine to threonine at position 437 in the deduced amino acid sequence. This substitution is located on the NA surface, relatively close to the active site (Fig. 8A). A conserved NA epitope that is recognized by HCA-2 under denaturing conditions (65, 66) and localized within the NA catalytic site, and the amino acids necessary for binding a recently reported set of MAbs that recognize H1N1 and H5N1 NA (67) do not overlap this N1-5-VHHm escape mutation (Fig. 8A). Sequence alignment of a set of representative N1 IAVs, including human H1N1, other H5N1 viruses, and an avian H6N1 virus, showed that I437 is conserved in these NA sequences (Fig. 8B). Binding studies with immobilized virus revealed that the I437T escape virus was no longer bound by N1-3-VHHm, N1-3-VHHb, N1-5-VHHm, and N1-5-VHHb (Fig. 8C). This result is in accordance with the Biacore data, which showed that N1-3-VHHm and N1-5-VHHm compete for binding to recombinant H5N1 NA (Fig. 1F) and suggests that I437 is part of the VHH epitope in NA or stabilizes the conformation of that epitope.
FIG 8.
Epitope mapping on the NA of N1-5-VHHm escape NIBRG-14ma mutants. MDCK cells were infected with NIBRG-14ma in the presence of 3.7 μM N1-5-VHHm. Evidence of resistant viruses was based on a cytopathic effect on infected cell cultures and was monitored by plaque purification of candidate escape viruses. The NA coding sequence of the resultant escape viruses was amplified by RT-PCR and sequenced. One selected escape virus carried an Ile-to-Thr change at position 437. The deduced amino acid sequence in NA was modeled with PyMol (Delano Scientific, http://www.pymol.org) with the H5N1 NA structure derived from A/Vietnam/1194/2004 (PDB code 2HTY). (A) Top (left) and lateral (right) views of the surface-exposed amino acid residues of the NA dimer. The I437T amino acid substitution of the NA of the N1-5-VHHm escape mutant is surface exposed and marked in yellow. Residues that form the catalytic-site framework are depicted in red. The N1 “150 loop” is shown in green. For comparison, the residues involved in MAb HCA-2 binding are shown in dark blue (residues 222 to 230) and those involved in MAb 3H10 binding are shown in orange (N272, V339, and S340). (B) Sequence alignment of relevant N1 NAs. The I437T change in one of the N1-5-VHHm escape viruses is highlighted in yellow. The residues highlighted in red are important for MAb 3H10 binding. (C) Fluorogram showing the T-to-C substitution in the cloned cDNA of the N1-5-VHHm escape virus (left). Mono- and bivalent NA-specific VHHs bind to NIBRG-14 virus but not to I437T escape virus (right). ELISA plates were coated with 8 HA units of NIBRG-14 or I437T virus and, after blocking, incubated with different concentrations of N1-3-VHHm, N1-3-VHHb, N1-5-VHHm, or N1-5-VHHb. N1-VHH binding was revealed with an anti-His tag MAb.
Interestingly, the other three plaque-purified N1-5-VHHm escape viruses and all three plaque-purified viruses that grew in the presence of N1-3-VHHm contained wild-type NA. Possibly, these viruses carry compensatory mutations in other viral proteins such as HA, as was described before for IAV passaged in vitro in the presence of small-molecule inhibitors of NA (68). Taken together, the results show that H5N1 viruses that overcome inhibition by N1-3-VHHm and N1-5-VHHm can be selected in vitro and one way of escape is through the acquisition of a mutation in NA that results in loss of binding to the VHHs.
DISCUSSION
NA Abs usually do not neutralize virus and are thought to exert their antiviral effects further downstream in the IAV infection process. Still, NA-specific Abs can significantly reduce viral replication and disease severity (69, 70). Titers of Ab against N1 or N2 have been associated with high NA enzymatic activity in vaccine preparations, suggesting that the NA content and activity of IAV vaccines predict immunogenicity (71). NA of human influenza viruses is subject to antigenic drift at a pace that is comparable to that of HA (72). This suggests that NA is subject to intense immune selection pressure, which is surprising given that Abs directed against NA do not neutralize the virus in vitro and that during natural infections immune responses against HA are more pronounced than those against NA, presumably because HA molecules outnumber NA molecules on influenza virions. Biologically, HA and NA cooperate and antigenic variation in NA is, in part, driven by antigenic escape selection pressure on HA (73). Nevertheless, Abs directed against NA remain underexplored as a potential influenza treatment.
Our N1-VHHs are not the first inhibitory MAbs against H5N1 NA described. Recently, IgG2bκ MAb 2B9 directed against clade 1 A/Vietnam/1203/04 NA showed interclade and oseltamivir-resistant mutant NAI activity in vitro (74). In NAI assays with the 4-MUNANA and NA-Star substrates, the 2B9 MAb IC50s against H5N1 clade 1 and clade 2.1 virions and against an oseltamivir-resistant H274Y and N294S double mutant ranged from 1 to 8 μg/ml. In another recent report on IgG2b MAbs targeting multiple N1 NAs, intrasubtype MAbs showed relatively high H1N1 NAI potency (IC50s of 23.6 to 68.1 ng/ml), which was even lower when A/Vietnam/1203/04 H5N1 NAI potency was assessed (IC50s of 1.41 to 2.42 μg/ml) (67). We used two inhibitory N1-VHH formats to assess NA-inhibitory and antiviral potentials. Results of our N1rec inhibition assays showed that the N1-3-VHHm and N1-5-VHHm IC50s were 425 nM (6.3 μg/ml) and 374 nM (5.6 μg/ml), respectively, and the introduction of bivalency (N1-VHHb and N1-VHH-Fc) resulted in 2- to 3-log-fold further increases in their inhibitory potential (5.2 to 125 ng/ml) (Table 2). The above-mentioned conventional MAbs, which are bivalent, have H5N1 NAI activity that is similar to that of our N1-VHHm. We hypothesize that the much smaller size of VHH molecules than that of conventional Abs compensates for the lack of avidity present in conventional Abs to explain this.
Harmsen et al. reported the first VHH targeting NA (39). In contrast to our work, where we used recombinant NA protein to immunize an alpaca, those authors obtained VHH by immunizing llamas with whole IAV. This strategy yielded mainly HA and NP binders and required a strategy of cross-selection with homologous NA, followed by further selection with recombinant NA, to obtain NA binders. This approach resulted in VHH binding to homologous and nonhomologous IAV, but only some of the NA cross-binding VHHs showed NA inhibition activity (fetuin substrate). It is also important to point out that the recombinant NA activities used by Harmsen et al. were 316 to 13.5 times lower than our N1rec specific activity (57 μmol/mg/min), while their VHH NA inhibition IC50s ranged from 124 to 2 ng/ml, with homologous NA. The apparently lower IC50s of the Harmsen VHHs (3- to 200-fold lower than that of our N1-VHHm) could therefore be explained by a 1- to 2-log lower NA-specific subtype activity. All together, these results suggest that a similar NAI potency range was obtained with our monovalent N1-VHHm. In vitro, monovalent N1-3-VHHm and N1-5-VHHm had an IC50 that was almost identical to that of oseltamivir for N1rec and for the two oseltamivir-sensitive H5N1 viruses we assayed (Table 2).
In the TMPRSS2-expressing MDCK-based assay, which allowed us to assess multiple rounds of viral replication in the absence of exogenous trypsin, the antiviral effect of bivalent VHH formats (N1-VHHb and N1-VHH-Fc) was clearly greater than that of monovalent N1-VHHm molecules (Table 2). For NIBRG-14ma, the N1-VHH concentrations necessary to reduce the plaque size by 50% (plaque assay IC50) for N1-3-VHHb and N1-5-VHHb were 7.6 nM (243 ng/ml) and 14.5 nM (464 ng/ml), respectively, which are 240- and 58-fold lower than those of their monovalent formats. In fact, we noticed a consistent 30-fold or higher inhibitory activity of bivalent N1-3-VHHb and N1-3-VHH-Fc than of monovalent VHHs in a biochemical assay with N1rec and against the three H5N1 viruses tested in vitro. The H5N1 H274Y mutant was an exception in that the N1-5-VHH mono- and bivalent formats failed to inhibit this oseltamivir-resistant virus (Table 2). These results also suggest that the epitope recognized by N1-3-VHHm and N1-5-VHHm is conserved between the NAs of clade 1 and 2 H5N1 viruses.
Intranasal administration of a relatively high dose of N1-3-VHHb or N1-5-VHHb (60 μg; 3.3 mg/kg) 24 h before an H5N1 challenge protected mice against death, but significant body weight was still lost (Fig. 5). In this challenge model, the N1-VHH-Fc formats protected better than the N1-VHHb formats. A single dose of 17 μg (1.1 mg/kg, N1-3-VHH-Fc) or 3.5 μg (0.23 mg/kg, N1-5-VHH-Fc) resulted in 100% survival of mice and less morbidity than in control-treated animals (Fig. 6 and 7). When comparing the molar ratios of N1-VHH-Fc (N1-3-VHH-Fc and N1-5-VHH-Fc) with N1-VHHb (N1-3-VHHb or N1-5-VHHb), we found that approximately 10 to 50 times smaller amounts of the N1-VHH-Fc formats were necessary for 100% survival of H5N1-challenged mice. For comparison, also in a mouse model, conventional MAb 2B9 resulted in only partial (50%) protection against an H5N1 A/Vietnam/1194/2004 challenge (74). Even so, this required the intravenous administration of 500 μg (33.3 mg/kg) of 2B9 1 h before 10 LD50s of HPAIV H5N1, with daily boosts for 5 days. Furthermore, when we compared the 2B9 treatment with our single-dose treatment with N1-VHHb or N1-VHH-Fc, a 10- or 150-fold smaller amount of MAb, respectively, was required to rescue 100% of the H5N1-challenged mice without any boost. Compared with a study by Wan et al., the smallest prophylactic single dose of a conventional IgG2a MAb necessary to fully rescue mice from a 10-LD50 H5N1 challenge was 15 mg/kg, whereas 0.23 mg/kg of our N1-5-VHH-Fc protected all of the mice from death after a 4-LD50 challenge with NIBRG-14ma (67). The target NA of NIBRG-14 (derived from A/Vietnam/1194/2004) is almost identical to that used in the Wan study (A/Vietnam/1203/2004). In both of the reports on NA MAbs discussed above (67, 74), the route of administration (intravenous or intraperitoneal) could have been an important factor and it might help explain the apparent superior protection of our N1-VHH formats, which we administered intranasally, since VHHs withstand nebulization. Moreover, it has been shown that anti-RSV Nanobodies give extended protection against viral infection in vivo when administered via a nebulizer. It may be expected that optimization of its delivery will further improve the efficacy of N1-VHH. N1-VHHm treatments did not significantly reduce the virus titers or viral genomic loads in lung homogenates 4 days after infection (Fig. 4C). The weak, direct antiviral activity of our NA-specific VHHs in the in vivo model is in line with the paradigm that NA Abs or NA-inhibitory drugs do not prevent infection.
The observation that N1-5-VHHm, N1-5-VHHb, and N1-5-VHH-Fc did not have any antiviral activity against the oseltamivir-resistant H5N1 H274Y virus in vitro was surprising since in the SPR assay, N1-3-VHHm and N1-5-VHHm bound to overlapping epitopes in NA or sterically hindered each other's binding (Fig. 1F). This discrepancy was also observed in vitro for N1-7-VHHm, whereas N1-7-VHH-Fc did protect mice against a challenge with the H5N1 H274Y mutant virus (Fig. 7). The latter observation indicates that protection by NA-based Abs does not require the NAI activity of these Abs, provided that an Fc domain is present to contribute Fc receptor-dependent effector functions. The finding that N1-7-VHH-Fc also had in vivo antiviral activity despite a lack of NAI activity is in line with this (Fig. 7B and D). Mouse IgG2a is one of the most potent Ab isotypes to induce Ab-dependent cellular cytotoxicity or complement-dependent cellular cytotoxicity, because of its high affinity for FcγRIII and -IV and especially for FcγRI, which binds exclusively to IgG2a. IgG2a has an intermediate affinity for FcγRIV that is 1 order of magnitude higher than its affinity for the inhibitor receptor FcγRIIB (75).
VHH CDR3 is a major region responsible for the contact of VHH with antigen (25, 76). All of the NA-inhibitory N1-VHHs reported here (N1-1-VHHm, N1-3-VHHm, N1-5-VHHm, and N1-6-VHHm) have a predicted electronegatively charged protruding paratope that corresponds to their CDR3 (Fig. 1C and D). The CDR3 sequences of N1-3-VHHm (HGXXHGHGXHX, 10 residues) and N1-5-VHHm (HXHXHHXHXH, 12 residues), where H is a hydrophobic residue and X is an aspartic or glutamic acid, might be involved in the direct contact with their epitope or epitopes. When comparing N1-3-VHHm and N1-5-VHHm CDR3 sequences with the most potent NA-inhibitory VHHs reported by Harmsen et al., we found similarities in their CDR3 sequences. For example, the CDR3 sequences of one N8-binding VHH (UHXHHHGHGUHHXUPHXH, 18 residues) and one N2-binding VHH (HHXHGHHGGHGHUHXH, 16 residues), where U is a polar uncharged residue, showed similarities to the N1-3-VHHm and N1-5-VHHm CDR3s.
The available crystal structures of MAbs in complex with NA are all for the group 2 NA, specifically, N2 and N9 (77–80). These structures showed that these MAb epitopes rarely include conserved amino acid residues from the catalytic site; rather, they target amino acids in loops surrounding the NA catalytic site, also described as the major antigenic regions of NA. It seems that the N1-5-VHHm epitope is partially shared with other N1 NA molecules, as the pandemic H1N1 NAI suggests (Fig. 3).
Plant-produced Abs have been considered a promising and effective alternative for Ab production by recombinant yeast or mammalian cell culture systems. Our N1-VHH-Fc is the first reported plant-produced Ab against an influenza virus antigen. Their scalability, robustness, and speed of production are major advantages, even though the glycosylation patterns of the plant-produced Abs (rich in high-mannose residues) are different from those of the Abs produced with mammalian expression platforms (81). The use of plants for Ab production also presents an interesting approach for developing countries; long storage life, relative ease of purification or delivery, and the use of established agricultural techniques are very attractive. To our knowledge, only three single-domain Abs produced in plants have been reported as stably transformed lines (82–84), but their yields were very low. Results from our lab demonstrated that the Fc moiety significantly improved VHH accumulation in seeds (compared to the monomeric VHH alone) with expression levels of 10 to 20% of the total seed proteins (54, 85).
HPAIV H5N1 remains a public health threat. The experimental adaptation of H5N1 virus for airborne transmission in ferrets, with mutations in HA and PB2, warns of the possibility of human-to-human transmission of zoonotic HPAIV (1, 2). Should HPAIV H5N1 become transmissible among humans, NA targeting could be part of an effective strategy to mitigate disease caused by such a virus. In addition, an interesting prophylactic and therapeutic approach that deserves further validation is the combination of VHH-targeting IAV NA, HA, or M2 proteins in cocktails or biparatopic bivalent VHH constructs that could diminish the antigenic drift in IAV and lead to much higher selective pressure against IAV VHH.
ACKNOWLEDGMENTS
We gratefully appreciate the excellent technical support of Tine Ysenbaert, Frederick Vervallle, and Jonah Nolf. We thank Thierry Van den Bergh of the Veterinary and Agrochemical Research Centre in Brussels (Belgium) for providing cDNA of NA of A/crested eagle/Belgium/01/2004 (H5N1) influenza virus; Wolgang Garten, Philipps-Universität Marburg, Marburg, Germany, for TMPRSS2-expressing MDCK cells; John Wood of the UK National Institute for Biological Standards and Control for providing the NIBRG-14 and -23 viruses; Alan Hay (MRC National Institute for Medical Research, London, United Kingdom) for providing the A/New Caledonia/20/99 virus; and St. Jude Children's Research Hospital for providing the A/PR/8/34-based eight-plasmid system for generating recombinant IAVs.
F.M.C. was funded by a predoctoral grant from the Biotechnology Institute of Flanders (VIB) and supported by IUAP BELVIR project p7/45.
Footnotes
Published ahead of print 14 May 2014
REFERENCES
- 1.Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336:1534–1541. 10.1126/science.1213362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y. 2012. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486:420–428. 10.1038/nature10831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tran TH, Nguyen TL, Nguyen TD, Luong TS, Pham PM, Nguyen vV, Pham TS, Vo CD, Le TQ, Ngo TT, Dao BK, Le PP, Nguyen TT, Hoang TL, Cao VT, Le TG, Nguyen DT, Le HN, Nguyen KT, Le HS, Le VT, Christiane D, Tran TT, de Jong M, Schultsz C, Cheng P, Lim W, Horby P, Farrar J, World Health Organization International Avian Influenza Investigative Team 2004. Avian influenza A (H5N1) in 10 patients in Vietnam. N. Engl. J. Med. 350:1179–1188. 10.1056/NEJMoa040419 [DOI] [PubMed] [Google Scholar]
- 4.Trifonov V, Khiabanian H, Rabadan R. 2009. Geographic dependence, surveillance, and origins of the 2009 influenza A (H1N1) virus. N. Engl. J. Med. 361:115–119. 10.1056/NEJMp0904572 [DOI] [PubMed] [Google Scholar]
- 5.Gao HN, Lu HZ, Cao B, Du B, Shang H, Gan JH, Lu SH, Yang YD, Fang Q, Shen YZ, Xi XM, Gu Q, Zhou XM, Qu HP, Yan Z, Li FM, Zhao W, Gao ZC, Wang GF, Ruan LX, Wang WH, Ye J, Cao HF, Li XW, Zhang WH, Fang XC, He J, Liang WF, Xie J, Zeng M, Wu XZ, Li J, Xia Q, Jin ZC, Chen Q, Tang C, Zhang ZY, Hou BM, Feng ZX, Sheng JF, Zhong NS, Li LJ. 2013. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N. Engl. J. Med. 368:2277–2285. 10.1056/NEJMoa1305584 [DOI] [PubMed] [Google Scholar]
- 6.Roose K, Fiers W, Saelens X. 2009. Pandemic preparedness: toward a universal influenza vaccine. Drug News Perspect. 22:80–92. 10.1358/dnp.2009.22.2.1334451 [DOI] [PubMed] [Google Scholar]
- 7.de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN, Hoang DM, Chau NVV, Khanh TH, Dong VC, Qui PT, Cam BV, Ha DQ, Guan Y, Peiris JS, Chinh NT, Hien TT, Farrar J. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 12:1203–1207. 10.1038/nm1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. 2004. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J. Virol. 78:12665–12667. 10.1128/JVI.78.22.12665-12667.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johansson BE, Matthews JT, Kilbourne ED. 1998. Supplementation of conventional influenza A vaccine with purified viral neuraminidase results in a balanced and broadened immune response. Vaccine 16:1009–1015. 10.1016/S0264-410X(97)00279-X [DOI] [PubMed] [Google Scholar]
- 10.Kilbourne ED, Pokorny BA, Johansson B, Brett I, Milev Y, Matthews JT. 2004. Protection of mice with recombinant influenza virus neuraminidase. J. Infect. Dis. 189:459–461. 10.1086/381123 [DOI] [PubMed] [Google Scholar]
- 11.Marcelin G, Sandbulte MR, Webby RJ. 2012. Contribution of antibody production against neuraminidase to the protection afforded by influenza vaccines. Rev. Med. Virol. 22:267–279. 10.1002/rmv.1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goto H, Kawaoka Y. 1998. A novel mechanism for the acquisition of virulence by a human influenza A virus. Proc. Natl. Acad. Sci. U. S. A. 95:10224–10228. 10.1073/pnas.95.17.10224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li S, Schulman J, Itamura S, Palese P. 1993. Glycosylation of neuraminidase determines the neurovirulence of influenza A/WSN/33 virus. J. Virol. 67:6667–6673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuoka Y, Swayne DE, Thomas C, Rameix-Welti MA, Naffakh N, Warnes C, Altholtz M, Donis R, Subbarao K. 2009. Neuraminidase stalk length and additional glycosylation of the hemagglutinin influence the virulence of influenza H5N1 viruses for mice. J. Virol. 83:4704–4708. 10.1128/JVI.01987-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang IC, Li W, Sui J, Marasco W, Choe H, Farzan M. 2008. Influenza A virus neuraminidase limits viral superinfection. J. Virol. 82:4834–4843. 10.1128/JVI.00079-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ilyushina NA, Bovin NV, Webster RG. 2012. Decreased neuraminidase activity is important for the adaptation of H5N1 influenza virus to human airway epithelium. J. Virol. 86:4724–4733. 10.1128/JVI.06774-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi Y, Hasegawa H, Hara Y, Ato M, Ninomiya A, Takagi H, Odagiri T, Sata T, Tashiro M, Kobayashi K. 2009. Protective immunity afforded by inactivated H5N1 (NIBRG-14) vaccine requires antibodies against both hemagglutinin and neuraminidase in mice. J. Infect. Dis. 199:1629–1637. 10.1086/598954 [DOI] [PubMed] [Google Scholar]
- 18.Gubareva LV, Nedyalkova MS, Novikov DV, Murti KG, Hoffmann E, Hayden FG. 2002. A release-competent influenza A virus mutant lacking the coding capacity for the neuraminidase active site. J. Gen. Virol. 83:2683–2692 [DOI] [PubMed] [Google Scholar]
- 19.Mitnaul LJ, Matrosovich MN, Castrucci MR, Tuzikov AB, Bovin NV, Kobasa D, Kawaoka Y. 2000. Balanced hemagglutinin and neuraminidase activities are critical for efficient replication of influenza A virus. J. Virol. 74:6015–6020. 10.1128/JVI.74.13.6015-6020.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nedyalkova MS, Hayden FG, Webster RG, Gubareva LV. 2002. Accumulation of defective neuraminidase (NA) genes by influenza A viruses in the presence of NA inhibitors as a marker of reduced dependence on NA. J. Infect. Dis. 185:591–598. 10.1086/339358 [DOI] [PubMed] [Google Scholar]
- 21.Murphy BR, Kasel JA, Chanock RM. 1972. Association of serum anti-neuraminidase antibody with resistance to influenza in man. N. Engl. J. Med. 286:1329–1332. 10.1056/NEJM197206222862502 [DOI] [PubMed] [Google Scholar]
- 22.Vanlandschoot P, Stortelers C, Beirnaert E, Ibañez LI, Schepens B, Depla E, Saelens X. 2011. Nanobodies®: new ammunition to battle viruses. Antiviral Res. 92:389–407. 10.1016/j.antiviral.2011.09.002 [DOI] [PubMed] [Google Scholar]
- 23.Greenberg AS, Avila D, Hughes M, Hughes A, McKinney EC, Flajnik MF. 1995. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature 374:168–173. 10.1038/374168a0 [DOI] [PubMed] [Google Scholar]
- 24.Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N, Hamers R. 1993. Naturally occurring antibodies devoid of light chains. Nature 363:446–448. 10.1038/363446a0 [DOI] [PubMed] [Google Scholar]
- 25.Coppieters K, Dreier T, Silence K, de Haard H, Lauwereys M, Casteels P, Beirnaert E, Jonckheere H, Van de Wiele C, Staelens L, Hostens J, Revets H, Remaut E, Elewaut D, Rottiers P. 2006. Formatted anti-tumor necrosis factor alpha VHH proteins derived from camelids show superior potency and targeting to inflamed joints in a murine model of collagen-induced arthritis. Arthritis Rheum. 54:1856–1866. 10.1002/art.21827 [DOI] [PubMed] [Google Scholar]
- 26.Saerens D, Ghassabeh GH, Muyldermans S. 2008. Single-domain antibodies as building blocks for novel therapeutics. Curr. Opin. Pharmacol. 8:600–608. 10.1016/j.coph.2008.07.006 [DOI] [PubMed] [Google Scholar]
- 27.Decanniere K, Desmyter A, Lauwereys M, Ghahroudi MA, Muyldermans S, Wyns L. 1999. A single-domain antibody fragment in complex with RNase A: non-canonical loop structures and nanomolar affinity using two CDR loops. Structure 7:361–370. 10.1016/S0969-2126(99)80049-5 [DOI] [PubMed] [Google Scholar]
- 28.Decanniere K, Muyldermans S, Wyns L. 2000. Canonical antigen-binding loop structures in immunoglobulins: more structures, more canonical classes? J. Mol. Biol. 300:83–91. 10.1006/jmbi.2000.3839 [DOI] [PubMed] [Google Scholar]
- 29.Ward ES, Gussow D, Griffiths AD, Jones PT, Winter G. 1989. Binding activities of a repertoire of single immunoglobulin variable domains secreted from Escherichia coli. Nature 341:544–546. 10.1038/341544a0 [DOI] [PubMed] [Google Scholar]
- 30.Desmyter A, Spinelli S, Payan F, Lauwereys M, Wyns L, Muyldermans S, Cambillau C. 2002. Three camelid VHH domains in complex with porcine pancreatic alpha-amylase. Inhibition and versatility of binding topology. J. Biol. Chem. 277:23645–23650. 10.1074/jbc.M202327200 [DOI] [PubMed] [Google Scholar]
- 31.Lauwereys M, Arbabi Ghahroudi M, Desmyter A, Kinne J, Holzer W, De Genst E, Wyns L, Muyldermans S. 1998. Potent enzyme inhibitors derived from dromedary heavy-chain antibodies. EMBO J. 17:3512–3520. 10.1093/emboj/17.13.3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, Devree BT, Rosenbaum DM, Thian FS, Kobilka TS, Schnapp A, Konetzki I, Sunahara RK, Gellman SH, Pautsch A, Steyaert J, Weis WI, Kobilka BK. 2011. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature 469:175–180. 10.1038/nature09648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Genst E, Silence K, Decanniere K, Conrath K, Loris R, Kinne J, Muyldermans S, Wyns L. 2006. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc. Natl. Acad. Sci. U. S. A. 103:4586–4591. 10.1073/pnas.0505379103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hultberg A, Temperton NJ, Rosseels V, Koenders M, Gonzalez-Pajuelo M, Schepens B, Ibañez LI, Vanlandschoot P, Schillemans J, Saunders M, Weiss RA, Saelens X, Melero JA, Verrips CT, Van Gucht S, de Haard HJ. 2011. Llama-derived single domain antibodies to build multivalent, superpotent and broadened neutralizing anti-viral molecules. PLoS One 6:e17665. 10.1371/journal.pone.0017665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ibañez LI, De Filette M, Hultberg A, Verrips T, Temperton N, Weiss RA, Vandevelde W, Schepens B, Vanlandschoot P, Saelens X. 2011. Nanobodies with in vitro neutralizing activity protect mice against H5N1 influenza virus infection. J. Infect. Dis. 203:1063–1072. 10.1093/infdis/jiq168 [DOI] [PubMed] [Google Scholar]
- 36.Schepens B, Ibañez LI, De Baets S, Hultberg A, Bogaert P, De Bleser P, Vervalle F, Verrips T, Melero J, Vandevelde W, Vanlandschoot P, Saelens X. 2011. Nanobodies® specific for respiratory syncytial virus fusion protein protect against infection by inhibition of fusion. J. Infect. Dis. 204:1692–1701. 10.1093/infdis/jir622 [DOI] [PubMed] [Google Scholar]
- 37.Tillib SV, Ivanova TI, Vasilev LA, Rutovskaya MV, Saakyan SA, Gribova IY, Tutykhina IL, Sedova ES, Lysenko AA, Shmarov MM, Logunov DY, Naroditsky BS, Gintsburg AL. 2013. Formatted single-domain antibodies can protect mice against infection with influenza virus (H5N2). Antiviral Res. 97:245–254. 10.1016/j.antiviral.2012.12.014 [DOI] [PubMed] [Google Scholar]
- 38.Wei G, Meng W, Guo H, Pan W, Liu J, Peng T, Chen L, Chen CY. 2011. Potent neutralization of influenza A virus by a single-domain antibody blocking M2 ion channel protein. PLoS One 6:e28309. 10.1371/journal.pone.0028309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harmsen M, Pritz-Verschuren BJS, Bartenlink W, Van der Burg H, Koch G. 2013. Isolation of panels of llama single-domain antibody fragments binding all nine neuraminidase subtypes of influenza A virus. Antibodies 2:168–192. 10.3390/antib2020168 [DOI] [Google Scholar]
- 40.Van Borm S, Thomas I, Hanquet G, Lambrecht B, Boschmans M, Dupont G, Decaestecker M, Snacken R, van den Berg T. 2005. Highly pathogenic H5N1 influenza virus in smuggled Thai eagles, Belgium. Emerg. Infect. Dis. 11:702–705. 10.3201/eid1105.050211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffmann E, Krauss S, Perez D, Webby R, Webster RG. 2002. Eight-plasmid system for rapid generation of influenza virus vaccines. Vaccine 20:3165–3170. 10.1016/S0264-410X(02)00268-2 [DOI] [PubMed] [Google Scholar]
- 42.Schotsaert M, Ysenbaert T, Neyt K, Ibañez LI, Bogaert P, Schepens B, Lambrecht BN, Fiers W, Saelens X. 2013. Natural and long-lasting cellular immune responses against influenza in the M2e-immune host. Mucosal Immunol. 6:276–287. 10.1038/mi.2012.69 [DOI] [PubMed] [Google Scholar]
- 43.Reed LJ, Muench H. 1938. A simple method for estimating fifty percent endpoints. Am. J. Hyg. 27:493–497 [Google Scholar]
- 44.De Filette M, Martens W, Roose K, Deroo T, Vervalle F, Bentahir M, Vandekerckhove J, Fiers W, Saelens X. 2008. An influenza A vaccine based on tetrameric ectodomain of matrix protein 2. J. Biol. Chem. 283:11382–11387. 10.1074/jbc.M800650200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harbury PB, Zhang T, Kim PS, Alber T. 1993. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262:1401–1407. 10.1126/science.8248779 [DOI] [PubMed] [Google Scholar]
- 46.Hassantoufighi A, Zhang H, Sandbulte M, Gao J, Manischewitz J, King L, Golding H, Straight TM, Eichelberger MC. 2010. A practical influenza neutralization assay to simultaneously quantify hemagglutinin and neuraminidase-inhibiting antibody responses. Vaccine 28:790–797. 10.1016/j.vaccine.2009.10.066 [DOI] [PubMed] [Google Scholar]
- 47.Arbabi Ghahroudi M, Desmyter A, Wyns L, Hamers R, Muyldermans S. 1997. Selection and identification of single domain antibody fragments from camel heavy-chain antibodies. FEBS Lett. 414:521–526. 10.1016/S0014-5793(97)01062-4 [DOI] [PubMed] [Google Scholar]
- 48.Kang AS, Jones TM, Burton DR. 1991. Antibody redesign by chain shuffling from random combinatorial immunoglobulin libraries. Proc. Natl. Acad. Sci. U. S. A. 88:11120–11123. 10.1073/pnas.88.24.11120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Meyer T, Eeckhout D, De Rycke R, De Buck S, Muyldermans S, Depicker A. 2014. Generation of VHH antibodies against the Arabidopsis thaliana seed storage proteins. Plant Mol. Biol. 84:83–93. 10.1007/s11103-013-0118-0 [DOI] [PubMed] [Google Scholar]
- 50.Lambert C, Leonard N, De Bolle X, Depiereux E. 2002. ESyPred3D: prediction of proteins [sic] 3D structures. Bioinformatics 18:1250–1256. 10.1093/bioinformatics/18.9.1250 [DOI] [PubMed] [Google Scholar]
- 51.Els Conrath K, Lauwereys M, Wyns L, Muyldermans S. 2001. Camel single-domain antibodies as modular building units in bispecific and bivalent antibody constructs. J. Biol. Chem. 276:7346–7350. 10.1074/jbc.M007734200 [DOI] [PubMed] [Google Scholar]
- 52.Conrath KE, Lauwereys M, Galleni M, Matagne A, Frere JM, Kinne J, Wyns L, Muyldermans S. 2001. β-Lactamase inhibitors derived from single-domain antibody fragments elicited in the Camelidae. Antimicrob. Agents Chemother. 45:2807–2812. 10.1128/AAC.45.10.2807-2812.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16:735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- 54.De Buck S, Virdi V, De Meyer T, De Wilde K, Piron R, Nolf J, Van Lerberge E, De Paepe A, Depicker A. 2012. Production of camel-like antibodies in plants. Methods Mol. Biol. 911:305–324. 10.1007/978-1-61779-968-6_19 [DOI] [PubMed] [Google Scholar]
- 55.Böttcher E, Freuer C, Steinmetzer T, Klenk HD, Garten W. 2009. MDCK cells that express proteases TMPRSS2 and HAT provide a cell system to propagate influenza viruses in the absence of trypsin and to study cleavage of HA and its inhibition. Vaccine 27:6324–6329. 10.1016/j.vaccine.2009.03.029 [DOI] [PubMed] [Google Scholar]
- 56.Matrosovich M, Matrosovich T, Garten W, Klenk HD. 2006. New low-viscosity overlay medium for viral plaque assays. Virol. J. 3:63. 10.1186/1743-422X-3-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stech J, Stech O, Herwig A, Altmeppen H, Hundt J, Gohrbandt S, Kreibich A, Weber S, Klenk HD, Mettenleiter TC. 2008. Rapid and reliable universal cloning of influenza A virus genes by target-primed plasmid amplification. Nucleic Acids Res. 36:e139. 10.1093/nar/gkn646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Jaeger G, Scheffer S, Jacobs A, Zambre M, Zobell O, Goossens A, Depicker A, Angenon G. 2002. Boosting heterologous protein production in transgenic dicotyledonous seeds using Phaseolus vulgaris regulatory sequences. Nat. Biotechnol. 20:1265–1268. 10.1038/nbt755 [DOI] [PubMed] [Google Scholar]
- 59.Van Droogenbroeck B, Cao J, Stadlmann J, Altmann F, Colanesi S, Hillmer S, Robinson DG, Van Lerberge E, Terryn N, Van Montagu M, Liang M, Depicker A, De Jaeger G. 2007. Aberrant localization and underglycosylation of highly accumulating single-chain Fv-Fc antibodies in transgenic Arabidopsis seeds. Proc. Natl. Acad. Sci. U. S. A. 104:1430–1435. 10.1073/pnas.0609997104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.WHO/OIE/FAO H5N1 Evolution Working Group. 2012. Continued evolution of highly pathogenic avian influenza A (H5N1): updated nomenclature. Influenza Other Respir. Viruses 6:1–5. 10.1111/j.1750-2659.2011.00298.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kandun IN, Wibisono H, Sedyaningsih ER, Yusharmen Hadisoedarsuno W, Purba W, Santoso H, Septiawati C, Tresnaningsih E, Heriyanto B, Yuwono D, Harun S, Soeroso S, Giriputra S, Blair PJ, Jeremijenko A, Kosasih H, Putnam SD, Samaan G, Silitonga M, Chan KH, Poon LL, Lim W, Klimov A, Lindstrom S, Guan Y, Donis R, Katz J, Cox N, Peiris M, Uyeki TM. 2006. Three Indonesian clusters of H5N1 virus infection in 2005. N. Engl. J. Med. 355:2186–2194. 10.1056/NEJMoa060930 [DOI] [PubMed] [Google Scholar]
- 62.Wang MZ, Tai CY, Mendel DB. 2002. Mechanism by which mutations at His274 alter sensitivity of influenza A virus N1 neuraminidase to oseltamivir carboxylate and zanamivir. Antimicrob. Agents Chemother. 46:3809–3816. 10.1128/AAC.46.12.3809-3816.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brandes M, Klauschen F, Kuchen S, Germain RN. 2013. A systems analysis identifies a feedforward inflammatory circuit leading to lethal influenza infection. Cell 154:197–212. 10.1016/j.cell.2013.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Govorkova EA, Leneva IA, Goloubeva OG, Bush K, Webster RG. 2001. Comparison of efficacies of RWJ-270201, zanamivir, and oseltamivir against H5N1, H9N2, and other avian influenza viruses. Antimicrob. Agents Chemother. 45:2723–2732. 10.1128/AAC.45.10.2723-2732.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Doyle TM, Jaentschke B, Van Domselaar G, Hashem AM, Farnsworth A, Forbes NE, Li C, Wang J, He R, Brown EG, Li X. 2013. The universal epitope of influenza A viral neuraminidase fundamentally contributes to enzyme activity and viral replication. J. Biol. Chem. 288:18283–18289. 10.1074/jbc.M113.468884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gravel C, Li C, Wang J, Hashem AM, Jaentschke B, Xu KW, Lorbetskie B, Gingras G, Aubin Y, Van Domselaar G, Girard M, He R, Li X. 2010. Qualitative and quantitative analyses of virtually all subtypes of influenza A and B viral neuraminidases using antibodies targeting the universally conserved sequences. Vaccine 28:5774–5784. 10.1016/j.vaccine.2010.06.075 [DOI] [PubMed] [Google Scholar]
- 67.Wan H, Gao J, Xu K, Chen H, Couzens LK, Rivers KH, Easterbrook JD, Yang K, Zhong L, Rajabi M, Ye J, Sultana I, Wan XF, Liu X, Perez DR, Taubenberger JK, Eichelberger MC. 2013. Molecular basis for broad neuraminidase immunity: conserved epitopes in seasonal and pandemic H1N1 as well as H5N1 influenza viruses. J. Virol. 87:9290–9300. 10.1128/JVI.01203-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McKimm-Breschkin JL, Blick TJ, Sahasrabudhe A, Tiong T, Marshall D, Hart GJ, Bethell RC, Penn CR. 1996. Generation and characterization of variants of NWS/G70C influenza virus after in vitro passage in 4-amino-Neu5Ac2en and 4-guanidino-Neu5Ac2en. Antimicrob. Agents Chemother. 40:40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johansson BE, Moran TM, Bona CA, Popple SW, Kilbourne ED. 1987. Immunologic response to influenza virus neuraminidase is influenced by prior experience with the associated viral hemagglutinin. II. Sequential infection of mice simulates human experience. J. Immunol. 139:2010–2014 [PubMed] [Google Scholar]
- 70.Schulman JL, Khakpour M, Kilbourne ED. 1968. Protective effects of specific immunity to viral neuraminidase on influenza virus infection of mice. J. Virol. 2:778–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cate TR, Rayford Y, Nino D, Winokur P, Brady R, Belshe R, Chen W, Atmar RL, Couch RB. 2010. A high dosage influenza vaccine induced significantly more neuraminidase antibody than standard vaccine among elderly subjects. Vaccine 28:2076–2079. 10.1016/j.vaccine.2009.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Air GM. 2012. Influenza neuraminidase. Influenza Other Respir. Viruses 6:245–256. 10.1111/j.1750-2659.2011.00304.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hensley SE, Das SR, Gibbs JS, Bailey AL, Schmidt LM, Bennink JR, Yewdell JW. 2011. Influenza A virus hemagglutinin antibody escape promotes neuraminidase antigenic variation and drug resistance. PLoS One 6:e15190. 10.1371/journal.pone.0015190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shoji Y, Chichester JA, Palmer GA, Farrance CE, Stevens R, Stewart M, Goldschmidt L, Deyde V, Gubareva L, Klimov A, Mett V, Yusibov V. 2011. An influenza N1 neuraminidase-specific monoclonal antibody with broad neuraminidase inhibition activity against H5N1 HPAI viruses. Hum. Vaccin. 7(Suppl):199–204. 10.4161/hv.7.0.14595 [DOI] [PubMed] [Google Scholar]
- 75.Nimmerjahn F, Ravetch JV. 2008. Fcgamma receptors as regulators of immune responses. Nat. Rev. Immunol. 8:34–47. 10.1038/nri2206 [DOI] [PubMed] [Google Scholar]
- 76.Desmyter A, Transue TR, Ghahroudi MA, Thi MH, Poortmans F, Hamers R, Muyldermans S, Wyns L. 1996. Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme. Nat. Struct. Biol. 3:803–811. 10.1038/nsb0996-803 [DOI] [PubMed] [Google Scholar]
- 77.Colman PM, Laver WG, Varghese JN, Baker AT, Tulloch PA, Air GM, Webster RG. 1987. Three-dimensional structure of a complex of antibody with influenza virus neuraminidase. Nature 326:358–363. 10.1038/326358a0 [DOI] [PubMed] [Google Scholar]
- 78.Malby RL, McCoy AJ, Kortt AA, Hudson PJ, Colman PM. 1998. Three-dimensional structures of single-chain Fv-neuraminidase complexes. J. Mol. Biol. 279:901–910. 10.1006/jmbi.1998.1794 [DOI] [PubMed] [Google Scholar]
- 79.Malby RL, Tulip WR, Harley VR, McKimm-Breschkin JL, Laver WG, Webster RG, Colman PM. 1994. The structure of a complex between the NC10 antibody and influenza virus neuraminidase and comparison with the overlapping binding site of the NC41 antibody. Structure 2:733–746. 10.1016/S0969-2126(00)00074-5 [DOI] [PubMed] [Google Scholar]
- 80.Tulip WR, Varghese JN, Laver WG, Webster RG, Colman PM. 1992. Refined crystal structure of the influenza virus N9 neuraminidase-NC41 Fab complex. J. Mol. Biol. 227:122–148. 10.1016/0022-2836(92)90687-F [DOI] [PubMed] [Google Scholar]
- 81.De Muynck B, Navarre C, Boutry M. 2010. Production of antibodies in plants: status after twenty years. Plant Biotechnol. J. 8:529–563. 10.1111/j.1467-7652.2009.00494.x [DOI] [PubMed] [Google Scholar]
- 82.Ismaili A, Jalali-Javaran M, Rasaee MJ, Rahbarizadeh F, Forouzandeh-Moghadam M, Memari HR. 2007. Production and characterization of anti-(mucin MUC1) single-domain antibody in tobacco (Nicotiana tabacum cultivar Xanthi). Biotechnol. Appl. Biochem. 47:11–19. 10.1042/BA20060071 [DOI] [PubMed] [Google Scholar]
- 83.Jobling SA, Jarman C, Teh MM, Holmberg N, Blake C, Verhoeyen ME. 2003. Immunomodulation of enzyme function in plants by single-domain antibody fragments. Nat. Biotechnol. 21:77–80. 10.1038/nbt772 [DOI] [PubMed] [Google Scholar]
- 84.Winichayakul S, Pernthaner A, Scott R, Vlaming R, Roberts N. 2009. Head-to-tail fusions of camelid antibodies can be expressed in planta and bind in rumen fluid. Biotechnol. Appl. Biochem. 53:111–122. 10.1042/BA20080076 [DOI] [PubMed] [Google Scholar]
- 85.De Buck S, Nolf J, De Meyer T, Virdi V, De Wilde K, Van Lerberge E, Van Droogenbroeck B, Depicker A. 2013. Fusion of an Fc chain to a VHH boosts the accumulation levels in Arabidopsis seeds. Plant Biotechnol. J. 11:1006–1016. 10.1111/pbi.12094 [DOI] [PubMed] [Google Scholar]