ABSTRACT
Kaposi's sarcoma-associated herpesvirus (KSHV) ORF6 is homologous to the herpes simplex virus 1 (HSV-1) ICP8 and Epstein-Barr virus (EBV) BALF2 proteins. Here, we describe its single-stranded DNA (ssDNA) binding properties. Based on previous findings with ICP8 and BALF2, a 60-amino-acid C-terminal deletion mutant of Orf6 was generated, and the protein was purified to explore the function of the C terminus in ssDNA binding. We showed that full-length ORF6 binds cooperatively to M13 ssDNA, disrupting its secondary structure and extending it to a length equivalent to that of duplex M13 DNA. The width of the ORF6-ssDNA filament is 9 nm, and a 7.3-nm repeat can be distinguished along the filament axis. Fluorescence polarization analysis revealed that the wild-type and C-terminal mutant ORF6 proteins bind equally well to short ssDNA substrates, with dissociation constant (Kd) values of 2.2 × 10−7M and 1.5 × 10−7M, respectively. These values were confirmed by electrophoretic mobility shift assay (EMSA) analysis, which also suggested that binding by the full-length protein may involve both monomers and small multimers. While no significant difference in affinities of binding between full-length ORF6 and the C-terminal deletion mutant were observed with the short DNAs, binding of the C-terminal mutant protein to M13 ssDNA showed a clear lack of cooperativity as seen by electron microscopy (EM). Incubation of a duplex DNA containing a long single-stranded tail with double-helical ORF6 protein filaments revealed that the ssDNA segment can be enveloped within the protein filament without disrupting the filament structure.
IMPORTANCE This work describes the biochemical characterization of the single-stranded DNA binding protein of KSHV, ORF6, central to viral DNA replication in infected cells. A C-terminal deletion mutant protein was generated to aid in understanding the role of the C terminus in DNA binding. Here we analyze the binding of the wild-type and mutant proteins to short oligomeric and longer genomic ssDNA substrates. Although it is capable of interacting with the short substrates, the inability of mutant ORF6 to form oligomers in solution hindered it from fully covering the long genomic substrates. We previously showed that ORF6 forms long filaments in solution, and we showed here that these can absorb ssDNA without disruption of the filament structure. This work will provide an important basis for future studies by us and/or others.
INTRODUCTION
Kaposi's sarcoma-associated herpesvirus (KSHV) is a member of the gammaherpesvirus family (1) and the causative agent of primary effusion lymphoma, Kaposi's sarcoma, and multicentric Castleman's disease (2). Similar to herpes simplex virus 1 (HSV-1) and Epstein-Barr virus (EBV), the life cycle of KSHV consists of a latent phase with periodic episodes of lytic replication. During the lytic phase, replication is driven by a set of 6 core replication factors (3), which for KSHV have been identified as ORF9 (DNA polymerase), ORF59 (polymerase processivity factor), ORF56 (primase), ORF44 (helicase), ORF40/41 (primase-associated factor), and ORF6 (DNA binding protein), based on homology with the HSV-1 and EBV proteins. This identification was bolstered by the finding that when the KSHV proteins are expressed in Vero cells, they will catalyze replication from the EBV lytic origin (4). ORF6 is homologous to the HSV-1 ICP8 and EBV BALF2 proteins (5), both of which were first identified as single-stranded DNA (ssDNA) binding proteins. Interestingly, in murine gammaherpesvirus 68 (MHV68), a gammaherpesvirus related to EBV and KSHV, deletion of the MHV68 Orf6 homolog resulted in a virus that could establish latency in vivo but was defective for viral replication (6).
Most of what is known about these single-stranded binding proteins comes from studies of ICP8. ICP8 was first shown to be required for in vitro replication reactions, suggesting that it might be the herpesvirus equivalent of the Escherichia coli single-stranded DNA binding protein (SSB) protein or eukaryotic replication protein A (RPA) (7). Further studies, however, revealed that in the absence of DNA, it forms long double-helical protein filaments (8, 9), a property shared with the RecA recombinase but not with SSB or RPA. Deletion of the C-terminal 60 amino acids of ICP8 was found to render it unable to form the double-helical filaments and also reduced the cooperative nature of its ssDNA binding (10). It was then shown that ICP8 is able to catalyze the annealing of homologous ssDNAs (11, 12), placing it in the annealase category, which contains numerous proteins, including Redβ. Finally, when ICP8 is incubated with a linear double-stranded DNA (dsDNA) containing a resected tail and a homologous ssDNA circle, it catalyzes DNA strand transfer over distances of >7 kb (13–15). These varied properties suggest that ICP8 has multiple functions in replication and recombination and does not fit into just one protein family. Little information is currently available as to whether ORF6 also shares these properties. However, we recently showed that ORF6 is able to form double-helical protein filaments in the absence of DNA (16).
ICP8 (17) and BALF2 (18) are abundantly expressed throughout lytic replication and are highly concentrated in nuclear replication bodies where viral DNA replication occurs. These bodies contain many cellular replication and repair factors (19). During KSHV lytic infection, ORF6 has been shown to be present in replication bodies as well. The six core replication proteins together with two other viral proteins, K8 and RTA, form a prereplication complex, which loads onto ori-Lyt DNA and becomes a replication initiation complex (20). Fluorescence microscopy and coimmunoprecipitation assays have shown that certain cellular DNA repair proteins, such as MSH2/6 and DNA-PK/Ku86/70 complexes, topoisomerase I, and topoisomerase II, accumulate in the viral replication bodies. Furthermore, their expression levels elevate dramatically during viral lytic DNA replication (21). The toposiomerases were of particular importance, since inhibition of their activity abolished lytic KSHV DNA replication altogether.
In this study, we have continued our biochemical characterization of the ORF6 protein by examining its ssDNA binding properties. To understand the role of oligomerization in ORF6-catalyzed reactions, we generated a C-terminal deletion mutant. We have employed electrophoretic mobility shift assay (EMSA), fluorescence polarization, and electron microscopic (EM) assays to examine the structure of the ssDNA-protein complexes and the effects of oligomerization on DNA binding kinetics.
MATERIALS AND METHODS
Expression constructs.
The Orf6 gene was cloned into the pFastBac HTa plasmid for baculovirus expression as described previously (16). The last 60 amino acids from the C terminus of the Orf6 gene were removed by PCR using the following primers: forward, 5′-GGC TAT GGA TCC GAT GGCGCAAA GGG ACC ACA-3′; reverse, 5′-GCT TTC AAA GCT TCT AAC CCT CGG CCG TCC A-3′. The mutant Orf6 [Orf6(ΔC)] gene was subcloned into the pFastBac HTa plasmid at the HindIII and BamHI restriction sites and contains a 6× His tag on the N terminus. The integrity of the mutant Orf6 gene was confirmed by sequencing. The baculovirus for protein expression was generated as described by the manufacturer (Invitrogen, Carlsbad, CA). The incorporation of mutant Orf6 into the viral genome was verified by PCR.
ORF6(ΔC) expression and purification.
The wild-type ORF6 protein was expressed and purified as previously described (16). ORF6(ΔC) was expressed and purified as follows. Sf21 cells (250 ml) were grown in suspension in Sf-900 II SFM and were inoculated with the virus at a multiplicity of infection (MOI) of 10. The cells were incubated for 48 h at 27°C and then pelleted at 1,500 × g for 10 min and washed once with ice-cold 1× phosphate-buffered saline (PBS) (Gibco BRL, Carlsbad, CA). After pelleting again, the cells were frozen and kept at −80°C until further use. Batch purification was used by following the Qiagen (Valencia, CA) nickel resin protocol with minor modifications. After thawing the cell pellet on ice, the cells were resuspended in cold lysis buffer (20 mM Tris, pH 7.4, 300 mM NaCl, 0.1% NP-40, 8 mM β-mercaptoethanol, and EDTA-free protease cocktail tablets [Roche, Indianapolis, IN]) and incubated on ice for 20 min. The cells were then sonicated (0.5-s pulses) for 2 min on ice. The cell extract was clarified by spinning at 18,000 × g for 60 min at 4°C. Qiagen nickel resin (1 ml) was equilibrated with the lysis buffer and applied to the clarified cell extract. The protein was allowed to bind to the resin by rocking in the cold room overnight. The resin was then washed several times with lysis buffer containing 5% glycerol and 20 mM Imidazole (pH 8.0). The ORF6(ΔC) protein was eluted with elution buffer (20 mM Tris, pH 7.4, 200 mM NaCl, and 250 mM imidazole) and dialyzed overnight against 20 mM HEPES, pH 7.4, 150 mM NaCl, 0.1 mM EDTA, and 20% glycerol and stored at −80°C. The purity of the protein was determined by SDS-PAGE and Coomassie staining.
DNA substrates.
For EMSA, homopolymers of [dT]14, [dT]28, and [dT]45 were labeled with fluorescein at the 5′ end. The DNA was visualized by exciting the fluorophore with 494-nm-wavelength light and recording its emission at 521 nm. For the electron microscopy (EM) experiments, we used M13mp18 ssDNA (NEB, Ipswich, MA) and a circular dsDNA plasmid with a 400-nucleotide (nt)-long ssDNA tail prepared as described before (22).
Fluorescence polarization.
Solutions (70 μl) containing 0.15 μM fluorescein isothiocyanate (FITC)-labeled [dT]14 or [dT]28 with varied amounts of ORF6 or ORF6(ΔC) in 20 mM HEPES, 50 mM NaCl, and 1 mM dithiothreitol (DTT) were prepared for the assays. The mixtures were incubated at room temperature for 20 min. The fluorescence anisotropies were measured on a Fluorolog-3 instrument (Horiba Jobin Yvon, Edison, NJ) with an excitation wavelength of 494 nm and an emission wavelength of 515 nm. The statistical analysis of the data and the graphs were generated using the Prism 6 software program (GraphPad, La Jolla, CA).
EMSA.
Binding reactions were carried out in a final volume of 20 μl. The reaction mixtures contained 20 nM DNA in 20 mM HEPES (pH 7.9), 50 mM NaCl, and 1 mM DTT with appropriate amounts of the ORF6 and ORF6(ΔC) proteins. The mixtures were incubated at room temperature for 20 min, and the complexes were resolved on 5% nondenaturing polyacrylamide gels in Tris-borate-EDTA (TBE) buffer run at 50 V at 4°C. The DNA was visualized by using a Typhoon 9400 instrument (GE Life Sciences, Piscataway, NJ) to record the emission signal of the excited fluorescein labels. The images were adjusted for contrast using Adobe Photoshop software (Adobe Systems, San Jose, CA). Statistical analysis of the data and the graphs were generated using Prism 6 software.
Electron microscopy.
To image the ORF6 filaments by negative staining, ORF6 or the C-terminal mutant protein was incubated at room temperature for 3 h in a buffer containing 20 mM HEPES (pH 7.9), 50 mM NaCl, and 1 mM DTT as described previously (16). Aliquots were then absorbed to copper grids covered by thin glow-charged carbon foils and stained for 5 min with 2% (wt/vol) uranyl acetate in water. Samples were examined in a Tecnai 12 transmission electron microscopy (TEM) (FEI, Hillsboro, OR) at 80 kV, and images were captured on a Gatan Orius charge-coupled device (CCD) camera programmed with Digital Micrograph software (Gatan, Warrendale, PA).
To visualize DNA-protein complexes, binding mixtures were prepared as described for the EMSAs. For rotary metal shadowcasting, aliquots of the DNA-protein binding mixtures were cross-linked with 0.6% glutaraldehyde (final concentration) for 5 min at room temperature and then chromatographed through - ml Biogel A-5m columns equilibrated with 10 mM Tris (pH 7.4)–0.1 mM EDTA buffer. The samples were mixed with a buffer containing spermidine hydrochloride (final concentration of 2 mM) and applied to copper grids covered by thin glow-charged carbon foils, washed, dehydrated, and rotary shadowcast with tungsten as described previously (23). The grids were examined in an FEI Tecnai 12 TEM at 40 kV, and the images were captured as described above. Contrast in the final images was adjusted using Photoshop software (Adobe Systems, San Jose, CA). For negative staining, binding reactions were carried out in a final volume of 10 μl. The reaction mixtures contained 20 nM M13mp18 ssDNA in 20 mM HEPES (pH 7.9), 50 mM NaCl, and 1 mM DTT with various amounts of the ORF6 and ORF6(ΔC) proteins. The mixtures were incubated at room temperature or at 37°C for 20 min. Aliquots were then absorbed to copper grids covered by thin glow-charged carbon foils, stained for 5 min with 2% (wt/vol) uranyl acetate in water, and examined as described above.
RESULTS
Expression and purification of the ORF6ΔC protein.
The objective of this study was to examine the binding of the ORF6 protein to ssDNA and to further examine the properties of the double-helical protein filaments formed in the absence of DNA. We previously reported that deletion of the last 60 amino acids of ICP8 abolishes its ability to form double-helical protein filaments (24). Similar results were obtained with the C-terminus deletion mutant of BALF2 (24). Thus, we constructed a similar deletion mutant of ORF6. We successfully overexpressed the C-terminal deletion mutant (ORF6ΔC) in insect cells as a recombinant N-terminal 6×His-tagged protein and purified it to homogeneity (see Materials and Methods). The expression levels of ORF6ΔC were very high, with almost 3 mg of protein obtained from 500 ml of insect cell culture. Wild-type ORF6 was expressed as described previously (16). The purities of both wild-type and mutant ORF6 were determined by SDS-PAGE and Coomassie staining (see Fig. S1 in the supplemental material).
ORF6ΔC does not form double-helical protein filaments.
Under reducing conditions, ORF6 forms long filamentous structures (16), easily visualized by negative staining with 2% uranyl acetate (Fig. 1). All fields examined showed many of these filaments with >90% of the protein being present in filaments as contrasted to single particles on the background. These large protein oligomers elute in the void volume upon size exclusion chromatography on Superdex 200 columns, using UV absorption at 280 nm and SDS-PAGE staining to monitor the location of the protein (see Fig. S2 in the supplemental material). Omission of DTT did not completely eliminate filament formation, however, since a few shorter filaments and a few large unstructured aggregates were present in addition to a large amount of single Orf6 particles as observed by EM. The ORF6ΔC protein, however, eluted at the position expected for monomers when it was applied to Superdex 200 columns just after the purification. However, if the protein was incubated at 4°C or room temperature for several hours, then the bulk of the mutant protein eluted in the void volume (data not shown). Negative staining of ORF6ΔC from the void volume revealed the presence of large, unstructured protein aggregates (see Fig. S3), similar to those observed with the full-length protein in the absence of DTT. These results demonstrate that the mutant protein is blocked from forming long uniform filaments, in agreement with previous studies of ICP8 (8, 25).
FIG 1.
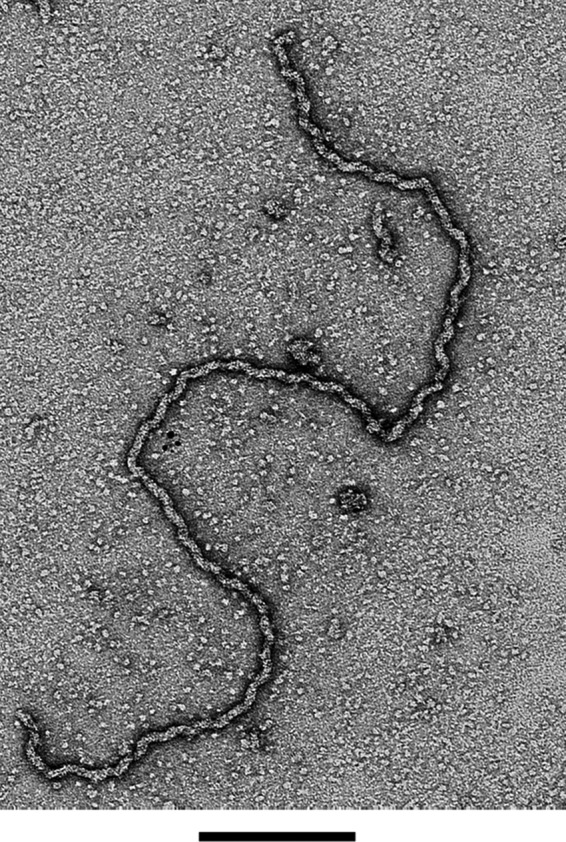
Visualization of ORF6 filaments by electron microscopy. The samples were adsorbed to glow discharge-treated thin carbon foils and stained with 2% uranyl acetate (see Materials and Methods). Bar = 200 nm.
Wild-type and mutant ORF6 bind ssDNA as shown using biochemical assays.
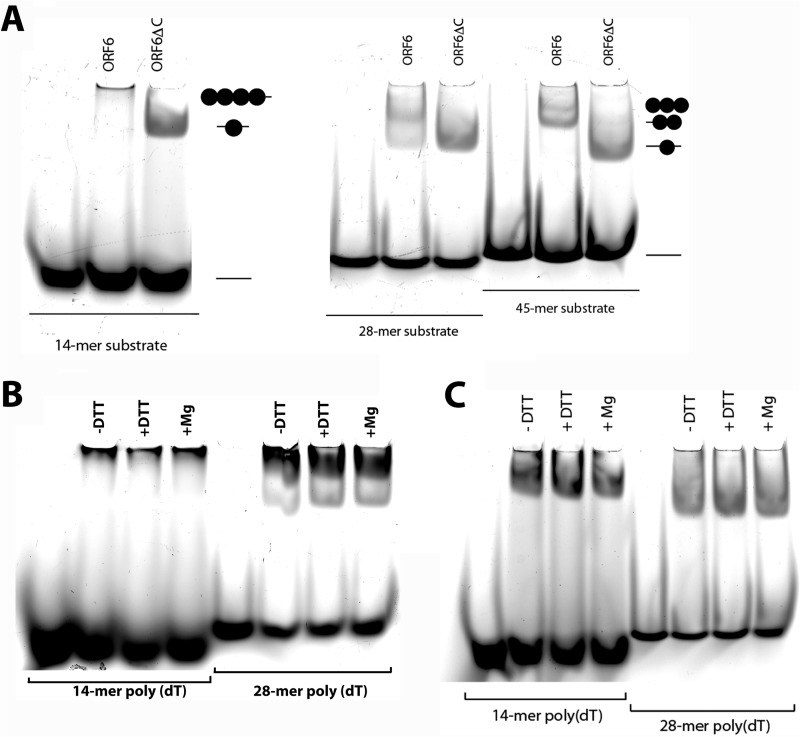
To examine the binding of full-length ORF6 and ORF6ΔC to ssDNA, we employed electrophoretic mobility shift assays (EMSA). The assays were performed with poly(dT) oligonucleotides, 14, 28, and 45 nt in length, labeled with 5′-fluorescein. The fluorescent tag made it possible to measure fluorescence polarization and follow the position of the DNA on the gels in the EMSAs by monitoring fluorescence at 515 nm with an excitation at 494 nm. With the 28-nt substrate, full-length ORF6 formed two supershifted nucleoprotein species in the gel, while ORF6ΔC formed only a single supershifted band (Fig. 2A). Incubation of full-length ORF6 with the 14-mer resulted in a single, slow-moving complex, which barely penetrated the gel, while ORF6ΔC formed a faster-moving complex that easily penetrated the gel (Fig. 2A). Previously, it has been shown that a 14-nt-long substrate can accommodate only a single ICP8 molecule, and this is clearly what we observed with ORF6ΔC. However, wild-type ORF6 formed a much larger complex with this substrate, suggesting that in the presence of short stretches of ssDNA, the protein tends to bind in an oligomeric form. ORF6ΔC bound only as a monomer to each substrate, from 14 to 45 nt. ORF6, however, showed two bands, likely a dimer and possibly a trimer with the 45-nt template (Fig. 2A). These results are different from previously published data with ICP8. First, by deleting the C terminus of ORF6, we eliminated its binding cooperativity, whereas ICP8 displayed some cooperativity since it was capable of binding as a dimer to longer substrates (10). Second, ORF6 formed large nucleoprotein complexes with the short 14-mer substrate, whereas ICP8 bound as a monomer. Negative staining of full-length ORF6 showed that in 20 min at room temperature, it had already started to form protein-only filaments, but most of the protein was still present as monomers. Similar examination of ORF6ΔC showed that it was predominantly monomeric with occasional aggregate forms. It is quite possible that wild -type ORF6 binds as a monomer and then starts forming oligomers. Similarly, when we increased the protein concentration in the presence of the 28-mer, ORF6 started to lose the monomeric species, and a new, heavier complex, which had difficulty penetrating the gel, appeared (see Fig. 4A). Mg2+ ions have been shown to be important for ICP8 function (8, 25). Even though we have shown previously (16) that Mg2+ ions do not affect ORF6 filament formation, we asked whether they would alter ORF6 interaction with small oligonucleotide substrates (Fig. 2B and C). Neither protein was affected by the presence of magnesium in the reaction as observed with the EMSA, nor did omission of DTT alter the results. This indicates that although DTT is critical for the formation of long double-helical filaments in solution, it does not affect the binding properties of ORF6 for small oligonucleotide DNAs.
FIG 2.
Binding of ORF6 and ORF6ΔC to ssDNA. (A) ORF6 and ORF6ΔC were incubated with 5′ fluorescein-labeled [dT]14, [dT]28, and d[T]45 single-stranded DNA, and the complexes were separated on 5% nondenaturing polyacrylamide gels (see Materials and Methods). The gel was imaged with a Typhoon 9400 scanner. The effect of reducing conditions and Mg2+ ions on the activity of ORF6 (B) or ORF6ΔC (C) is shown.
FIG 4.
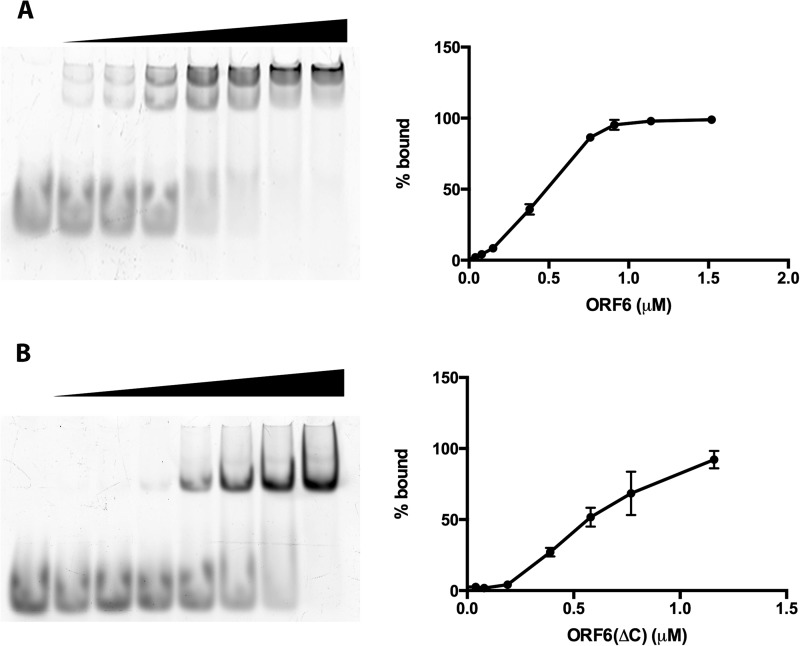
EMSA analysis of the binding of ORF6 (A) or ORF6ΔC (B) to ssDNAs. The proteins at the indicated concentrations were incubated with 0.2 μM 28-nt-long ssDNA, and the protein-DNA complexes were separated on 5% nondenaturing polyacrylamide gels and subjected to image analysis with a Typhoon 9400 scanner. Left panels show representative gel images, and right panels show quantitative analysis of data from three independent experiments. The bars indicate standard errors.
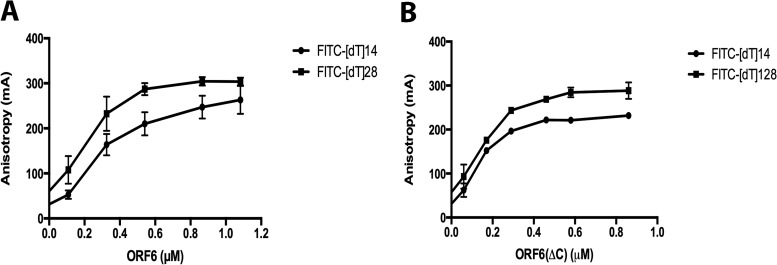
To further investigate the nature of binding of ORF6 to ssDNA, we carried out protein titrations and fluorescence polarization analysis. The fluorescein-labeled oligonucleotides were diluted to 0.15 µM concentrations in the binding buffer as described in Materials and Methods. The proteins were added at the needed concentrations, and the reactions were allowed to proceed for 20 min at room temperature. The fluorescence anisotropy of each reaction was measured by monitoring emission at 515 nm with excitation at 494 nm. The equilibrium binding data were then analyzed using nonlinear regression. Based on the results (Fig. 3) with both 14- and 28-nt substrates, the dissociation constant (Kd) values obtained for the proteins were similar, 2.2 × 10−7M for ORF6 and 1.5 × 10−7M for ORF6ΔC. Next, we repeated the protein titration assays with the 28-mer substrate using EMSA. The Kd values obtained from EMSAs, 4.6 × 10−7M for full-length ORF6 and 6.3 × 10−7M for ORF6ΔC (Fig. 4), indicate that the affinities of the two forms of the protein for these short DNAs are similar. These values are in good agreement with the results obtained from the fluorescence polarization analysis of the same binding experiments (Fig. 3). Both sets of experiments show that deletion of the C-terminal 60 amino acids has a minimal effect on the protein's binding affinity for short ssDNA.
FIG 3.
Fluorescence polarization analysis of binding of ORF6 and ORF6ΔC to ssDNAs. ORF6 (A)or ORF6ΔC (B), at the indicated concentrations, was incubated with 0.15 μM (each) FITC-labeled poly[dT] substrate for 20 min, and fluorescence polarization was measured. Bars indicate standard errors.
Visualization of binding of ORF6 and ORF6ΔC to M13 ssDNA reveals major differences.
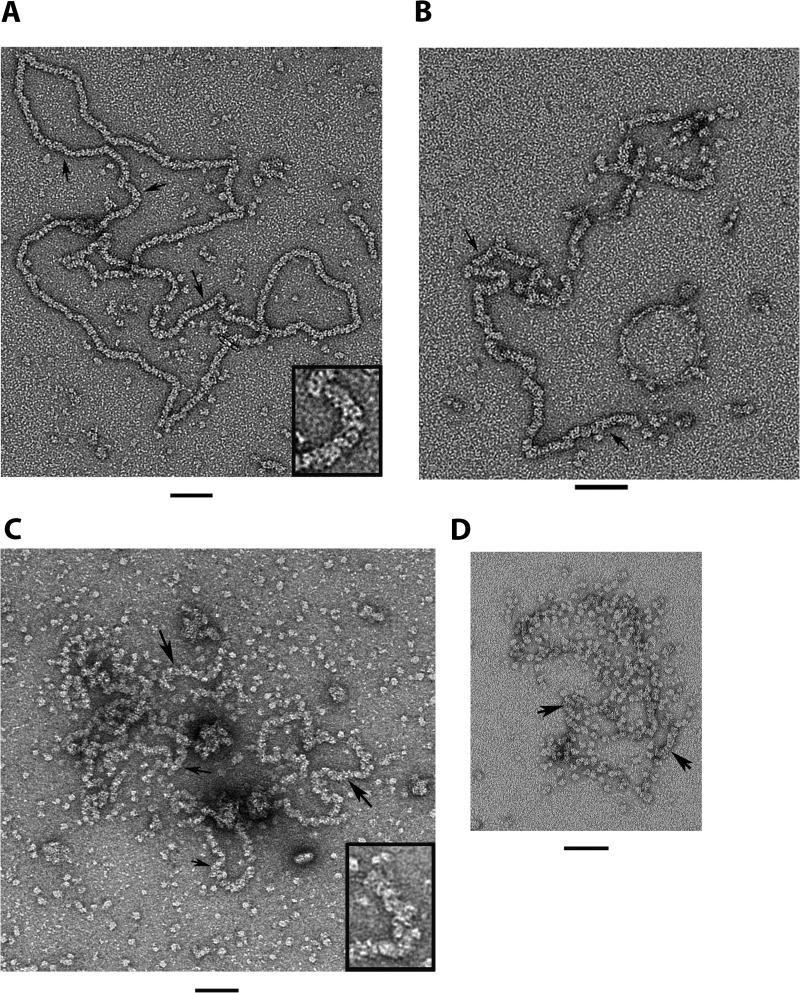
While it is essential to know how ORF6 interacts with small, defined substrates, it is also important to examine its interaction with longer substrates typical of DNA segments present during viral replication. We thus employed M13mp18 ssDNA and visualized the ORF6-ssDNA complexes using EM. The ssDNA was incubated with ORF6 or ORF6ΔC in a buffer containing 20 mM HEPES, 50 mM NaCl, and 1 mM DTT at room temperature. We varied the mass ratio of protein to DNA from 4:1 to 38:1 (micrograms of ORF6 per microgram of ssDNA). This corresponds to ratios from 1 monomer per 100 nt (4:1) to 1 monomer per 10 nt (38:1). The higher mass ratios are typical of complexes formed by ICP8, RecA, UvsX, and other proteins that form helical filaments on ssDNA. The samples were then stained with 2% uranyl acetate (without fixation) and visualized by EM at 80 kV. In Fig. 5, we show a typical field seen by EM. At higher ratios of ORF6 to ssDNA (Fig. 5A), uniform-appearing circular nucleoprotein filaments were observed, with all fields examined showing similar structures. The filaments showed a repeat along the filament axis of 7.3 ± 0.8 nm (n = 13), suggestive of single protein particles stacked side by side (inset, Fig. 5A). These ORF6-ssDNA filaments were very different from the double-helical protein filaments which ORF6 forms in the absence of ssDNA. The ORF6-ssDNA filament measured 9 ± 0.3 nm in width (n = 206 measurements). The ORF6 self-filaments are thicker in diameter and have a helical twist (16), similar to ICP8 self-filaments formed in the presence of Mg2+. The length of the circular ORF6-M13mp18 ssDNA filaments measured 2,060 ± 180 nm (n = 6). Preparation of M13mp18 dsDNA for EM by spreading it on an air-water interface with cytochrome c yielded a value of 2,330 nm ± 55 nm (n = 15). This suggests that when ssDNA is fully bound by the ORF6 protein, the ssDNA thread is extended to a value of 0.28 nm per nt, a value close to what it would be were it in a duplex conformation (0.34 nm/bp). This is similar to the extension of ssDNA by the T4 gene 32 protein (26) but not by the E. coli SSB protein, where the ssDNA is extended only to 0.3 times the length of an equivalent dsDNA (27).
FIG 5.
Visualization of the binding of ORF6 and ORF6ΔC to M13 ssDNA. ORF6 (A and B) was incubated with M13 ssDNA (see Materials and Methods). The complexes were visualized by staining with 2% uranyl acetate. In panel A, the monomer/nucleotide ratio is 10:1, and in panel B, 1:10. (C and D) ORF6ΔC with M13 ssDNA. In panel C, the reaction was carried out for 10 min at 37°C with a monomer/nucleotide ratio of 10:1. In panel D, the reaction was carried out with a monomer/nucleotide ratio of 5:1 and performed at 20°C. Arrows indicate nucleoprotein filaments. The insets in panels A and C are close-ups of the nucleoprotein filaments. Bar = 50 nm for all panels.
To estimate the number of nucleotides covered by a single ORF6 protein particle along the filament axis, we measured the lengths of segments of the ORF6-M13ssDNA filaments where there was a clear string of repeating particles. The segment lengths were divided by the number of particles and converted to nucleotides based on the value of 0.28 nm/nt determined above. This revealed that a single particle covers 22 nt. Based on the dimensions of the axial particles (average diameter of 11 nm [16]) and comparison with the sizes of single ORF6 particles present on the background, we assume that these axial particles represent single protein monomers. This is in agreement with the dimensions of ICP8 from the crystal studies (10.9 nm by 14.6 by 16.8 nm [28]). This represents a somewhat longer segment of ssDNA covered by ORF6 than had been reported for ICP8 and ICP8ΔC (24, 29) (15.8 and 14.6 nt, respectively). However, as discussed below, our estimate is expected to be biased toward a higher value.
The C-terminal deletion of ORF6 eliminates cooperative binding to ssDNA.
When the ratio of full-length ORF6 monomers per nucleotide was reduced stepwise below 1 monomer per 20 nt, the resulting nucleoprotein filaments showed long tracts of ORF6 spaced by segments of unbound ssDNA (Fig. 5B), reflecting the highly cooperative assembly of ORF6 onto ssDNA. When we increased the temperature to 37°C with wild-type ORF6, the M13 ssDNA molecules appeared fully covered by protein even after a 2-min incubation. In contrast, when the same titrations were repeated with ORF6ΔC, the protein was unable to cover the M13 molecules fully even at ratios of 1 monomer per 10 nt. As seen by EM, examining many fields of complexes, only short stretches of nucleoprotein filaments had formed, and most of the ORF6ΔC was visible in the background of the EM grids as monomers or larger particles (Fig. 5D), which are probably just protein aggregates since we see only monomers bound to short oligomeric substrates in our EMSAs. Increasing the temperature to 37°C slightly improved the ability of ORF6ΔC to interact with ssDNA, but it was still unable to fully cover the DNA (Fig. 5C). ICP8ΔC, however, had been shown to form filaments with ssDNA, which also had a larger diameter (24). The ORF6ΔC particles did not appear to be tightly stacked as in the case with wild-type ORF6. Thus, while the full-length and C-terminally deleted ORF6 proteins show similar binding to very short substrates, on long natural DNA, the full-length protein binds in a highly cooperative manner, forming stable nucleoprotein filaments, while ORF6ΔC appears to have lost its cooperativity and is unable to fully cover the long ssDNA.
ORF6 double-helical filaments can take up ssDNA without losing their structure.
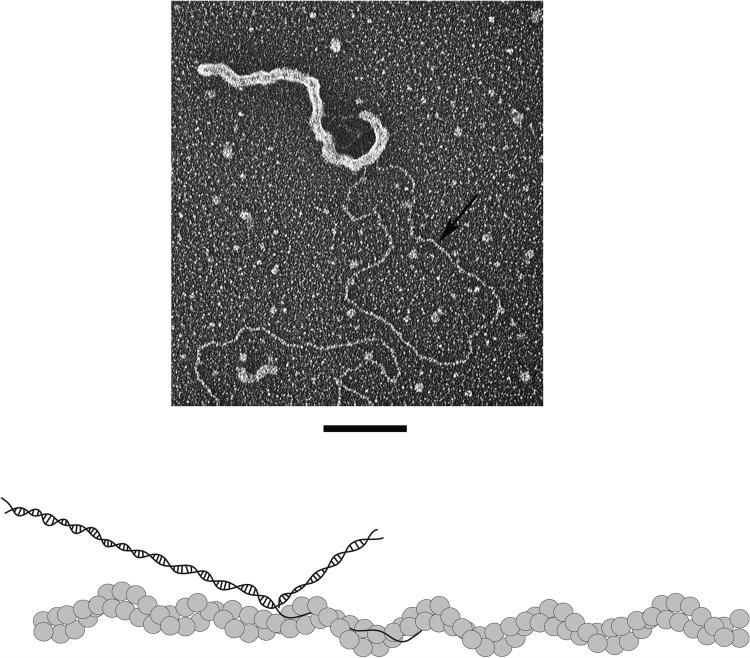
While we have suggested that in vivo the double-helical ORF6 or ICP8 protein filaments might organize the viral replication bodies, there has been no evidence for this proposal, nor indeed that these filaments can even bind DNA. Thus, it is possible that incubation of the double-helical protein filaments with ssDNA would lead to their rapid disintegration and concomitant assembly of ORF6 (or ICP8) onto the ssDNA. To examine this question, a circular double-stranded DNA with a long single-strand tail was prepared as described previously (22). ORF6 double-helical filaments were generated by incubating the protein for 3 h at room temperature, followed by addition of the tailed dsDNA circles to the protein filaments. Examination of the mixture by EM (Fig. 6; see also Fig. S4 in the supplemental material) revealed that the ssDNA tails of the plasmid circles had become engulfed within the double-helical filaments without visibly altering the filament structure. The duplex portion of the plasmid, however, remained unbound. Similar experiments with ORF6ΔC showed that the protein can bind to the ssDNA tails of the plasmid circles, but none of the filamentous structures were detected, as expected (see Fig. S5). This demonstrates that dsDNA is not a substrate for ORF6 and supports our proposal, discussed below, that the double-helical ORF6 filaments can bind ssDNA.
FIG 6.
Binding of ORF6 to dsDNA containing ssDNA tails. The ORF6 protein was incubated at room temperature for 3 h in the absence of DNA to form protein filaments. A circular dsDNA with a single-stranded tail was then added to the protein filaments. The sample was adsorbed to a charged carbon foil grid, followed by rotary tungsten shadowcasting (see Materials and Methods). Bar = 100 nm. The model represents the ORF6 filament interaction with the substrate. The arrow points to dsDNA. The single-stranded tail is buried within the ORF6 filament.
DISCUSSION
Orf6 was originally identified as the KSHV gene with considerable homology to the genes for the EBV BALF2 and HSV ICP8 proteins, both of which are well-characterized ssDNA binding proteins. In our previous study, we purified the ORF6 protein to homogeneity and showed that it shares a unique property with ICP8 in forming long, highly regular double-helical protein filaments in the absence of DNA (16). There, we hypothesized that these filaments might organize the replication bodies and provide a scaffold for the replication and repair factors, increasing their effective concentration and facilitating their function. This established the first structural similarity between ORF6 and ICP8. In this new study, we examined the ability of ORF6 to bind to ssDNA using biochemical assays for short DNA substrate binding and EM to visualize binding to long natural ssDNA. A 60-amino-acid (aa) C-terminal deletion mutant of the Orf6 gene was constructed, and the protein was purified to localize the region responsible for cooperative binding. Design of this mutant was based on our previous studies of ICP8 and BALF2. As described here, full-length ORF6 and ORF6ΔC bound to 14- and 28-nt poly(dT) with similar efficiencies and Kds. When bound to M13 ssDNA, ORF6 disrupted the secondary structure, showed highly cooperative binding, covered ∼22 nt/protein monomer, and extended the ssDNA filament to 0.28 nm/nt. ORF6ΔC, in contrast, bound poorly and showed little evidence of cooperative binding.
The Kd values obtained for binding of ORF6 and ORF6ΔC to short poly(dT) oligonucleotides were close to each other as determined by both EMSAs (4.6 × 10−7M for ORF6 and 6.3 × 10−7M for ORF6ΔC) and fluorescence polarization methods (2.2 × 10−7M for ORF6 and 1.5 × 10−7M for ORF6ΔC). The latter method provided values that reflect a somewhat tighter binding to the substrates. Because fluorescence polarization measures binding in solution as contrasted to a gel matrix, we would place more confidence in those values. Extensive analysis of the binding of ICP8 to short DNA molecules has shown that one monomer covers 15.8 nt (24, 28, 29). Our estimate of 22 nt bound by a single monomer of ORF6 is greater than the ICP8 value even though ORF6 is slightly smaller than ICP8. However, our estimate derived here was obtained by searching for segments of the ORF6-M13 ssDNA filaments, where the number of individual protein particles could be clearly counted. This required that each particle was separated from its neighbors, and thus the particles would not be as densely packed as they would be in regions where there were no ssDNA gaps. Given this caveat, our measurement of 22 nt/monomer should be taken as being somewhat of an overestimate and likely reflecting a real value closer to that for ICP8 (15.8 nt/monomer) (29). It does, nonetheless, emphasize the similarity between the filaments formed by the two proteins.
Examination of the EMSA patterns produced by binding of full-length ORF6 and ORF6ΔC to the 14-mer, 28-mer, and 45-mer substrates revealed major differences. ORF6ΔC formed a single slow-migrating complex with each substrate. The C-terminal deletion completely eliminated the ability of ORF6 to cooperatively bind to ssDNA. Full-length ORF6, however, formed different complexes with each substrate. It formed a single slower-migrating nucleoprotein complex with the 14-mer but two supershifted nucleoprotein species with the 28-mer substrate. In the presence of the 45-nt substrate, ORF6 bound only as a dimer or trimer. This suggests that the 14-mer was able to engage only oligomers, whereas longer substrates can engage monomers. This was different from findings in a previous study of ICP8 (10) binding to similar substrates. In that report, the authors concluded that the 14-mer will accommodate only a single ICP8 monomer, while the 28-mer binds monomers and dimers of ICP8, and they concluded that the 14-mer was too short to accommodate two ICP8 molecules. Here, we observed that ORF6ΔC forms a single faster-moving band with all three substrates, suggesting that DNAs were bound by protein monomers. This was in contrast to the ICP8 study (10), where the C-terminally truncated ICP8 generated two supershifted bands with the 28-nt substrate.
The RecA protein exists in solution in an equilibrium between monomers, dimers, and short rings and rods (30). Given that both RecA and ORF6 form self-filaments in solution, it is likely that ORF6 is also present as a spectrum of monomeric and oligomeric species. This may help explain the EMSA results summarized above. We suggest that full-length ORF6 is able to bind to the 28-mer as both monomers and small oligomers, the latter generating the slow-moving species. On the other hand, the 14-mer is so short that monomer binding is much less stable than oligomer binding, leading to a single slow-moving band in the gels. This model is supported by the observation that ORF6ΔC does not form the self-filaments and hence the only species available to bind to the 14- or 28- and 45-mer may be protein monomers, and indeed only a single fast-moving band was observed with either DNA.
Our demonstration that preformed double-helical filaments of ORF6 are able to take up ssDNA and that this occurs without any obvious disruption of the protein filament structure has implications for the way in which ORF6 may function in vivo. ORF6 is essential for viral DNA replication and is highly expressed, based on immunostaining assays where it is located in the nuclear replication compartments, which also contain other viral and host replication, recombination and repair factors (21). In the case of HSV-1, many of these other factors have been shown to bind ICP8 (31). Currently, there are no structural data regarding the oligomerization status of ORF6 in vivo, but we would suggest that it is organized into a network of double-helical protein filaments upon which other factors required for viral replication are assembled. Such a preformed replication factory would aid in arranging the replicating DNA and concentrating the other viral and host replication factors. This model is the converse of conventional thinking, in which the replicating DNA provides the scaffold for protein assembly. Given our finding that the ORF6ΔC mutant retains simple DNA binding but has lost the ability to form protein filaments, it will be of great interest to examine the phenotype of this mutant in the environment of a viral infection in vivo.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health, CA19014 and GM31819.
We appreciate helpful discussion from Sandra Weller.
Footnotes
Published ahead of print 21 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03652-13.
REFERENCES
- 1.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865–1869. 10.1126/science.7997879 [DOI] [PubMed] [Google Scholar]
- 2.Staskus KA, Sun R, Miller G, Racz P, Jaslowski A, Metroka C, Brett-Smith H, Haase AT. 1999. Cellular tropism and viral interleukin-6 expression distinguish human herpesvirus 8 involvement in Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. J. Virol. 73:4181–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fixman ED, Hayward GS, Hayward SD. 1992. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J. Virol. 66:5030–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu FY, Ahn JH, Alcendor DJ, Jang WJ, Xiao J, Hayward SD, Hayward GS. 2001. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 75:1487–1506. 10.1128/JVI.75.3.1487-1506.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicholas J, Ruvolo V, Zong J, Ciufo D, Guo HG, Reitz MS, Hayward GS. 1997. A single 13-kilobase divergent locus in the Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genome contains nine open reading frames that are homologous to or related to cellular proteins. J. Virol. 71:1963–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H, Ikuta K, Sixbey JW, Tibbetts SA. 2008. A replication-defective gammaherpesvirus efficiently establishes long-term latency in macrophages but not in B cells in vivo. J. Virol. 82:8500–8508. 10.1128/JVI.00186-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skaliter R, Lehman IR. 1994. Rolling circle DNA replication in vitro by a complex of herpes simplex virus type 1-encoded enzymes. Proc. Natl. Acad. Sci. U. S. A. 91:10665–10669. 10.1073/pnas.91.22.10665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Donnell ME, Elias P, Funnell BE, Lehman IR. 1987. Interaction between the DNA polymerase and single-stranded DNA-binding protein (infected cell protein 8) of herpes simplex virus 1. J. Biol. Chem. 262:4260–4266 [PubMed] [Google Scholar]
- 9.Makhov AM, Sen A, Yu X, Simon MN, Griffith JD, Egelman EH. 2009. The bipolar filaments formed by herpes simplex virus type 1 SSB/recombination protein (ICP8) suggest a mechanism for DNA annealing. J. Mol. Biol. 386:273–279. 10.1016/j.jmb.2008.12.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mapelli M, Muhleisen M, Persico G, van Der Zandt H, Tucker PA. 2000. The 60-residue C-terminal region of the single-stranded DNA binding protein of herpes simplex virus type 1 is required for cooperative DNA binding. J. Virol. 74:8812–8822. 10.1128/JVI.74.19.8812-8822.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dutch RE, Lehman IR. 1993. Renaturation of complementary DNA strands by herpes simplex virus type 1 ICP8. J. Virol. 67:6945–6949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makhov AM, Griffith JD. 2006. Visualization of the annealing of complementary single-stranded DNA catalyzed by the herpes simplex virus type 1 ICP8 SSB/recombinase. J. Mol. Biol. 355:911–922. 10.1016/j.jmb.2005.11.022 [DOI] [PubMed] [Google Scholar]
- 13.Bortner C, Hernandez TR, Lehman IR, Griffith J. 1993. Herpes simplex virus 1 single-strand DNA-binding protein (ICP8) will promote homologous pairing and strand transfer. J. Mol. Biol. 231:241–250. 10.1006/jmbi.1993.1279 [DOI] [PubMed] [Google Scholar]
- 14.Reuven NB, Staire AE, Myers RS, Weller SK. 2003. The herpes simplex virus type 1 alkaline nuclease and single-stranded DNA binding protein mediate strand exchange in vitro. J. Virol. 77:7425–7433. 10.1128/JVI.77.13.7425-7433.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reuven NB, Willcox S, Griffith JD, Weller SK. 2004. Catalysis of strand exchange by the HSV-1 UL12 and ICP8 proteins: potent ICP8 recombinase activity is revealed upon resection of dsDNA substrate by nuclease. J. Mol. Biol. 342:57–71. 10.1016/j.jmb.2004.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ozgur S, Damania B, Griffith J. 2011. The Kaposi's sarcoma-associated herpesvirus ORF6 DNA binding protein forms long DNA-free helical protein filaments. J. Struct. Biol. 174:37–43. 10.1016/j.jsb.2010.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burkham J, Coen DM, Weller SK. 1998. ND10 protein PML is recruited to herpes simplex virus type 1 prereplicative sites and replication compartments in the presence of viral DNA polymerase. J. Virol. 72:10100–10107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amon W, White RE, Farrell PJ. 2006. Epstein-Barr virus origin of lytic replication mediates association of replicating episomes with promyelocytic leukaemia protein nuclear bodies and replication compartments. J. Gen. Virol. 87:1133–1137. 10.1099/vir.0.81589-0 [DOI] [PubMed] [Google Scholar]
- 19.Wilcock D, Lane DP. 1991. Localization of p53, retinoblastoma and host replication proteins at sites of viral replication in herpes-infected cells. Nature 349:429–431. 10.1038/349429a0 [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Tang Q, Maul GG, Yuan Y. 2006. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: dual role of replication and transcription activator. J. Virol. 80:12171–12186. 10.1128/JVI.00990-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Li H, Tang Q, Maul GG, Yuan Y. 2008. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: involvement of host cellular factors. J. Virol. 82:2867–2882. 10.1128/JVI.01319-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Compton SA, Tolun G, Kamath-Loeb AS, Loeb LA, Griffith JD. 2008. The Werner syndrome protein binds replication fork and Holliday junction DNAs as an oligomer. J. Biol. Chem. 283:24478–24483. 10.1074/jbc.M803370200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffith JD, Christiansen G. 1978. Electron microscope visualization of chromatin and other DNA-protein complexes. Annu. Rev. Biophys. Bioeng. 7:19–35. 10.1146/annurev.bb.07.060178.000315 [DOI] [PubMed] [Google Scholar]
- 24.Mumtsidu E, Makhov AM, Konarev PV, Svergun DI, Griffith JD, Tucker PA. 2008. Structural features of the single-stranded DNA-binding protein of Epstein-Barr virus. J. Struct. Biol. 161:172–187. 10.1016/j.jsb.2007.10.014 [DOI] [PubMed] [Google Scholar]
- 25.Makhov AM, Taylor DW, Griffith JD. 2004. Two-dimensional crystallization of herpes simplex virus type 1 single-stranded DNA-binding protein, ICP8, on a lipid monolayer. Biochim. Biophys. Acta 1701:101–108. 10.1016/j.bbapap.2004.06.006 [DOI] [PubMed] [Google Scholar]
- 26.Delius H, Mantell NJ, Alberts B. 1972. Characterization by electron microscopy of the complex formed between T4 bacteriophage gene 32-protein and DNA. J. Mol. Biol. 67:341–350. 10.1016/0022-2836(72)90454-8 [DOI] [PubMed] [Google Scholar]
- 27.Chrysogelos S, Griffith J. 1982. Escherichia coli single-strand binding protein organizes single-stranded DNA in nucleosome-like units. Proc. Natl. Acad. Sci. U. S. A. 79:5803–5807. 10.1073/pnas.79.19.5803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mapelli M, Panjikar S, Tucker PA. 2005. The crystal structure of the herpes simplex virus 1 ssDNA-binding protein suggests the structural basis for flexible, cooperative single-stranded DNA binding. J. Biol. Chem. 280:2990–2997. 10.1074/jbc.M406780200 [DOI] [PubMed] [Google Scholar]
- 29.Makhov AM, Boehmer PE, Lehman IR, Griffith JD. 1996. Visualization of the unwinding of long DNA chains by the herpes simplex virus type 1 UL9 protein and ICP8. J. Mol. Biol. 258:789–799. 10.1006/jmbi.1996.0287 [DOI] [PubMed] [Google Scholar]
- 30.Brenner SL, Zlotnick A, Griffith JD. 1988. RecA protein self-assembly. Multiple discrete aggregation states. J. Mol. Biol. 204:959–972 [DOI] [PubMed] [Google Scholar]
- 31.Taylor TJ, Knipe DM. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 78:5856–5866. 10.1128/JVI.78.11.5856-5866.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.