ABSTRACT
Activation of the ATM (ataxia telangiectasia-mutated kinase)-dependent DNA damage response (DDR) is necessary for productive replication of human papillomavirus 31 (HPV31). We previously found that DNA repair and homologous recombination (HR) factors localize to sites of HPV replication, suggesting that ATM activity is required to recruit factors to viral genomes that can productively replicate viral DNA in a recombination-dependent manner. The Mre11-Rad50-Nbs1 (MRN) complex is an essential component of the DDR that is necessary for ATM-mediated HR repair and localizes to HPV DNA foci. In this study, we demonstrate that the HPV E7 protein is sufficient to increase levels of the MRN complex and also interacts with MRN components. We have found that Nbs1 depletion blocks productive viral replication and results in decreased localization of Mre11, Rad50, and the principal HR factor Rad51 to HPV DNA foci upon differentiation. Nbs1 contributes to the DDR by acting as an upstream activator of ATM in response to double-strand DNA breaks (DSBs) and as a downstream effector of ATM activity in the intra-S-phase checkpoint. We have found that phosphorylation of ATM and its downstream target Chk2, as well as SMC1 (structural maintenance of chromosome 1), is maintained upon Nbs1 knockdown in differentiating cells. Given that ATM and Chk2 are required for productive replication, our results suggest that Nbs1 contributes to viral replication outside its role as an ATM activator, potentially through ensuring localization of DNA repair factors to viral genomes that are necessary for efficient productive replication.
IMPORTANCE The mechanisms that regulate human papillomavirus (HPV) replication during the viral life cycle are not well understood. Our finding that Nbs1 is necessary for productive replication even in the presence of ATM (ataxia telangiectasia-mutated kinase) and Chk2 phosphorylation offers evidence that Nbs1 contributes to viral replication downstream of facilitating ATM activation. Nbs1 is required for the recruitment of Mre11 and Rad50 to viral genomes, suggesting that the MRN complex plays a direct role in facilitating productive viral replication, potentially through the processing of substrates that are recognized by the key homologous recombination (HR) factor Rad51. The discovery that E7 increases levels of MRN components, and MRN complex formation, identifies a novel role for E7 in facilitating productive replication. Our study not only identifies DNA repair factors necessary for HPV replication but also provides a deeper understanding of how HPV utilizes the DNA damage response to regulate viral replication.
INTRODUCTION
Human papillomaviruses (HPVs) are small double-stranded DNA viruses that exhibit a strict tropism for epithelial cells (1). A subset of HPV types (termed high-risk HPVs) are the causative agents of cervical cancer and are also associated with other genital malignancies, as well as an increasing number of head and neck cancers (2). The life cycle of HPV is dependent upon the differentiation of its host cell, the keratinocyte. There are three phases of viral replication that characterize the viral life cycle (3). Upon infection of basal keratinocytes, the virus transiently amplifies to 50 to 100 episomal copies per cell. In undifferentiated cells, the virus is maintained at a low copy number by replicating once per cell cycle along with cellular DNA (4). In contrast, upon keratinocyte differentiation, the productive phase of the viral life cycle is activated, resulting in late gene expression, viral genome amplification to thousands of copies per cell, and virion assembly and release (1). Viral genome amplification is thought to occur through multiple rounds of replication following cellular DNA synthesis in cells arrested in an S- or G2-like environment (5–8), with some evidence indicating that this occurs through a switch to rolling circle replication (9). Although it is well established that the viral E7 protein promotes S-phase reentry of differentiating cells to provide cellular factors necessary for productive replication (10), the mechanisms that regulate the switch to viral genome amplification in differentiating cells are not well understood.
Over the past several years, it has become evident that DNA and RNA viruses facilitate replication by targeting the DNA damage response (DDR) (11). Previously, we showed that high-risk HPV31 induces constitutive activation of an ATM (ataxia telangiectasia-mutated kinase)-dependent DNA damage response throughout the viral life cycle (12). ATM is a member of the phosphatidylinositol 3-kinase-like kinase (PIK) family of kinases, which along with ATR (ataxia telangiectasia and Rad3-related protein) and DNA-PK, respond to certain types of DNA damage (13). ATM and DNA-PK are typically activated in response to double-strand DNA breaks (DSBs), while ATR is activated in response to single-stranded DNA breaks, as well as replication stress. Our previous studies demonstrated that HPV31 requires ATM kinase activity for productive replication upon differentiation but not for episomal maintenance in undifferentiated cells (12). In HPV31-positive cells, the ATM response was characterized by phosphorylation of downstream targets, including Chk2, Nbs1, and Brca1 (12). Similarly to inhibition of ATM, Chk2 inhibition also blocked productive replication, indicating an important role for ATM signaling specifically during the differentiation-dependent phase of the viral life cycle. How HPV activates ATM is currently unclear, though we have found that E7 expression alone is sufficient to induce activation of ATM targets (12), possibly through the induction of replication stress and DNA damage (14, 15). Recent studies by Hong and Laimins demonstrated that E7-induced STAT5 activation is necessary for ATM activation, possibly through peroxisome proliferator-activated receptor γ (PPARγ) expression (16). We, as well as others, have shown that expression of viral helicase E1 can also stimulate ATM activation (8, 17). Why HPV requires ATM activity for productive replication, as well as which ATM effectors contribute to viral DNA synthesis, is not well understood.
In more recent studies, we demonstrated that multiple components of the ATM DNA damage response pathway localize to sites of HPV replication, including γH2AX, Chk2, and 53BP1 and components of the MRN complex (Mre11, Rad50, and Nbs1) (18). In addition, we found that Rad51 and Brca1, two proteins necessary for the repair of DSBs through homologous recombination (HR) (19), are increased in expression in HPV-positive cells and localize to viral DNA foci. The localization of cellular replication factors, PCNA and RPA32, to HPV DNA foci indicated that these were sites of viral DNA synthesis. In addition, we observed that RPA32 is phosphorylated at sites of HPV replication, which is thought to redirect the function of replication protein A (RPA) from DNA replication to repair synthesis and has been linked to DSB resection (20–22). This suggests that HPV may utilize ATM signaling to recruit DNA repair machinery to viral genomes to promote replication through DNA repair. In support of this, we found that γH2AX is bound to viral chromatin throughout the viral life cycle, with binding increasing upon productive replication (18).
Increasing evidence suggests a role for recombination in HPV replication (23). Indeed, the localization of Rad51 and Brca1 to sites of HPV DNA synthesis suggests that productive replication may result in structures that require HR for processing. HR is a high-fidelity repair mechanism that requires ATM activity and functions to rejoin DSBs and restart broken replication forks (19, 24). Replication-coupled recombination is thought to play a role in the life cycles of several viruses, including simian virus 40 (SV40) (25), herpes simplex virus 1 (HSV-1) (26–28), Epstein-Barr virus (EBV) (29, 30), and Kaposi's sarcoma-associated herpesvirus (KSHV) (31). The MRN complex is also essential to homology-directed repair (32). The MRN complex serves as a sensor of DNA damage that also controls the DNA damage response (DDR) through activation of ATM (33–35). ATM is recruited to sites of DSBs by directly binding Nbs1, where ATM activates the DNA damage checkpoint and regulates DNA repair by phosphorylating specific substrates (36–38). In addition to facilitating ATM activation and recruitment to DSBs, Nbs1 also acts downstream as an effector of ATM activity and initiates HR with Mre11, a nuclease involved in resection of DNA ends (22, 39). The importance of Nbs1 in facilitating DNA repair is evident in patients with Nijmegen breakage syndrome (NBS), a disorder due to hypomorphic mutations in the Nbs1 gene, which is characterized by cellular radiosensitivity, cell cycle abnormalities, and a defective response to DNA damage (40–42). Several viruses have been shown to relocalize and/or degrade components of the MRN complex to facilitate viral replication (11). In addition, the SV40 large T antigen (43), as well as the HSV protein UL12 (44), has been shown to interact with MRN components. Our previous studies demonstrated that HPV-positive cells exhibit high levels of MRN components throughout the viral life cycle (12), which may be important in HPV's ability to activate ATM, and therefore contribute to efficient viral replication upon differentiation.
In this study, we investigate whether Nbs1 and maintenance of the MRN complex have an impact on the ability of HPV to efficiently replicate. We report here that HPV31 and HPV16 E7 bind to the MRN components Nbs1 and Rad50 but not Mre11. However, formation of the MRN complex is not disrupted and rather is increased compared to uninfected keratinocytes. We have found that Nbs1 is required for productive viral replication but not episomal maintenance. Depletion of Nbs1 results in a loss of Mre11 and Rad50 as well as Rad51 from sites of viral replication upon differentiation, suggesting that productive replication may occur through a mechanism dependent on recombination. Although phosphorylation of ATM and Chk2 is decreased in the absence of Nbs1, relatively high levels remain during the productive phase of the viral life cycle. Importantly, our results indicate that Nbs1 does not contribute to productive viral replication solely as an upstream regulator of ATM activity but rather has functions downstream as well, with Nbs1 potentially acting as an effector of ATM activity and/or ensuring efficient viral DNA synthesis through a recombination-dependent mechanism.
MATERIALS AND METHODS
Cell culture.
Human foreskin keratinocytes (HFKs) were collected from neonatal foreskin tissue as described previously (45) and were maintained in Dermalife keratinocyte growth medium (KGM; Lifeline Cell Technology). The human cervical carcinoma cell line C33A was grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% bovine growth serum (BGS) (Life Technologies). CIN612 9E cells, which are derived from a CIN1 biopsy specimen and stably maintain HPV31 genomes, were grown in E medium supplemented with 5 ng/ml mouse epidermal growth factor (BD Biosciences) and cocultured with mitomycin C-treated J2 3T3 fibroblasts, as described previously (45). The NBS-ILB1 fibroblast cell line was a generous gift from K. Cerosaletti and was described previously (46). NBS-ILB1 cells were maintained in DMEM with sodium pyruvate supplemented with 10% BGS. Generation and maintenance of cells stably expressing pLXSN or pLXSN-HPV31 E7 or pLXSN-HPV16 E7 through retroviral transduction have been previously described (47). When necessary, J2 feeders were removed from HPV-positive cells by incubation with 1 mM EDTA in phosphate-buffered saline (PBS). U20S and 293T cells were grown in DMEM supplemented with 10% BGS.
Plasmids and chemicals.
The pBR322 min-HPV31 plasmid has been described previously (48). The hemagglutinin (HA)-tagged HPV31 E7 proteins were previously described (49) and are as follows: HA-E7 ΔLHCYE contains an in-frame deletion of the Rb binding domain, and the HA-E7 L67R construct contains a point mutation in the histone deacetylase (HDAC) binding site, converting a leucine to an arginine. The TAP-tagged HPV16 E7 construct was generously provided by J. Bodily and was described previously (50). The retroviral plasmids pLXIN and pLXIN-Nbs1 and those expressing Nbs1 truncation mutants (Nbs1 652 and Nbs1 ΔATM) were kind gifts from P. Concannon and were described previously (51, 52). The pLXIN-Nbs1 FR5 deletion construct was obtained from K. Cerosaletti and was described previously (52). The Mre11 binding mutant was also obtained from K. Cerosaletti and was generated by site-directed mutagenesis (QuikChange; Agilent Technologies) based on the identification of the Mre11 binding domain by You et al. (53), resulting in an NFKK684-687AAAA conversion. KU-55933 was obtained from Calbiochem, and Mirin was obtained from Tocris.
Generation of HPV31-positive HFKs.
HFKs stably maintaining HPV31 episomes were created as previously described (45). Briefly, HPV31 genomes were excised from the pBR322 plasmid using HindIII (New England BioLabs) and religated using T4 DNA ligase (Life Technologies). Primary HFKs were transfected with 1 μg of the ligated genomes and 1 μg pSV2-Neo using FuGene 6 according to the manufacturer's instructions (Promega). Stable cell lines were generated through neomycin selection (Sigma-Aldrich). After selection was complete, pooled populations were expanded for further analysis.
Induction of keratinocyte differentiation.
For differentiation, 1.5% methylcellulose was used as described previously (54). Cells were harvested at T0 (undifferentiated), as well as 24 and 48 h postsuspension. High-calcium medium was also used to induce differentiation as previously described (7). Cells were harvested at T0, as well as 48, 72, or 96 h after exposure to high calcium. For both methylcellulose and calcium, at each time point DNA was harvested from one half of the cells, and protein was harvested from the other half. For every experiment, viral genome amplification was measured by Southern blotting to ensure activation of the productive phase of the viral life cycle.
Generation of lentivirus.
Lentivirus was produced as previously described (55). Plasmids carrying an Nbs1 short hairpin RNA (shRNA) (TRCN0000010393) or a Scramble nontarget control shRNA in the pLKO background were obtained from Open Biosystems (Pittsburgh, PA). Each of these plasmids (5 μg) was cotransfected with 1.6 μg vesicular stomatitis virus G plasmid DNA and 3.37 μg Gag-Pol-Tet-Rev plasmid DNA into 293T cells using polyethyleneimine (PEI) (VWR). Supernatants containing lentivirus were harvested 3 days posttransfection, filter sterilized, and stored at −80°C until use. CIN612 9E and HFK-31 cells were transduced with 5 ml viral supernatant consisting of Scramble or Nbs1 shRNA lentivirus particles in the presence of 4.8 μg/ml hexadimethrine bromide (Polybrene) (Sigma-Aldrich) for 3 days, followed by selection in puromycin to generate stable cell lines. Knockdown of Nbs1 was confirmed for each experiment by Western blotting.
Nuclear/cytoplasmic fractionation.
Fractionation was carried out according to the methods of Schreiber et al. (56). Briefly, after being washed in cold PBS, cells were resuspended in 400 μl Schreiber buffer A (10 mM HEPES, 0.4 M NaCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride [PMSF]) and swollen on ice for 15 min. The cells were then lysed by the addition of 25 μl 10% NP-40 and vortexing for 10 s. The nuclei were subsequently pelleted by centrifugation at 4°C. The supernatant containing the cytoplasmic extract was removed, and the nuclei were lysed by addition of 50 μl Schreiber buffer C (20 mM HEPES, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, 0.5 mM PMSF) and shaking for 15 min at 4°C. The soluble nuclear fraction was separated by centrifugation for 5 min at 4°C. Purity of nuclear and cytoplasmic fractions was determined by Western blotting using antibodies to lamin A/C (GeneTex) and tubulin (Sigma-Aldrich), respectively.
Western blot analysis/immunoprecipitation.
For Western blotting, whole-cell lysates were harvested in radioimmunoprecipitation assay (RIPA) lysis buffer supplemented with Complete Mini and PhosSTOP tablets (Roche). Total protein levels were determined by the Bio-Rad protein assay. Western blot analysis was performed as described previously (49). Equal protein amounts were electrophoresed on SDS-polyacrylamide gels and transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore). For immunoprecipitations, cells were harvested in 1× cell lysis buffer (Cell Signaling), as described previously (12). The following primary antibodies were used: anti-mouse HA, anti-rabbit HA, anti-goat Nbs1, anti-mouse involucrin, anti-mouse Ku70, anti-mouse CDC25C, anti-mouse cyclin A, anti-rabbit cyclin B, anti-rabbit RPA32, and anti-mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Santa Cruz); anti-mouse cyclin E (Pharmingen); anti-mouse HPV16 E7 (Life Technologies); anti-rabbit Nbs1 and phospho-Nbs1 S343 (Novus Biologicals); anti-rabbit phospho-ATM S1981, anti-mouse CDK2, and anti-rabbit PCNA (Abcam); anti-rabbit phospho-Chk2 Thr68 and Chk2, anti-rabbit CDK1, and anti-rabbit SMC1 (Cell Signaling Technologies); anti-rabbit ATM and anti-rabbit phospho-SMC1 Ser966 (Bethyl Laboratories); anti-mouse lamin A/C, anti-mouse Mre11, and anti-rabbit Rad50 (GeneTex); anti-mouse tubulin (Sigma-Aldrich). Secondary antibodies used were horseradish peroxidase (HRP)-conjugated anti-goat (Santa Cruz), HRP-conjugated anti-rabbit (Cell Signaling Technologies), and HRP-conjugated anti-mouse (GE Life Sciences) antibodies.
Southern blot analysis.
DNA isolation and Southern blotting were performed as previously described (54). Briefly, cells were harvested in buffer containing 400 mM NaCl, 10 mM Tris (pH 7.5), and 10 mM EDTA. Cells were lysed by the addition of 30 μl of 20% SDS and subsequently treated with 15 μl of 10-mg/ml proteinase K overnight at 37°C. DNA was then extracted by phenol chloroform and precipitated using sodium acetate and ethanol. Resultant DNAs were digested with BamHI (which does not cut the HPV31 genome) or HindIII (which cuts the HPV31 genome once). DNAs were resolved on a 0.8% agarose gel and transferred to a positively charged nylon membrane (GeneScreen Plus; PerkinElmer). Hybridization was performed using 32P-labeled HPV31 genome as a probe.
IF and FISH.
CIN612 9E cells stably expressing the Scramble or Nbs1 shRNA were grown on coverslips and harvested at T0 or after 72 h of differentiation in high-calcium medium. At the indicated times, the cells were fixed with 4% paraformaldehyde (PFA) for 10 min and then permeabilized with 1% Triton X-100 in phosphate-buffered saline (PBS) for 10 min, followed by three washes with PBS. Coverslips were blocked with 3% bovine serum albumin (BSA) in PBS for 30 min and incubated with primary antibodies in 3% BSA-PBS overnight at 4°C in a humidified chamber. Coverslips were washed three times with PBS and incubated with secondary antibodies for 1 h, followed by three washes with PBS. Coverslips were cross-linked with cold methanol-acetic acid (3:1) at −20°C for 10 min, followed by fixation with 4% PFA for 10 min at room temperature. Coverslips were then analyzed for HPV DNA by fluorescence in situ hybridization (FISH) using tyramide-enhanced fluorescence (Invitrogen) as previously described (12, 57). The coverslips were mounted using Vectashield containing 4′,6-diamidino-2-phenylindole (DAPI) to counterstain the cellular DNA. Images were captured using a Zeiss CLSM 710 spectral confocal laser scanning microscope. For immunofluorescence (IF), the following antibodies were used: mouse monoclonal anti-Mre11 (1:200; GeneTex), mouse monoclonal anti-Rad50 (1:200; GeneTex), and rabbit polyclonal Rad51 (1:200; Santa Cruz). For the secondary antibody, Alexa Fluor 568 goat anti-mouse antibody was used (Life Technologies). IF-FISH was carried out for each repair factor on three independent experiments. The number of foci positive for both HPV DNA and each repair factor was quantified, with 25 to 40 FISH-positive cells being counted for each experiment.
Real-time PCR.
RNA was isolated from HFKs, as well as HFKs stably expressing pLXSN-HPV31 or -HPV16 E7 using RNA Stat 60 (Tel-Test), followed by treatment with DNase (Promega) according to the manufacturer's instructions. RNA was reverse transcribed using the iScript reverse transcription kit (Bio-Rad). Fifty nanograms of cDNA was analyzed in triplicate reactions using quantitative PCR (qPCR) with 375 nM primers and iTaq Universal SYBR green Supermix (Bio-Rad) in a total reaction volume of 20 μl. Reactions were carried out in an ABI 7500 thermal cycler with a thermal profile of 3 min at 95°C, 40 cycles of 95°C for 15 s, and then 30 s at 60°C, followed by a melting curve to ensure proper annealing. The results were analyzed using version 2.0.5 of the ABI 7500 software application. The following gene-specific primer sequences were used: for Mre11, forward, 5′-GCCTTCCCGAAATGTCACTA-3′, and reverse, 5′-TTCAAAATCAACCCCTTTCG-3′; for Rad50, forward, 5′-GGAAGAGCAGTTGTCCAGTTACG-3′, and reverse, 5′-GAGTAAACTGCTGTGGCTCCAG-3′; for Nbs1, forward, 5′-CACCTCCAAAGACAACTGCGGA-3′, and reverse, 5′-TCTGTCAGGACGGCAGGAAAGA-3′; for GAPDH, forward, 3′-CTGTTGCTGTAGCCAAATTCGT-5′, and reverse, 3′-ACCCACTCCACCTTTGAC-5′. Relative transcript amounts were calculated using the threshold cycle (ΔΔCT) method using GAPDH as a reference gene.
RESULTS
Expression of high-risk HPV E7 increases levels of repair factors, including the MRN complex.
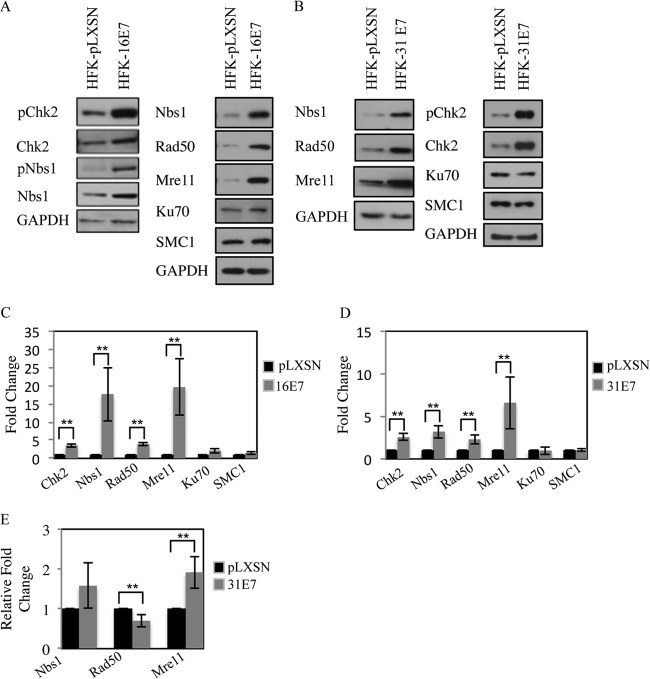
Previously, we demonstrated that HPV31 E7 is sufficient to induce phosphorylation of the ATM target Chk2 (12). To determine if this observation extends to HPV16 E7, we examined Chk2 phosphorylation in human foreskin keratinocytes (HFKs) stably expressing pLXSN-HPV16 E7 or vector control. As shown in Fig. 1A, we found that the expression of HPV16 E7, similarly to HPV31 E7 (Fig. 1B), is sufficient to induce Chk2, as well as Nbs1 phosphorylation, and also significantly increases total levels of Chk2 (Fig. 1C and D). Our previous studies also revealed that the MRN components Mre11, Rad50, and Nbs1 are expressed at increased levels in HPV-positive cells compared to uninfected keratinocytes (12). To determine if E7 could be responsible for increased levels of the MRN components in infected cells, we examined the levels of Mre11, Rad50, and Nbs1 in keratinocytes stably expressing HPV16 E7 or HPV31 E7. As shown in Fig. 1, expression of both HPV16 E7 and HPV31 E7 resulted in a significant increase in protein levels of Mre11, Rad50, and Nbs1; however, the repair proteins Ku70 and SMC1 were not affected (Fig. 1A to D). To determine if the increase in MRN components was regulated transcriptionally, we performed quantitative real-time PCR on RNA extracted from HFKs stably expressing HPV31 E7 or vector control. As shown in Fig. 1E, although expression of HPV 31E7 resulted in increased transcript levels of Mre11 and Nbs1, only Mre11 was significantly changed. In contrast, Rad50 was significantly decreased at the transcript level in HPV31 E7 cells compared to the vector control. Since Mre11 stabilizes Rad50 and Nbs1 through binding (58, 59), it is possible that the E7-mediated increase in Mre11 transcript and protein levels is sufficient to maintain elevated levels of Rad50 and Nbs1. Overall, these results suggest that in addition to activation of ATM signaling, E7 may be necessary to provide essential DNA repair factors for viral replication.
FIG 1.
Expression of HPV E7 increases levels of proteins associated with detection and repair of DNA damage. (A) Whole-cell extracts were harvested from HFKs stably expressing pLXSN vector control or pLXSN-HPV16 E7. Immunoblotting was performed using antibodies to phosphorylated Chk2 (Thr68) (pChk2), total Chk2, phosphorylated Nbs1 S343 (pNbs1), total Nbs1, Rad50, Ku70, and SMC1. (B) Lysates harvested from HFK-pLXSN or HFK-pLXSN HPV31 E7 cells were analyzed by Western blotting using antibodies to pChk2, Chk2, Nbs1, Rad50, Mre11, Ku70, and SMC1. GAPDH served as a loading control. (C and D) Bar graphs demonstrate the average expression level of target proteins, normalized to GAPDH, in at least four independent Western blot analyses, including data from panels A and B. Densitometry was performed using ImageJ software. The statistical analysis was assayed by 2-tailed t test. Data are means ± standard errors. **, P value less than 0.05. (E) Quantitative real-time PCR of gene expression analysis in HFK-pLXSN and HFK-pLXSN-31E7 lines. Expression levels are shown relative to HFK-pLXSN cells and were calculated using GAPDH as a reference gene. Shown is the relative fold change in gene expression over three independent experiments. The statistical analysis was assayed by 2-tailed t test. Data are means ± standard errors. **, P value less than 0.05.
E7 binds to the MRN components Nbs1 and Rad50.
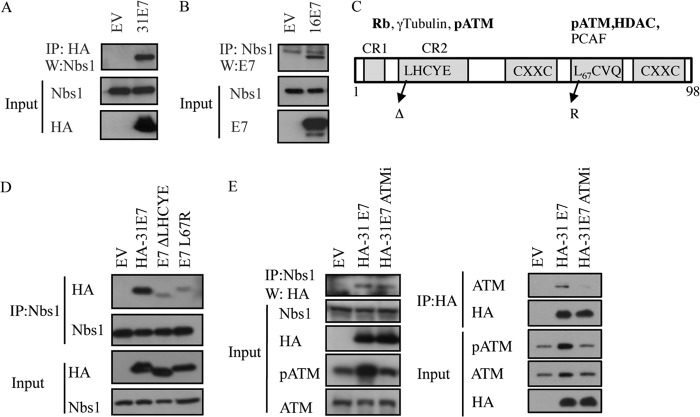
In previous studies, we found that HPV31 E7 binds preferentially to the phosphorylated form of ATM (12), potentially due to the exposure of the E7 binding site upon dissociation of inactive ATM dimers to active monomers. Since Nbs1 binds ATM for recruitment to DSBs, we wanted to determine if E7 also binds Nbs1. For this, we immunoprecipitated HA-tagged HPV31 E7 from transiently transfected U20S cells and performed Western blotting for endogenous Nbs1. As shown in Fig. 2A, Nbs1 coimmunoprecipitates with HPV31 E7, and we found similar results when endogenous Nbs1 was immunoprecipitated, followed by Western blotting for HPV16 E7 (Fig. 2B) or HPV31 E7 (Fig. 2D and E). The E7-Nbs1 interaction does not appear to occur nonspecifically through DNA, as immunoprecipitation in the presence of ethidium bromide did not affect the ability of E7 to bind Nbs1 (data not shown).
FIG 2.
HPV E7 interacts with Nbs1 independently of ATM. (A) Whole-cell lysates of U20S cells transiently transfected with HA-HPV31 E7 or empty vector (EV) were immunoprecipitated with antibodies to HA or Nbs1, followed by immunoblotting with an antibody to Nbs1 or HA. (B) Whole-cell lysates of U20S cells transiently transfected with TAP-HPV16 E7 or empty vector (EV) were immunoprecipitated using an anti-Nbs1 antibody, followed by immunoblotting with an HPV16 E7 antibody. Inputs were analyzed using antibodies to Nbs1 and HA (A) and Nbs1 and HPV16E7 (B). (C) Structure of E7 and mutations examined in this study. Indicated are cellular targets that have been shown to interact with the Rb (LHCYE) and HDAC (L67) binding domains. Targets in bold have been identified as binding to HPV31 E7 (97). (D) Whole-cell lysates of U20S cells transiently transfected with EV alone, HA-HPV31 E7, HA-HPV31 E7 ΔLHCYE, or HA-HPV31 E7 L67R were immunoprecipitated using an anti-Nbs1 antibody and subsequently immunoblotted using an anti-HA or anti-Nbs1 antibody. Input lysates were analyzed by Western blotting using antibodies to HA and Nbs1. (E) Whole-cell lysates of U20S cells expressing EV alone or HA-HPV31 E7 in the presence or absence of 10 μM ATM inhibitor KU-55933 were immunoprecipitated using an anti-Nbs1 antibody and immunoblotted using an anti-HA antibody (left panel). Input lysates were immunoblotted with antibodies to Nbs1 and HA as well as phosphorylated ATM (Ser1981) and total ATM to demonstrate ATM inhibition. Lysates were also subjected to immunoprecipitation with an antibody to HA, followed by immunoblotting to ATM and HA. Inputs were analyzed using an antibody to HA, as well as phosphorylated and total ATM to demonstrate ATM inhibition. Ab, antibody; IP, immunoprecipitation; W, immunoblotting; ATMi, KU-55933. All results are representative of observations from three or more independent experiments.
In order to map the Nbs1 interaction domain on HPV31 E7, we used previously characterized mutants that block the binding of either Rb (ΔLHCYE) or histone deacetylases (HDACs) (L67R) (Fig. 2C) (49). As shown in Fig. 2D, both the E7 Rb binding mutant and the HDAC binding mutant were deficient in immunoprecipitating Nbs1. Since we previously found that ATM also binds E7 through these two domains (12), we wanted to next determine if E7 interacts with Nbs1 indirectly through its binding to ATM. For this, we treated cells with the small-molecule inhibitor of ATM, KU-55933, which inhibits ATM phosphorylation and ablates the ability of E7 to bind ATM (Fig. 2E) (12). However, we found that Nbs1 was still able to immunoprecipitate E7, even when the E7-ATM interaction was disrupted (Fig. 2E), indicating that E7 binds Nbs1 independently of ATM.
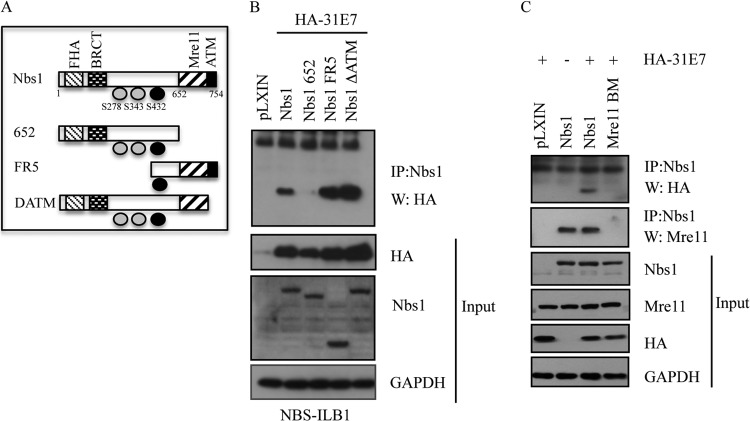
To map the HPV31 E7 interaction site on Nbs1, we utilized a series of previously characterized Nbs1 deletion mutants (Fig. 3A) (51, 52). For these studies, we used the NBS-ILB1 fibroblast cell line that is hypomorphic for Nbs1 and produces undetectable levels of a 70-kDa C-terminal polypeptide (46, 60). NBS-ILB1 cells were retrovirally transduced with wild-type (WT) Nbs1, a mutant that lacks the C terminus (Nbs1 652), a mutant that lacks the N terminus (FR5), or a mutant that lacks the ATM binding site (ΔATM), and stable cell lines were generated through neomycin selection. Immunoprecipitation of Nbs1 from lysates of cells containing the pLXIN vector, WT Nbs1, or the mutants revealed that the C terminus of Nbs1 is required to immunoprecipitate E7 (Fig. 3B). Importantly, we found that E7 was still able to coimmunoprecipitate with the Nbs1 mutant lacking the ATM binding domain, confirming our results with the ATM inhibitor (Fig. 3B). The C terminus of Nbs1 contains an Mre11 binding domain (52, 53), in addition to an ATM binding site (38, 53, 61). To determine if the Mre11 binding domain of Nbs1 is required for immunoprecipitation of E7, we utilized a construct that is mutated in the Mre11 binding site. As shown in Fig. 3C, mutation of the Mre11 binding site abrogated the ability of E7 to immunoprecipitate with Nbs1, suggesting that E7 interacts with Nbs1 through the Mre11 binding domain, though it is currently unclear if this occurs in a direct or indirect manner.
FIG 3.
HPV31 E7 interacts with Nbs1 through the Mre11 binding domain. (A) Schematic of Nbs1 constructs depicting relevant binding domains and phosphorylation sites. (B) NBS-ILB1 cells stably expressing pLXIN vector alone or the indicated Nbs1 mutants were transiently transfected with HA-HPV31 E7. Immunoprecipitations were performed using an antibody to Nbs1, followed by Western blotting using an antibody to HA. Western blot analysis was performed on the indicated input lysates using antibodies to HA, Nbs1, and GAPDH. (C) NBS-ILB1 cells stably expressing pLXIN vector alone, pLXIN-Nbs1, or a pLXIN-Nbs1 Mre11 binding mutant (Mre11 BM) were transiently transfected with HA-HPV31 E7. Immunoprecipitation of whole-cell lysates was performed using an antibody to Nbs1 followed by Western blotting with an antibody to HA and Mre11. Western blot analysis was performed on the indicated input lysates using antibodies to HA, Nbs1, Mre11, and GAPDH. IP, immunoprecipitation; W, immunoblotting. All results are representative of observations of three independent experiments.
To form the MRN complex, Nbs1 binds Mre11, which binds to Rad50 (40, 62). Since we were able to immunoprecipitate Nbs1 with E7, we next wanted to determine if E7 interacts with other components of the MRN complex. For this, we immunoprecipitated HA-tagged HPV31 E7 or TAP-tagged HPV16 E7 from lysates harvested from U20S cells and performed Western blot analysis for Mre11 and Rad50. Interestingly, as shown in Fig. 4A, we found that both HPV31 E7 and HPV16 E7 could immunoprecipitate with Rad50, but neither interacted with Mre11. However, we found that the presence of E7 did not disrupt formation of the MRN complex, as Nbs1 was still able to interact with Rad50 and Mre11 (Fig. 4B). Rather, our results indicate that the presence of E7 increases the formation of the MRN complex, which may result from increased Mre11 expression. We also found that the formation of the MRN complex is not disrupted in HPV-infected cells (see Fig. 8B) or cells stably expressing HPV31 E7 (data not shown).
FIG 4.
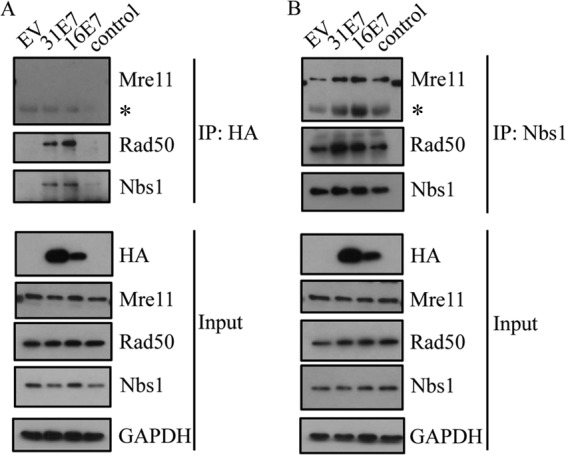
HPV E7 interacts with Nbs1 and Rad50 but not Mre11. (A) U20S cells were transiently transfected with vector alone (EV), HA-HPV31 E7, and TAP-HPV16 E7, and immunoprecipitations of whole-cell lysates were performed using an HA antibody, followed by immunoblotting with Mre11, Rad50, and Nbs1 antibodies. (B) U20S cells were transiently transfected with EV alone, HA-HPV31 E7, or TAP-HPV16 E7. Immunoprecipitations were performed on lysates using an antibody to Nbs1, followed by Western blotting using antibodies to Mre11, Rad50, and Nbs1 antibodies. For panels A and B, input lysates were analyzed by Western blotting using antibodies to HA, Mre11, Rad50, and Nbs1. GAPDH served as a loading control. IP, immunoprecipitation; *, antibody heavy chain. All results are representative of observations from three independent experiments.
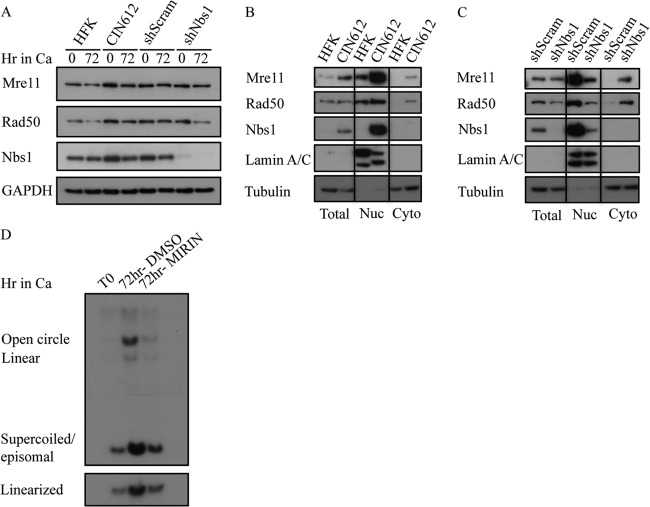
FIG 8.
Nbs1 knockdown disrupts MRN complex formation. (A) Whole-cell lysates were harvested from HFK, CIN612, and CIN612 9E cells stably expressing shScramble or shNBS1 at T0 and after 72 h of differentiation in high-calcium medium (Ca). Immunoblotting was performed using Mre11, Rad50, and Nbs1 antibodies. GAPDH was used as a loading control. (B) Total, nuclear (Nuc), and cytoplasmic (Cyto) lysates were harvested from HFK and CIN612 cells. Immunoblotting was performed using Mre11, Rad50, and Nbs1 antibodies. (C) Total, nuclear (Nuc), and cytoplasmic (Cyto) lysates were harvested from stable CIN612 9E shScramble and CIN612 9E shNBS1 cells. Immunoblotting was performed using Mre11, Rad50, and Nbs1 antibodies. For panels B and C, lamin A/C and tubulin were used to confirm nuclear and cytoplasmic fractionation, respectively. (D) DNA was harvested from CIN612 9E cells at T0 and after 72 h of differentiation in high-calcium medium with dimethyl sulfoxide (DMSO) as a vehicle control or 50 μM Mre11 inhibitor Mirin. Southern blot analysis was performed to analyze viral genome amplification of DNA digested with BamHI (nonviral genome cutter; upper panel) or HindIII (cuts viral genome once; lower panel). All results are representative of observations of two or more independent experiments.
Nbs1 is necessary for productive viral replication.
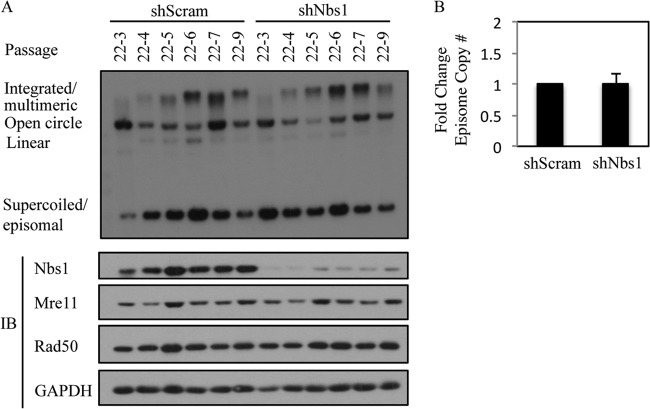
Previously, we demonstrated that inhibition of ATM kinase activity has minimal effect on the ability of HPV to be maintained as an episome (12). However, in addition to being essential for ATM activation in response to DSBs, Nbs1 can also mediate ATR activation (63, 64), which could potentially be important for episomal maintenance. To examine the effect of Nbs1 depletion on episomal maintenance, we transduced HPV31-positive CIN612 9E cells with a Scramble control shRNA or a previously validated Nbs1 shRNA (65) and generated stable cell lines. The cells were routinely passaged, and DNA and protein were harvested at every passage. Southern blot analysis was performed to examine the status of the episomal viral DNA. As shown in Fig. 5 (see also Fig. 12A), in two independently derived CIN612 9E lines, there was no significant difference in the ability of viral episomes to be maintained across passages in cells containing either the Scramble control or the Nbs1 shRNA. Similar results were observed with four independent experiments (Fig. 5B). These results mirror what we previously observed upon inhibition of ATM activity and indicate that Nbs1 is not necessary for episomal maintenance. In addition, we observed no effect of Nbs1 knockdown on Chk1 phosphorylation (data not shown).
FIG 5.
Nbs1 is not necessary for HPV31 genome maintenance. (A) DNA was isolated at the indicated passages from CIN612 9E cells stably maintaining a Scramble shRNA (shScram) or Nbs1 shRNA (shNbs1) and analyzed by Southern blotting. Passage 22 (p22) represents the passage at which the CIN612 9E cells were transduced with the respective shRNAs. Each passage following transduction is represented by p22-#. Western blot analysis was performed on lysates harvested at each passage using an antibody to Nbs1 to demonstrate knockdown, as well as antibodies to Mre11 and Rad50. GAPDH served as a loading control. IB, immunoblotting. (B) Bar graph represents episome copy number quantified and averaged across the passages of cells containing Scramble shRNA (set at 1) compared to passages of Nbs1 shRNA-containing cells. The data represent the averages of four independent experiments. Densitometry was performed using ImageJ software. The statistical analysis was assayed by 2-tailed t test. Data are means ± standard errors.
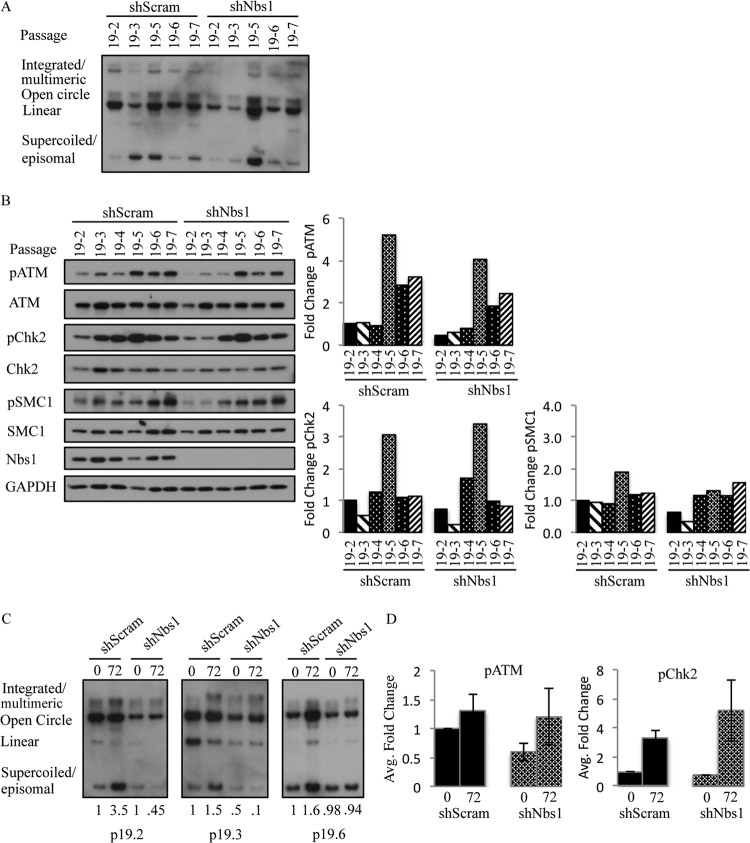
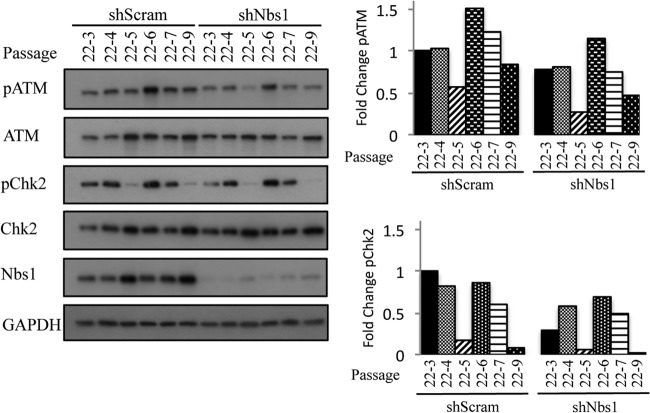
FIG 12.
Levels of phosphorylated ATM and Chk2 do not influence viral genome amplification. (A) DNA was harvested from CIN612 9E cells stably expressing shScramble and shNbs1 at the indicated passages and digested with BamHI (nonviral genome cutter). HPV genomes were visualized via Southern blot analysis. Passage 19 (p19) represents the passage at which the CIN612 9E cells were transduced with the indicated shRNAs. p19-# indicates that passage at which DNA and protein were harvested following transduction. (B) Whole-cell lysates were harvested from CIN612 9E shScramble and shNbs1 cells at the indicated passages, and immunoblotting was performed with antibodies to phosphorylated ATM (Ser1981) (pATM), phosphorylated Chk2 (Thr68) (pChk2), phosphorylated SMC1 (Ser966) (pSMC1), and total ATM, Chk2, SMC1, and Nbs1. GAPDH was used as a loading control. Protein levels were quantified using ImageJ, with phosphorylated protein levels normalized first to total levels and then to GAPDH. Levels are graphed as fold change compared to the first passage (p19-2) of the shScramble cells, which is set to 1. (C) DNA was harvested from p19-2, p19-3, and p19-6 CIN612 9E shScramble and shNbs1 cells at T0 and after differentiation for 72 h in high-calcium medium. Southern blot analysis was performed to analyze viral genome amplification. Levels of HPV episomes were quantified using ImageJ; episome amounts are shown relative to the T0 shScramble, which is set to 1. (D) Protein was harvested from p19-2, p19-3, and p19-6 CIN612 9E shScramble and shNbs1 cells at T0 as well as after 72 h of differentiation in high-calcium medium. Western blot analysis was performed using antibodies to phosphorylated ATM (S1981) (pATM) and phosphorylated Chk2 (Thr68) (pChk2). GAPDH served as a loading control. Protein levels were quantified using ImageJ, as described above. The average fold change in pATM and pChk2 levels across the three passages in undifferentiated shScram and shNbs1 cells compared to cells differentiated in high-calcium medium for 72 h is graphed. Fold change is shown relative to shScram T0, which is set to 1. Results shown are representative of observations of three or more independent experiments.
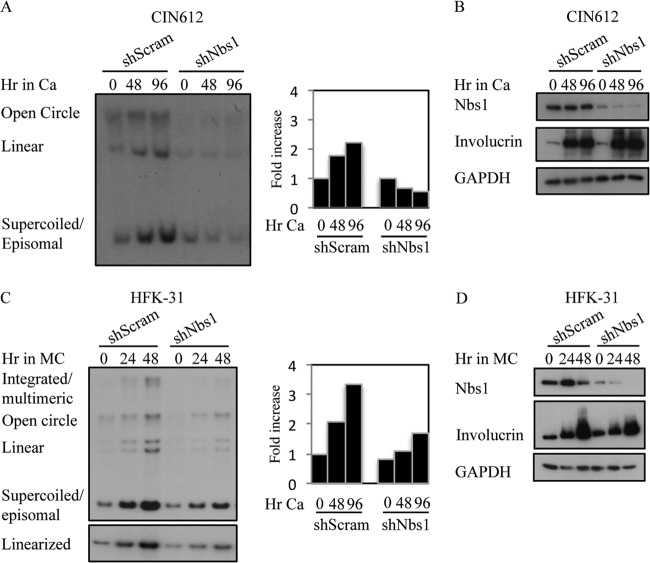
Since we previously found that ATM activity is necessary for productive viral replication, and given that Nbs1 is necessary for ATM activation in response to DSBs (32, 35, 36), we next wanted to determine the effect of Nbs1 depletion on productive replication. For this, CIN612 9E cells stably maintaining the Scramble or Nbs1 shRNAs were induced to differentiate in high-calcium medium, which activates the productive phase of the viral life cycle by 48 h postexposure (66). As shown in Fig. 6A, Southern blot analysis for HPV DNA demonstrated that cells containing the Nbs1 shRNA were greatly inhibited in their ability to undergo viral genome amplification compared to the Scramble shRNA control, exhibiting a decrease in episomal copy number upon differentiation. This experiment was performed at least three times with three independently derived CIN612 9E-Scramble and Nbs1 shRNA lines with similar results. In addition, we found that Nbs1 knockdown also decreased productive replication of human foreskin keratinocytes stably maintaining HPV31 genomes (HFK-31) after differentiation in methylcellulose (Fig. 6C). Importantly, Nbs1 knockdown did not inhibit the ability of HPV-positive cells to differentiate in high-calcium medium or methylcellulose as indicated by the expression of the differentiation-specific marker involucrin (Fig. 6B and D). In addition, as shown in Fig. 7, Nbs1 knockdown had minimal effect on the level of cellular factors involved in replication and cell cycle regulation, including the S-phase cyclins (cyclin A and cyclin E) and CDK (CKD2) (Fig. 7A), as well as mitotic cyclin B, the M-phase CDK (CDK1), and the CDC25C phosphatase (Fig. 7B). In addition, both RPA and PCNA were maintained at levels in the Nbs1 shRNA cells similar to those in the Scramble control (Fig. 7C). These results suggest that the block in productive viral replication observed in response to Nbs1 knockdown was not due to alterations in cell cycle control or the lack of cellular factors directly required for viral replication. We also observed similar effects on productive viral replication following transient knockdown of Nbs1 for 3 days (data not shown). Overall, these results indicate that Nbs1 is specifically necessary for differentiation-dependent viral replication.
FIG 6.
Nbs1 is necessary for productive viral replication. (A) DNA was harvested from CIN612 9E cells stably expressing a Scramble shRNA or Nbs1 shRNA at T0 (undifferentiated) or after 48 and 96 h of differentiation in high-calcium medium. Southern blot analysis was performed to analyze viral genome amplification. The bar graph represents quantification of the episome copy number present at each time point, relative to T0 shScramble, which was set to 1. Densitometry was performed using ImageJ. Ca, calcium. (B) Total protein was harvested from CIN612 9E shScramble and shNbs1 cells at T0 or after 48 and 96 h of differentiation in high-calcium medium. Western blot analysis was performed using antibodies to Nbs1 and involucrin, and GAPDH as a loading control. (C) DNA was harvested from human foreskin keratinocytes stably maintaining HPV31 genomes (HFK-31) at T0 or after 24 and 48 h of differentiation in methylcellulose (MC) and analyzed by Southern blotting for amplification of viral genomes. DNA samples were digested with BamHI (does not cut the viral genome; upper panel) or with HindIII to linearize viral genomes. The bar graph represents quantification of the episome copy number present at each time point, relative to T0 shScramble (set to 1). Densitometry was performed using ImageJ. (D) Western blot analysis was performed on lysates harvested from HFK-31 cells at T0 or after 24 and 48 h of differentiation in methylcellulose using antibodies to Nbs1 and involucrin. GAPDH was used as a loading control. All results are representative of observations of four or more independent experiments.
FIG 7.
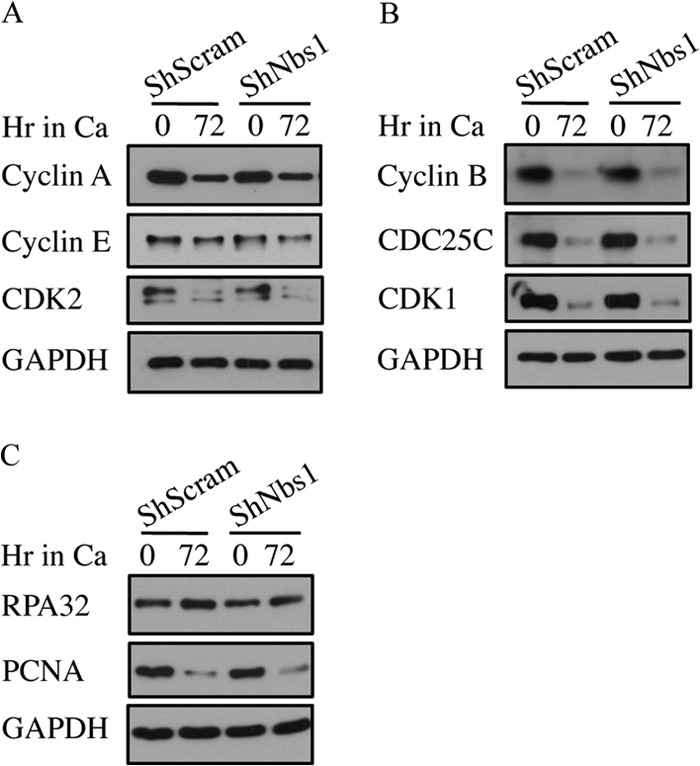
Levels of cell cycle or replication proteins are not affected by Nbs1 knockdown. Western blot analysis was performed on lysates harvested from CIN612 9E cells stably maintaining the Scramble (shScram) or Nbs1 shRNAs at T0, or after 72 h of differentiation in high-calcium medium using antibodies to cyclin A, cyclin E, and CDK2 (A); cyclin B, CDC25C, and CDK1 (B); and RPA (C), as well as PCNA. GAPDH served as a loading control. All results are representative observations of at least three independent experiments. Ca, calcium.
The MRN complex is disrupted upon Nbs1 knockdown.
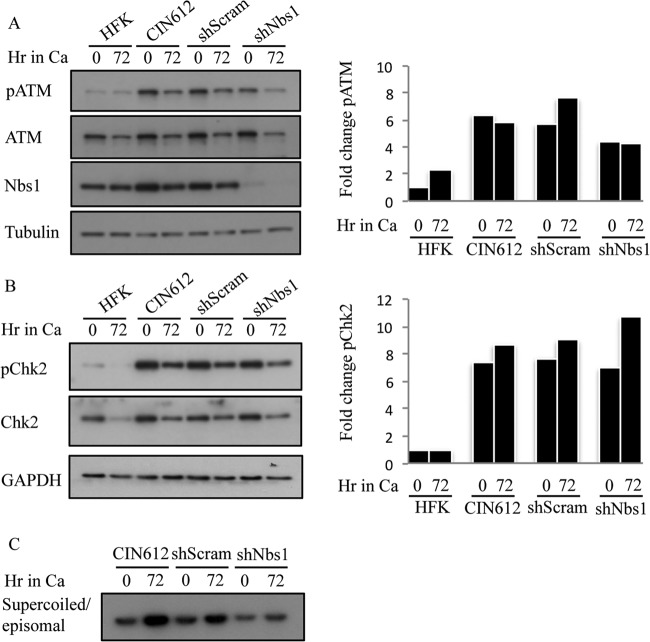
As Nbs1 functions in a complex with Mre11 and Rad50, we next wanted to determine if Nbs1 depletion affected the levels of Mre11 and Rad50. As shown in Fig. 5A and 8A, Nbs1 knockdown had little effect on the levels of Mre11 or Rad50, as measured by Western blotting, consistent with previous studies (58). Although Rad50 levels appear to decrease after 72 h in high-calcium medium (Fig. 8A), this result was not consistently observed. Since Nbs1 contains a nuclear localization signal and is necessary for the localization of Mre11 and Rad50 to the nucleus (52, 67), we next wanted to determine if Nbs1 depletion affected Mre11 and Rad50 localization. As shown in Fig. 8B, CIN612 9E cells exhibited increased levels of Mre11, Rad50, and Nbs1 in the nucleus compared to uninfected HFKs, indicating increased MRN complex formation, which is consistent with our immunoprecipitation results (Fig. 4B). However, upon depletion of Nbs1, we observed a dramatic decrease in levels of Mre11 and Rad50 in the nucleus of CIN612 9E cells and a marked relocalization to the cytoplasm (Fig. 8C), indicating disruption of the MRN complex, as previously shown in NBS fibroblasts (40). These results raise the possibility that formation of the MRN complex, rather than Nbs1 alone, is crucial to productive viral replication. To test this, we examined the effect of Mirin, an inhibitor of Mre11 nuclease activity, on productive replication of CIN612 9E cells. As shown in Fig. 8D, inhibition of Mre11 by Mirin resulted in a decreased ability of viral episomes to amplify upon differentiation, similarly to Nbs1 depletion. Similar results were observed in three independent experiments. Overall, these results indicate that the MRN complex is required for productive viral replication.
Nbs1 knockdown affects the localization of Mre11, Rad50, and the HR factor Rad51 to HPV DNA foci.
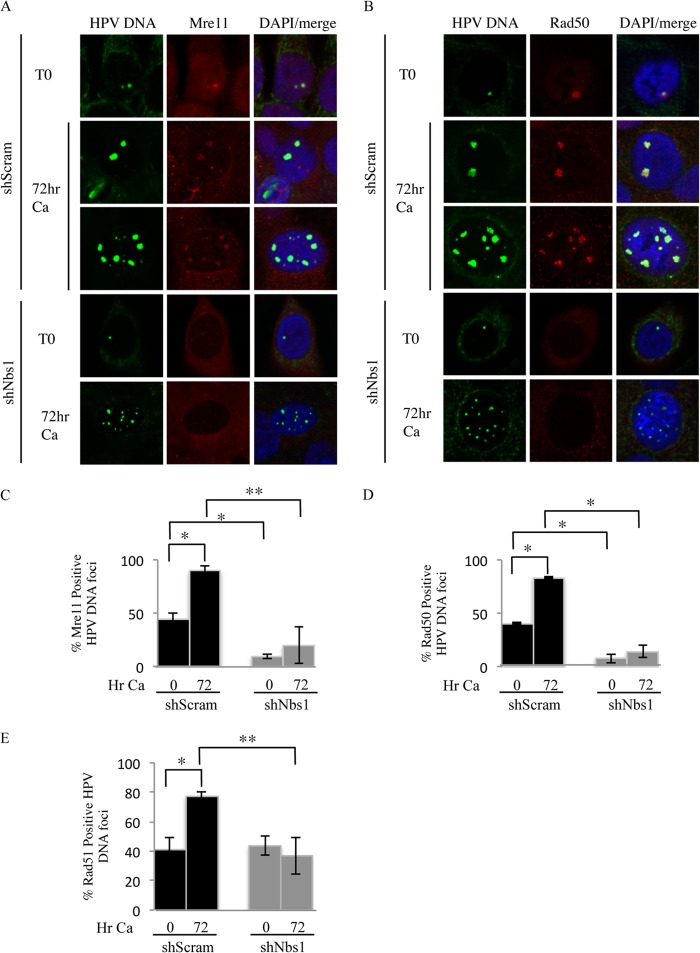
Since we previously observed that MRN components colocalize with HPV DNA foci (18), we next wanted to determine if Nbs1 knockdown affected the localization of Mre11 and Rad50 to sites of HPV DNA replication. We performed fluorescence in situ hybridization (FISH) for HPV DNA coupled with immunofluorescence for Mre11 and Rad50, as described previously (3, 18). As shown in Fig. 9, in undifferentiated CIN612 9E cells containing the Scramble shRNA, we found that Mre11 and Rad50 colocalized with viral genomes in ∼44% ± 5.6% and 41% ± 0.78% of cells containing HPV DNA foci, respectively (Fig. 9A to D). Upon differentiation in high-calcium medium, this number significantly increased, with Mre11 and Rad50 colocalizing with viral genomes in 90% ± 5.3% and 86% ± 0.98% of cells positive for HPV DNA foci, respectively. Consistent with viral genome amplification, the size of the Mre11/Rad50/HPV DNA foci increased upon differentiation. In contrast, Nbs1 knockdown resulted in a significant decrease in the localization of Mre11 and Rad50 to HPV DNA foci in undifferentiated cells, with Mre11 and Rad50 localized to viral genomes in 9% ± 1.3% and 8% ± 4.3% of HPV DNA focus-positive cells, respectively (Fig. 9A to D). Although an increase in Mre11 and Rad50 localization to viral genomes was observed upon differentiation, this was significantly less compared to the Scramble control cells (Mre11, 20% ± 17%; Rad50, 14% ± 5.5%). In addition, consistent with a block in viral genome amplification, the size of the HPV DNA/Mre11/Rad50 foci did not increase upon differentiation. These results suggest that Nbs1 may contribute to productive replication through the localization of MRN components to viral genomes.
FIG 9.
Nbs1 knockdown results in decreased localization of Mre11, Rad50, and Rad51 to viral genomes. (A and B) Immunofluorescence (IF) for Mre11 (red) (A) and Rad50 (red) (B), followed by fluorescence in situ hybridization (FISH) for HPV DNA (green), was performed on CIN612 9E cells stably maintaining the Scramble or Nbs1 shRNAs at T0 (undifferentiated), as well as after 72 h of differentiation in high-calcium medium. Cellular DNA was counterstained with DAPI. (C and D) Bar graphs represents quantification of percentages of HPV DNA focus-positive cells that were also positive for Mre11 (C) and Rad50 (D). (E) Quantification of percentage of cells positive for HPV DNA foci by FISH that were also positive for Rad51 by IF in CIN612 9E-shScram and -shNbs1 cells at T0, as well as after 72 h of differentiation in high-calcium medium. The data represent the averages of at least three independent experiments. The statistical analysis was assayed by 2-tailed t test. Data are means ± standard errors. **, P value less than 0.05; *, P value less than 0.01; Ca, calcium.
Since the MRN complex is important in facilitating ATM-mediated HR repair (22, 39), we examined the effect of Nbs1 knockdown on the localization of the principal HR factor Rad51, a recombinase that we previously showed localized to HPV replication foci (18). Interestingly, in undifferentiated cells, we found no difference in Rad51 localization to HPV DNA foci between the Scramble control (∼41% ± 8.4%) and Nbs1 knockdown (∼44% ± 6.25%) (Fig. 9E). Upon differentiation, however, while Rad51 localization to HPV DNA foci significantly increased in the Scramble control cells (∼77% ± 3.4%), no increase was observed in the Nbs1 knockdown cells, with Rad51 localizing to ∼37% ± 12.8% of cells containing HPV DNA foci. Overall, these results indicate that Nbs1 is required for the localization of Mre11 and Rad50 to viral genomes and suggest that productively replicating genomes require Rad51 activity, which may rely on an intact MRN complex.
Nbs1 knockdown moderately affects phosphorylation of ATM and Chk2 in HPV-positive cells.
Nbs1, as part of the MRN complex, is an essential part of the DDR, acting upstream to activate ATM, as well as downstream in the intra-S-phase checkpoint, and in the repair of DSBs through HR (13). To determine if Nbs1 contributes to productive viral replication through facilitating ATM activation, we examined the effect of Nbs1 depletion on ATM and Chk2 phosphorylation in CIN612 9E cells. We first examined the effect of stable Nbs1 knockdown on ATM pSer1981 (pATM) and Chk2 pThr68 (pChk2) in undifferentiated cells over several passages. As shown in Fig. 10, pATM and pChk2 were both decreased to a certain extent in response to Nbs1 depletion. While the decrease in pATM relative to total ATM ranged from 1.3- to 2-fold over the indicated passages, the decrease in Chk2 phosphorylation relative to Chk2 was more dramatic, ranging from a 1.2- to a 8.3-fold difference in this particular line (Fig. 10). We previously showed that Chk2 activation is dependent on ATM activity in HPV-positive cells (12), indicating that a small decrease in ATM phosphorylation is sufficient to induce significant changes in downstream targets. These results suggest that Nbs1 may affect viral genome amplification through its effects on ATM and Chk2, both of which are necessary for productive replication.
FIG 10.
Phosphorylated ATM and Chk2 levels are decreased variably in response to Nbs1 knockdown. Whole-cell lysates were harvested from stable CIN612 9E shScramble and shNbs1 cells at the indicated passages. Passage 22 (p22) represents the passage at which the CIN612 9E cells were transduced with the respective shRNAs. Each passage following transduction is represented by p22-#. Lysates were immunoblotted with antibodies to phosphorylated ATM (Ser1981) (pATM) and Chk2 (Thr68) (pChk2), as well as total ATM, Chk2, and Nbs1. GAPDH served as a loading control. Protein levels were quantified using ImageJ, with phosphorylated protein levels first normalized to total levels and then GAPDH. Bar graphs represent the fold changes compared to the first passage of the shScramble cells for this representative experiment, which is set at 1. Results shown are representative of observations of three independent experiments.
We next determined if Nbs1 knockdown results in a further decrease in pATM and pChk2 upon differentiation. As shown in Fig. 11A and B, in this particular passage (p22-6) at time zero (undifferentiated), the levels of pATM in CIN612 9E-shNbs1 cells were decreased only 1.3-fold compared to Scramble control, while pChk2 remained essentially unchanged. Upon differentiation, although Nbs1 levels decreased in CIN612-shNbs1 cells, there was no further decrease in pATM levels compared to total, and these cells exhibited only a moderate 1.6-fold decrease in pATM upon differentiation compared to Scramble control. Despite the relatively high level of pATM, however, viral genome amplification was still diminished in shNbs1 cells (Fig. 11C). Interestingly, the decrease in pATM levels in shNbs1 cells upon differentiation did not correlate with decreased pChk2 levels. Rather, pChk2 levels increased relative to total Chk2 in CIN612-shNbs1 cells, as well as in CIN612-shScramble and CIN612 control cells (Fig. 11B). An increase in pChk2 was observed upon differentiation across multiple passages (Fig. 12D), as well as with HFK-31–shScramble and –shNbs1 cells suspended in methylcellulose (data not shown). While the decrease in pATM and pChk2 observed in undifferentiated cells suggests that Nbs1 contributes to their activation in HPV-positive cells, the relatively high levels of pATM and pChk2 observed upon differentiation in shNbs1 cells potentially indicate an alternative mechanism for ATM activation, independent of the MRN complex. Overall, these data offer support for the idea that Nbs1 contributes to productive replication at least in part through a mechanism that is independent of promoting ATM activation. Nbs1 may serve as a downstream effector of ATM activity through phosphorylation and/or ensure the localization of repair factors to viral genomes that are required for efficient DNA synthesis, which could potentially occur in a recombination-dependent manner.
FIG 11.
Phosphorylation of ATM and Chk2 is maintained with Nbs1 knockdown upon differentiation. (A) Whole-cell lysates were harvested from HFKs and CIN612 9E cells, as well as CIN612 9E cells stably expressing shScramble or shNBS1 cells at T0 and 72 h after differentiation in high-calcium medium. Immunoblotting was performed using antibodies to phosphorylated ATM (Ser1981) (pATM), total ATM, and Nbs1. Tubulin was used as a loading control. Protein levels were quantified using ImageJ, with phosphorylated protein levels normalized first to total levels and then to tubulin. Levels for this representative experiment are graphed as fold change compared to the T0 HFK sample, which is set to 1. (B) Whole-cell lysates were harvested from HFK, CIN612 9E, CIN612 9E shScramble, and CIN612 9E shNBS1 cells at T0 and 72 h after differentiation in high-calcium medium. Immunoblotting was performed using antibodies to phosphorylated Chk2 (Thr68) (pChk2), total Chk2, and Nbs1. GAPDH was used as a loading control. Protein levels were quantified using ImageJ as indicated above. Shown is a representative experiment where levels are graphed as fold change compared to the T0 HFK sample, which is set at 1. (C) DNA was harvested from CIN612 9E, CIN612 9E shScramble, and CIN612 9E shNBS1 cells at T0 and after 72 h of differentiation in high-calcium medium and linearized by digestion with HindIII. HPV episomes were visualized via Southern blot analysis. Results shown are representative observations of four or more independent experiments.
Levels of pATM and pChk2 do not influence productive viral replication.
While we previously found that ATM and Chk2 activity is necessary for productive replication (12), it is unclear if higher levels of pATM and/or pChk2 could influence the ability of HPV to productively replicate. As shown in Fig. 10 and 12B, in two independently derived CIN612-shScramble and -shNbs1 lines, we have found that the levels of pATM and pChk2 fluctuate over time, with or without depletion of Nbs1. Similar results were observed for phosphorylated SMC1 S966 (pSMC1), a target of ATM that requires Nbs1 for phosphorylation and is involved in the intra-S-phase checkpoint (Fig. 12B) (68–70). To determine if the levels of pATM and pChk2 in undifferentiated cells affect productive replication, and to further examine the effect of Nbs1 depletion on pATM and pChk2 levels upon differentiation, we examined viral genome amplification in three different passages of CIN612 9E cells stably expressing the Scramble or Nbs1 shRNA (Fig. 12C). We compared passages 19-2, 19-3, and 19-6, in which, in undifferentiated cells, Nbs1 knockdown resulted in a 2.2-, 1.75-, and 1.53-fold decrease in pATM, respectively, and a 1.4-, 2.2-, and 1.1-fold decrease in pChk2, respectively (Fig. 12B). We found that even though the p19-6 Scramble control cells (undifferentiated) exhibited higher levels of pATM than did p19-2 and p19-3 (Fig. 12B), productive viral replication was not enhanced (Fig. 12C). Likewise, in the Nbs1 shRNA cells, the higher level of pATM and pChk2 present in the p19-6 undifferentiated cells was not sufficient to allow for productive viral replication. In addition, as shown in Fig. 12D, Nbs1 knockdown had no significant effect on pATM and pChk2 levels upon differentiation when averaged across these three passages, and we observed similar results for pSMC1 (data not shown). The lack of productive replication in Nbs1 knockdown cells, despite maintenance of pATM and pChk2, furthers suggests that Nbs1 has a multifaceted role in promoting productive replication, one that is more complex than functioning solely to activate ATM.
DISCUSSION
Previously, we demonstrated that MRN components localize to sites of HPV DNA synthesis (18). We have now found that Nbs1 is necessary for productive replication, though exactly how Nbs1 contributes to efficient viral DNA synthesis is unclear. Nbs1, as part of the MRN complex, acts as a sensor of DNA breaks and also regulates activation of ATM (32, 39). We have found that depletion of Nbs1 disrupts formation of the MRN complex, resulting in relocalization of Mre11 and Rad50 to the cytoplasm. This, coupled with the finding that the nuclease activity of Mre11 is also required for productive replication, suggests a key role for the MRN complex during the productive phase of the viral life cycle.
One of the simplest explanations for how Nbs1 contributes to productive viral replication is through facilitating activation of ATM. However, while Nbs1 knockdown did affect the levels of pATM and pChk2 to some extent, pATM and pChk2 remained quite high for most passages, especially upon differentiation. Despite the presence of pATM and pChk2, however, a defect in HPV genome amplification was consistently observed. These results suggest that Nbs1, while certainly influencing activation of ATM in HPV-infected cells, is likely playing a role outside its function as an upstream regulator of ATM activation to drive productive viral replication.
There are several possible ways by which Nbs1 could contribute to viral replication independently of activating ATM. Nbs1 is a target for phosphorylation by ATM on Ser343, and this phosphorylation is necessary for activation of the intra-S-phase checkpoint (71), which serves to slow down cellular DNA replication in response to DNA damage (70). However, while phosphorylation of SMC1, a key regulator in this checkpoint (68, 69), was decreased upon Nbs1 knockdown, pSMC1 levels exhibited little change upon differentiation, suggesting that a block in the intra-S-phase checkpoint was not responsible for the abrogation of productive replication. Nbs1 has also been implicated in DNA damage-induced apoptosis (72). While we have shown that caspase activation is necessary for productive viral replication (66), Nbs1 knockdown had no effect on caspase activation in differentiating cells (data not shown). This is not surprising, as pChk2 levels were not affected upon differentiation by Nbs1 depletion, and we previously found Chk2 activation to be necessary for caspase activation (66).
As mentioned, downstream of ATM activation, Nbs1 and the MRN complex also assist in the repair of DNA breaks, primarily through homologous recombination (HR) when cells are in the S or G2 phase of the cell cycle (32). Nbs1, as part of this complex, has specific functions that contribute to efficient HR repair. Nbs1 contains protein-protein interaction motifs important for the localization of MRN to nuclear foci (52, 73, 74), as well as the recruitment of CtIP (C-terminal-binding-protein-interacting protein) to DNA DSB ends for end processing and HR (22, 75–77). In addition to two ATM phosphorylation sites (S278 and S343) (71, 78, 79), Nbs1 is also phosphorylated by CDK2 on Ser432, which stimulates MRN-dependent conversion of DSBs into structures recognized by HR for repair (80, 81). Furthermore, recent studies indicate that Nbs1 may be required for Rad51 localization to RPA-coated DNA and the restart of stalled replication forks (81, 82).
Our finding that productive viral replication requires ATM activity and Nbs1, coupled with the localization of the MRN complex and the HR factors Rad51 and Brca1 to sites of viral genome synthesis (18, 83), suggests that recombination-dependent repair may play a role in productive viral replication. We have found that Mre11, Rad50, and Rad51 increase in localization to viral genomes upon differentiation and that this is abrogated in the absence of Nbs1. These results suggest that these repair and recombination factors are required at viral DNA to ensure efficient productive replication, possibly through facilitating HR. In support of this, studies from our lab indicate that Rad51 is necessary for productive viral replication (W. Chappell and C. Moody, unpublished data). Whether Nbs1 recruits Rad51 to viral DNA, or if productive replication results in HR structures that are processed by MRN and then bound by Rad51, is currently unclear. The massive amplification of HPV genomes upon differentiation could result in DSBs or stalled replication forks that require ATM-mediated HR to restart (84), and it will be important to examine the effect of ATM inhibition, as well as depletion of Nbs1, on the formation of replication intermediates to assess this possibility, as has been done recently for SV40 (85).
Curiously, we found that Mre11 and Rad50 localize to viral genomes in ∼40% of cells containing HPV DNA foci in undifferentiated Scramble control cells, despite Nbs1 not being required for episomal maintenance. The possibility exists that the MRN complex contributes to viral replication in undifferentiated cells but that, in its absence, replication proceeds efficiently enough such that episomal copy number and maintenance are not affected at a discernible level. This may also apply to Rad51, which we found localized to viral genomes in a similar percentage of undifferentiated HPV DNA focus-positive cells. In contrast to Mre11 and Rad50, though, Nbs1 knockdown did not affect Rad51 localization to viral genomes in undifferentiated cells. However, recent studies have shown that in the absence of Nbs1, Rad51 foci serve as markers of HR substrates, rather than reflecting HR activity (86), and could potentially account for Rad51 localization to viral genomes in these cells. Understanding the contribution of Rad51 and recombination to maintenance replication, in addition to productive replication, will be important areas of future research.
The finding that ATM and Chk2 are phosphorylated at high levels in differentiating cells upon Nbs1 knockdown suggests either that ATM activation occurs in an MRN-independent manner or that an alternative mode of ATM activation is stimulated in the presence of decreased Nbs1. The ATM interactor (ATMIN) protein is required for ATM signaling in an Nbs1-independent manner in response to hypotonic stress, as well as inhibitors of replication such as hydroxyurea (87). Nbs1 knockdown could potentially increase the flux through the ATMIN-dependent arm of the ATM signaling pathway (87, 88), maintaining phosphorylation of ATM targets. The DNA repair protein 53BP1, which is expressed at high levels in HPV-positive cells (18), has also been shown to be a mediator of ATM function (89, 90), with its effects on ATM activity enhanced in situations in which the MRN complex is present at low levels (90). Activated STAT5 has been shown to be necessary for ATM activation in HPV-positive cells (16), though whether this occurs in an MRN-dependent manner is unclear. Regardless, the maintenance of ATM signaling upon differentiation is not sufficient to drive productive viral replication, as the decreased levels of Nbs1 still act as a barrier to viral replication. Understanding the role of the MRN complex in the regulation of ATM activity in HPV-infected cells will be the focus of future investigations.
Our studies indicate that E7 expression is sufficient to increase protein levels of MRN components, as well as those of other repair factors, such as Chk2. Interestingly, E7 significantly increased only the transcript levels of Mre11, which may in turn serve to stabilize protein levels of Nbs1 and Rad50 in infected cells. We previously showed that HPV-positive cells exhibit increased levels of key proteins involved in HR repair (18), including Rad51 and Brca1, and preliminary studies indicate that this occurs in an E7- and E2F-dependent manner (W. Chappell, B. Johnson, and C. Moody, unpublished data). Taken together, these results suggest that E7 may increase levels of proteins involved in the DNA damage response to support viral replication. Recent studies by Hong and Laimins demonstrated that the protein levels of ATM, Chk2, Brca1, and Rad51 decreased upon STAT5 depletion (16). Deregulation of E2F and STAT5 activity may be necessary to ensure sufficient levels of DNA repair genes that drive viral replication in response to E7-, or potentially E6- and E1-induced, DNA damage (8, 15, 17, 83).
We have found that E7 immunoprecipitates with Nbs1, but in a manner independent of ATM. Interestingly, we found that E7 also immunoprecipitates with Rad50 but not Mre11. Nbs1 has been shown to be capable of forming a complex with Rad50 (91), and it will be important to determine if E7 binds directly with either of these MRN components, as has been shown for HSV UL12 (44). We have found that immunoprecipitation of Mre11 does not pull down E7 (data not shown), suggesting that at least two, possibly three, complexes exist in HPV-infected cells: Mre11/Rad50/Nbs1 and E7/Nbs1/+/−Rad50. E7 may form a complex with Nbs1 in the nucleus, regulating its stability when not bound to Mre11. Several studies indicate that the activity of Nbs1 is regulated by posttranslational modifications, including acetylation in addition to phosphorylation, and E7 may contribute to this regulation through multiple mechanisms. E7 increases levels of SIRT1 (92), which deacetylates Nbs1 to allow its phosphorylation by ATM on S343 (93). E7 decreases the acetyltransferase activity of PCAF (94), which has been shown to acetylate Nbs1 and block phosphorylation (93). In addition, E7 maintains CDK2 activity (95, 96) and may target Nbs1 for phosphorylation through binding, potentially regulating HR repair in HPV-infected cells.
In summary, our studies demonstrate that HPV utilizes Nbs1 and the MRN complex for efficient productive replication. Nbs1 knockdown does not significantly affect the phosphorylation of ATM and Chk2 upon differentiation, suggesting that Nbs1 may contribute to efficient productive replication outside its ability to facilitate ATM activation, potentially through the localization of Mre11, Rad50, and Rad51 to viral genomes. In addition, we have shown that E7 affects expression of DNA repair factors that are required for productive replication. E7 may further modulate the DNA damage response and drive viral replication through interactions with components of the MRN complex.
ACKNOWLEDGMENTS
C.A.M. was supported by a Pathway to Independence award from the National Cancer Institute (4-R00-CA137160-03) and by a grant from the American Cancer Society (RSG-13-229-01).
We thank members of the laboratory of Nathanial Moorman for helpful discussions and Blossom Damania for critical reading of the manuscript. We thank Jason Bodily, Pat Concannon, and Karen Cerosaletti for their generous gifts of reagents.
Footnotes
Published ahead of print 21 May 2014
REFERENCES
- 1.Longworth MS, Laimins LA. 2004. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol. Mol. Biol. Rev. 68:362–372. 10.1128/MMBR.68.2.362-372.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.zur Hausen H. 2009. Papillomaviruses in the causation of human cancers—a brief historical account. Virology 384:260–265. 10.1016/j.virol.2008.11.046 [DOI] [PubMed] [Google Scholar]
- 3.Kadaja M, Silla T, Ustav E, Ustav M. 2009. Papillomavirus DNA replication—from initiation to genomic instability. Virology 384:360–368. 10.1016/j.virol.2008.11.032 [DOI] [PubMed] [Google Scholar]
- 4.Hoffmann R, Hirt B, Bechtold V, Beard P, Raj K. 2006. Different modes of human papillomavirus DNA replication during maintenance. J. Virol. 80:4431–4439. 10.1128/JVI.80.9.4431-4439.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fradet-Turcotte A, Moody C, Laimins LA, Archambault J. 2010. Nuclear export of human papillomavirus type 31 E1 is regulated by Cdk2 phosphorylation and required for viral genome maintenance. J. Virol. 84:11747–11760. 10.1128/JVI.01445-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerjee NS, Wang HK, Broker TR, Chow LT. 2011. Human papillomavirus (HPV) E7 induces prolonged G2 following S phase reentry in differentiated human keratinocytes. J. Biol. Chem. 286:15473–15482. 10.1074/jbc.M110.197574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang HK, Duffy AA, Broker TR, Chow LT. 2009. Robust production and passaging of infectious HPV in squamous epithelium of primary human keratinocytes. Genes Dev. 23:181–194. 10.1101/gad.1735109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fradet-Turcotte A, Bergeron-Labrecque F, Moody CA, Lehoux M, Laimins LA, Archambault J. 2011. Nuclear accumulation of the papillomavirus E1 helicase blocks S-phase progression and triggers an ATM-dependent DNA damage response. J. Virol. 85:8996–9012. 10.1128/JVI.00542-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flores ER, Lambert PF. 1997. Evidence for a switch in the mode of human papillomavirus type 16 DNA replication during the viral life cycle. J. Virol. 71:7167–7179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munger K, Baldwin A, Edwards KM, Hayakawa H, Nguyen CL, Owens M, Grace M, Huh K. 2004. Mechanisms of human papillomavirus-induced oncogenesis. J. Virol. 78:11451–11460. 10.1128/JVI.78.21.11451-11460.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weitzman MD, Lilley CE, Chaurushiya MS. 2010. Genomes in conflict: maintaining genome integrity during virus infection. Annu. Rev. Microbiol. 64:61–81. 10.1146/annurev.micro.112408.134016 [DOI] [PubMed] [Google Scholar]
- 12.Moody CA, Laimins LA. 2009. Human papillomaviruses activate the ATM DNA damage pathway for viral genome amplification upon differentiation. PLoS Pathog. 5:e1000605. 10.1371/journal.ppat.1000605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciccia A, Elledge SJ. 2010. The DNA damage response: making it safe to play with knives. Mol. Cell 40:179–204. 10.1016/j.molcel.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach DS, Kerem B. 2011. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 145:435–446. 10.1016/j.cell.2011.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duensing S, Munger K. 2002. The human papillomavirus type 16 E6 and E7 oncoproteins independently induce numerical and structural chromosome instability. Cancer Res. 62:7075–7082 [PubMed] [Google Scholar]
- 16.Hong S, Laimins LA. 2013. The JAK-STAT transcriptional regulator, STAT-5, activates the ATM DNA damage pathway to induce HPV 31 genome amplification upon epithelial differentiation. PLoS Pathog. 9:e1003295. 10.1371/journal.ppat.1003295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakakibara N, Mitra R, McBride AA. 2011. The papillomavirus E1 helicase activates a cellular DNA damage response in viral replication foci. J. Virol. 85:8981–8995. 10.1128/JVI.00541-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillespie KA, Mehta KP, Laimins LA, Moody CA. 2012. Human papillomaviruses recruit cellular DNA repair and homologous recombination factors to viral replication centers. J. Virol. 86:9520–9526. 10.1128/JVI.00247-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.San Filippo J, Sung P, Klein H. 2008. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77:229–257. 10.1146/annurev.biochem.77.061306.125255 [DOI] [PubMed] [Google Scholar]
- 20.Nuss JE, Patrick SM, Oakley GG, Alter GM, Robison JG, Dixon K, Turchi JJ. 2005. DNA damage induced hyperphosphorylation of replication protein A. 1. Identification of novel sites of phosphorylation in response to DNA damage. Biochemistry 44:8428–8437. 10.1021/bi0480584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zernik-Kobak M, Vasunia K, Connelly M, Anderson CW, Dixon K. 1997. Sites of UV-induced phosphorylation of the p34 subunit of replication protein A from HeLa cells. J. Biol. Chem. 272:23896–23904. 10.1074/jbc.272.38.23896 [DOI] [PubMed] [Google Scholar]
- 22.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. 2007. Human CtIP promotes DNA end resection. Nature 450:509–514. 10.1038/nature06337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakakibara N, Chen D, McBride AA. 2013. Papillomaviruses use recombination-dependent replication to vegetatively amplify their genomes in differentiated cells. PLoS Pathog. 9:e1003321. 10.1371/journal.ppat.1003321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moynahan ME, Jasin M. 2010. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 11:196–207. 10.1038/nrm2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boichuk S, Hu L, Hein J, Gjoerup OV. 2010. Multiple DNA damage signaling and repair pathways deregulated by simian virus 40 large T antigen. J. Virol. 84:8007–8020. 10.1128/JVI.00334-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schumacher AJ, Mohni KN, Kan Y, Hendrickson EA, Stark JM, Weller SK. 2012. The HSV-1 exonuclease, UL12, stimulates recombination by a single strand annealing mechanism. PLoS Pathog. 8:e1002862. 10.1371/journal.ppat.1002862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilkinson DE, Weller SK. 2004. Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J. Virol. 78:4783–4796. 10.1128/JVI.78.9.4783-4796.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilkinson DE, Weller SK. 2003. The role of DNA recombination in herpes simplex virus DNA replication. IUBMB Life 55:451–458. 10.1080/15216540310001612237 [DOI] [PubMed] [Google Scholar]
- 29.Kudoh A, Iwahori S, Sato Y, Nakayama S, Isomura H, Murata T, Tsurumi T. 2009. Homologous recombinational repair factors are recruited and loaded onto the viral DNA genome in Epstein-Barr virus replication compartments. J. Virol. 83:6641–6651. 10.1128/JVI.00049-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dheekollu J, Deng Z, Wiedmer A, Weitzman MD, Lieberman PM. 2007. A role for MRE11, NBS1, and recombination junctions in replication and stable maintenance of EBV episomes. PLoS One 2:e1257. 10.1371/journal.pone.0001257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dheekollu J, Chen HS, Kaye KM, Lieberman PM. 2013. Timeless-dependent DNA replication-coupled recombination promotes Kaposi's sarcoma-associated herpesvirus episome maintenance and terminal repeat stability. J. Virol. 87:3699–3709. 10.1128/JVI.02211-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams RS, Williams JS, Tainer JA. 2007. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 85:509–520. 10.1139/O07-069 [DOI] [PubMed] [Google Scholar]
- 33.Lavin MF. 2007. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene 26:7749–7758. 10.1038/sj.onc.1210880 [DOI] [PubMed] [Google Scholar]
- 34.Lee JH, Paull TT. 2007. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene 26:7741–7748. 10.1038/sj.onc.1210872 [DOI] [PubMed] [Google Scholar]
- 35.Lee JH, Paull TT. 2005. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308:551–554. 10.1126/science.1108297 [DOI] [PubMed] [Google Scholar]
- 36.Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. 2003. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 22:5612–5621. 10.1093/emboj/cdg541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316:1160–1166. 10.1126/science.1140321 [DOI] [PubMed] [Google Scholar]
- 38.Falck J, Coates J, Jackson SP. 2005. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434:605–611. 10.1038/nature03442 [DOI] [PubMed] [Google Scholar]
- 39.Lamarche BJ, Orazio NI, Weitzman MD. 2010. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 584:3682–3695. 10.1016/j.febslet.2010.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, III, Hays L, Morgan WF, Petrini JH. 1998. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93:477–486. 10.1016/S0092-8674(00)81175-7 [DOI] [PubMed] [Google Scholar]
- 41.Digweed M, Sperling K. 2004. Nijmegen breakage syndrome: clinical manifestation of defective response to DNA double-strand breaks. DNA Repair (Amst.) 3:1207–1217. 10.1016/j.dnarep.2004.03.004 [DOI] [PubMed] [Google Scholar]
- 42.Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, Saar K, Beckmann G, Seemanova E, Cooper PR, Nowak NJ, Stumm M, Weemaes CM, Gatti RA, Wilson RK, Digweed M, Rosenthal A, Sperling K, Concannon P, Reis A. 1998. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell 93:467–476. 10.1016/S0092-8674(00)81174-5 [DOI] [PubMed] [Google Scholar]
- 43.Wu X, Avni D, Chiba T, Yan F, Zhao Q, Lin Y, Heng H, Livingston D. 2004. SV40 T antigen interacts with Nbs1 to disrupt DNA replication control. Genes Dev. 18:1305–1316. 10.1101/gad.1182804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balasubramanian N, Bai P, Buchek G, Korza G, Weller SK. 2010. Physical interaction between the herpes simplex virus type 1 exonuclease, UL12, and the DNA double-strand break-sensing MRN complex. J. Virol. 84:12504–12514. 10.1128/JVI.01506-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson R, Laimins LA. 2005. Differentiation of HPV-containing cells using organotypic “raft” culture or methylcellulose. Methods Mol. Med. 119:157–169. 10.1385/1-59259-982-6:157 [DOI] [PubMed] [Google Scholar]
- 46.Kraakman-van der Zwet M, Overkamp WJ, Friedl AA, Klein B, Verhaegh GW, Jaspers NG, Midro AT, Eckardt-Schupp F, Lohman PH, Zdzienicka MZ. 1999. Immortalization and characterization of Nijmegen breakage syndrome fibroblasts. Mutat. Res. 434:17–27. 10.1016/S0921-8777(99)00009-9 [DOI] [PubMed] [Google Scholar]
- 47.Hebner CM, Wilson R, Rader J, Bidder M, Laimins LA. 2006. Human papillomaviruses target the double-stranded RNA protein kinase pathway. J. Gen. Virol. 87:3183–3193. 10.1099/vir.0.82098-0 [DOI] [PubMed] [Google Scholar]
- 48.Hubert WG, Laimins LA. 2002. Human papillomavirus type 31 replication modes during the early phases of the viral life cycle depend on transcriptional and posttranscriptional regulation of E1 and E2 expression. J. Virol. 76:2263–2273. 10.1128/jvi.76.5.2263-2273.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Longworth MS, Laimins LA. 2004. The binding of histone deacetylases and the integrity of zinc finger-like motifs of the E7 protein are essential for the life cycle of human papillomavirus type 31. J. Virol. 78:3533–3541. 10.1128/JVI.78.7.3533-3541.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bodily JM, Mehta KP, Laimins LA. 2011. Human papillomavirus E7 enhances hypoxia-inducible factor 1-mediated transcription by inhibiting binding of histone deacetylases. Cancer Res. 71:1187–1195. 10.1158/0008-5472.CAN-10-2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cerosaletti KM, Desai-Mehta A, Yeo TC, Kraakman-Van Der Zwet M, Zdzienicka MZ, Concannon P. 2000. Retroviral expression of the NBS1 gene in cultured Nijmegen breakage syndrome cells restores normal radiation sensitivity and nuclear focus formation. Mutagenesis 15:281–286. 10.1093/mutage/15.3.281 [DOI] [PubMed] [Google Scholar]
- 52.Desai-Mehta A, Cerosaletti KM, Concannon P. 2001. Distinct functional domains of nibrin mediate Mre11 binding, focus formation, and nuclear localization. Mol. Cell. Biol. 21:2184–2191. 10.1128/MCB.21.6.2184-2191.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.You Z, Chahwan C, Bailis J, Hunter T, Russell P. 2005. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell. Biol. 25:5363–5379. 10.1128/MCB.25.13.5363-5379.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fehrmann F, Klumpp DJ, Laimins LA. 2003. Human papillomavirus type 31 E5 protein supports cell cycle progression and activates late viral functions upon epithelial differentiation. J. Virol. 77:2819–2831. 10.1128/JVI.77.5.2819-2831.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mighty KK, Laimins LA. 2011. p63 is necessary for the activation of human papillomavirus late viral functions upon epithelial differentiation. J. Virol. 85:8863–8869. 10.1128/JVI.00750-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schreiber E, Matthias P, Muller MM, Schaffner W. 1989. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 17:6419. 10.1093/nar/17.15.6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kadaja M, Isok-Paas H, Laos T, Ustav E, Ustav M. 2009. Mechanism of genomic instability in cells infected with the high-risk human papillomaviruses. PLoS Pathog. 5:e1000397. 10.1371/journal.ppat.1000397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM. 1999. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99:577–587. 10.1016/S0092-8674(00)81547-0 [DOI] [PubMed] [Google Scholar]
- 59.Takemura H, Rao VA, Sordet O, Furuta T, Miao ZH, Meng L, Zhang H, Pommier Y. 2006. Defective Mre11-dependent activation of Chk2 by ataxia telangiectasia mutated in colorectal carcinoma cells in response to replication-dependent DNA double strand breaks. J. Biol. Chem. 281:30814–30823. 10.1074/jbc.M603747200 [DOI] [PubMed] [Google Scholar]
- 60.Maser RS, Zinkel R, Petrini JH. 2001. An alternative mode of translation permits production of a variant NBS1 protein from the common Nijmegen breakage syndrome allele. Nat. Genet. 27:417–421. 10.1038/86920 [DOI] [PubMed] [Google Scholar]
- 61.Cerosaletti K, Wright J, Concannon P. 2006. Active role for nibrin in the kinetics of atm activation. Mol. Cell. Biol. 26:1691–1699. 10.1128/MCB.26.5.1691-1699.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stracker TH, Petrini JH. 2011. The MRE11 complex: starting from the ends. Nat. Rev. Mol. Cell Biol. 12:90–103. 10.1038/nrm3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duursma AM, Driscoll R, Elias JE, Cimprich KA. 2013. A role for the MRN complex in ATR activation via TOPBP1 recruitment. Mol. Cell 50:116–122. 10.1016/j.molcel.2013.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shiotani B, Nguyen HD, Hakansson P, Marechal A, Tse A, Tahara H, Zou L. 2013. Two distinct modes of ATR activation orchestrated by Rad17 and Nbs1. Cell Rep. 3:1651–1662. 10.1016/j.celrep.2013.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rodier F, Coppe JP, Patil CK, Hoeijmakers WA, Munoz DP, Raza SR, Freund A, Campeau E, Davalos AR, Campisi J. 2009. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 11:973–979. 10.1038/ncb1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moody CA, Fradet-Turcotte A, Archambault J, Laimins LA. 2007. Human papillomaviruses activate caspases upon epithelial differentiation to induce viral genome amplification. Proc. Natl. Acad. Sci. U. S. A. 104:19541–19546. 10.1073/pnas.0707947104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tseng SF, Chang CY, Wu KJ, Teng SC. 2005. Importin KPNA2 is required for proper nuclear localization and multiple functions of NBS1. J. Biol. Chem. 280:39594–39600. 10.1074/jbc.M508425200 [DOI] [PubMed] [Google Scholar]
- 68.Kim ST, Xu B, Kastan MB. 2002. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 16:560–570. 10.1101/gad.970602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kitagawa R, Bakkenist CJ, McKinnon PJ, Kastan MB. 2004. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes Dev. 18:1423–1438. 10.1101/gad.1200304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bartek J, Lukas C, Lukas J. 2004. Checking on DNA damage in S phase. Nat. Rev. Mol. Cell Biol. 5:792–804. 10.1038/nrm1493 [DOI] [PubMed] [Google Scholar]
- 71.Lim DS, Kim ST, Xu B, Maser RS, Lin J, Petrini JH, Kastan MB. 2000. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature 404:613–617. 10.1038/35007091 [DOI] [PubMed] [Google Scholar]
- 72.Iijima K, Muranaka C, Kobayashi J, Sakamoto S, Komatsu K, Matsuura S, Kubota N, Tauchi H. 2008. NBS1 regulates a novel apoptotic pathway through Bax activation. DNA Repair (Amst.) 7:1705–1716. 10.1016/j.dnarep.2008.06.013 [DOI] [PubMed] [Google Scholar]
- 73.Cerosaletti KM, Concannon P. 2003. Nibrin forkhead-associated domain and breast cancer C-terminal domain are both required for nuclear focus formation and phosphorylation. J. Biol. Chem. 278:21944–21951. 10.1074/jbc.M211689200 [DOI] [PubMed] [Google Scholar]
- 74.Zhao S, Renthal W, Lee EY. 2002. Functional analysis of FHA and BRCT domains of NBS1 in chromatin association and DNA damage responses. Nucleic Acids Res. 30:4815–4822. 10.1093/nar/gkf612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen L, Nievera CJ, Lee AY, Wu X. 2008. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J. Biol. Chem. 283:7713–7720. 10.1074/jbc.M710245200 [DOI] [PubMed] [Google Scholar]
- 76.Yuan J, Chen J. 2009. N terminus of CtIP is critical for homologous recombination-mediated double-strand break repair. J. Biol. Chem. 284:31746–31752. 10.1074/jbc.M109.023424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang H, Shi LZ, Wong CC, Han X, Hwang PY, Truong LN, Zhu Q, Shao Z, Chen DJ, Berns MW, Yates JR, III, Chen L, Wu X. 2013. The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS Genet. 9:e1003277. 10.1371/journal.pgen.1003277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu X, Ranganathan V, Weisman DS, Heine WF, Ciccone DN, O'Neill TB, Crick KE, Pierce KA, Lane WS, Rathbun G, Livingston DM, Weaver DT. 2000. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature 405:477–482. 10.1038/35013089 [DOI] [PubMed] [Google Scholar]
- 79.Gatei M, Young D, Cerosaletti KM, Desai-Mehta A, Spring K, Kozlov S, Lavin MF, Gatti RA, Concannon P, Khanna K. 2000. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat. Genet. 25:115–119. 10.1038/75508 [DOI] [PubMed] [Google Scholar]
- 80.Wohlbold L, Merrick KA, De S, Amat R, Kim JH, Larochelle S, Allen JJ, Zhang C, Shokat KM, Petrini JH, Fisher RP. 2012. Chemical genetics reveals a specific requirement for Cdk2 activity in the DNA damage response and identifies Nbs1 as a Cdk2 substrate in human cells. PLoS Genet. 8:e1002935. 10.1371/journal.pgen.1002935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Falck J, Forment JV, Coates J, Mistrik M, Lukas J, Bartek J, Jackson SP. 2012. CDK targeting of NBS1 promotes DNA-end resection, replication restart and homologous recombination. EMBO Rep. 13:561–568. 10.1038/embor.2012.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yata K, Lloyd J, Maslen S, Bleuyard JY, Skehel M, Smerdon SJ, Esashi F. 2012. Plk1 and CK2 act in concert to regulate Rad51 during DNA double strand break repair. Mol. Cell 45:371–383. 10.1016/j.molcel.2011.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sakakibara N, Chen D, Jang MK, Kang DW, Luecke HF, Wu SY, Chiang CM, McBride AA. 2013. Brd4 is displaced from HPV replication factories as they expand and amplify viral DNA. PLoS Pathog. 9:e1003777. 10.1371/journal.ppat.1003777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Petermann E, Helleday T. 2010. Pathways of mammalian replication fork restart. Nat. Rev. Mol. Cell Biol. 11:683–687. 10.1038/nrm2974 [DOI] [PubMed] [Google Scholar]
- 85.Sowd GA, Li NY, Fanning E. 2013. ATM and ATR activities maintain replication fork integrity during SV40 chromatin replication. PLoS Pathog. 9:e1003283. 10.1371/journal.ppat.1003283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bruhn C, Zhou ZW, Ai H, Wang ZQ. 2014. The essential function of the MRN complex in the resolution of endogenous replication intermediates. Cell Rep. 6:182–195. 10.1016/j.celrep.2013.12.018 [DOI] [PubMed] [Google Scholar]
- 87.Kanu N, Behrens A. 2007. ATMIN defines an NBS1-independent pathway of ATM signalling. EMBO J. 26:2933–2941. 10.1038/sj.emboj.7601733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang T, Penicud K, Bruhn C, Loizou JI, Kanu N, Wang ZQ, Behrens A. 2012. Competition between NBS1 and ATMIN controls ATM signaling pathway choice. Cell Rep. 2:1498–1504. 10.1016/j.celrep.2012.11.002 [DOI] [PubMed] [Google Scholar]
- 89.Mochan TA, Venere M, DiTullio RA, Jr, Halazonetis TD. 2003. 53BP1 and NFBD1/MDC1-Nbs1 function in parallel interacting pathways activating ataxia-telangiectasia mutated (ATM) in response to DNA damage. Cancer Res. 63:8586–8591 [PubMed] [Google Scholar]
- 90.Lee JH, Goodarzi AA, Jeggo PA, Paull TT. 2010. 53BP1 promotes ATM activity through direct interactions with the MRN complex. EMBO J. 29:574–585. 10.1038/emboj.2009.372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.van der Linden E, Sanchez H, Kinoshita E, Kanaar R, Wyman C. 2009. RAD50 and NBS1 form a stable complex functional in DNA binding and tethering. Nucleic Acids Res. 37:1580–1588. 10.1093/nar/gkn1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Allison SJ, Jiang M, Milner J. 2009. Oncogenic viral protein HPV E7 up-regulates the SIRT1 longevity protein in human cervical cancer cells. Aging (Albany NY) 1:316–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E. 2007. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol. Cell 27:149–162. 10.1016/j.molcel.2007.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Avvakumov N, Torchia J, Mymryk JS. 2003. Interaction of the HPV E7 proteins with the pCAF acetyltransferase. Oncogene 22:3833–3841. 10.1038/sj.onc.1206562 [DOI] [PubMed] [Google Scholar]
- 95.Nguyen CL, Munger K. 2008. Direct association of the HPV16 E7 oncoprotein with cyclin A/CDK2 and cyclin E/CDK2 complexes. Virology 380:21–25. 10.1016/j.virol.2008.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moody CA, Laimins LA. 2010. Human papillomavirus oncoproteins: pathways to transformation. Nat. Rev. Cancer 10:550–560. 10.1038/nrc2886 [DOI] [PubMed] [Google Scholar]
- 97.Roman A, Munger K. 2013. The papillomavirus E7 proteins. Virology 445:138–168. 10.1016/j.virol.2013.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]