Abstract
Mammalian cells have the ability to recognize virus infection and mount a powerful antiviral transcriptional response that provides an initial barrier to replication and impacts both innate and adaptive immune responses. Retinoic acid-inducible gene I (RIG-I)-like receptor (RLR) proteins mediate intracellular virus recognition and are activated by viral RNA ligands to induce antiviral signal transduction. While the mechanisms of RIG-I regulation are already well understood, less is known about the more enigmatic melanoma differentiation-associated 5 (MDA5) and laboratory of genetics and physiology 2 (LGP2). Emerging evidence suggests that these two RLRs are intimately associated as both accomplices and antagonists of antiviral signal transduction.
INTRODUCTION
Cellular antiviral signaling is initiated following recognition of virus-encoded molecular signatures, often in the form of nucleic acids. Infection by RNA viruses results in cytosolic accumulation of double-stranded RNA (dsRNA) or otherwise chemically distinct, non-self RNA species. Sentry proteins in the cytoplasm recognize characteristics of non-self RNAs and can trigger downstream signal transduction pathways that culminate in activated antiviral transcription regulators (1–3). These factors accumulate in the nucleus, where they drive the expression of virus-induced genes, including the primary antiviral cytokine, beta interferon (IFN-β), and diverse direct and indirect antiviral effectors (4).
One group of intracellular responders, the retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), includes the proteins RIG-I (5), melanoma differentiation-associated 5 (MDA5) (6), and laboratory of genetics and physiology 2 (LGP2) (7). Similar in structure and function, these proteins share significant sequence homologies that define them as the products of an evolutionarily conserved gene family (8) (Fig. 1A). The RLR proteins are thought to share the ability to detect molecular signatures of virus infection and activate antiviral signaling cascades, but they differ in both their RNA recognition capacities and signaling properties (9, 10). RIG-I, MDA5, and LGP2 share homologous DECH box helicase domain regions that have intrinsic dsRNA binding and ATP hydrolysis functions and a C-terminal domain that has been implicated in binding to RNA termini and autoregulation (11–13). RIG-I and MDA5 both have tandem N-terminal caspase activation and recruitment domain (CARD) motifs, protein interaction domains that mediate associations with upstream and downstream regulatory machinery. The CARDs are regulated by posttranslational modifications, including ubiquitination and phosphorylation (14–16). Current evidence indicates that RNA recognition by RIG-I and MDA5 is accompanied by CARD dephosphorylation, enabling their productive interaction with signaling machinery. Much of the knowledge of the RLR signaling pathway was developed by detailed study of RIG-I, the prototype RLR (17), and investigations of MDA5 have contributed greatly to a general paradigm for RLR signaling (Fig. 1B). Interaction with non-self-ligand RNAs leads to exposure of active RLR CARDs, enabling association with the CARD of an essential mitochondrial antiviral signaling protein, IPS-1/MAVS (18–21). IPS-1/MAVS is thought to act as a polymeric signaling scaffold that facilitates the assembly and activation of signaling proteins, including TRAF2, TRAF5, and TRAF6, and associated serine kinases, including IκB kinase alpha (IKKα), IKKβ, IKKγ, IKKε, and TANK-binding kinase 1 (TBK1) (22). These kinases activate the master transcription regulators, interferon regulatory factor 3 (IRF3) and NF-κB, that induce the production of IFN-β and diverse antiviral target genes (23, 24). Secreted IFN-β amplifies the antiviral response via IFNAR-JAK-STAT signaling, inducing the expression of many more effectors that together produce a cellular antiviral state that provides a broadly effective barrier against virus replication (25, 26).
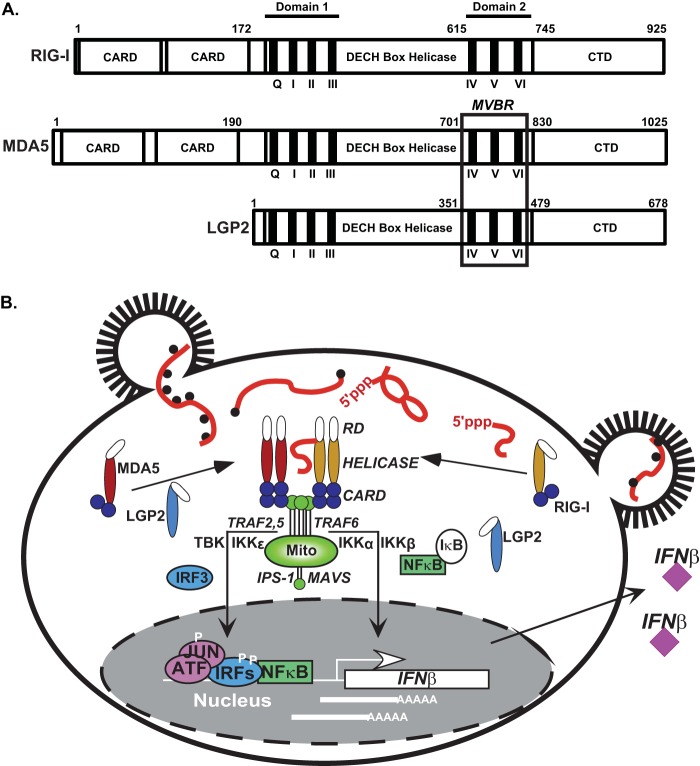
FIG 1.
RLR family proteins in antiviral signaling. (A) Diagram illustrating key features of RIG-I, MDA5, and LGP2 mentioned in the text. The three RLRs are composed of a central DECH box helicase domain containing conserved domain 1 (encompassing helicase motifs Q, I, II, and III) and domain 2 (encompassing helicase motifs IV, V, and VI), both of which are essential to coordinate RNA binding and ATP hydrolysis activities. A C-terminal domain (CTD) is required for RNA terminus recognition and is involved in autoregulation of RIG-I. RIG-I and MDA5 contain tandem caspase activation and recruitment domain (CARD) regions at their N termini, which are essential for downstream signaling activity. The position of the minimal V protein binding region (MVBR), the target site for paramyxovirus V protein antagonism of MDA5 and LGP2, coincides with the boundaries of domain 2. (B) Simplified overview of RLR signal transduction leading to IFN-β gene transcription. Virus infection causes accumulation of cytosolic RNA species with non-self features, including 5′-triphosphates and double-stranded regions (for RIG-I recognition) or long RNAs with structural features (for MDA5 recognition). These RNAs trigger derepression of RIG-I and activate exposure of RLR CARDs to allow interaction with the CARD of the signaling adaptor, IPS-1/MAVS, that is localized on the mitochondria (Mito). Activation of IPS-1/MAVS polymerization stimulates assembly of a signaling scaffold for TRAF2, TRAF5, and TRAF6 and their associated kinases (e.g., TBK, IKKα, IKKβ, and IKKε) that result in phosphorylation-mediated activation of latent IRF3 and NF-κB transcription factors. These factors, along with ATF/JUN, assemble on the IFN-β gene proximal enhancer, resolve nucleosome-mediated repression, and recruit RNA polymerase to induce transcription. Transcribed IFN-β mRNA is translated and subsequently secreted from the cell.
While MDA5 has been the subject of many investigations, its redundant and nonredundant functions with respect to RIG-I are only beginning to be unraveled. The study of natural and synthetic RNAs has yet to reveal precise features that clearly define an MDA5-specific ligand or reveal the basis for non-self-recognition capacity, contributing to a paucity of mechanistic details regarding the basis for MDA5 non-self RNA recognition leading to signal transduction. For LGP2, the absence of CARDs or other recognizable signaling domains has impeded drawing clear comparisons with the functions of the other RLR proteins. Furthermore, the apparently antithetical reported functions of LGP2 as both a positive and negative regulator of antiviral responses have been difficult to reconcile. These mechanistic details leave a large gap in our understanding of RLR signaling and contribute to contradictory reports that implicate MDA5 and LGP2 as either collaborators or competitors in the antiviral system. The complex relationship of MDA5 and LGP2 is described here, and a unifying model is presented to help clarify their roles in the regulation of antiviral signaling.
RIG-I
RIG-I, the prototypic RLR, is the first recognized and best characterized family member (17). As RIG-I has been reviewed extensively (e.g., in references 1, 2, 9, and 3), it is discussed here cursorily, primarily to provide a basis of comparison to contrast with MDA5 and LGP2. RIG-I is autoinhibited at steady state by CARD interactions with the helicase domain, and it can be conformationally activated by engaging an appropriate RNA ligand (11, 27, 28). RNA recognition by RIG-I involves helicase domain-mediated interaction with dsRNA, ATP-powered translocation along the dsRNA (29), and C-terminal domain (CTD)-mediated recognition of tri- and diphosphorylated RNA ends (28, 30). RIG-I is responsive to short or long dsRNA or single-stranded RNA (ssRNA) with 5′-triphosphate and base-paired ends, including poly(I·C), viral hairpins and defective interfering RNAs, and hepatitis C virus (HCV) poly(U)-rich untranslated region (UTR) RNA. These diverse ligands reflect the broad range of RNA viruses that have been demonstrated to be susceptible to detection by RIG-I. RIG-I deficiency in mice results in high susceptibility to RNA virus infections, including several negative-strand RNA genome viruses, such as Newcastle disease virus, vesicular stomatitis virus, influenza A virus, and Sendai virus. Cells derived from these mice fail to initiate antiviral signaling programs, resulting in reduced production of IFN-β (32). RIG-I has been linked to many additional regulatory pathways that have been characterized to positively or negatively regulate RIG-I signaling capacity, including ubiquitination, phosphorylation, and association with antiviral mediators (e.g., see references 2, 33, 34, and 35). Moreover, several viral IFN antagonists have been described to disrupt RIG-I directly or indirectly (36, 37), confirming its role as a critical component of the RNA-induced antiviral system.
MDA5
MDA5 shares the overall domain structure of RIG-I with tandem CARDs fused to homologous helicase and CTD regions (Fig. 1A) and is thought to signal through a similar CARD-mediated, IPS-1/MAVS-dependent system to activate antiviral gene expression. Unlike the autoinhibited RIG-I, the expression of MDA5 alone is sufficient to activate the IFN-β gene in the absence of specific RNA recognition (38, 39), though its activity is regulated in vivo by CARD ubiquitination, phosphorylation, and direct or indirect association with other antiviral mediators (2, 16, 33–35). MDA5 deficiency in mice created a defect in the response to poly(I·C) and greater susceptibility to certain positive-sense single-stranded RNA viruses, specifically, the picornaviruses poliovirus and encephalomyocarditis virus (EMCV) and murine norovirus (32, 40, 41). All of these viruses have a protein covalently attached to the RNA 5′ end, effectively preventing their genome RNA termini from recognition by RIG-I. However, the simplistic distinction between negative-sense and positive-sense single-stranded RNA virus recognition by RIG-I or MDA5 cannot be generalized. For example, flaviviruses are more potent IFN activators in wild-type than in RIG-I-deficient mice (32, 42), but further work shows that dengue virus and West Nile virus are detected by both RIG-I and MDA5 (32, 43, 44). Similarly, although Sendai virus is thought to be recognized by RIG-I during early infection, the importance of MDA5 becomes more apparent during later phases of infection in vivo (40, 45, 46).
There is a relative lack of detailed information regarding MDA5 RNA recognition substrates, in part due to the apparently poor RNA binding activity of MDA5. However, a few studies have elucidated potential RNA features or modifications or specific viral RNA regions that are discriminated by MDA5. MDA5 was found to be activated by enzymatically digested or sheared populations of RNA longer than 2 kbp (47), and high-molecular-weight RNAs extracted from virus-infected cells were shown to preferentially activate MDA5-mediated signaling (48). It was proposed that structural features such as RNA branches found in RNAs with both single-stranded and double-stranded regions might be required for recognition by MDA5. This is consistent with the observation that poly(I·C), a synthetic dsRNA analogue, is able to activate MDA5 in vitro and in vivo (32, 40). MDA5 may also be able to discriminate some features specific to virus-derived mRNAs, including 2′-O-methylation or primary and secondary structures (49, 50, 76). EMCV is a virus that effectively escapes RIG-I detection by masking its RNA 5′ ends and replicates more efficiently in the absence of either MDA5 or LGP2. A region of the EMCV negative-strand RNA that was found to copurify with LGP2 acts as a physiological agonist of MDA5-dependent signaling (51). This RNA, derived from the antisense RNA complementary to the EMCV L gene, is necessary and sufficient for antiviral responses to EMCV mediated by MDA5. While the identification of this virus-origin MDA5 agonist RNA failed to yield information regarding specific RNA features that are required for MDA5 responses, it adds to the increasing evidence implicating LGP2 as a collaborator for MDA5 RNA recognition (more below).
Despite its relatively low solution RNA binding affinity compared to that of either RIG-I or LGP2, electron microscopy has revealed that MDA5 is able to assemble into filaments on dsRNA, with ring-like asymmetric units that form helical twists (52–57). Data indicate that MDA5 initially binds slowly and with low affinity to dsRNA as a monomer or dimer and, under controlled conditions, can be observed to assemble onto dsRNAs to form long head-to-tail filaments. Curiously, these structures are destabilized by ATP hydrolysis (56–59) but are clearly visible in the presence of the ATP transition state analogue, ADP-aluminum tetrafluoride (AlF4). Physiological ATP levels promote MDA5 dissociation from RNA, and these filaments have yet to be observed inside living cells. In light of evidence that, while MDA5 does require ATP hydrolysis for its activity, catalytically inactive MDA5 mutants are capable of constitutive signaling (38), as well as the discovery of a similar signaling-competent, ATPase-deficient MDA5 mutant in mammalian autoimmunity (60), the exact reason for filaments in MDA5 signaling remains to be determined. The observed long MDA5 filaments may represent a captured transition state formed during MDA5 signaling rather than a stable intracellular structure. Interestingly, RIG-I also forms filamentous assemblies on RNA, albeit with clear mechanistic distinctions from MDA5 (61), possibly indicating that oligomerization is a conserved feature of RLR signaling that engages or induces IPS-1/MAVS polymers.
LGP2
LGP2 shares sequence conservation with the other RLRs within the helicase domain and the C terminus but lacks a CARD region entirely (Fig. 1A). LGP2 is present at low levels in the uninfected cell but accumulates in response to virus infection or antiviral mediators, including poly(I·C) and IFNs (13, 62, 63). Due to the absence of a CARD or other known signaling interaction domain, the functions of LGP2 in antiviral signaling have been difficult to generalize, and its precise roles in innate antiviral immunity remain to be fully elucidated. Three LGP2 knockout mice were reported with disparate conclusions (64–66), and different experimental strategies have demonstrated seemingly antithetic biological activities. While the exact roles for LGP2 in RNA recognition and antiviral signaling remain to be resolved, evidence is accumulating that it can impact a wide range of cellular responses.
LGP2 AS A NEGATIVE REGULATOR
LGP2 expressed from plasmid vectors can function as a negative regulator of RLR signaling (13, 62, 63). This observation, combined with the fact that LGP2 expression is induced during antiviral signaling, supports the characterization of LGP2 as a feedback inhibitor of antiviral responses. Several potential mechanisms for LGP2-mediated feedback inhibition have been proposed. The fact that LGP2 has a stronger RNA binding affinity than MDA5 or RIG-I was interpreted as evidence for RNA sequestration as an inhibitory mechanism (63). LGP2 was proposed to act as a sponge that soaks up excess RIG-I ligands. The characterization of a C-terminal regulatory domain in RIG-I that mediates autoinhibition in cis and in trans via interaction with the CARD domains was interpreted as another potential source of LGP2-mediated interference. It was proposed by analogy that LGP2's C-terminal domain could also act as a regulatory domain to mediate RIG-I interference in trans (13). A third report demonstrated that LGP2 could inhibit antiviral signaling independent of dsRNA or virus infection by engaging in a protein complex with IPS-1/MAVS (62). Experiments indicated that LGP2 interaction with IPS-1/MAVS could interfere with kinase recruitment that is required for antiviral signaling. Based on currently available evidence, these negative regulatory mechanisms are not necessarily mutually exclusive, but evidence indicates that the negative regulatory activity of LGP2 remains intact in the absence of RNA binding ability or ATP hydrolysis activity (38, 39). This suggests that LGP2 uses an interference mechanism distinct from that used for MDA5 coactivation, which requires both ATPase and RNA interactions (39). Further research will be necessary to fully appreciate the basis for LGP2-mediated negative regulation and the relative importance of these mechanisms to antiviral signaling.
LGP2 AS A POSITIVE REGULATOR
Mice with a targeted disruption in the LGP2 locus are more susceptible to certain virus infections than heterozygous littermates and have defects in generating antiviral responses, supporting a positive role for LGP2 in antiviral signaling (64). LGP2 deficiency reduces IFN-β production and other host responses to several RNA viruses, notably including the picornaviruses EMCV and poliovirus that had been previously linked to detection by MDA5 (32, 64). The effect of LGP2 deficiency extends to cytokine responses triggered by cytosolic dsDNA and DNA-genome pathogens, which are impaired in the absence of LGP2 (67). Experiments in cells lacking both LGP2 and MDA5 revealed a synergistic signal transduction activity resulting from coexpression of LGP2 with MDA5 that was not attributable to either RLR alone (64), suggesting that LGP2 may synergize with MDA5 to promote efficient signal transduction. Replacing LGP2 with an enzymatically inactive mutant did not reconstitute defective positive signaling responses in vivo, indicating the importance of ATP hydrolysis in LGP2 positive regulation of antiviral signaling. This requirement distinguishes positive regulation by LGP2 from its negative regulation, which is independent of enzymatic activity (38, 39).
Biochemical analysis of ATP hydrolysis was combined with single-molecule dsRNA binding and antiviral signaling assays to study LGP2's physical properties related to RNA interaction and signal transduction (39). The results indicate that LGP2 uses ATP hydrolysis to enhance its ability to scan the cytoplasm and efficiently engage diverse dsRNA species. This ATP-enhanced RNA interaction is connected to the ability of LGP2 to potentiate MDA5-mediated signal transduction, as specific mutations that eliminate LGP2 basal ATP hydrolysis also prevent its ability to enhance MDA5 signaling. LGP2 positive regulation of antiviral signaling through MDA5 is further supported by the aforementioned identification of an EMCV-derived MDA5 agonist RNA. The MDA5 agonist was not identified based on association with MDA5 but through its association with LGP2 (51). Together, these features of LGP2 have greatly expanded our understanding of this important innate immune sensor and enable the generation of new models to reconcile its dual roles in RLR regulation (38, 45, 68).
LGP2's positive contributions to antiviral signaling are also highlighted by the actions of virus-encoded IFN antagonists, the paramyxovirus V proteins. V proteins can bind directly to the LGP2 helicase domain, disrupting its ATP hydrolysis activity (Fig. 1A) (69). The V proteins are known to disrupt MDA5 but not RIG-I signaling, and this interference is mediated by a discrete binding site in the helicase domain (69–72). The ∼130-amino-acid minimal V protein binding region (MVBR) encompasses helicase domain 2, an evolutionarily conserved domain within both MDA5 and LGP2 that is more divergent in RIG-I (69). The common viral antagonism supports a positive role for LGP2 and a connection with MDA5. LGP2 has also been linked to more unexpected tissue-specific functions in adaptive immune responses and cancer. The investigation of adaptive immune responses in a line of LGP2-deficient mice indicated a role in promoting the survival of West Nile virus-specific CD8+ T cells (65). LGP2 has also been identified as a critical component of cancer cell resistance to ionizing radiation that is associated with clinical outcomes (73). It is unclear how these LGP2-driven phenomena relate to its roles in antiviral signaling, but these findings demonstrate the importance of further studies to fully understand the mechanisms of LGP2 function in a variety of contexts.
LGP2 AS A CONCENTRATION-DEPENDENT BIPHASIC SWITCH
Combining information from a variety of sources has suggested a theoretical foundation for understanding the mechanisms of LGP2 as both a positive and negative regulator of RLR signaling (Fig. 2). The evidence supports a model in which LGP2 functions as a concentration-dependent switch between MDA5-specific enhancement and a more general RLR interference (68). It has been observed that MDA5 is able to drive transcription from the IFN-β promoter, and titrating LGP2 expression demonstrates that low levels of LGP2 are synergistic with MDA5. LGP2 can produce a concentration-dependent increase in MDA5 signaling activity, referred to as enhancement, optimal activation, or sensitization (39, 74, 75). This activity of LGP2 requires both ATP hydrolysis and RNA binding activities. Further titration of LGP2 expression ultimately achieves a concentration that inhibits MDA5 signaling activity, driving IFN-β expression back toward baseline (39, 75). The MDA5-coactivating activity of LGP2 is revealed in a narrow stoichiometric range, and experiments with single high doses of LGP2 invariably reveal only the inhibitory activity (13, 62, 63).
FIG 2.
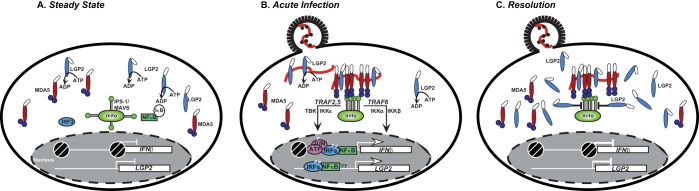
Unifying model to reconcile dual LGP2 functions in antiviral signaling. The panels illustrate known aspects of MDA5 and LGP2 before and after virus infection. For greater clarity, RIG-I is not depicted. (A) Depiction of an uninfected cell at steady state. Blue spheres represent MDA5 CARD motifs, colored ellipses represent MDA5 (red) and LGP2 (light blue) helicase domains, and open ellipses represent CTDs. Both RLRs are present at steady state at modest concentrations, and LGP2 uses its RNA ligand-independent ATP hydrolysis to scan the cytoplasm for dsRNA molecules. IFN-β gene expression is in the repressed, nucleosome (white-hatched black circle)-occupied state, and basal LGP2 transcription is low. In the absence of signaling, IPS-1/MAVS is inactive, and transcription factors IRF3 and NF-κB are in their quiescent, latent cytoplasmic state. (B) Depiction of an acutely infected cell. After virus entry, potential dsRNA ligands accumulate (red line) and are bound by LGP2, which facilitates MDA5 recognition, triggering MDA5-RNA association and seeding filament formation. In turn, this activates IPS-1/MAVS polymerization and downstream signal transduction that induces IFN-β and other antiviral genes, including the LGP2 gene itself. (C) Depiction of an infected cell during the resolution of RLR signaling. LGP2 accumulation as a result of new protein synthesis reaches sufficient concentration to negatively regulate RLR signaling, which occurs downstream from both MDA5 and RIG-I by ATP- and RNA-independent mechanisms. Interference with IPS-1/MAVS signaling by LGP2 is illustrated as a block to downstream signaling, allowing the IFN-β promoter and LGP2 transcription to return to their steady-state configurations.
Titrating LGP2 with RIG-I instead of MDA5 illustrates a key difference between the two RLR responses to LGP2 (62, 75). RIG-I signaling is not enhanced by low LGP2 concentrations, but increasing LGP2 expression results in concentration-dependent inhibition of RIG-I signaling. LGP2 acts as a negative regulator for both RIG-I and MDA5. However, LGP2 is only able to enhance MDA5 signaling. LGP2 mutants with defective ATP hydrolysis and/or RNA binding are unable to stimulate MDA5 signaling but retain negative regulation (39). This difference suggests that the two regulatory actions of LGP2 target different cellular effectors or pathways.
A kinetic model of RLR signaling is proposed to unify these ostensibly opposing actions of LGP2 in MDA5 and RIG-I responses (Fig. 2). At steady state, low concentrations of LGP2 constantly hydrolyze ATP to scan the cytoplasm and sample any dsRNA species (Fig. 2A). During acute infection, LGP2 can mediate viral recognition and communicate with MDA5 to facilitate CARD-mediated signal transduction. LGP2 may function by assisting MDA5 RNA recognition, regulating filament assembly, or otherwise enabling MDA5 communication with IPS-1/MAVS, inducing its polymerization and activation of associated signaling apparatus. The intracellular LGP2 concentration increases as a result of new mRNA and protein synthesis stimulated by the antiviral response (Fig. 2B). As new LGP2 accumulates, ATP- and RNA-independent negative regulatory phenomena cause it to become refractory toward signaling by mechanisms that are directed downstream from both MDA5 and RIG-I (Fig. 2C). This model would support the concept that viral evasion mechanisms could evolve to inactivate positive signaling effects of LGP2 in the MDA5 axis, as has been accomplished by paramyxovirus V proteins that antagonize both MDA5 and LGP2. By disrupting LGP2 ATP hydrolysis, V proteins prevent LGP2 RNA recognition, but negative regulatory effects remain intact (77), providing the virus with a double benefit in overcoming host immunity. Although this hypothetical model is currently based on the idea that the concentration of LGP2 determines its differential activity, it is possible and likely that additional research will reveal posttranslational modifications to LGP2 or MDA5 that regulate the switch between positive and negative actions or control more nuanced signaling activities.
ACKNOWLEDGMENTS
We are grateful to Jean-Patrick Parisien for his comments on the manuscript.
Research on MDA5 and LGP2 in the Horvath laboratory is supported by NIH grants AI073919 and AI50707 to C.M.H. A.M.B. was supported in part by a predoctoral fellowship from the NIH Cellular and Molecular Basis of Disease Training grant T32GM008061.
Footnotes
Published ahead of print 21 May 2014
REFERENCES
- 1.Goubau D, Deddouche S, Reis ESC. 2013. Cytosolic sensing of viruses. Immunity 38:855–869. 10.1016/j.immuni.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacobs JL, Coyne CB. 2013. Mechanisms of MAVS regulation at the mitochondrial membrane. J. Mol. Biol. 425:5009–5019. 10.1016/j.jmb.2013.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rathinam VA, Fitzgerald KA. 2011. Cytosolic surveillance and antiviral immunity. Curr. Opin. Virol 1:455–462. 10.1016/j.coviro.2011.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kato H, Takahasi K, Fujita T. 2011. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol. Rev. 243:91–98. 10.1111/j.1600-065X.2011.01052.x [DOI] [PubMed] [Google Scholar]
- 5.Sun YW. 1997. RIG-I, a human homolog gene of RNA helicase, is induced by retinoic acid during the differentiation of acute promyelocytic leukemia cell. Ph.D. dissertation Shanghai Second Medical University, Shanghai, China [Google Scholar]
- 6.Kang DC, Gopalkrishnan RV, Wu Q, Jankowsky E, Pyle AM, Fisher PB. 2002. MDA-5: an interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc. Natl. Acad. Sci. U. S. A. 99:637–642. 10.1073/pnas.022637199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui Y, Li M, Walton KD, Sun K, Hanover JA, Furth PA, Hennighausen L. 2001. The Stat3/5 locus encodes novel endoplasmic reticulum and helicase-like proteins that are preferentially expressed in normal and neoplastic mammary tissue. Genomics 78:129–134. 10.1006/geno.2001.6661 [DOI] [PubMed] [Google Scholar]
- 8.Cagliani R, Forni D, Tresoldi C, Pozzoli U, Filippi G, Rainone V, De Gioia L, Clerici M, Sironi M. 2014. RIG-I-like receptors evolved adaptively in mammals, with parallel evolution at LGP2 and RIG-I. J. Mol. Biol. 426:1351–1365. 10.1016/j.jmb.2013.10.040 [DOI] [PubMed] [Google Scholar]
- 9.Ramos HJ, Gale M., Jr 2011. RIG-I like receptors and their signaling crosstalk in the regulation of antiviral immunity. Curr. Opin. Virol. 1:167–176. 10.1016/j.coviro.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Jr, Akira S, Yonehara S, Kato A, Fujita T. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851–2858. 10.4049/jimmunol.175.5.2851 [DOI] [PubMed] [Google Scholar]
- 11.Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M, Jr, Patel SS, Marcotrigiano J. 2011. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature 479:423–427. 10.1038/nature10537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, Ranjith-Kumar CT, Brooks MT, Dharmaiah S, Herr AB, Kao C, Li P. 2009. The RIG-I-like receptor LGP2 recognizes the termini of double-stranded RNA. J. Biol. Chem. 284:13881–13891. 10.1074/jbc.M900818200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M., Jr 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. U. S. A. 104:582–587. 10.1073/pnas.0606699104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maelfait J, Beyaert R. 2012. Emerging role of ubiquitination in antiviral RIG-I signaling. Microbiol. Mol. Biol. Rev. 76:33–45. 10.1128/MMBR.05012-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsumiya T, Stafforini DM. 2010. Function and regulation of retinoic acid-inducible gene-I. Crit. Rev. Immunol. 30:489–513. 10.1615/CritRevImmunol.v30.i6.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wies E, Wang MK, Maharaj NP, Chen K, Zhou S, Finberg RW, Gack MU. 2013. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity 38:437–449. 10.1016/j.immuni.2012.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730–737. 10.1038/ni1087 [DOI] [PubMed] [Google Scholar]
- 18.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981–988. 10.1038/ni1243 [DOI] [PubMed] [Google Scholar]
- 19.Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C, Yamamoto M, Uematsu S, Ishii KJ, Takeuchi O, Akira S. 2006. Essential role of IPS-1 in innate immune responses against RNA viruses. J. Exp. Med. 203:1795–1803. 10.1084/jem.20060792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seth RB, Sun L, Ea CK, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669–682. 10.1016/j.cell.2005.08.012 [DOI] [PubMed] [Google Scholar]
- 21.Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ. 2006. The specific and essential role of MAVS in antiviral innate immune responses. Immunity 24:633–642. 10.1016/j.immuni.2006.04.004 [DOI] [PubMed] [Google Scholar]
- 22.Liu S, Chen J, Cai X, Wu J, Chen X, Wu YT, Sun L, Chen ZJ. 2013. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. Elife 2:e00785. 10.7554/eLife.00785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freaney JE, Kim R, Mandhana R, Horvath CM. 2013. Extensive cooperation of immune master regulators IRF3 and NFkappaB in RNA Pol II recruitment and pause release in human innate antiviral transcription. Cell Rep 4:959–973. 10.1016/j.celrep.2013.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Honda K, Taniguchi T. 2006. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 6:644–658. 10.1038/nri1900 [DOI] [PubMed] [Google Scholar]
- 25.Au-Yeung N, Mandhana R, Horvath CM. 2013. Transcriptional regulation by STAT1 and STAT2 in the interferon JAK-STAT pathway. JAKSTAT 2:e23931. 10.4161/jkst.23931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fink K, Grandvaux N. 2013. STAT2 and IRF9: beyond ISGF3. JAKSTAT 2:e27521. 10.4161/jkst.27521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. 2011. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell 147:423–435. 10.1016/j.cell.2011.09.039 [DOI] [PubMed] [Google Scholar]
- 28.Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM. 2011. Structural insights into RNA recognition by RIG-I. Cell 147:409–422. 10.1016/j.cell.2011.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. 2009. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science 323:1070–1074. 10.1126/science.1168352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP. 2008. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol. Cell 29:169–179. 10.1016/j.molcel.2007.10.032 [DOI] [PubMed] [Google Scholar]
- 31. Reference deleted.
- 32.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105. 10.1038/nature04734 [DOI] [PubMed] [Google Scholar]
- 33.Belgnaoui SM, Paz S, Hiscott J. 2011. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr. Opin. Immunol. 23:564–572. 10.1016/j.coi.2011.08.001 [DOI] [PubMed] [Google Scholar]
- 34.Hiscott J, Ware C. 2011. Cytokines. Curr. Opin. Immunol. 23:561–563. 10.1016/j.coi.2011.09.001 [DOI] [PubMed] [Google Scholar]
- 35.Paz S, Vilasco M, Werden SJ, Arguello M, Joseph-Pillai D, Zhao T, Nguyen TL, Sun Q, Meurs EF, Lin R, Hiscott J. 2011. A functional C-terminal TRAF3-binding site in MAVS participates in positive and negative regulation of the IFN antiviral response. Cell Res. 21:895–910. 10.1038/cr.2011.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leung DW, Basler CF, Amarasinghe GK. 2012. Molecular mechanisms of viral inhibitors of RIG-I-like receptors. Trends Microbiol. 20:139–146. 10.1016/j.tim.2011.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loo YM, Gale M., Jr 2011. Immune signaling by RIG-I-like receptors. Immunity 34:680–692. 10.1016/j.immuni.2011.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bamming D, Horvath CM. 2009. Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I, and LGP2. J. Biol. Chem. 284:9700–9712. 10.1074/jbc.M807365200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bruns AM, Pollpeter D, Hadizadeh N, Myong S, Marko JF, Horvath CM. 2013. ATP hydrolysis enhances RNA recognition and antiviral signal transduction by the innate immune sensor, laboratory of genetics and physiology 2 (LGP2). J. Biol. Chem. 288:938–946. 10.1074/jbc.M112.424416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. 2006. Essential role of MDA-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. U. S. A. 103:8459–8464. 10.1073/pnas.0603082103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCartney SA, Thackray LB, Gitlin L, Gilfillan S, Virgin HW, Colonna M. 2008. MDA-5 recognition of a murine norovirus. PLoS Pathog. 4:e1000108. 10.1371/journal.ppat.1000108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sumpter R, Jr, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M., Jr 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79:2689–2699. 10.1128/JVI.79.5.2689-2699.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fredericksen BL, Keller BC, Fornek J, Katze MG, Gale M., Jr 2008. Establishment and maintenance of the innate antiviral response to West Nile Virus involves both RIG-I and MDA5 signaling through IPS-1. J. Virol. 82:609–616. 10.1128/JVI.01305-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, Carney DS, Wang T, Ishida H, Yoneyama M, Fujita T, Saito T, Lee WM, Hagedorn CH, Lau DT, Weinman SA, Lemon SM, Gale M., Jr 2006. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc. Natl. Acad. Sci. U. S. A. 103:6001–6006. 10.1073/pnas.0601523103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gitlin L, Benoit L, Song C, Cella M, Gilfillan S, Holtzman MJ, Colonna M. 2010. Melanoma differentiation-associated gene 5 (MDA5) is involved in the innate immune response to Paramyxoviridae infection in vivo. PLoS Pathog. 6:e1000734. 10.1371/journal.ppat.1000734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yount JS, Gitlin L, Moran TM, Lopez CB. 2008. MDA5 participates in the detection of paramyxovirus infection and is essential for the early activation of dendritic cells in response to Sendai Virus defective interfering particles. J. Immunol. 180:4910–4918. 10.4049/jimmunol.180.7.4910 [DOI] [PubMed] [Google Scholar]
- 47.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 205:1601–1610. 10.1084/jem.20080091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pichlmair A, Schulz O, Tan CP, Rehwinkel J, Kato H, Takeuchi O, Akira S, Way M, Schiavo G, Reis e Sousa C. 2009. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J. Virol. 83:10761–10769. 10.1128/JVI.00770-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luthra P, Sun D, Silverman RH, He B. 2011. Activation of IFN expression by a viral mRNA through RNase L and MDA5. Proc. Natl. Acad. Sci. U. S. A. 108:2118–2123. 10.1073/pnas.1012409108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, Siddell SG, Ludewig B, Thiel V. 2011. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 12:137–143. 10.1038/ni.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deddouche S, Goubau D, Rehwinkel J, Chakravarty P, Begum S, Maillard PV, Borg A, Matthews N, Feng Q, van Kuppeveld FJ, Reis e Sousa C. 2014. Identification of an LGP2-associated MDA5 agonist in picornavirus-infected cells. Elife. 3:e01535. 10.7554/eLife.01535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berke IC, Li Y, Modis Y. 2013. Structural basis of innate immune recognition of viral RNA. Cell. Microbiol. 15:386–394. 10.1111/cmi.12061 [DOI] [PubMed] [Google Scholar]
- 53.Berke IC, Modis Y. 2012. MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA. EMBO J. 31:1714–1726. 10.1038/emboj.2012.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berke IC, Yu X, Modis Y, Egelman EH. 2012. MDA5 assembles into a polar helical filament on dsRNA. Proc. Natl. Acad. Sci. U. S. A. 109:18437–18441. 10.1073/pnas.1212186109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Motz C, Schuhmann KM, Kirchhofer A, Moldt M, Witte G, Conzelmann KK, Hopfner KP. 2013. Paramyxovirus V proteins disrupt the fold of the RNA sensor MDA5 to inhibit antiviral signaling. Science 339:690–693. 10.1126/science.1230949 [DOI] [PubMed] [Google Scholar]
- 56.Peisley A, Jo MH, Lin C, Wu B, Orme-Johnson M, Walz T, Hohng S, Hur S. 2012. Kinetic mechanism for viral dsRNA length discrimination by MDA5 filaments. Proc. Natl. Acad. Sci. U. S. A. 109:E3340–E3349. 10.1073/pnas.1208618109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu B, Peisley A, Richards C, Yao H, Zeng X, Lin C, Chu F, Walz T, Hur S. 2013. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell 152:276–289. 10.1016/j.cell.2012.11.048 [DOI] [PubMed] [Google Scholar]
- 58.Feng Q, Hato SV, Langereis MA, Zoll J, Virgen-Slane R, Peisley A, Hur S, Semler BL, van Rij RP, van Kuppeveld FJ. 2012. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Rep. 2:1187–1196. 10.1016/j.celrep.2012.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peisley A, Lin C, Wu B, Orme-Johnson M, Liu M, Walz T, Hur S. 2011. Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proc. Natl. Acad. Sci. U. S. A. 108:21010–21015. 10.1073/pnas.1113651108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Funabiki M, Kato H, Miyachi Y, Toki H, Motegi H, Inoue M, Minowa O, Yoshida A, Deguchi K, Sato H, Ito S, Shiroishi T, Takeyasu K, Noda T, Fujita T. 2014. autoimmune disorders associated with gain of function of the intracellular sensor MDA5. Immunity 40:199–212. 10.1016/j.immuni.2013.12.014 [DOI] [PubMed] [Google Scholar]
- 61.Peisley A, Wu B, Yao H, Walz T, Hur S. 2013. RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Mol. Cell 51:573–583. 10.1016/j.molcel.2013.07.024 [DOI] [PubMed] [Google Scholar]
- 62.Komuro A, Horvath CM. 2006. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J. Virol. 80:12332–12342. 10.1128/JVI.01325-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rothenfusser S, Goutagny N, DiPerna G, Gong M, Monks BG, Schoenemeyer A, Yamamoto M, Akira S, Fitzgerald KA. 2005. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 175:5260–5268. 10.4049/jimmunol.175.8.5260 [DOI] [PubMed] [Google Scholar]
- 64.Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O. 2010. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc. Natl. Acad. Sci. U. S. A. 107:1512–1517. 10.1073/pnas.0912986107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suthar MS, Ramos HJ, Brassil MM, Netland J, Chappell CP, Blahnik G, McMillan A, Diamond MS, Clark EA, Bevan MJ, Gale M., Jr 2012. The RIG-I-like receptor LGP2 controls CD8(+) T cell survival and fitness. Immunity 37:235–248. 10.1016/j.immuni.2012.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Venkataraman T, Valdes M, Elsby R, Kakuta S, Caceres G, Saijo S, Iwakura Y, Barber GN. 2007. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J. Immunol. 178:6444–6455. 10.4049/jimmunol.178.10.6444 [DOI] [PubMed] [Google Scholar]
- 67.Pollpeter D, Komuro A, Barber GN, Horvath CM. 2011. Impaired cellular responses to cytosolic DNA or infection with Listeria monocytogenes and vaccinia virus in the absence of the murine LGP2 protein. PLoS One 6:e18842. 10.1371/journal.pone.0018842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bruns AM, Horvath CM. 2012. Activation of RIG-I-like receptor signal transduction. Crit. Rev. Biochem. Mol. Biol. 47:194–206. 10.3109/10409238.2011.630974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parisien JP, Bamming D, Komuro A, Ramachandran A, Rodriguez JJ, Barber G, Wojahn RD, Horvath CM. 2009. A shared interface mediates paramyxovirus interference with antiviral RNA helicases MDA5 and LGP2. J. Virol. 83:7252–7260. 10.1128/JVI.00153-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, Randall RE. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, MDA-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. U. S. A. 101:17264–17269. 10.1073/pnas.0407639101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Childs K, Stock N, Ross C, Andrejeva J, Hilton L, Skinner M, Randall R, Goodbourn S. 2007. MDA-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology 359:190–200. 10.1016/j.virol.2006.09.023 [DOI] [PubMed] [Google Scholar]
- 72.Ramachandran A, Horvath CM. 2010. Dissociation of paramyxovirus interferon evasion activities: universal and virus-specific requirements for conserved V protein amino acids in MDA5 interference. J. Virol. 84:11152–11163. 10.1128/JVI.01375-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Widau RC, Parekh AD, Ranck MC, Golden DW, Kumar KA, Sood RF, Pitroda SP, Liao Z, Huang X, Darga TE, Xu D, Huang L, Andrade J, Roizman B, Weichselbaum RR, Khodarev NN. 2014. RIG-I-like receptor LGP2 protects tumor cells from ionizing radiation. Proc. Natl. Acad. Sci. U. S. A. 111:E484–E491. 10.1073/pnas.1323253111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Childs KS, Randall RE, Goodbourn S. 2013. LGP2 plays a critical role in sensitizing MDA-5 to activation by double-stranded RNA. PLoS One 8:e64202. 10.1371/journal.pone.0064202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pippig DA, Hellmuth JC, Cui S, Kirchhofer A, Lammens K, Lammens A, Schmidt A, Rothenfusser S, Hopfner KP. 2009. The regulatory domain of the RIG-I family ATPase LGP2 senses double-stranded RNA. Nucleic Acids Res. 37:2014–2025. 10.1093/nar/gkp059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Runge S, Sparrer KM, Lässig C, Hembach K, Baum A, García-Sastre A, Söding J, Conzelmann KK, Hopfner KP. 2014. In vivo ligands of MDA5 and RIG-I in measles virus-infected cells. PLoS Pathog. 10:e1004081. 10.1371/journal.ppat.1004081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rodriguez KR, Horvath CM. 14 May 2014. Paramyxovirus V protein interaction with the antiviral sensor LGP2 disrupts MDA5 signaling enhancement but is not relevant to LGP2-mediated RLR signaling inhibition. J Virol. 10.1128/JVI.00737-14 [DOI] [PMC free article] [PubMed] [Google Scholar]