ABSTRACT
The Bunyaviridae constitute a large family of enveloped animal viruses, many of which are important emerging pathogens. How bunyaviruses enter and infect mammalian cells remains largely uncharacterized. We used two genome-wide silencing screens with distinct small interfering RNA (siRNA) libraries to investigate host proteins required during infection of human cells by the bunyavirus Uukuniemi virus (UUKV), a late-penetrating virus. Sequence analysis of the libraries revealed that many siRNAs in the screens inhibited infection by silencing not only the intended targets but additional genes in a microRNA (miRNA)-like manner. That the 7-nucleotide seed regions in the siRNAs can cause a perturbation in infection was confirmed by using synthetic miRNAs (miRs). One of the miRs tested, miR-142-3p, was shown to interfere with the intracellular trafficking of incoming viruses by regulating the v-SNARE VAMP3, a strong hit shared by both siRNA screens. Inactivation of VAMP3 by the tetanus toxin led to a block in infection. Using fluorescence-based techniques in fixed and live cells, we found that the viruses enter VAMP3+ endosomal vesicles 5 min after internalization and that colocalization was maximal 15 min thereafter. At this time, LAMP1 was associated with the VAMP3+ virus-containing endosomes. In cells depleted of VAMP3, viruses were mainly trapped in LAMP1-negative compartments. Together, our results indicated that UUKV relies on VAMP3 for penetration, providing an indication of added complexity in the trafficking of viruses through the endocytic network.
IMPORTANCE Bunyaviruses represent a growing threat to humans and livestock globally. Unfortunately, relatively little is known about these emerging pathogens. We report here the first human genome-wide siRNA screens for a bunyavirus. The screens resulted in the identification of 562 host cell factors with a potential role in cell entry and virus replication. To demonstrate the robustness of our approach, we confirmed and analyzed the role of the v-SNARE VAMP3 in Uukuniemi virus entry and infection. The information gained lays the basis for future research into the cell biology of bunyavirus infection and new antiviral strategies. In addition, by shedding light on serious caveats in large-scale siRNA screening, our experimental and bioinformatics procedures will be valuable in the comprehensive analysis of past and future high-content screening data.
INTRODUCTION
With over 350 identified isolates worldwide, bunyaviruses represent a global threat to public health, livestock, and agricultural productivity (1). Many cause health problems in humans, such as fatal pulmonary syndromes, encephalitis, and hemorrhagic fever. Severe fever with thrombocytopenia syndrome virus in China (2009), Rift Valley fever virus (RVFV) in South Africa (2010), Schmallenberg virus in Europe (2011), and hantavirus in California (2012) are recent examples of outbreaks demonstrating that bunyaviruses must be taken seriously as emerging agents of disease. Currently, there are no approved vaccines or treatments to protect humans from infection.
Bunyaviruses are enveloped, negative-strand RNA viruses that are roughly spherical (∼100 nm), with a trisegmented genome that is replicated in the cytosol. The envelope glycoproteins GN and GC form spike-like projections responsible for virus attachment to host cells and penetration by membrane fusion. Receptors, cellular factors, and entry pathways remain poorly characterized. However, many bunyaviruses use the C-type lectin CD209 as a receptor to enter dendritic cells (DCs), and some members of the hantavirus subfamily have been shown to use β3-integrin to infect endothelial cells (2, 3). Heparan sulfate has been implicated in RVFV attachment (4). Sensitivity to clathrin and dynamin-2 perturbation implies a role for clathrin-mediated endocytosis in uptake and internalization (5–9).
Several lines of evidence suggest that many bunyaviruses are late-penetrating viruses (L-PVs), a large group of viruses that depends on trafficking to late endosomes (LE) for productive infection (10, 11). Inhibitor studies have shown that infection relies on late endosomal maturation and vacuolar acidification (11, 12). Weak bases that neutralize the endosomal pH, such as ammonium chloride (NH4Cl), inhibit infection by most bunyaviruses (1). Acid-activated viral membrane fusion, which results in the release of the virus genome in the cytosol, occurs typically at pH levels below 5.8, which are characteristic of late endosomal vacuoles (5, 11, 13, 14).
To further investigate the life cycle of bunyaviruses, we used two separate libraries of small interfering RNAs (siRNAs) against the whole human genome to screen for host factors involved in infection by Uukuniemi virus (UUKV), a bunyavirus of the Phlebovirus genus. Since UUKV is not pathogenic to humans, it enables state-of-the-art experimental approaches that are nearly impossible for pathogenic bunyaviruses, most of which must be contained within high-level biosafety laboratories. Together, the screens revealed 562 cellular factors with a potential role in UUKV endocytosis and replication. Further analysis, however, showed that many of the identified candidates were caused by off-target effects due to microRNA (miRNA)-like functions of the siRNAs. When the effects of a synthetic miRNA (miR) that mimicked the endogenous miRNA-142-3p were analyzed in detail, we observed that miR-142-3p inhibited UUKV entry and penetration. The v-SNARE VAMP3, a hit shared between both of the siRNA screens, was identified as an important target of miR-142-3p and a critical player in the late penetration of the virus.
MATERIALS AND METHODS
Cells and viruses.
All products used for cell culture were from Life Technologies. Human epithelial cells (HeLa) that stably express the human C-type lectin CD209 were cultured according to ATCC recommendations and used as the cell model (3). The prototype strains of Uukuniemi virus S23, influenza A virus X-31 (IAV; Virapur), and green fluorescent protein (GFP)-recombinant Semliki Forest virus (SFV; a kind gift from the University of Helsinki) have been described previously (15–17). Production, purification, and titration of UUKV were performed in BHK-21 cells as described elsewhere (11). Labeling of UUKV with AF-succinimidyl esters and Bodipy TR-thiosulfates (Invitrogen) was performed in HEPES (20 mM) as recently described (11). Alternatively, UUKV (109 focus-forming units [FFU/ml]) was labeled with the lipid dye R18 (20 μM; Life Technologies) according to a similar protocol. The multiplicity of infection (MOI) is given according to the titer determined on BHK-21 cells.
siRNAs, miRs, and plasmids.
siRNAs (siR-VAMP3#1, CAGCAUGUUUCUGAUAAUUAU [SI00759227]; siR-VAMP3#2, CCCAAAUAUGAAGAUAAACUA [SI00759248]; siR-N, GGAGAUCUUGGAUGCCAAUGA), AllStars negative-control siRNA (siR-Ctrl), and miRs were all from Qiagen. siR-N has been described previously (3). The plasmid carrying the gene for enhanced GFP (EGFP)-VAMP3 was generated using the human open reading frame library V3.1 (hORFeome v3.1) and Gateway cloning technology according to the manufacturer's instructions (18). Plasmids carrying genes for red fluorescent protein (RFP)-LAMP1 and EGFP-LAMP1 were kindly provided by J. Gruenberg (University of Geneva, Geneva, Switzerland). The catalytic chain of tetanus toxin (TeTx) was a kind gift from G. Coppolino (University of Guelph, Guelph, Ontario, Canada).
Antibodies and reagents.
The mouse monoclonal antibody (MAb) 8B11A3 (kind gift from Ludwig Institute for Cancer Research, Stockholm, Sweden) and HB-65 (ATCC) are directed against the nucleoproteins of UUKV and IAV, respectively (19). The phycoerythrin (PE)-conjugated fragment antigen binding (Fab) 1621P (R&D Systems) was used to detect CD209 at the cell surface. The mouse MAb against α-tubulin and the polyclonal rabbit antibody (Ab) against VAMP3 were purchased from Abcam and Thermo Scientific, respectively. An AF647-conjugated anti-mouse antibody (Life Technologies) was used as a secondary Ab in a fluorescence-activated cell sorting (FACS)-based infection assay. NH4Cl (Sigma) stock was dissolved in water.
High-throughput siRNA screening procedures. (i) Human genome-wide siRNA libraries.
The human genome-wide siRNA libraries employed in the screens were from Qiagen and Dharmacon. The library of Qiagen consists of four independent siRNAs against each gene, while that of Dharmacon involves pools of four different siRNAs per gene. The Qiagen library (OnGuard) was comprised of the druggable genome version 3, the genome supplement version 1, and the predicted gene set version 1. The Dharmacon library (On-TargetPlus) included the G-104655-02 OTP drug targets, the G-104675-02 OTP druggable subsets, and the G-105005-02 OTP genome. The gene information released by the National Center for Biotechnology Information (NCBI) in November 2012 was used for the annotation of libraries.
Several siRNAs in each plate were used to control the successful conduct of the general screening procedures. The AllStarsNegative and scrambled siRNAs from Qiagen and Dharmacon, respectively, as well as mock-transfected cells were used as negative controls. siRNA silencing of proteins known to be essential for UUKV infection, such as the vacuolar-type H+-ATPase V1 subunit A1 and the virus receptor CD209, but also against the viral genome, such as siR-N, were used as positive controls (3, 11). To assess the efficiency of transfection, cell death was induced with siRNAs that silence proteins critical for cell viability, such as the kinesin family member 11.
(ii) Liquid handling procedure.
All liquid handling steps, including preparation of cell plates, reverse transfection, virus infection, cell fixation, and immunofluorescence staining, were performed with an EL406 microplate washer and dispenser as well as with a microplate Biostak stacker (BioTek).
(iii) siRNA screen reverse transfection.
RNA interference screenings were achieved in single runs (Qiagen) or in triplicate (Dhamarcon). Experiments were conducted in a 384-well plate format, amounting to 296 and 171 plates for the Qiagen and Dharmacon screens, respectively. Reverse transfection was performed by seeding 400 cells in 50 μl of culture medium into a well containing 30 μl of transfection mix (24.9 μl of Dulbecco's modified Eagle's medium [DMEM], 0.1 μl of RNAiMax [LifeTechnologies], and 1.6 pmol siRNA in 5 μl [final siRNA concentration of ∼20 nM]). Cells were then left at 37°C for 72 h prior to infection.
(iv) Infection, immunofluorescence staining, and imaging.
Transfected cells were washed with complete medium before infection with UUKV (15,000 FFU [MOI, ∼5], diluted in DMEM containing 20 mM HEPES and 0.2% bovine serum albumin [BSA; Life Technologies]) at 37°C for 1 h. After exposition, the virus inoculum was replaced by culture medium. Infected cells were fixed with 4% formaldehyde 19 h later and immunostained with the mouse MAb 8B11A3 against N (diluted 1:500 in phosphate-buffered saline [PBS], 0.1% Triton X-100, 0.2% BSA) at room temperature (RT) for 1.5 h. The primary Ab and nuclei were then detected with a mix containing an anti-mouse Alexa Fluor 488-conjugated secondary Ab (1:1,000; Life Technologies) and Hoechst 33258 (1 μg ml−1; Life Technologies) in PBS with 0.2% BSA at RT in the dark for 1.5 h. After washing with 0.04% NaN3, plates were sealed and imaged by using automated inverted epifluorescence microscopes (ImageXpress; Molecular Devices) with a 10× objective, 0.3 numerical aperture (NA) Plan Fluor lens.
Image analysis. (i) Imaging error detection (out of focus or illumination problems).
As the first step, image quality was checked. Out-of-focus and illumination problems were automatically detected. Eight global image properties were extracted based on image intensities and their gradients and total variation. There was not a single property that clearly indicated whether an image was in focus or had to be reimaged. Therefore, we used all global parameters and a supervised machine learning algorithm based on the Random Forest classifier (20). Manually selected in-focus, out-of-focus, and illumination problem-affected images were used to train the model. The recognition accuracy was higher than 99%. Plates with out-of-focus wells were reacquired.
(ii) Image segmentation and feature extraction.
The next step of the image analysis was the identification of the cells, followed by the segmentation of their nuclear and cytoplasmic regions. We used a custom modified version of the CellProfiler program (21). The extraction started with an adaptive threshold determination on the Hoechst channel and followed by a morphological filter to eliminate objects that were too small or too large. Cells were kept in areas between 176 and 1,963 pixels (73.2 μm2 and 816.7 μm2). To perform measurements of the cytoplasm, we extended the nuclear mask by a 12-pixel-wide (7.7-μm-wide) ring. The extension value was deduced from the homogeneous fluorescence throughout the cytoplasm of infected cells. Using the detected objects, 37 cellular properties were measured for every single cell. The properties were based on intensity, texture, and morphology. They were used to further characterize the cellular phenotypes. The CellProfiler pipeline, the refactoring function, and a detailed list of features can be found at www.highcontentanalysis.org.
(iii) Phenotypic classification.
Infected cells were visualized using immunofluorescence staining against the viral protein N, which is expressed in the cytoplasm. Using simple fluorescence intensity-based threshold determinations resulted in a 20% infection recognition error. This error could be reduced to less than 4% by using a multiparametric supervised machine learning algorithm (22). We tested several models successfully applied earlier for high-content screening for phenotypic discovery (the analyzed classifiers are described in reference 23). By combining a committee of such classifiers using a majority voting method (20), the accuracy of the recognition rate was 96%.
Screen analysis and hit definition. (i) Transformation and normalization.
While the number of cells seemed to be homogenous within plates over all the screens, from raw data infections in outer wells were higher than inner wells for most of the plates (data not shown). Consequently, the systematic infection edge effect was normalized by using an adapted plate-wise local polynomial regression fitting Loess function (24). To this end, the sigmoidal distribution of the infection indices (i.e., values in the range 0 to 1) was first linearized with empirical logit transformation. From these processed values, the predicted values obtained with the Loess fit function were subtracted, and the mean of the predicted values was finally added (data not shown). In addition, plate-to-plate variations were corrected with a robust Z* score normalization function, and data were back-transformed by using the equation eZ* score/(1 + eZ* score) and normalized to the median of the overall negative siRNA controls (25).
(ii) Quality control.
After transformation and normalization, the data quality of individual plates was controlled with the strictly standardized means difference (SSMD) method applied to each positive control within a plate (26). The criteria for quality control were fixed according to the SSMD definition for a “very strong control” (data not shown). Only plates with at least 75% of positive controls considered good or excellent were further processed for the hit analysis. Other parameters that may reflect experimental problems were also considered, such as the infection index and siRNA cytotoxicity. Plates with median infection indices below 10% in the negative siRNA controls were removed to avoid increased errors on potential inhibitory effects. Data for wells of the Qiagen screen were removed when the number of cells was less than 75% of the median. As the Dharmacon screen was performed in triplicate, data were removed when the mean cell number for a gene was 3 standard deviations away from the mean observed for the siRNA negative controls.
(iii) Hit definition.
Plates and wells that successfully passed the quality control step were further processed to quantitatively score and rank genes according to the impact of their depletion on UUKV infection. The redundant siRNA activity (RSA) method, which is based on a hypergeometric distribution formula, was employed to rank siRNA efficiency and define hits in the Qiagen screen (27). This method allowed generation of two ranked gene lists according to the overall rank of each siRNA against the same gene, for either inhibition or enhancement siRNA strength effects. Subsequently, siRNAs were ranked following the Z* score method, and inhibitor and enhancer hits were defined by P values of 0.01 and 0.001, respectively. Different thresholds were used because the siRNA effects on infection were asymmetrically distributed. The SSMD method combined with fold change approaches was utilized to rank genes in the Dharmacon screen, as that screen was performed in triplicate. SSMD cutoffs of less than −3 and greater than 2 with changes of 0.5-fold and 1.5-fold were used for definitions of inhibitors and enhancers, respectively.
FACS-based infection and plasma membrane-virus fusion assays.
Cells, transfected or not, were infected at the indicated MOIs as previously described (11). After fixation and permeabilization with 0.1% saponin, UUKV and IAV infections were detected by standard immunofluorescence staining against the nucleoprotein N with the mouse MAbs 8B11A3 and HB-65, respectively, and quantified by flow cytometry. In the case of SFV, GFP expression was used to monitor infection. For siRNA- and miR-based infection assays, transfected cells were washed 1 h before infection. For the VAMP3 inactivation assay, cells were transfected with the plasmid carrying the TeTx gene, washed 6 h later, and exposed to viruses 18 h posttransfection. Virus fusion at the surface of siRNA- and miR-transfected cells was performed as recently reported (11).
siRNA- and miR-mediated knockdown confirmation.
siRNA and miR reverse transfections were performed using Lipofectamine RNAiMAX transfection reagent according to the manufacturer's reverse protocol (Life Technologies). Briefly, 40,000 cells were transfected with a 20 nM final concentration of siRNAs or miRs in fetal calf serum-free medium and seeded in a 24-well dish 3 days before infection or virus fusion.
Surface immunofluorescence staining.
The expression of CD209 was assessed at the surface of cells (not permeabilized) by FACS, using the PE-conjugated Fab 1621P (1:50) according to a standard procedure (3).
DNA transfection.
Cells were transfected with plasmids carrying genes for TeTx, EGFP-VAMP3, RFP-LAMP1, or EGFP-LAMP1 by using Lipofectamine 2000 (Life Technologies) according to the manufacturer's instructions.
Binding and internalization assays.
Viruses were bound to the cells for up to 2 h on ice at the indicated MOIs in binding buffer (DMEM [pH ∼7.4] containing 0.2% BSA, 1 mM CaCl2, and 2 mM MgCl2) as described before (3). In internalization assays, fluorescent virus-bound cells were rapidly warmed to 37°C for up to 40 min to trigger endocytosis. Binding and internalization were quantified by flow cytometry or analyzed by confocal microscopy. For FACS analysis, to distinguish between internalized and external particles, samples were treated with trypan blue (0.01%; Sigma) for 15 s at RT (3). To assess the role of VAMP3 in UUKV intracellular trafficking by using confocal microscopy, cells were transfected with the plasmids carrying genes for EGFP-VAMP3 and RFP-LAMP1, washed 6 h later, and incubated at 37°C for 12 h prior to the exposure to the virus. To further investigate the function of VAMP3 in UUKV late penetration, cells were first transfected with the control siRNA, siR-VAMP3#1, and miR-142-3p, and 48 h later with the plasmid harboring the gene for EGFP-LAMP1, before exposure to UUKV. Cells were fixed and analyzed by using a Zeiss LSM510 Meta microscope and a 63×, 1.4 NA Plan-Apochromat immersion oil objective lens.
R18-based virus fusion.
Transfected cells were collected from the culture surface with PBS-EDTA (0.5 mM), washed, and exposed to R18-labeled UUKV (UUKV-R18; MOI, ∼5) in binding buffer (DMEM [pH ∼7.4] containing 0.2% BSA and 20 mM HEPES) on ice for 1 h. Virus internalization was triggered by rapid warming at 37°C, in the presence or absence of NH4Cl (50 mM), directly inside a Varian Cary Eclipse fluorimeter (Agilent Technologies), and the fluorescence was measured for 1 h.
Microarray gene expression analysis.
Microarray experiments were conducted using Agilent Human Gene Expression 8×60k chips according to the manufacturer's recommendations. Briefly, total mRNA was isolated from cells transfected with siR-Ctrl by using TRIzol reagent, digested with DNase I (Qiagen), and purified. When RNA samples passed quality controls, i.e., when the A260/A280 ratio was above 2 and no RNA degradation was detected, microarray analysis was performed. Data were processed with the quantile normalization method and further analyzed with a two-group-based statistical method in combination with a fold change approach using the Limma package under the open source software R (28). Down- and upregulated genes were defined by a false-discovery rate better than 0.01 and a change below 0.5-fold or above 1.5-fold, respectively.
Protein analysis.
Proteins were analyzed by SDS-PAGE and transferred to polyvinylidene difluoride membranes (iBlot transfer stacks; Life Technologies). Incubation with primary Abs against α-tubulin (1:5,000) and VAMP3 (1:500) was followed by incubation with anti-rabbit or anti-mouse horseradish peroxidase-conjugated secondary Abs (1:5,000).
RESULTS
Genome-wide siRNA screens to elucidate bunyavirus-host cell interactions in human cells.
We developed an automated, high-content siRNA screening procedure with fluorescence microscopy as a readout to elucidate host cell factors and processes important during early UUKV-host cell interactions. As a cellular system, we used HeLa cells expressing the surface lectin CD209. Given that CD209 serves as a receptor for UUKV in DCs, these HeLa cells provided a model system to investigate the early stages of infection, including cell attachment, endocytosis, and translation of the viral proteins (3).
Each gene in the human genome was silenced by using two independent siRNA libraries, as illustrated in Fig. 1A. While one of the libraries (Qiagen) contained four unpooled, single siRNAs targeting distinct regions of each gene, the second (Dharmacon) relied on pools of four different siRNAs against each gene. The screens were carried out as detailed in Fig. 1A and in Materials and Methods. Automated transfection, infection, and immunostaining against newly synthesized nucleoprotein N as well as automated fluorescence microscopy and image analysis were employed to analyze cell factors required during the replication cycle, up to and including translation (Fig. 1A).
FIG 1.
UUKV siRNA screens. (A) Outlines of screening and bioinformatics procedures. (B) Z* score distribution of infection indices obtained for each siRNA in the Qiagen screen regarding inhibitor candidate genes. A redundant siRNA activity (RSA) P value below 0.01 defined potential hits. (C) Z* score distribution, as described for panel B but for enhancer candidate genes. An RSA P value below 0.001 defined potential hits. (D) Dual flash plot for Dharmacon siRNAs, showing selected candidates based on SSMD criteria and the fold change. Inhibitor genes were defined by SSMD and change values below −3-fold and 0.5-fold, respectively, and enhancers were defined as those with values above 2-fold and 1.5-fold (indicated by the gray horizontal lines). (E) Overlaps of candidate genes between the two siRNA screens. The numbers of potential hits that were specific to the Dharmacon and Qiagen screens are indicated in the top and bottom circles, respectively. The total numbers of potential hits for the Dharmacon and Qiagen screens are indicated next to the top and bottom circles, respectively. The P values, based on Fisher's exact statistical tests, indicate that potential hits obtained for both libraries were not independent.
Before potential hits were defined, systematic errors, such as batch-to-batch, plate-to-plate, or well-to-well differences and plate-wise spatial effects, generally caused by the experimental procedure itself, were normalized by computer-based mathematical procedures (data not shown). Parameters reflecting experimental problems were also considered, such as siRNA cytotoxicity and a variable infection index (i.e., the fraction of infected cells). Plates with median infection indices lower than 10% in the negative siRNA controls (compared to the median values of 20% and 34% for all plates in the Qiagen and Dharmacon screens, respectively) were removed to avoid any misinterpretation. Of the plates and wells, 10 to 20% were not taken into consideration for further analysis. This represented 28 plates out of 296 for Qiagen and 0 for Dharmacon, or 21,514 and 1,453 wells out of 94,720 and 18,003 for Qiagen and Dharmacon, respectively.
Because the intrinsic features of the siRNA libraries were different (single unpooled versus pooled siRNAs), we used distinct statistical analyses to define potential hits. Next, we determined the transcriptome of our HeLa cell line by using an Agilent-based microarray approach. Consistent with previous transcriptome studies conducted in HeLa strains, we found that the cells expressed some 13,000 different mRNA transcripts (see Table S1 at http://www.highcontentanalysis.org/download/Table_S1.xlsx). As expected from previous HeLa transcriptome studies, G-protein-coupled receptor signaling and immune defense factors were poorly represented (29, 30). The set of repressed genes was therefore excluded from the analysis, to minimize false-positive hits. For complete details on the screen procedures and analysis as well as the hit definition, refer to Materials and Methods.
siRNA screens identified 562 candidate genes with a role in UUKV infection.
The Qiagen screen led to the identification of 370 genes with a potential role in UUKV entry and replication, 331 inhibitors and 39 enhancers (Fig. 1B, C, and E; see also Tables S2 and S3 at http://www.highcontentanalysis.org/download/Table_S2.xlsx and http://www.highcontentanalysis.org/download/Table_S3.xlsx, respectively). The Dharmacon screen revealed 212 candidate genes, 186 inhibitors and 26 enhancers (Fig. 1D and E; see also Table S4 at http://www.highcontentanalysis.org/download/Table_S4.xlsx). Twenty candidates were present in both the Qiagen and Dharmacon screens (Fig. 1E). Although the direct overlap was modest, Fisher's exact test clearly established that the results of both screens were not statistically independent. Bioinformatics analysis via open source software like STRING, DAVID, and Reactome showed, moreover, that the potential hits complemented each other with regard to cell functions (data not shown). Consequently, we combined the results from both siRNA libraries. The new list of candidates (shown in Table S5 at http://www.highcontentanalysis.org/download/Table_S5.xlsx) contained 562 host factors representing a broad range of cell functions. The majority of genes in the list had no prior connection to bunyavirus infection.
Some of the identified cell functions would be expected for any RNA virus, such as those related to transcription. Others were specific to viruses that depend on endosomal acidification and membrane trafficking for infection, as illustrated by the presence of many subunits of the vacuolar-type H+-ATPases and several proteins involved in intracellular protein transport (e.g., AP2M1 and CLTCL1). The largest and most significant groups of potential hits represented vacuolar acidification and other membrane-related functions. Cell functions also included signaling, protein localization, cellular homeostasis, and others that are likely to lay the basis for future research into bunyaviruses.
siRNAs can silence genes through their seed regions in a miRNA-like manner.
When siRNA screens have been performed in different laboratories to identify host factors against a virus, the genes in published hit lists have shown poor overlap, suggesting serious inherent problems in the approach. This is in part explained by different screening procedures and analysis, as well as by siRNA off-target effects (31–33). When our analysis was limited to the 13,000 transcribed genes, the number of candidates was reduced to about 70% of the original lists independently of the Qiagen or Dharmacon screens. This implied that at least 30% of the potential hits in both the Qiagen and Dharmacon original lists were likely to represent false positives, since the corresponding genes were not expressed in our cell model. More specifically, it has been shown that siRNA antisense seed regions of 7 nucleotides (nt) regulate the expression of many genes through mechanisms identical to those involved in the silencing of mRNAs by miRNAs (Fig. 2A) (34–36). This may result in off-target effects.
FIG 2.
miRs, with sequences identical to those of siRNA seeds that inhibit the UUKV cycle, block infection. (A) Principles of gene silencing by siRNAs through on- and off-target mechanisms. The siRNA against epoxide hydrolase 1 (EPHX1), the seed region of which is identical to that of miR-142-3p, is shown as an example. VAMP3, vesicular-associated membrane protein 3; RAC1, ras-related C3 botulinum toxin substrate 1. (B) Cells treated with miRs (20 nM), which have identical seeds to those of siRNAs that blocked UUKV infection in the two screens, were infected with UUKV (MOI, ∼5) and harvested 7 h later. Infection was quantified via FACS after immunostaining against the viral nucleoprotein N, and results were normalized to the infection of cells treated with a negative-control siRNA (siR-Ctrl). An siRNA directed against N was used as a positive-control siRNA (siR-N). The light gray graphs indicate the impact predicted for the corresponding siRNA seeds. The value for the correlation between predicted and measured effects was 0.7, according to the Pearson statistical test. Error bars indicate the standard errors (SE) of the means of three independent experiments. (C) To bypass the need of virus endocytosis for infection, cells treated with miRs (20 nM) were exposed to UUKV (MOI, ∼0.5) in the cold, washed, and treated at the indicated pHs at 37°C for 1.5 min. Infected cells were then incubated for 8 h in the presence of NH4Cl (50 mM) to block virus penetration from endosomes. Consequently, only the release of viral genomes from the plasma membrane was monitored. Infection was analyzed via FACS, following immunostaining against the protein N. siR-Ctrl and siR-N were used as negative- and positive-control siRNAs, respectively. Error bars indicate SE of the means of three independent experiments.
To investigate the possibility that siRNAs acted like miRNAs in our screens, we first classified all the siRNAs in each library according to their 7-nt seed regions, i.e., we sorted them into groups according to the nucleotide sequences from position 2 to 8 in the 5′ end of the antisense siRNA strand (data not shown). We could identify 9,541 and 11,890 different seed sequences among the 72,773 and 71,557 siRNAs in the Qiagen and Dharmacon libraries, respectively (data not shown). By applying the SSMD and fold change approach to the seed sequences that were present in a minimum of 3 different siRNAs (totals of 5,658 and 7,622 seeds in the Qiagen and Dharmacon libraries, respectively), we found that 610 seeds in the Qiagen screen conferred the ability to block UUKV infection. Only 6 such sequences were identified in the Dharmacon screen. In other words, 5,257 siRNAs out of the 64,793 in the Qiagen screen and 54 siRNAs out of the 65,936 in the Dharmacon screen had the potential to inhibit the UUKV life cycle through miRNA-like mechanisms in addition to the intended siRNA effect. Although this phenomenon may appear relatively limited, the fraction of siRNAs that potentially acted like miRNAs was dramatically higher within lists of potential hits (see Tables S2 to S4 at http://www.highcontentanalysis.org). This implied that miRNA-like mechanisms caused by seed regions are likely to play a major role in the generation of off-target effects.
miRs bearing seed sequences that were predicted to impact infection had adverse effects on the virus life cycle.
Next, we selected 10 seed sequences predicted to affect UUKV and analyzed their potential miR functions in more detail. In addition, we selected 5 seed sequences predicted to have no effect on UUKV infection as controls. To predict whether a seed region has the capacity to impair the UUKV life cycle, SSMD and fold change values inferior to −3 and superior to 0.5 were used as cutoffs, respectively. When miRs with these seed sequences were tested for their impact on the UUKV life cycle in the HeLa CD209 cell line, we observed that the effect on infection strongly correlated with the predicted seed effects (correlation coefficient of 0.7) (Fig. 2B). Interestingly, in the lung epithelium cell line A549, which lacks endogenous CD209 expression, similar results were obtained for many of the miRs, including miR-142-3p, miR-921, miR-1303, and miR-369-3p (data not shown). This suggested that UUKV uses mechanisms for penetration and infection that are conserved in cell lines, beyond receptor-mediated virus uptake.
To determine whether the miRs that blocked UUKV infection affected the viral entry process (endocytosis and penetration into the cytosol), we used a bypass approach (11, 37). Bypass allows for the delivery of viral genomes into the cytosol of cells without endocytosis and endosomal trafficking. After allowing the virus to bind to the cell surface, the pH in the medium was briefly lowered to induce fusion of the viral envelope with the plasma membrane. Of the six miRs tested, one, miR-142-3p, failed to reduce infection after bypass, independently of the MOI (Fig. 2C). This was consistent with an effect on prepenetration steps of UUKV entry.
To define which entry steps were affected by miR-142-3p, the major stages in the entry program, from virus attachment to fusion, were analyzed. The surface expression of the virus receptor CD209 was unaffected by miR-142-3p-transfection (Fig. 3A). Using Alexa Fluor (AF) 488-labeled viral particles (UUKV-AF488), we found that virus binding to the plasma membrane was normal (Fig. 3B). To monitor virus endocytosis, a flow cytometry-based assay was used to discriminate between virus particles on the cell surface and those which had internalized (3). This assay relies on a membrane-impermeable dye, trypan blue, that quenches the fluorescence emitted by surface-exposed UUKV-AF488 while leaving intracellular viruses unquenched. When the amount of trypan blue-resistant fluorescence of cell-associated AF488-conjugated UUKV was analyzed 25 min after warming, no significant difference was observed between cells transfected with miR-142-3p and control RNA (Fig. 3C). This indicated that virus internalization by endocytosis was functional.
FIG 3.
miR-142-3p regulates UUKV intracellular trafficking. (A) Cells transfected with a negative-control siRNA (siR-Ctrl) or miR-142-3p (20 nM) were immunostained for CD209 expression before FACS analysis. The data shown are representative of three independent experiments. (B) AF488-labeled UUKV (UUKV-AF488) was bound to cells treated with siR-Ctrl or miR-142-3p for 1 h on ice before fixation and FACS analysis. The results are representative of three independent experiments. (C) UUKV-AF488 (MOI, ∼2) was bound to siR-Ctrl- or miR-142-3p-transfected cells on ice before warming to 37°C for 25 min. Internalization was analyzed by FACS after cell fixation and trypan blue treatment to quench fluorescence due to cell surface-bound viruses. Internalization was normalized to internalization in cells treated with siR-Ctrl. The mean value for the siR-Ctrl samples was 20%. Error bars indicate standard errors (SE) of three independent experiments. (D) R18-labeled UUKV (MOI, ∼5) was bound to siR-Ctrl- or miR-142-3p-transfected cells on ice before warming to 37°C for 1 h. The fluorescence increase corresponded to the dequenching of the lipid dye R18, following virus fusion within endosomes in living cells, and was measured with a fluorimeter. NH4Cl was used to block the virus fusion by raising the endosomal pH and thus to determine the fluorescence background due to spontaneous flip-flop of the dye R18 between the virus envelope and the adjacent cell membrane. RFI, relative fluorescence intensity. The data are representative of three independent experiments. (E) Results of an experiment similar to that shown in panel D, but for two time points and with data normalized to the fusion in siR-Ctrl-transfected cells, as follows: 100 × [(fluorescence in miR-142-3p-transfected cells) − (fluorescence in siR-Ctrl-transfected cells in the presence of NH4Cl)]/[(fluorescence in siR-Ctrl-transfected cells) – (fluorescence in siR-Ctrl-transfected cells in the presence of NH4Cl)]. Error bars indicate SE of the means of three independent experiments.
To analyze acid-activated membrane fusion in late endosomal vacuoles, we relied on the autoquenching of the fluorescence of a lipid dye, R18 (38). At high concentrations, the dye molecules in the virus envelope of labeled UUKV resulted in the autoquenching of the fluorescence signal. Fusion with cellular membranes allowed the release of R18 in the target membrane, resulting in dilution and dequenching. When a fluorimeter was used to monitor living cells, we observed that virus fusion was strongly impaired in cells transfected with miR-142-3p (Fig. 3D). The extent of fusion was 25% of control values (Fig. 3E).
These results indicated that in cells transfected with miR-142-3p, virus binding and endocytosis were normal but the viruses failed to fuse. The steps affected by miR-142-3p could therefore include intracellular trafficking of the virus to a fusion-competent compartment, acidification, or the fusion event itself. Of these, the last one, fusion, was less likely, because acid-activated fusion at the plasma membrane took place normally in bypass experiments.
miR-142-3p regulates the expression of the v-SNARE VAMP3 and 283 other genes.
With a sequence of only 7 nt, the seed sequences are so short that they are likely to match sequences in many mRNAs. Thus, a single miRNA can have many mRNA targets and can influence a broad spectrum of cellular functions (39). To identify which of the 13,000 genes expressed in our HeLa CD209 cell line were silenced by miR-142-3p, we used Agilent microarrays to compare the level of each mRNA in miR-142-3p-transfected cells with control cells. We found that 172 genes were upregulated and 112 were downregulated (Fig. 4A and B; see also Table S6 at http://www.highcontentanalysis.org/download/Table_S6.xlsx). The identity of the genes indicated that miR-142-3p had effects on many cellular functions, ranging from cell death to leukocyte activation. Many genes related to endocytosis, the cytoskeleton, and membrane trafficking were also included.
FIG 4.
miR-142-3p regulates VAMP3. (A) Microarray chips were used to define the genes regulated by miR-142-3p. The x axis of the volcano plot indicates the magnitude of gene expression changes, on a log2 scale, and the y axis shows the corrected P value for multiple comparisons. The correction was performed using the false-discovery rate (FDR) method, and values were log transformed and multiplied by −1. Data represent the combination of four independent experiments. (B) Overlaps of potential hits between microarray data obtained for miR-142-3p and both siRNA screens.
It was of interest to compare this list of genes with those obtained for UUKV in the siRNA screens and determine whether any of the genes were shared. If such genes existed, they would probably represent strong candidates. As shown in Fig. 4B, there was in fact one such gene, the vesicle-associated membrane protein 3 (VAMP3). VAMP3 is an integral membrane protein in the v-SNARE family of membrane fusion proteins with a role in late vesicular trafficking (40). Since UUKV uses late endosomal vesicles for penetration into host cells, it seemed worthwhile to follow up on VAMP3.
The v-SNARE VAMP3 is a key player in UUKV late penetration.
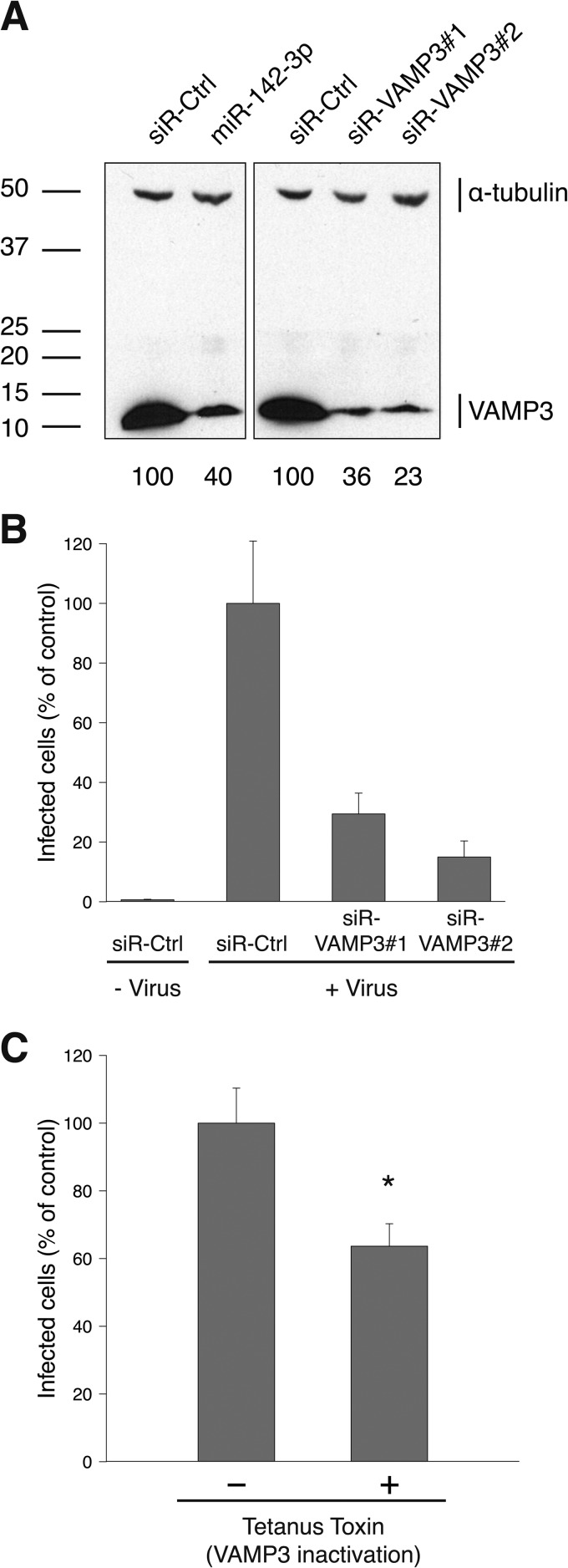
We first depleted HeLa CD209 cells of VAMP3 by using miR-142-3p and two nonoverlapping siRNAs. The level of VAMP3 protein was reduced by 60% with the miR-142-3p and by up to 75% with the siRNAs, as assayed by Western blotting (Fig. 5A). In the miR-142-3p-transfected cells, infection was inhibited by 70%. In the siRNA-treated cells, the inhibition was 70 to 80% of that in cells transfected with a control siRNA (Fig. 5B).
FIG 5.
VAMP3 depletion inhibits UUKV infection. (A) Efficiency of VAMP3 knockdown, assayed by Western blotting. VAMP3 protein levels are expressed as the percentage of VAMP3 levels in cells treated with interfering RNAs against VAMP3 (miR-142-3p and siR-VAMP3#1 and -#2) normalized to levels of α-tubulin and VAMP3 in cells treated with negative-control siRNAs (siR-Ctrl). (B) Cells were treated with siRNAs against VAMP3 (20 nM) and detached with PBS-EDTA 72 h posttransfection, prior to the exposure to UUKV (MOI, ∼2). Infection was analyzed via FACS, after immunostaining against N, 8 h later. The values were normalized to the infection level in samples treated with siR-Ctrl. Error bars indicate the standard error (SE) of the means of three independent experiments. (C) TeTx was transiently expressed in cells. The cells were then infected with UUKV (MOI, ∼2), and infection was quantified via FACS; data were normalized to results with cells transfected with the empty control plasmid. Error bars indicate SE of the means of three independent experiments. *, P ≤ 0.05.
This indicated that VAMP3 was, indeed, needed for efficient UUKV infection. To confirm its specific role in the UUKV infection cycle, we assessed infection in cells expressing the catalytic light chain of TeTx, a protease that cleaves and inactivates VAMP molecules, including VAMP3 (41). The expression of the toxin resulted in a significant decrease in UUKV infection (about 40%) (Fig. 5C).
We next determined whether UUKV enters VAMP3-positive vesicles during its journey through the endocytic network. Bodipy Texas Red-conjugated viral particles (UUKV-TR) were prepared as previously described (3). They were allowed to attach to cells expressing EGFP-VAMP3 (EGFP-tagged VAMP3) on ice. The temperature was then rapidly shifted to 37°C for periods of up to 40 min. Forty minutes is the time needed for UUKV to penetrate by membrane fusion from endosomes (3, 11).
While a fraction of the particles remained at the plasma membrane, as expected and previously reported (3, 11), confocal microscopy 5 min postwarming showed some colocalizations between internalized UUKV and EGFP-VAMP3 in vesicles located in the peripheral cytosol (Fig. 6A). Colocalization reached a maximum 20 min postwarming, when UUKV-positive vacuoles were mainly in the nuclear periphery, a typical location for late endosomal vesicles (Fig. 6A and B). In addition, confocal microscopy in live cells revealed that UUKV-TR actually moved in the cytoplasm together with the EGFP-VAMP3 in the same vesicles (see Movie S1 in the supplemental material).
FIG 6.
UUKV associates with VAMP3 and LAMP1. (A) Bodipy Texas Red-conjugated UUKV (UUKV-TR; MOI, ∼10) was bound to EGFP-VAMP3-expressing cells on ice. Cells were washed, rapidly warmed for up to 40 min, and fixed. UUKV (red) and EGFP-VAMP3 (green) were imaged in one focal plane by confocal microscopy. Magnifications of the association between UUKV and green vesicles (yellow squares) or noncolocalizing, internalized particles (white squares) are shown on the bottom. Bars, 10 μm. (B) The percentage of viruses colocalizing with the marker EGFP-VAMP3 at different times postwarming was calculated from series of z stacks, as described for those shown in panel A, and merged to one plane. Cells (n = 10) were counted in 10 independent fields for each time point. Error bars indicate the standard error (SE) of the means of the counted cells. The data are representative of three independent experiments. (C) Entry of prebound AF647-labeled UUKV (UUKV-AF647; MOI, ∼10) into cells expressing EGFP-VAMP3 and RFP-LAMP1, based on confocal microscopy. Viruses were internalized at 37°C for 30 min. One focal plane is show; UUKV is seen in blue, cell vesicles containing EGFP-VAMP3 are green, and RFP-LAMP1 is red. Magnifications of association between VAMP3+ LAMP1+ vesicles and internalized UUKV are shown on the bottom (yellow squares). Bar, 5 μm.
Thirty minutes after warming, several endocytosed viral particles were seen associated with large EGFP-VAMP3 and RFP-LAMP1 containing vacuoles in the nuclear periphery (Fig. 6C). LAMP1 is a marker for LEs and lysosomes. This indicated that UUKV and VAMP3 were present in the same vacuoles and that these most likely corresponded to LEs or endolysosomes. UUKV intracellular trafficking was then assessed by confocal microscopy in EGFP-LAMP1-expressing cells transfected with an siRNA against VAMP3 or miR-142-3p. Both RNA interferences resulted in a significant decrease in the colocalizations between LAMP1 and UUKV (Fig. 7A and B). This suggests that the viral particles could not reach LAMP1-positive late endosomal vesicles. These results highlight the functional importance of VAMP3 in UUKV late penetration.
FIG 7.
Depletion of VAMP3 blocks UUKV intracellular trafficking. (A) Cells transfected with miR-142-3p or siR-VAMP3#1 were exposed to AF594-labeled UUKV (UUKV-AF594; MOI, ∼5) on ice for 2 h, then washed and rapidly warmed to 37°C to allow virus endocytosis for 30 min. UUKV (red) and EGFP-LAMP1 (green) were imaged in one focal plane by confocal microscopy. Magnifications of association between internalized UUKV and LAMP1+ vesicles (yellow squares) or noncolocalizing, internalized particles (white squares) are shown on the bottom. Bars, 5 μm. (B) The percentage of viruses colocalizing with EGFP-LAMP1 was calculated from series of z stacks, as described for those shown in panel A. Cells (n ≥ 10) were counted in 10 independent fields, at least. Error bars indicate standard errors of the means of the counted cells. ***, P ≤ 0.001. The data are representative of two independent experiments.
Silencing of VAMP3 by using siRNA or miR-142-3p had no significant effect on infection by SFV. SFV is a virus that penetrates from early endosomes (37, 42). Interestingly, we also observed no effect on IAV, another L-PV (Fig. 8A and B). This implied that UUKV and IAV differently exploit late endocytic functions for entry.
FIG 8.
IAV does not rely on VAMP3 for penetration. (A) Cells were treated with siRNAs and then exposed to SFV, UUKV, or IAV as described in the legend for Fig. 5B. Infection was quantified by FACS 8 h later, and data were normalized to infection of cells treated with siR-Ctrl. **, P ≤ 0.01. Error bars indicate standard errors (SE) of the means of three independent experiments. (B) Cells transfected with miR-142-3p were exposed to SFV, UUKV, or IAV as described for Fig. 2B. Infection was quantified by FACS 8 h later, and data were normalized to infection of cells treated with siR-Ctrl. Error bars indicate SE of the means of three independent experiments. **, P ≤ 0.01; ns, nonsignificant.
DISCUSSION
In the last few years, large-scale siRNA screens have become a common tool for the analysis of virus-host cell interactions. For bunyaviruses, a Drosophila melanogaster genome-wide siRNA screen was used to show that mRNA uncapping restricts bunyavirus replication (43). We describe here the results from the first human genome-wide siRNA screen for a bunyavirus and report the identity of 562 cellular factors in human cells with a potential role in UUKV entry and replication. We were able to demonstrate that of the candidate genes, a large fraction was included because of off-target effects. Some of the siRNAs used could be shown to interfere with infection through an miRNA-like mechanism. In follow-up studies, we establish that one of the miRs, miR-142-3p, interfered with intracellular trafficking of the incoming UUKV and prevented its penetration into the cytosol. The identification and validation of the v-SNARE VAMP3 as one of the critical genes affected by miR-142-3p, as well as a true hit in the two siRNA screens, allowed us to define the role of this cell protein as a late endosomal factor required for UUKV infection.
Large-scale siRNA screens have in the last few years been applied to numerous viruses (31, 32). The information gained has led to the potential identification of hundreds of host cell factors with a role in the replication cycle of these viruses. The discrepancy in the hit lists reported for the same virus in different laboratories has, however, been alarmingly high (31, 32). Not more than 10% overlap was observed between the hits reported between IAV screens, with similar numbers observed for human immunodeficiency virus (HIV) screens (31, 32). The poor reproducibility has undermined confidence in the screening approach.
The comparison of two genome-wide siRNA libraries undertaken by us provides further clarification. Here, the screens were performed with the same virus stocks, cells, assays, and instruments, in an attempt to eliminate any trivial reasons for discrepancies. Still, the overlap between the lists of candidate genes obtained with Qiagen and Dharmacon libraries was not significantly higher than that seen in the IAV and HIV studies. It was apparent that the roots of false-negative and -positive hits were inherent in the design of the siRNA libraries and in the individual siRNA sequences that they contained.
The magnitude of the problem of false positives became evident when our potential hits were compared with the transcriptome. Nearly one-third of the candidates in our initial, uncorrected lists corresponded to genes in the human genome that were not transcribed in detectable amounts in the HeLa cells used. This allowed us to purge the initial 827 identified cellular factors of 265 false-positive candidates. It is clear that this represents a major problem inherent in the siRNA screening approach. It may explain many of the divergent results observed in previous screens. The extent of the problem probably also depends on factors such as the readout, the cellular processes involved, and the threshold settings.
One reason for the false positives was no doubt the large number of siRNAs with miRNA activity (34–36). We estimated that more than 5,200 of the 64,793 siRNAs in the Qiagen screen and 54 siRNAs of the 65,936 in the Dharmacon library were likely to affect UUKV infection through a miRNA-like mechanism. For a proof of principle, we experimentally tested 10 miRNAs that shared 7-nt seed sequences with those present among siRNAs that blocked UUKV infection, and we observed similar inhibition in infection. This indicated that many siRNAs in the libraries affected UUKV infection through a miRNA mechanism, or through combined siRNA and miRNA mechanisms.
Our results indicated that miRNA effects were less pronounced in the Dharmacon screen with pooled libraries than in the Qiagen screen, where cells were transfected with single siRNAs. However, the use of the siRNA pools in the Dharmacon library makes the analysis and correction of false-positive effects more difficult. Anyhow, it is unlikely that the nature of libraries, pooled versus unpooled, is by itself responsible for the off-target effect (44).
To increase the reliability of siRNA-based screens, there are several options. A tighter control of the specificity of siRNAs is a first obvious step. In this sense, the recent method developed by Buehler and colleagues (called control C911) may represent an important achievement (45). New approaches with built-in advantages for large loss-of-function screens also include the use of large libraries of short hairpin RNAs (shRNAs) in combination with the improvements in the plate design and the analysis of population context-regulated cell-to-cell variability (46–48).
Despite the problems, it is evident that the siRNA screens can provide valuable new information about the role of cellular factors in virus infection when carefully performed and validated. By focusing on gene clusters and performing stringent validation, it has been possible to identify legitimate hits of value (49, 50). With regard to UUKV infection, the screens did help us to identify candidate genes that made sense and brought our understanding of UUKV entry forward. Some of the potential hits confirmed cellular processes already described in the literature, and others were obvious and expected for RNA viruses. Among the unexpected findings was the v-SNARE VAMP3, a hit derived by combining miRNA (miR-142-3p) and siRNA screening data.
VAMP3 was the only cellular factor shared as an inhibitor hit between the miRNA screen and both siRNA screens. It is unlikely that the block in infection was caused by an adverse effect of VAMP3 depletion on the luminal pH of endosomes. Infection by SFV and IAV, which both rely on endosomal acidification for penetration, was not impaired in cells depleted of VAMP3. Follow-up studies showed that VAMP3 was, rather, needed for intracellular trafficking of UUKV to late endosomal compartments competent to support acid-activated membrane fusion between the virus and cellular membranes. About 20 min after internalization, the virus was maximally present in vacuoles containing LAMP1 and VAMP3. In cells depleted of VAMP3, viruses did not reach LAMP1+ late endosomal vesicles, suggesting that UUKV remained trapped in upstream endosomal compartments.
Our data support the view that the v-SNARE VAMP3 plays an active role in the maturation of the endosomal vacuoles used by UUKV during its journey within the endocytic network. That the intracellular trafficking of UUKV was abrogated in cells transfected with miR-142-3p supported this model, although we cannot completely exclude that genes other than VAMP3 that are targeted by miR-142-3p participate in UUKV infection. The role of VAMP3 was not restricted to cells expressing CD209. Interestingly, the synaptosome-associated protein 23 (SNAP23) was also a strong candidate. Consistent with the late timing for UUKV penetration, SNAP23 is part of the t-SNARE complexes that mediate fusion of LEs with each other and with other vacuoles (51). Together, these observations suggested that VAMP3 may have a role in late endosomal maturation and penetration of UUKV.
The observation that IAV was not dependent on VAMP3 indicated that the pathways of entry of the two L-PVs might diverge. Rather than mediating endosome-plasma membrane fusion, VAMP3 is actually thought to be involved in autophagosome-endosome fusion (40, 41, 52, 53). It is present on autophagosomes. This suggests that UUKV fusion may depend on the intersection between the endocytic and autophagocytic pathways, with VAMP3 acting like a bridge between both routes. Among the other potential hits, FIP200 (also known as RB1CC) and Rab1B also have functions related to autophagy and autophagosome maturation (54, 55). It has been recently proposed that high levels of autophagy increase adenovirus type 2 penetration and that echovirus 7 entry involves some cell factors of the autophagy machinery (56, 57). Though it is likely that some viruses induce and subvert autophagy for their own purposes, further investigations will be needed to clarify whether UUKV and other bunyaviruses depend on autophagy for productive entry.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from InfectX to C.V.M., from the European Research Council, the Swiss National Foundation for Research, ETH Zurich, and LipidX to A.H., and from the Natural Sciences and Engineering Research Council of Canada and the Banting Research Foundation to P.Y.L.
We thank M. Krzyzaniak, J. Mercer, L. Burleigh, S. Noerrelykke, and N. Tischler for proofreading. We also acknowledge S. Wirth and H. Rehrauer from the Functional Genomics Center Zurich, as well as G. Csúcs, A. Kaufman, T. Schwarz, and M. Stebler from the ScopeM ETH Zurich and A. Vonderheit from the IMB, Mainz, Germany.
Footnotes
Published ahead of print 21 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00388-14.
REFERENCES
- 1.Walter CT, Barr JN. 2011. Recent advances in the molecular and cellular biology of bunyaviruses. J. Gen. Virol. 92:2467–2484. 10.1099/vir.0.035105-0 [DOI] [PubMed] [Google Scholar]
- 2.Gavrilovskaya IN, Shepley M, Shaw R, Ginsberg MH, Mackow ER. 1998. β3 integrins mediate the cellular entry of hantaviruses that cause respiratory failure. Proc. Natl. Acad. Sci. U. S. A. 95:7074–7079. 10.1073/pnas.95.12.7074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lozach PY, Kuhbacher A, Meier R, Mancini R, Bitto D, Bouloy M, Helenius A. 2011. DC-SIGN as a receptor for phleboviruses. Cell Host Microbe 10:75–88. 10.1016/j.chom.2011.06.007 [DOI] [PubMed] [Google Scholar]
- 4.de Boer SM, Kortekaas J, de Haan CA, Rottier PJ, Moormann RJ, Bosch BJ. 2012. Heparan sulfate facilitates Rift Valley fever virus entry into the cell. J. Virol. 86:13767–13771. 10.1128/JVI.01364-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Boer SM, Kortekaas J, Spel L, Rottier PJ, Moormann RJ, Bosch BJ. 2012. Acid-activated structural reorganization of the Rift Valley fever virus Gc fusion protein. J. Virol. 86:13642–13652. 10.1128/JVI.01973-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hollidge BS, Nedelsky NB, Salzano MV, Fraser JW, Gonzalez-Scarano F, Soldan SS. 2012. Orthobunyavirus entry into neurons and other mammalian cells occurs via clathrin-mediated endocytosis and requires trafficking into early endosomes. J. Virol. 86:7988–8001. 10.1128/JVI.00140-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santos RI, Rodrigues AH, Silva ML, Mortara RA, Rossi MA, Jamur MC, Oliver C, Arruda E. 2008. Oropouche virus entry into HeLa cells involves clathrin and requires endosomal acidification. Virus Res. 138:139–143. 10.1016/j.virusres.2008.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simon M, Johansson C, Mirazimi A. 2009. Crimean-Congo hemorrhagic fever virus entry and replication is clathrin-, pH- and cholesterol-dependent. J. Gen. Virol. 90:210–215. 10.1099/vir.0.006387-0 [DOI] [PubMed] [Google Scholar]
- 9.Jin M, Park J, Lee S, Park B, Shin J, Song KJ, Ahn TI, Hwang SY, Ahn BY, Ahn K. 2002. Hantaan virus enters cells by clathrin-dependent receptor-mediated endocytosis. Virology 294:60–69. 10.1006/viro.2001.1303 [DOI] [PubMed] [Google Scholar]
- 10.Lozach PY, Huotari J, Helenius A. 2011. Late-penetrating viruses. Curr. Opin. Virol. 1:35–43. 10.1016/j.coviro.2011.05.004 [DOI] [PubMed] [Google Scholar]
- 11.Lozach PY, Mancini R, Bitto D, Meier R, Oestereich L, Overby AK, Pettersson RF, Helenius A. 2010. Entry of bunyaviruses into mammalian cells. Cell Host Microbe 7:488–499. 10.1016/j.chom.2010.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simon M, Johansson C, Lundkvist A, Mirazimi A. 2009. Microtubule-dependent and microtubule-independent steps in Crimean-Congo hemorrhagic fever virus replication cycle. Virology 385:313–322. 10.1016/j.virol.2008.11.020 [DOI] [PubMed] [Google Scholar]
- 13.Cifuentes-Munoz N, Barriga GP, Valenzuela PD, Tischler ND. 2011. Aromatic and polar residues spanning the candidate fusion peptide of the Andes virus Gc protein are essential for membrane fusion and infection. J. Gen. Virol. 92:552–563. 10.1099/vir.0.027235-0 [DOI] [PubMed] [Google Scholar]
- 14.Filone CM, Heise M, Doms RW, Bertolotti-Ciarlet A. 2006. Development and characterization of a Rift Valley fever virus cell-cell fusion assay using alphavirus replicon vectors. Virology 356:155–164. 10.1016/j.virol.2006.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pettersson R, Kaariainen L. 1973. The ribonucleic acids of Uukuniemi virus, a noncubical tick-borne arbovirus. Virology 56:608–619. 10.1016/0042-6822(73)90062-7 [DOI] [PubMed] [Google Scholar]
- 16.Kilbourne ED. 1969. Future influenza vaccines and the use of genetic recombinants. Bull. World Health Organ. 41:643–645 [PMC free article] [PubMed] [Google Scholar]
- 17.Spuul P, Balistreri G, Kaariainen L, Ahola T. 2010. Phosphatidylinositol 3-kinase-, actin-, and microtubule-dependent transport of Semliki Forest virus replication complexes from the plasma membrane to modified lysosomes. J. Virol. 84:7543–7557. 10.1128/JVI.00477-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamesch P, Li N, Milstein S, Fan C, Hao T, Szabo G, Hu Z, Venkatesan K, Bethel G, Martin P, Rogers J, Lawlor S, McLaren S, Dricot A, Borick H, Cusick ME, Vandenhaute J, Dunham I, Hill DE, Vidal M. 2007. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics 89:307–315. 10.1016/j.ygeno.2006.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Persson R, Pettersson RF. 1991. Formation and intracellular transport of a heterodimeric viral spike protein complex. J. Cell Biol. 112:257–266. 10.1083/jcb.112.2.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Witten IH, Frank E. 2005. Data mining: practical machine learning tools and techniques, 2nd ed. Elsevier, San Francisco, CA [Google Scholar]
- 21.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7:R100. 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horvath P, Wild T, Kutay U, Csucs G. 2011. Machine learning improves the precision and robustness of high-content screens: using nonlinear multiparametric methods to analyze screening results. J. Biomol. Screen. 16:1059–1067. 10.1177/1087057111414878 [DOI] [PubMed] [Google Scholar]
- 23.Banerjee I, Yamauchi Y, Helenius A, Horvath P. 2013. High-content analysis of sequential events during the early phase of influenza A virus infection. PLoS One 8:e68450. 10.1371/journal.pone.0068450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacoby WG. 2000. Loess: a nonparametric, graphical tool for depicting relationships between variables. Electoral Stud. 19:577–613 [Google Scholar]
- 25.Birmingham A, Selfors LM, Forster T, Wrobel D, Kennedy CJ, Shanks E, Santoyo-Lopez J, Dunican DJ, Long A, Kelleher D, Smith Q, Beijersbergen RL, Ghazal P, Shamu CE. 2009. Statistical methods for analysis of high-throughput RNA interference screens. Nat. Methods 6:569–575. 10.1038/nmeth.1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang XD. 2007. A pair of new statistical parameters for quality control in RNA interference high-throughput screening assays. Genomics 89:552–561. 10.1016/j.ygeno.2006.12.014 [DOI] [PubMed] [Google Scholar]
- 27.Konig R, Chiang CY, Tu BP, Yan SF, DeJesus PD, Romero A, Bergauer T, Orth A, Krueger U, Zhou Y, Chanda SK. 2007. A probability-based approach for the analysis of large-scale RNAi screens. Nat. Methods 4:847–849. 10.1038/nmeth1089 [DOI] [PubMed] [Google Scholar]
- 28.Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article3. 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- 29.Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, Paabo S, Mann M. 2011. Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 7:548. 10.1038/msb.2011.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landry JJ, Pyl PT, Rausch T, Zichner T, Tekkedil MM, Stutz AM, Jauch A, Aiyar RS, Pau G, Delhomme N, Gagneur J, Korbel JO, Huber W, Steinmetz LM. 2013. The genomic and transcriptomic landscape of a HeLa cell line. G3 3:1213–1224. 10.1534/g3.113.005777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bushman FD, Malani N, Fernandes J, D'Orso I, Cagney G, Diamond TL, Zhou H, Hazuda DJ, Espeseth AS, Konig R, Bandyopadhyay S, Ideker T, Goff SP, Krogan NJ, Frankel AD, Young JA, Chanda SK. 2009. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 5:e1000437. 10.1371/journal.ppat.1000437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cherry S. 2009. What have RNAi screens taught us about viral-host interactions? Curr. Opin. Microbiol. 12:446–452. 10.1016/j.mib.2009.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franceschini A, Meier R, Casanova A, Kreibich S, Daga N, Andritschke D, Dilling S, Ramo P, Emmenlauer M, Kaufmann A, Conde-Alvarez R, Low SH, Pelkmans L, Helenius A, Hardt WD, Dehio C, von Mering C. 2014. Specific inhibition of diverse pathogens in human cells by synthetic microRNA-like oligonucleotides inferred from RNAi screens. Proc. Natl. Acad. Sci. U. S. A. 111:4548–4553. 10.1073/pnas.1402353111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, Marshall WS, Khvorova A. 2006. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods 3:199–204. 10.1038/nmeth854 [DOI] [PubMed] [Google Scholar]
- 35.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. 2003. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21:635–637. 10.1038/nbt831 [DOI] [PubMed] [Google Scholar]
- 36.Lin X, Ruan X, Anderson MG, McDowell JA, Kroeger PE, Fesik SW, Shen Y. 2005. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 33:4527–4535. 10.1093/nar/gki762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Helenius A, Kartenbeck J, Simons K, Fries E. 1980. On the entry of Semliki Forest virus into BHK-21 cells. J. Cell Biol. 84:404–420. 10.1083/jcb.84.2.404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoekstra D, de Boer T, Klappe K, Wilschut J. 1984. Fluorescence method for measuring the kinetics of fusion between biological membranes. Biochemistry 23:5675–5681. 10.1021/bi00319a002 [DOI] [PubMed] [Google Scholar]
- 39.Pritchard CC, Cheng HH, Tewari M. 2012. MicroRNA profiling: approaches and considerations. Nat. Rev. Genet. 13:358–369. 10.1038/nrg3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fader CM, Sanchez DG, Mestre MB, Colombo MI. 2009. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim. Biophys. Acta 1793:1901–1916. 10.1016/j.bbamcr.2009.09.011 [DOI] [PubMed] [Google Scholar]
- 41.Galli T, Chilcote T, Mundigl O, Binz T, Niemann H, De Camilli P. 1994. Tetanus toxin-mediated cleavage of cellubrevin impairs exocytosis of transferrin receptor-containing vesicles in CHO cells. J. Cell Biol. 125:1015–1024. 10.1083/jcb.125.5.1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White J, Matlin K, Helenius A. 1981. Cell fusion by Semliki Forest, influenza, and vesicular stomatitis viruses. J. Cell Biol. 89:674–679. 10.1083/jcb.89.3.674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hopkins KC, McLane LM, Maqbool T, Panda D, Gordesky-Gold B, Cherry S. 2013. A genome-wide RNAi screen reveals that mRNA decapping restricts bunyaviral replication by limiting the pools of Dcp2-accessible targets for cap-snatching. Genes Dev. 27:1511–1525. 10.1101/gad.215384.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marine S, Bahl A, Ferrer M, Buehler E. 2012. Common seed analysis to identify off-target effects in siRNA screens. J. Biomol. Screen. 17:370–378. 10.1177/1087057111427348 [DOI] [PubMed] [Google Scholar]
- 45.Buehler E, Chen YC, Martin S. 2012. C911: a bench-level control for sequence specific siRNA off-target effects. PLoS One 7:e51942. 10.1371/journal.pone.0051942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bassik MC, Lebbink RJ, Churchman LS, Ingolia NT, Patena W, LeProust EM, Schuldiner M, Weissman JS, McManus MT. 2009. Rapid creation and quantitative monitoring of high coverage shRNA libraries. Nat. Methods 6:443–445. 10.1038/nmeth.1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snijder B, Sacher R, Ramo P, Damm EM, Liberali P, Pelkmans L. 2009. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature 461:520–523. 10.1038/nature08282 [DOI] [PubMed] [Google Scholar]
- 48.Collinet C, Stoter M, Bradshaw CR, Samusik N, Rink JC, Kenski D, Habermann B, Buchholz F, Henschel R, Mueller MS, Nagel WE, Fava E, Kalaidzidis Y, Zerial M. 2010. Systems survey of endocytosis by multiparametric image analysis. Nature 464:243–249. 10.1038/nature08779 [DOI] [PubMed] [Google Scholar]
- 49.Mercer J, Snijder B, Sacher R, Burkard C, Bleck CK, Stahlberg H, Pelkmans L, Helenius A. 2012. RNAi screening reveals proteasome- and Cullin3-dependent stages in vaccinia virus infection. Cell Rep. 2:1036–1047. 10.1016/j.celrep.2012.09.003 [DOI] [PubMed] [Google Scholar]
- 50.Sun E, He J, Zhuang X. 2013. Dissecting the role of COPI complexes in influenza virus infection. J. Virol. 87:2673–2685. 10.1128/JVI.02277-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valdez AC, Cabaniols JP, Brown MJ, Roche PA. 1999. Syntaxin 11 is associated with SNAP-23 on late endosomes and the trans-Golgi network. J. Cell Sci. 112:845–854 [DOI] [PubMed] [Google Scholar]
- 52.McMahon HT, Ushkaryov YA, Edelmann L, Link E, Binz T, Niemann H, Jahn R, Sudhof TC. 1993. Cellubrevin is a ubiquitous tetanus-toxin substrate homologous to a putative synaptic vesicle fusion protein. Nature 364:346–349. 10.1038/364346a0 [DOI] [PubMed] [Google Scholar]
- 53.Riggs KA, Hasan N, Humphrey D, Raleigh C, Nevitt C, Corbin D, Hu C. 2012. Regulation of integrin endocytic recycling and chemotactic cell migration by syntaxin 6 and VAMP3 interaction. J. Cell Sci. 125:3827–3839. 10.1242/jcs.102566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. 2008. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181:497–510. 10.1083/jcb.200712064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zoppino FC, Militello RD, Slavin I, Alvarez C, Colombo MI. 2010. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic 11:1246–1261. 10.1111/j.1600-0854.2010.01086.x [DOI] [PubMed] [Google Scholar]
- 56.Zeng X, Carlin CR. 2013. Host cell autophagy modulates early stages of adenovirus infections in airway epithelial cells. J. Virol. 87:2307–2319. 10.1128/JVI.02014-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim C, Bergelson JM. 2014. Echovirus 7 entry into polarized caco-2 intestinal epithelial cells involves core components of the autophagy machinery. J. Virol. 88:434–443. 10.1128/JVI.02706-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.