ABSTRACT
In recent decades, Raman spectroscopy has entered the biological and medical fields. It enables nondestructive analysis of structural details at the molecular level and has been used to study viruses and their constituents. Here, we used Raman spectroscopy to study echovirus 1 (EV1), a small, nonenveloped human pathogen, in two different uncoating states induced by heat treatments. Raman signals of capsid proteins and RNA genome were observed from the intact virus, the uncoating intermediate, and disrupted virions. Transmission electron microscopy data revealed general structural changes between the studied particles. Compared to spectral characteristics of proteins in the intact virion, those of the proteins of the heat-treated particles indicated reduced α-helix content with respect to β-sheets and coil structures. Changes observed in tryptophan and tyrosine signals suggest an increasingly hydrophilic environment around these residues. RNA signals revealed a change in the environment of the genome and in its conformation. The ionized-carbonyl vibrations showed small changes between the intact virion and the uncoating intermediate, which points to cleavage of salt bridges in the protein structure during the uncoating process. In conclusion, our data reveal distinguishable Raman signatures of the intact, intermediate, and disrupted EV1 particles. These changes indicate structural, chemical, and solute-solvent alterations in the genome and in the capsid proteins and lay the essential groundwork for investigating the uncoating of EV1 and related viruses in real time.
IMPORTANCE In order to combat virus infection, we need to know the details of virus uncoating. We present here the novel Raman signatures for opened and intact echovirus 1. This gives hope that the signatures may be used in the near future to evaluate the ambient conditions in endosomes leading to virus uncoating using, e.g., coherent anti-Stokes Raman spectroscopy (CARS) imaging. These studies will complement structural studies on virus uncoating. In addition, Raman/CARS imaging offers the possibility of making dynamic live measurements in vitro and in cells which are impossible to measure by, for example, cryo-electron tomography. Furthermore, as viral Raman spectra can be overwhelmed with various contaminants, our study is highly relevant in demonstrating the importance of sample preparation for Raman spectroscopy in the field of virology.
INTRODUCTION
Picornaviruses are small, nonenveloped human pathogens able to cause a wide range of illnesses (1). The subgroup enteroviruses include clinically important echoviruses, coxsackieviruses, and poliovirus. These viruses cause a variety of diseases varying from mild infections to aseptic meningitis, heart muscle damage, and paralysis. Enteroviruses have also recently been associated with chronic diseases such as type 1 diabetes, cardiomyopathies, and atherosclerosis (2, 3). Structural details of these viruses have been obtained by means of X-ray crystallography or at a lower resolution by cryo-electron microscopy (4). Various imaging techniques together with biochemical analysis have revealed important information about their entry into cells and the subsequent events leading to uncoating and virus replication (5, 6). However, information on the physicochemical details of genome release events of enteroviral particles is sparse. The uncoating process has been extensively studied by initiating the genome release using heat treatments where intact virions are irreversibly converted to subviral particles that resemble naturally occurring uncoating intermediates (7, 8, 9). Recently, by using such temperature-manipulated particles, a long-standing paradigm of the genome release model from the 5-fold axis was revised (10, 11). However, at present, information on the biochemical events leading to the genome release as well as the initiating environmental trigger of uncoating is lacking (1). Further studies are thus important for addressing one of the events in the enteroviral life cycle, the genome release, and also for the possibility of spotting new potential antiviral targets in related enteroviruses.
Echovirus 1 (EV1) is a picornavirus belonging to a structurally related group of enteroviruses. Common to all picornaviruses, EV1 is an icosahedron-shaped (T=1) assembly of 60 copies of identical protomers comprised of four viral proteins (VP1, VP2, VP3, and VP4), which encapsulate a positive-sense single-stranded RNA genome of approximately 7,500 nucleotides. A 14-carbon saturated fatty acid, myristate, is covalently attached to the N terminus of each picornavirus VP4 capsid protein (12), and it is thought to exit the capsid simultaneously with VP4 during the uncoating process (13). Additionally, studies on poliovirus have shown that the N terminus of VP1 is externalized to the virion surface during the uncoating process, increasing the overall hydrophobicity of the particle (14). Also, the release of the stability-mediating pocket factor, characterized as palmitic acid and located in the core of VP1, is thought to be a prerequisite in the uncoating process, as was shown for bovine enterovirus (15, 16).
Raman spectroscopy provides specific signatures of proteins, nucleic acids, and lipids. It can reveal vast amount of information about changes taking place in the chemical content of the particles, in the protein secondary structure, and in the physical environment of the particles, which undergo biologically important transformations (17). Raman spectroscopy is a noninvasive characterization technique which uses visible-light laser beams and thus enables microscopic mode with the same resolution as fluorescence microscopy. As the acquisition can be fast, processes in real time can be studied. As Raman spectra are only slightly disturbed by a water environment, information about molecules in their natural habitat or under a wide range of conditions can be obtained. This makes it an ideal probe to study detailed structural alterations and viral protein assembly, interactions, and dynamics, without any labels or other invasive sample preparation methods.
Raman spectroscopic studies on viruses such as turnip yellow mosaic virus (TYMV) (18), bean pod mottle virus (BPMV) (19, 20), and belladonna mottle virus (BDMV) (21) and bacteriophages such as filamentous bacteriophage Ff (22, 23), PRD1 virus (24), and P22 (25–27) have been reported. However, these viruses differ substantially from enteroviruses in several respects. In the plant viruses TYMV, BPMV, and BDMV, the genome is bound to the protein capsid in static form, whereas in the enteroviruses, the interior of the virion is spatially disordered with respect to the symmetric protein shell (28). In addition, the subunit packing is somewhat different between these bromoviruses and enteroviruses (28). The bacteriophage PRD1 is icosahedral but has a lipid envelope. Phage P22 is a nonenveloped, head-tail structured virus, with 6 prominent tail spikes. All such properties influence the Raman spectroscopic character of these viruses.
Detailed Raman spectroscopic signatures from the virus uncoating process of enteroviruses, both in vivo and in vitro, are missing. For such studies, linear Raman experiments of the initial and final states are a prerequisite. Here, we characterized vibrational spectroscopic properties of intact EV1 virions and investigated the spectral differences arising in the uncoating process of EV1 particles in vitro. First, we elucidated the Raman spectroscopic basis of recognition between the intact virion and the disrupted EV1 particle. Further, we determined the Raman signature of the uncoating intermediate, which is obtained by heating the intact, infectious particles at 50°C. Previous studies utilizing heat treatments in order to produce and to investigate uncoating intermediates of enteroviruses have produced a wealth of information. Heating enterovirus particles at 50 to 60°C for various amounts of time has been shown to be able to produce uncoating intermediates (135S) and result in genome ejecting, as well as producing empty (80S) particles indistinguishable from those observed in vivo (7, 10, 11). The Raman spectra of the particles that were heat treated at 50°C represent a state where the genome is partially ejected from the capsid, as confirmed by transmission electron microscopy (TEM) and thermal stability assays (29). We conclude that there are clear Raman markers which address the uncoating stage of the virion. The experiments reported here provide a framework for monitoring the sequence of chemical and conformational changes occurring during enterovirus uncoating in a time-resolved manner.
MATERIALS AND METHODS
Cell culture and virus purification.
Echovirus 1 (Farouk strain, obtained from the ATCC) was propagated in a monolayer of GMK cells and purified using an overall scheme similar to that described previously (30). To summarize, overnight (24-h)-infected cells were collected and repeatedly freeze-thawed (3 cycles) to lyse cellular structures. Bulk cell debris was removed by centrifugation (4,500 × g for 15 min). To precipitate protein, 8% (wt/vol) polyethylene glycol 6000 (PEG 6000) (Sigma-Aldrich) and 2.2% (wt/vol) NaCl were added to the supernatant and stirred overnight (24 h) at 4°C. Precipitated material was collected by centrifugation (8,000 × g for 40 min) and suspended in R buffer (10 mM Tris-HCl [pH 7.5], 200 mM NaCl, 50 mM MgCl2, 10% [wt/vol] glycerol). To disrupt membranous structures, 0.3% (wt/vol) sodium deoxycholate (Sigma-Aldrich) and 0.6% (vol/vol) Nonidet P-40 substitute (Sigma-Aldrich) were added to the suspension and incubated for 30 min at 4°C. The resulting mixture was clarified by centrifugation (3,500 × g for 15 min) and divided equally (<1.5 ml/gradient) on top of 10-ml linear density gradients of 10 to 40% (wt/vol) sucrose in R buffer. The gradients were subsequently ultracentrifuged (86,000 × g for 3 h at 4°C) and fractioned from the top into 500-μl aliquots. Based on optical density measurement at 260 nm (NanoDrop 1000; Thermo Scientific), three virion-containing fractions (fractions 11, 12, and 13) were pooled and diluted to 10 ml in phosphate-buffered saline (PBS) supplemented with 2 mM MgCl2. The diluted fractions were dialyzed three times against 1 liter of PBS supplemented with 2 mM MgCl2 using 50,000-molecular-mass-cutoff Spectra/Por Float-A-Lyzer cellulose ester membranes (Spectrum Laboratories). Finally, dialyzed virions were pelleted by ultracentrifugation (93,000 × g for 2 h at 4°C) and suspended in a small volume of PBS supplemented with 2 mM MgCl2. All measured samples were dissolved in the aforementioned PBS buffer. Virions were stored at −80°C.
Infectivity of the purified EV1 virions.
Infectivity of the purified virus batch was assayed using the endpoint titration method to determine the 50% tissue culture infective dose (TCID50). The TCID50 was calculated as described previously (31), positioning the TCID50 between the last infected and first noninfected well. Briefly, a monolayer of GMK cells seeded at 5 × 104 ml−1 on a 96-well plate were infected with serially diluted virus stock (starting from a 10−5 dilution). The progression of the infection was followed daily by light microscopy. After 72 h of incubation, the cells were stained using crystal violet supplemented with 10% formalin (for 10 min at room temperature [RT]). The detached cells were washed off with water, and remaining attached cells were counted as viable and noninfected. For the EV1 batches used in the experiments, the TCID50 was found to be 6.1 × 1011 to 1.5 × 1012 virus particles/ml.
Protein content and composition of purified EV1.
The protein content of purified EV1 was determined using a method described by Porterfield and Zlotnick (32), where an average value of molecular mass (23.5 kDa) and an average molar absorptivity value (ε280, 34,440 M−1 cm−1) for VP1 to -4 were used for the calculations. Protein concentrations of the batches were 0.9 to 3.8 mg ml−1 after subtraction of the scattering component (6.9% of the baseline subtracted absorption signal at 260 nm). The A260/A280 ratio was 1.65. The absorption spectrum was recorded with a PerkinElmer Lambda 850 spectrophotometer using a 10.0-mm optical path quartz cuvette (Hellma Analytics). The protein composition of purified EV1 was analyzed using 12% SDS-PAGE. The sample lane was loaded with 5 μg of EV1, and PageRuler Plus (Thermo Scientific) was used as a molecular weight marker.
Thermal stability.
The thermal stability of the purified EV1 particles was assayed using the method described by Walter et al. (29). The fluorescence signal was recorded using a Bio-Rad C1000 thermal cycler, and the final sample mixture contained 1 μg of EV1 and a 10× concentration of SYBR green II (Invitrogen). All samples were equilibrated at 20°C for 10 min before thermal stability measurements were started. For the full temperature range scan, the fluorescence signal was recorded at 10-s intervals with 0.5°C increments. In additional measurements, the designated temperature was kept constant for 3 min (50°C) or 10 min (60°C), and after heat treatment, the sample was cooled back to 20°C, at which point the fluorescence reading was taken or the sample was loaded to an EM grid.
Transmission electron microscopy.
The morphology of the heated and nonheated virions was visualized by negative staining with 1% (wt/vol) phosphotungstic acid (Sigma-Aldrich). The Formvar-coated EM grids were glow discharged (EMS/SC7620 mini-sputter coater), and samples were deposited on the grids for 15 s, after which the excess sample was blotted away (Whatman 3MM). Next, negative stain was added for 1 min and removed as before. Samples dried at least overnight were imaged with a JEM-1400 (JEOL) transmission electron microscope with a magnification of ×50,000 (80 kV).
Raman spectroscopy.
For all Raman viral experiments, aliquots of 6 to 10 μl with a sample concentration of 0.9 mg ml−1 for dried samples and 3.8 mg ml−1 for aqueous samples were either deposited on a CaF2 window or sealed in a glass capillary (SMI capillaries, P5070-902; American Dade). Heating of the aqueous samples was performed in a water bath, either at 50°C for 3 min or at 60°C for 10 min. The dried sample was allowed to dry for 15 min at ambient temperature. The Raman spectra were measured with a home-built Raman setup in a backscattering geometry. In this setup, a laser beam of solid-state laser (CNI) with an excitation wavelength of 532 nm was focused onto the sample with a spot diameter of 3 μm using a microscope objective (Plan Neofluar 10×/0.30; Zeiss). Raman scattering was recorded using a charge-coupled device (CCD) camera (Newton DU940P-BV; Andor) attached to an imaging spectrograph (Acton Spectra Pro 2500i) with an entrance slit of 50 μm and a grating of 600 grooves/mm. Raman spectra were accumulated using 30 10-s exposures. Laser powers of 45 mW for dried and 200 mW for aqueous samples were used. From each scan, a dark spectrum was subtracted. The reference spectra of water, buffer solution, empty capillary, and CaF2 window were detected with the same settings and subtracted from the sample spectra. The fluorescence background was defined by fitting a polynomial curve to the measured spectrum from the points where no Raman signal is expected to be present and subtracted from the data. After this procedure, no spectral signatures other than those of viral constituents were obtained.
Spectra of the aqueous samples at a concentration of 3.8 mg ml−1 represent means of measurements from three different EV1 batches normalized to the Phe peak at 1,003 cm−1 with a floating average with a span of three data points (2.7 cm−1). In addition, the differences between room temperature and heated samples were analyzed in four separated spectral sections, 610 to 945 cm−1, 945 to 1,145 cm−1, 1,145 to 1,520 cm−1, and 1,520 to 1,800 cm−1, which were normalized to the mean signals of each section and smoothed with a floating average with a span of seven data points (10 cm−1).
Radial distribution and positioning of salt bridges in EV1.
The atom coordinates were gathered from the EV1 protomer (PDB entry 1EV1) using UCSF Chimera (33) Build 1.8. Salt bridges were identified manually, scoring only arginine and lysine as salt bridge-forming residues with carboxylic oxygen of Glu and Asp. In the case of many possible connections (<3.5 Å), the closest was chosen, resulting in one connection per residue.
RESULTS
Sample purity and heat-induced uncoating of EV1.
The Raman analysis of viral particles imposes a demand for highly homogenous and pure samples. The sucrose gradient purification scheme used here resulted in a single, clearly visible band. The fractioned gradient showed a local absorbance maximum at 260 nm in fraction 12, while five top fractions also showed high optical density; no visible banding was observed (Fig. 1A). After dialysis, the collected fractions (fractions 11 to 13) were analyzed for protein composition using 12% SDS-PAGE. Figure 1C shows that the purified EV1 was contaminant-free in the molecular mass range of 250 to 10 kDa, while the prominent capsid proteins, VP1 to -3, banded next to 35- to 25-kDa markers, as expected. To quantify the protein concentration of a virus sample, a UV-visible (UV-Vis) spectroscopic method introduced by Porterfield and Zlotnick (32) was used. In our hands, the above-mentioned spectroscopic approach has been found to be in reasonable agreement with the more traditional Bradford assay (data not shown). The method incorporates scattering correction into the resulting absorption spectrum, with 6.9% of the total signal. This subsequently affects the A260/A280 ratio, which was 1.65 for dialyzed particles (Fig. 1B). In literature, the reported A260/A280 ratios for enteroviruses vary considerably (34–36), partially due to differences in genome contents but also resulting from uncorrected spectra. However, as the genome-to-protein ratio (i.e., the A260/A280 ratio) does not reflect virion integrity, EV1 was visualized by TEM and was found to consist of a highly homogenous population of intact particles approximately 25 nm in diameter (Fig. 1D).
FIG 1.
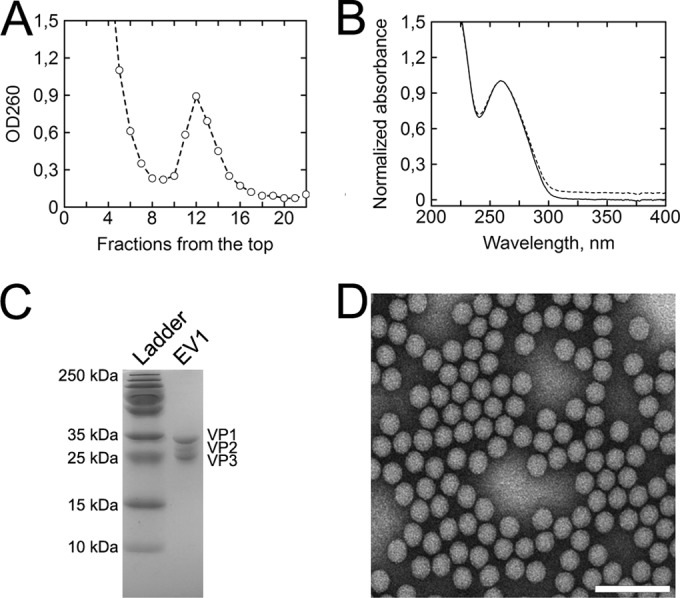
The scheme used to purify EV1 produces a homogenous population of intact virions. (A) A sucrose gradient fractioned from the top shows clear separation in optical density (OD260) between bulk, low-density material and EV1 virions. Fractions 11, 12, and 13 were collected for further purification. (B) Absorption spectrum of dialyzed EV1 (dashed line) and light scattering-corrected spectrum (solid line) normalized at 260 nm for better comparison between the two. The scattering component was calculated to be 6.9% of the total signal, and the A260/A280 ratio was 1.65. (C) SDS-PAGE analysis of the purified EV1 samples shows no protein contaminants in the range of 250 to 10 kDa, while bands of the prominent viral proteins VP1 to -3 appear between the 35-kDa and 25-kDa markers. (D) EV1 particles contrasted by negative staining and visualized by TEM show homogenous population of intact virions approximately 25 nm in diameter. Scale bar, 100 nm.
Since the EV1 capsid proved to be nonpermeable to the SYBR green II dye (Fig. 2A), thermal stability assay was used along with TEM to evaluate the extent of genome egress from intact EV1 particles. Figure 2A indicates that the melting temperature (Tm) for intact EV1 particles is 51°C. Noting that SYBR green II signal begins to increase around 40°C and accumulates over approximately 3 min of effective heating time prior to the 51°C Tm mark, 3 min of heating at 50°C was used to produce the so-called uncoating intermediate particles (Fig. 2B). The population of these genome-releasing virions was visualized by TEM (Fig. 3). After 3 min of heating at 50°C, the great majority of the visualized particles were connected to an electron-dense protrusion (Fig. 3B). Additionally, Fig. 3A and B show that both the intact (20°C) virions and partially disrupted virions (50°C) are structurally organized and symmetrical. Similar particles were obtained earlier using polioviruses (11). Heating the intact EV1 virions at 60°C resulted in a complete particle disruption (Fig. 3C). Overall, homogenously genome-deprived but structurally intact EV1 particles were challenging to produce by heat treatment alone (data not shown). The heat-induced chemical and structural alterations in the particles treated at 20°C, 50°C, and 60°C were then further probed by Raman spectroscopy.
FIG 2.
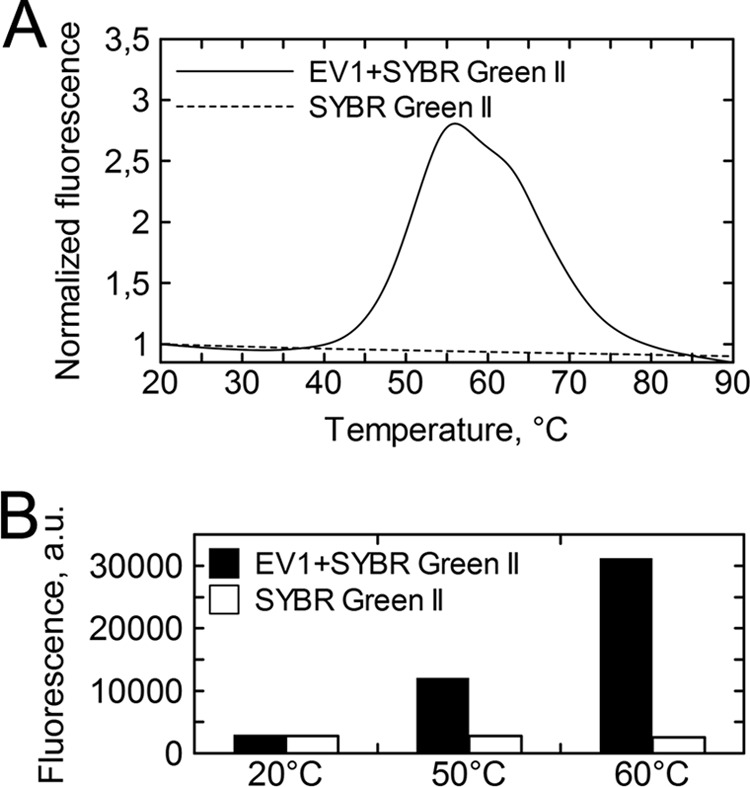
Thermal stability analysis of intact EV1 virions. (A) Fluorescence traces of SYBR green II in the presence of EV1 show a steep increase starting from around 40°C and a midpoint at 51°C (solid line). SYBR green II by itself shows no fluorescence increase as a function of temperature (dashed line). The fluorescence is normalized with respect to the initial fluorescence. (B) EV1 was incubated with SYBR green II at 50°C and 60°C for 3 and 10 min, respectively, and cooled back to 20°C for fluorescence measurement. At 20°C EV1 is not permeable to the dye, whereas temperature-induced genome egress is detected as an increase in fluorescence. The white bars represent the behavior of SYBR green II alone when subjected to the described treatment.
FIG 3.
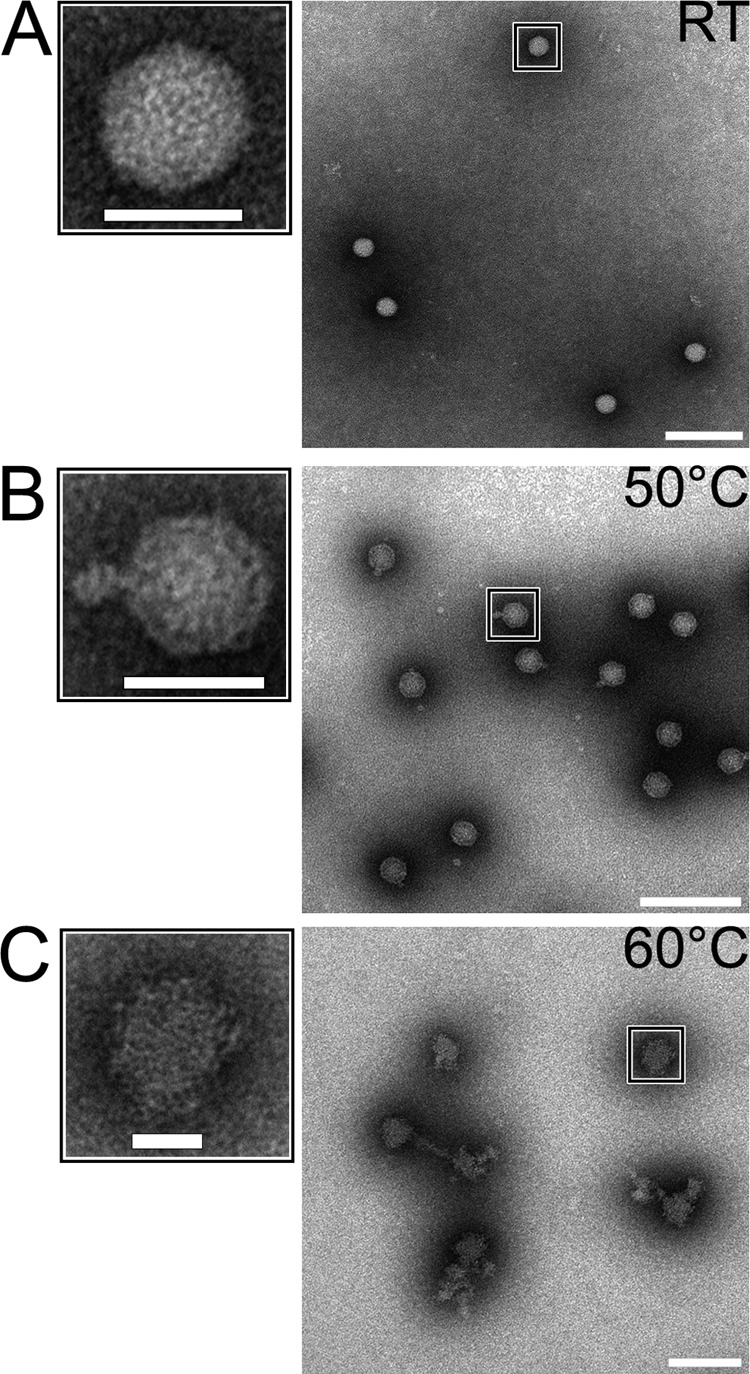
TEM images of negative stained EV1. (A) Intact EV1 virion. (B) EV1 heated at 50°C for 3 min shows an electron-dense protrusion from an otherwise structured particle. (C) Heating for 10 min at 60°C results in complete particle disruption. Scale bars, 25 nm and 100 nm (left and right images, respectively).
Raman analysis of viral particles.
The Raman spectrum of the intact virion (Fig. 4) shows a large number of vibrations that can be assigned reasonably well using values in the literature, as expressed in Tables 1 and 2 for proteins and RNA, respectively (17, 24, 37, 38, 39). The most prominent vibrational region at 2,900 cm−1 originates from aliphatic C-H vibrations. Here, however, we concentrated on the fingerprint region from 600 to 1,800 cm−1, where most previous biological applications of Raman spectroscopy have been performed, as it has higher sensitivity to the structural and chemical nature of particles (for examples, see references 18 and 23). The best-known spectral features found at typical positions (17) in the Raman spectra of the virus are the amide I (Am I) and Am III bands at around 1,670 cm−1 and in the region between 1,230 and 1,300 cm−1, respectively. The anti-symmetric C-C stretching modes of aromatic amino acids (Phe, Tyr, and Trp) locate in the region of 1,580 to 1,615 cm−1 and the ring breathing modes of aromatic amino acids at around 1,000 cm−1. Other side chain vibrations emerge at around 600 to 900 cm−1, and the methyl/methylene vibration bands are located at around 1,350 and 1,450 cm−1. Many RNA vibrations partially overlap the protein bands, although a large number of vibrations can still be specified. The ring breathing modes of RNA bases appear in the region below 800 cm−1, ribose and phosphate signals between 800 and 1,100 cm−1, and the stretching vibrations of the bases at higher wave numbers (1,200 to 1,600 cm−1) (39, 40).
FIG 4.
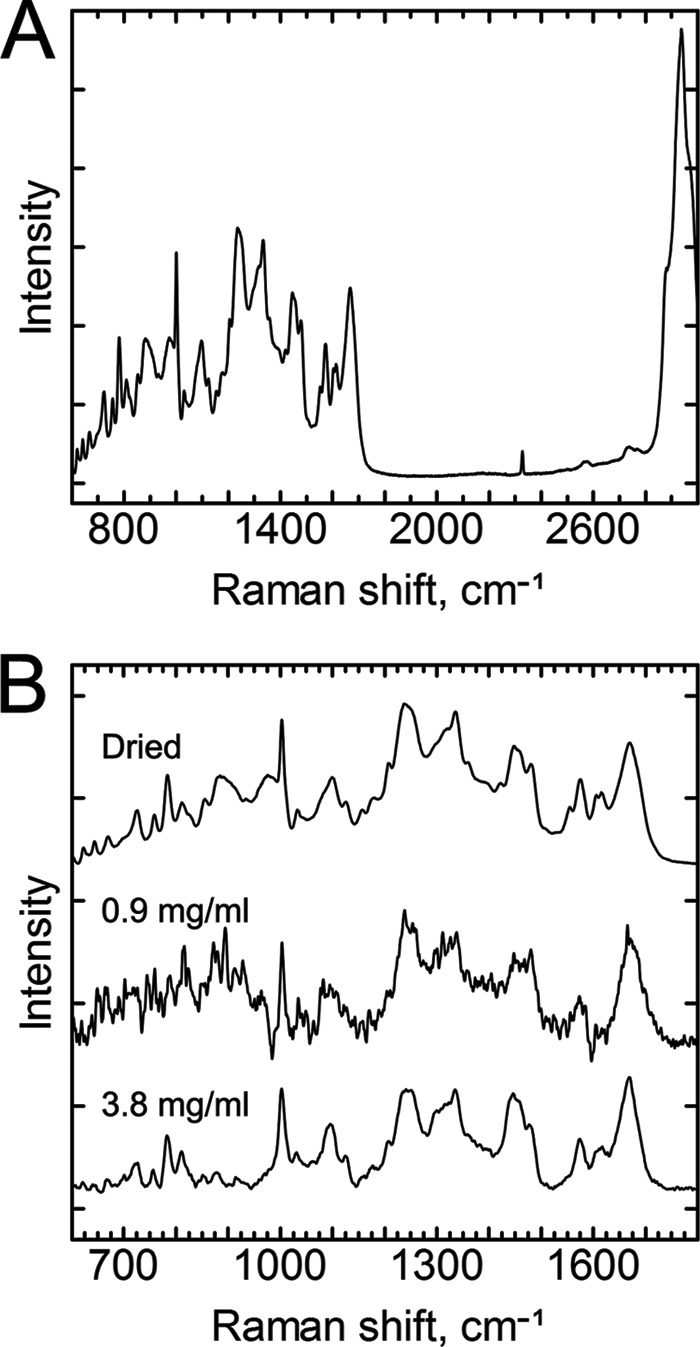
Comparison of the Raman spectra of dried and aqueous EV1 particles. (A) Full Raman spectra of dried intact EV1 virions. The most prominent vibrational region originates from the aliphatic C-H stretching vibrations shown at around 2,900 cm−1, which, however, has a low information content regarding the structural details of the virion particles. The fingerprint region is located between 700 and 1,800 cm−1. The sharp band at 2,331 cm−1 originates from N2. (B) Fingerprint regions of dried EV1 virion (top line) and of aqueous EV1 at 0.9 mg ml−1 and 3.8 mg ml−1 (middle and bottom lines, respectively). Laser excitation at 532 nm and laser powers of 45 mW for dried and 200 mW for the aqueous viral samples were used with 30 exposures of 10 s in each measurement. The spectra were normalized to that of the Phe vibration at 1,003 cm−1, and the baseline was shifted for better comparison of the spectra. All spectra were measured at ambient temperature.
TABLE 1.
Obtained frequencies of the vibrations of proteins found in this study
| Assignmenta | Frequency (cm−1) |
Heat-induced effect | |
|---|---|---|---|
| Dried sample | Aqueous sample | ||
| Phe | 623 | 620 | + |
| Tyr | 644 | 642 | |
| C-S | 670 | 668 | |
| Trp | 758 | 756 | + |
| Tyr | 826, 855 | 829, 854 | − (829)/+ (854) |
| Trp | 887 | 873–883 | − (60°C) |
| AmIII | 960–978 | 952–977 | |
| Phe | 1,003 | 1,003 | |
| Phe | 1,031 | 1,031 | |
| CC, CH | 1,071-1,096 | 1,073-1,098 | + |
| Nonaromatic side chains | 1,127 | 1,122-1,128 | + (1,120)/− (1,130) |
| CH3 | 1,156-1,176 | 1,160-1,178 | |
| Phe, Tyr | 1,208 | 1,205-1,210 | |
| Am III | 1,230-1,300 | 1,230-1,300 | + (1,240)/− (1,300) |
| Trp, CH3 | 1,318-1,378 | 1,322-1,383 | |
| Asp, Glu-COO− | 1,390-1,404 | 1,390-1,402 | + (60°C) |
| CH2 | 1,446-1,458 | 1,446-1,458 | − |
| Trp | 1,555, 1573 | 1,549, 1,572 | + |
| Phe | 1,588-1,594 | − | |
| Phe, Trp | 1,605 | 1,603 | |
| Tyr, Trp | 1,613, 1,618 | 1,612, 1,616 | |
| Am I | 1,669 | 1,668 | + |
| Asp, Glu-COOH | 1,710 | + (50°C) | |
TABLE 2.
Obtained frequencies assigned to the RNA molecule from the intact virus particles
| Assignmenta | Frequency (cm−1) |
Heat-induced effect | |
|---|---|---|---|
| Dried sample | Aqueous sample | ||
| A ring stretching (726) | 726 | 722-727 | |
| C/U breathing/stretching (787) | 783 | 783 | |
| O-P-O symmetric stretching (813) | 811 | 811 | + (60°C) |
| Ribose-phosphate, C-O stretching (919) | 914-921 | − | |
| Ribose-phosphate, C-O stretching (977) | 969-980 | − | |
| P-O stretch, sugar phosphate; C-O stretching (1,047) | 1,044 | 1,044shb | + |
| PO2− symmetric stretch (1,097) | 1,091-1,100 | 1,090-1,100 | |
| U/C ring stretching (1,253) | 1,252 | 1,250 | |
| C/A ring stretching (1,300) | 1,300 | 1,300 | − |
| A, G, U (1,336) | 1,336 | 1,335 | + |
| U/C ring stretching (1,460) | 1,460 | 1,458 | − |
| A/G ring stretching (1,485) | 1,480 | 1,478 | + |
| A/G ring stretching (1,578) | 1,576 | 1,578 | + |
The assignment is based on the study by Hobro et al. (39).
sh, shoulder.
Raman spectra of viral particles in the buffer environment show clear similarities with spectra of the dried particles (Fig. 4B), and basically all spectral features can be observed from the solution with a concentration as low as 0.9 mg ml−1. However, the details of the spectra were more clearly distinguished from the noise level of our experiment at a virus concentration of 3.8 mg ml−1, which was therefore used in the characterization of the heat-induced changes of the viral particles.
By detecting Raman spectra of the particles treated with different temperatures, we aimed to detect chemical and structural changes between the intact virus particle and an uncoating intermediate virion particle, as well as with an endpoint structure where the virion particle was disrupted by heat treatment (morphology of particles obtained with TEM [see above; Fig. 3]). As shown in Fig. 5, the Raman spectra of the particles that had been heat treated at 50°C and 60°C deviate clearly from that of the intact virion. As the heat treatment changes the form (Fig. 3) and possibly also the solvent shell of the particles, the scattering and fluorescence properties of the particles differ. This creates challenges in baseline determination and normalization of the spectra, and therefore, the Raman spectra were dissected in four individual spectral regions from 610 to 1,800 cm−1. Figure 5A to D show the differences between the particles. In most cases, a gradual change of Raman spectra from intact to 50°C-treated and then to 60°C-treated particles was observed. This effect can be expected according to the TEM analysis as well (Fig. 3), which shows basically a full disruption of the particles after 60°C heat treatment.
FIG 5.
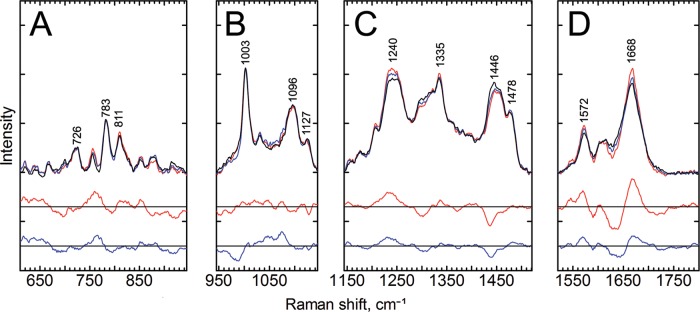
Raman spectra of aqueous EV1 particles at room temperature. The top lines show the Raman spectra of intact EV1 (RT, black line), intermediate particles (50°C, blue line), and disrupted particles (60°C, red line). Below are the 2-fold-magnified difference spectra corresponding to the temperature intervals between 60°C and RT (red) and between 50°C and RT (blue). The spectra were divided into individual sections and normalized to the mean signals of each section, and the baselines were shifted for better visualization. (A) 610 to 945 cm−1; (B) 945 to 1,245 cm−1; (C) 1,245 to 1,520 cm−1; (D) 1,520 to 1,800 cm−1. Marked vibrations from lower to higher frequency are as follows: 726 cm−1, RNA; 783 cm−1, RNA; 811 cm−1, RNA phosphate symmetric stretching, an A-form RNA helix marker; 1,003 cm−1, Phe; 1,096 cm−1, a complex region of CC and CH vibrations of lipid, RNA, and protein capsid; 1,127 cm−1, nonaromatic amino acids and lipids; 1,240 cm−1, Am III; 1,335 cm−1, RNA bases; 1,446 cm−1, C-H vibrations from the proteins; 1,478 cm−1, RNA bases; 1,572 cm−1, Trp/RNA; 1,668 cm−1, Am I, revealing information about the secondary structure of the proteins. Data acquisition was performed as described for Fig. 4, and the spectra represent mean signals of measurements from three different EV1 batches, all with a concentration of about 3.8 mg ml−1.
The largest changes in the spectra were observed in signals assigned to the amide bands, the methyl/methylene signal at 1,446 cm−1, and RNA signals at 750 to 800 cm−1, although many changes are located in regions where multiple factors affect the spectra. Interestingly, the 1,446 cm−1 band, reflecting C-H vibrations from the proteins, is more affected than the corresponding band for RNA molecules at 1,478 cm−1. Thus, the ratio between 1,446 and 1,478 cm−1 conveniently indicates changes of signals between proteins and genome, respectively, in the viral particles. The Am I and Am III regions, shown in Fig. 5, reveal information on the changes in the secondary structure of the coat proteins in the heat-treated particles. The differences between the spectra are the most pronounced at 1,630 to 1,650 cm−1 and 1,670 cm−1 (Fig. 5D). Broadening of the Am I peak is greatest in the spectrum of the intact virus. On the lower-energy side of the Am I vibration, the 1,572 cm−1 band, which originates from ring breathing modes, both from Trp as well as from adenine and guanine of RNA (39, 41), shows higher intensity in the heated particles, especially in the spectra of the particles treated at 60°C.
Most of the heat-induced changes grow gradually as the heat treatment becomes more intense. Some of the changes, however, are evident only after heat treatment at 50°C, namely, the decrease of signals at 990 and 1,650 cm−1. At around 990 cm−1, there are RNA vibrations from ribose phosphate (39) and protein-derived signals from proline, arginine, and tyrosine residues, as well as the nearby phenyl alanine signal at 1,003 cm−1 (37). At 1,650 cm−1, the signal originates from the amide bonds of proteins (17).
Apart from major differences in the Raman signals, subtle signal variations along the spectra can reveal information from important details of the EV1 uncoating process. These changes along with the assignments are indicated in Tables 1 and 2. For example, a slight change in the signal at around 1,710 cm−1 in the spectra of the particles treated at 50°C can be observed (Fig. 5D). This signal increase could imply protonation changes of carboxylic groups of Glu residues (42). In ideal case, such increase of signal of the protonated forms of Glu should lead to a decrease in the signals of the COO− population, found in the region between 1,390 cm−1 and 1,410 cm−1, which, however, is hard to detect in Fig. 5C from the spectra of viruses that were heat treated at 50°C. Another small change, a shift to lower wave numbers, or a decrease in the signal at the higher-frequency side of the band, is seen in the peak at 1,122 to 1,128 cm−1 (Fig. 5B). This peak represents nonaromatic amino acid residues, but it might also include some fatty acid vibrations. There are two kinds of fatty acids in the EV1 capsid, myristate and palmitic acids, the first being covalently attached to VP4 and the second being buried in a VP1-formed pocket. No obvious fatty acid peaks can be detected from the spectra, but there are small decreases in signals of heat-treated viruses at the fatty acid signal sites at 1,063 cm−1, 1,129 cm−1, and 1,296 cm−1 (37) that could be consistent with heat-induced changes in the local environment of the fatty acid molecules or reduction of fatty acid molecule amounts at the measured spot of the sample.
The Fermi doublets of the aromatic amino acids Trp and Tyr are often used as indicators of indole environment hydrophobicity and phenolic hydrogen bonding. Neither of these doublets stands out very clearly from the spectra, but they can be assigned to specific signals nonetheless. Small changes are seen in signals assigned to the Tyr Fermi doublet at 829 and 854 cm−1 (Fig. 5A), where heat treatments reduce the Raman signal ratio I829/I854, indicative of a greater hydrogen bond acceptor role, or exposure to the solvent, of the average phenoxyl group after heat treatments (43). The Trp Fermi doublet at 1,340 and 1,360 cm−1 shows a small increase in the signal at 1,340 cm−1 after heat treatment (Fig. 5C), indicating an increasingly hydrophilic environment of Trp residues (44, 45), although the overlapping RNA signal assigned to the larger peak at 1,335 cm−1 might affect this as well. A Trp band at around 880 cm−1 has been considered a marker for Trp hydrogen bonding, with signals at 882 to 883 cm−1 indicating Trp residues with no hydrogen bonding and a signal at 871 cm−1 indicating residues with strong hydrogen bonding (44). In intact viruses, this Trp signal is a single peak centered at 877 cm−1, and after heat treatments, the signal divides to two peaks at 873 and 881 cm−1, indicating changes in Trp hydrogen bonding status. The frequency of the Trp signal near 1,550 cm−1 has been reported to be indicative of changes in side chain conformation (46–48), and there are heat-induced changes in this region in our data (Fig. 5D). Particularly, in the spectrum of the intact virus, this signal overlaps with the peak at 1,572 cm−1, and after heat treatment, a new peak slightly under 1,550 cm−1 appears, suggesting a considerable change in conformation of some of the Trp residues. A similar change has been noticed for bovine enterovirus uncoating with resonance Raman spectroscopy (46). However, the RNA signal also appears relatively close at around 1,578 cm−1. Yet another Trp peak, at 756 cm−1, also shows apparent heat-induced changes, supporting the interpretation that Trp residues undergo significant changes upon virus uncoating.
Partially overlapping the Trp 756 cm−1 signal is an RNA signal at 783 cm−1, coming from the C/U ring breathing modes (Fig. 5A). This signal has been reported to change in intensity due to changes in the environment, particularly in nonhelical regions of RNA (39, 49). In our experiment, there were heat-induced changes around this peak. A small increase is also seen in the symmetric phosphate stretching vibration signal at 811 cm−1, considered a marker for the A-form RNA helix, suggesting a change in RNA backbone conformation. Most of the RNA signals somewhat overlap protein signals; however, RNA can be considered the main source of the peaks at 1,335 and 1,478 cm−1. More detailed assignments of signals to RNA vibrations are presented in Table 2 (39). Most RNA-assigned signals above 1,200 cm−1 slightly increase in intensity after heat treatments, with one exception. The decrease at 1,300 cm−1 could come from changes in cytosine or adenine ring stretching vibrations, although it could also be due to fatty acids or changes in protein secondary structures. A signal just below 1,050 cm−1 that can be assigned to RNA P-O and sugar phosphate C-O stretching appears only after heat treatments.
DISCUSSION
Virus particles of various species or in different assembling or uncoating states deviate from each other in terms of structure and chemical content. In this study, EV1 viral particles, in three different states, were characterized in terms of both structural and chemical information. The intact virion, the uncoating intermediate produced after heating the virion at 50°C for 3 min, and the disrupted viral particle treated at 60°C for 10 min, all showed different types of structures obtained by TEM analysis. Also, the Raman spectroscopy of different states produced unique spectral features.
The Raman spectrum of a complex biological sample is a mixture of spectral contributions from all Raman active molecular groups contained in the sample, and therefore it enables simultaneous studying of all sections taking part in the biological processes and ultimately determining the sequence of the changes taking place in the molecule. As a result, the Raman spectrum of a virus consists of numerous, overlapping bands and peak shoulders. The most prominent spectral features, seen in the spectra of large polypeptides, are Am I, originating from the polypeptide chain C-O stretching, and Am III, mainly resulting from the coupled C-N and N-H bending motions. These are widely used as indicators of the secondary structure of proteins (17). Other informative Raman marker bands originate from the aromatic amino acids. Although Raman spectra of biological material are usually interpreted mainly as indicators of secondary structure conformation, other factors in the local environment strongly influence the position, size, and shape of the Raman signal. Local electric fields, polarity, and protonation state of the vibrators (50) play an important role, which means that Raman signals are sensitive to ions, charged amino acids, and even charged phosphates in the protein or in the genome. As shown in this and other studies (18–20, 22–27), Raman spectroscopy can provide a vast amount of information about the character and status of virion particles. In many cases, however, it is difficult to distill all information from a single set of data. Below, we condense the essential features of the Raman fingerprint of EV1 and discuss some of the crucial factors that must be considered when one attempts to determine the genuine Raman spectra from viral samples in an aqueous environment.
As Raman signals are relatively weak, in comparison with, for example, fluorescence signals of fluorophores, relatively high sample concentrations are needed. Typically, the reported Raman spectra of viral particles are measured with concentrations up to 100 mg ml−1 (18–27). Low sample concentrations are preferable when the viral particles in nonaggregated form are being measured. On many occasions, the scattering intensity of viruses is small, and the changes at low concentrations are hidden inside the overruling background spectra. This leads to challenges in detecting the Raman signal of the sample and accurately subtracting the solvent and other background from the weak viral spectra (41). It is also important to keep in mind that some buffers include groups that have vibrations similar to those of many biological samples or, at worst, are quite reactive (51) and therefore can, if not obscure the spectra, at least skew the interpretation. Other informative vibrations may be overwhelmed in a heterogeneous sample containing leftovers, such as sugars, from the purification procedure. The material besides the sample may then result in the biggest changes in the Raman spectra, overlapping the small changes taking place in the actual sample. Alternatively, they may influence the behavior of the sample; for example, sucrose is known to stabilize protein structure (52). In our measurements, the viral particles were clearly dominated by sucrose contamination even after dialysis with a molecular weight cutoff of 2,000 (data not shown) and were properly purified only with a higher-molecular-weight cutoff of 50,000. Also, in a recently published article on virus-like particles, there are clear and identified sucrose leftovers in the Raman spectra (53). It is important to note that because Raman spectroscopy “sees everything,” the purity and homogeneity of the samples are crucial. Even smallest changes in the purification method used can cause additions to and interference with the spectra, which can lead to erroneous interpretations. This means that critical attention must be paid to the purification procedure, and as a result, better-quality spectra containing more information are achieved. Further, once a spectrum of a pure viral sample has been obtained, Raman spectroscopy could be used as a very sensitive probe for impurity studies for analytical purposes.
To the best of our knowledge, there are no previous Raman studies on EV1 and its close relatives. Bovine enterovirus and rhinovirus have been studied by means of surface enhanced Raman scattering (SERS) and UV resonance Raman spectroscopy (46, 54). It is notable that, in the case of SERS, the choice of the substrate material and, in the case of resonance Raman studies, the choice of the laser wavelength contribute to the intensity of each Raman transition. Our spectra of EV1 are especially similar to those obtained for the BPMV virion (20), which shares considerable structural similarity with picornaviruses (55). Li et al. also observed differences in the Raman spectra between crystalline and liquid BPMV viral particles and concluded that changes mainly originate from packed RNA molecules that have different electrostatic environments in the samples (56). It should be recognized, though, that one particular difference with most of the previously reported linear Raman data is that our Raman spectra were measured with much lower protein concentrations, i.e., less than 4 mg ml−1 in our study versus 80 mg ml−1 or more in other studies (for example, see references 19, 20, and 24). The amount of aggregation and interaction between viral particles at such concentrations could diminish the differences compared to the crystalline state. In our EV1 samples there are differences between spectra measured in solvated and dried states, for example, a broadening or a shift in the Am I band to higher wave numbers, indicating a larger amount of disordered secondary structures in the dried sample. On the other hand, the width of some bands originating from individual amino acids, like the Phe band at 1,003 cm−1, is similar to the width in spectra of dried and solvated samples. Drying removes the stabilizing hydration shell (57) and can influence stability, cause aggregation, and hamper at least certain parts of capsid structures. This also implies that the SERS technique (54), where viral particles are attached to metallic surfaces or nanoparticles, can reveal somewhat different vibrational spectroscopic information than measurement of viral particles normally diffusing freely in a buffer solution.
Care was taken to remove the contribution of the wagging mode of water molecules from the Raman data of the diluted samples, which has been determined to be essential for a proper analysis of the Am I modes (41). The Raman Am I band centered at 1,668 cm−1 indicates β-sheets as the predominant secondary structure of the capsid, which is consistent with the known structure of the major capsid proteins, VP1, VP2, and VP3, in EV1 (58). However, temperature-induced alterations around 1,653 cm−1 reveal a decrease in α-helical secondary structure due to the partial unfolding of the shell subunits, especially at the intermediate state. The decreasing signal in the Am III area at around 1,300 cm−1 could also be due to reduced α-helical content of proteins. This indicates that the heat-induced uncoating is dominating in the α-helical regions of the protein. The signals assigned to β-sheets and coiled structures in Am I and III regions, at 1,668 and 1,240 cm−1, respectively, increase in magnitude at the same time, suggesting that some of the α-helix structures are converted to β-sheets or coils. Flexible loops between structured domains are known to make protein dynamics less constrained and play a critical role in the uncoating process. For example, these mechanically uncoupled structures could enable the capsid expansion, which is clearly visible from the TEM image of the uncoated EV1 particle (Fig. 3C).
Signals of aromatic amino acids reveal information on hydrogen bonding, hydrophobicity, and side chain conformation of these residues (43–48). Many of these informative signals of EV1 change with heat treatments. Small changes are seen in the Fermi doublets of Tyr and Trp that change with hydrogen bonding of Tyr and hydrophobicity of Trp environment. Changes in the 877 cm−1 signal indicate variations in Trp hydrogen bonding status, and the appearance of a new Trp signal at around 1,550 cm−1 indicates changes in side chain conformations. All these signals point in the same direction as the changes seen in secondary structures of viral proteins and TEM-images, i.e., loosening of the capsid structure and subsequent exposure of these residues to water.
Obtaining Glu vibrations of protonated carboxyl group suggests that protonation processes take place in the heat-induced uncoating of EV1. In the unprotonated state, the negatively charged Glu may interact with positively charged residues (e.g., Lys and Arg) to form salt bridges, which have been shown to be important stabilizers of HIV, influenza virus, filamentous viruses, and bacteriophage hk97 (59–62). These salt bridges may be involved in a constrained network of interactions within one subunit or between two neighboring units, clamping the domains in a considerably more stable capsid structure. Our data suggest that salt bridges may be involved in fine-tuning the metastable capsid structure of EV1. By studying the EV1 structure, the distribution of possible salt bridges in the EV1 structure was shown to be in the interior of the capsid or buried within the capsid structure (Fig. 6A and B). Interestingly, the enterovirus uncoating intermediates reported were shown to have lost the internal protein VP4 (13), which mediates two interprotomer and one inter-VP salt bridge connection (Fig. 6C). It is thus tempting to speculate that the extraction of VP4 from the capsid particle might be controlled, at least partially, by salt bridges. In comparison to the native virion, the heat-induced uncoating intermediate of EV1 showed a small increase in protonated carboxylic acid stretching. Also, a small decrease in signals that could be assigned to fatty acids was detected. The disconnected salt bridges, together with the expelled fatty acid chains, are expected to shift the capsid dynamics into a more flexible motion as these constraints are removed.
FIG 6.
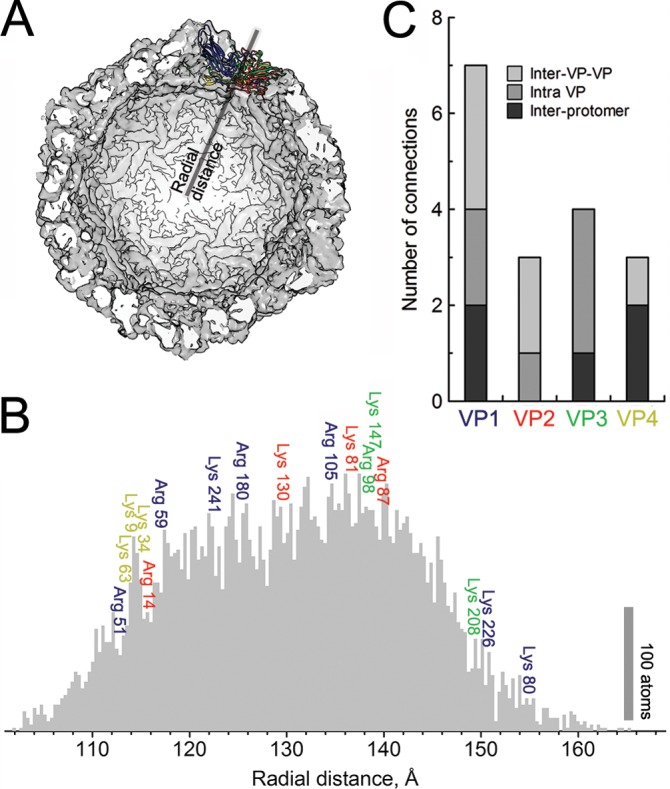
Radial distribution of salt bridge donors in EV1 protomer. (A) Single-protomer (colored) positioning with respect to the whole capsid (a half capsid is displayed). (B) The radial distribution of all atoms in EV1 protomer (PDB entry 1EV1) are depicted in the gray histogram to illustrate the density of the capsid shell. Colors indicate VP1 (blue), VP2 (red), VP3 (green), and VP4 (yellow), and the positioning of the salt bridge-forming amino acids (Lys and Arg) in a single protomer is shown, followed by their sequential positioning within these structural proteins. (C) The distribution of the salt bridge donors shows various degrees of connectivity within and between protomers. Interestingly, the inner protein VP4 forms connections mainly between protomers and one inner VP-VP connection.
In addition to the changes in the capsid proteins, several heat-induced changes in the RNA signals were observed. In general, the changes are difficult to interpret as particular structural changes but are more like a general change of the solute-solvent interactions of the RNA molecule. In particular, the signal variation at 783 cm−1 suggests that there are changes in the RNA solvation shell (39, 49). Both the ejection of genome and an increased permeability of the capsid structure could explain these changes in the signal sensitive to the environment of RNA. Still, a small increase in the A-form helix marker band at 811 cm−1 (39, 40) after heat treatment at 60°C also suggests that RNA is no longer bound by the intact capsid structure and refolds to the A-form double-helical structure.
In conclusion, we were able to acquire signature Raman spectra that distinguish between intact and uncoated EV1 diluted in buffer at relatively low concentrations. Very few vibrational spectroscopic data on the conformational changes during viral uncoating are available as yet. The observed Raman signatures are in good agreement with the present knowledge of enteroviral uncoating. For example, a UV resonance Raman study comparing intact and empty bovine enterovirus particles (46) revealed increasing hydrophobicity of the virus, or moving of Trp residues to increasingly hydrophilic environment during the viral opening process, consistent with our measurements. However, the detected differences between the intact virion in dried phase and in the liquid state indicate that the natural environment is essential in order to retain all the information from the uncoating process. Most biological macromolecules are physiologically active in aqueous solutions, and water molecules are thought to play a crucial role in the function and structure of biomolecules.
The Raman signatures of EV1 virion particles consist of numerous signals, with the most pronounced features being the following. The first is amide bands. The Am I became less broad after the uncoating process and revealed a decrease of α-helical structures and an increase of irregular structures or β-sheets. The second such feature is aromatic amino acids. The vibration modes of tryptophan and tyrosine residues suggest loosening of the capsid structure with increasing hydrophilicity around these residues. Third is chemical changes in the virion particles. The carbonyl vibrations showed small changes between the native virion and the heated particles, which indicates at least partial disruption of salt bridges. The fourth feature is genome signals. RNA signatures indicate both changes in the environment of the genome and a change in RNA conformation.
The observed differences in the Raman spectra between the intact and uncoated virions give novel insight into the structural changes occurring during virus opening. Most probably, findings would be similar for the enteroviruses that are close relatives of EV1. The prominent Raman marker bands of the intact virion, intermediate uncoating state of the virion, and disrupted virion particles presented here also enable in vivo studies of factors leading to viral uncoating in cellular structures with Raman mapping and coherent anti-Stokes Raman scattering microscopy.
ACKNOWLEDGMENTS
This research was supported by the Academy of Finland (134061 and 257125) and the National Doctoral Programme in Nanoscience (NGS).
We thank Alli Liukkonen, Eila Korhonen, and Arja Mansikkaviita for practical help, together with Tommi Isoniemi for supportive studies. Thomas Huser, Mika Pettersson, Olli Pentikäinen, and Vesa Aho are warmly appreciated for valuable discussions.
Footnotes
Published ahead of print 21 May 2014
REFERENCES
- 1.Tuthill TJ, Groppelli E, Hogle JM, Rowlands DJ. 2010. Picornaviruses. Curr. Top. Microbiol. Immunol. 343:43–89. 10.1007/82_2010_37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hober D, Sauter P. 2010. Pathogenesis of type 1 diabetes mellitus: interplay between enterovirus and host. Nat. Rev. Endocrinol. 6:279–289. 10.1038/nrendo.2010.27 [DOI] [PubMed] [Google Scholar]
- 3.Roivainen M, Alfthan G, Jousilahti P, Kimpimaki M, Hovi T, Tuomilehto J. 1998. Enterovirus infections as a possible risk factor for myocardial infarction. Circulation 98:2534–2537. 10.1161/01.CIR.98.23.2534 [DOI] [PubMed] [Google Scholar]
- 4.Fry EE, Stuart DI. 2010. Virion structure, p 59–71 In Ehrenfeld E, Domingo E, Roos RP. (ed), The picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 5.Brandenburg B, Lee LY, Lakadamyali M, Rust MJ, Zhuang X, Hogle JM. 2007. Imaging poliovirus entry in live cells. PLoS Biol. 5:1543–1555. 10.1371/journal.pbio.0050183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karjalainen M, Rintanen N, Lehkonen M, Kallio K, Mäki A, Hellström K, Siljamäki V, Upla P, Marjomäki V. 2011. Echovirus 1 infection depends on biogenesis of novel multivesicular bodies. Cell. Microbiol. 13:1975–1995. 10.1111/j.1462-5822.2011.01685.x [DOI] [PubMed] [Google Scholar]
- 7.Curry S, Chow M, Hogle JM. 1996. The poliovirus 135S particle is infectious. J. Virol. 70:7125–7131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bubeck D, Filman DJ, Cheng N, Steven AC, Hogle JM, Belnap DM. 2005. The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes. J. Virol. 79:7745–7755. 10.1128/JVI.79.12.7745-7755.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin J, Cheng N, Chow M, Filman DJ, Steven AC, Hogle JM, Belnap DM. 2011. An externalized polypeptide partitions between two distinct sites on genome-released poliovirus particles. J. Virol. 85:9974–9983. 10.1128/JVI.05013-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy HC, Bostina M, Filman DJ, Hogle JM. 2010. Catching a virus in the act of RNA release: a novel poliovirus uncoating intermediate characterized by cryo-electron microscopy. J. Virol. 84:4426–4441. 10.1128/JVI.02393-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bostina M, Levy H, Filman DJ, Hogle JM. 2011. Poliovirus RNA is released from the capsid near a twofold symmetry axis. J. Virol. 85:776–783. 10.1128/JVI.00531-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chow M, Newman JFE, Filman D, Hogle JM, Rowlands DJ, Brown F. 1987. Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature 327:482–486. 10.1038/327482a0 [DOI] [PubMed] [Google Scholar]
- 13.Fricks CE, Hogle JM. 1990. Cell-Induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J. Virol. 64:1934–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Racaniello VR. 1996. Early events in poliovirus infection: virus-receptor interactions. Proc. Natl. Acad. Sci. U. S. A. 93:11378–11381. 10.1073/pnas.93.21.11378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smyth M, Pettitt T, Symonds A, Martin J. 2003. Identification of the pocket factors in a picornavirus. Arch. Virol. 148:1225–1233. 10.1007/s00705-002-0974-4 [DOI] [PubMed] [Google Scholar]
- 16.Smyth MS, Martin JH. 2002. Picornavirus uncoating. Mol. Pathol. 55:214–219. 10.1136/mp.55.4.214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tuma R. 2005. Raman spectroscopy of proteins: from peptides to large assemblies. J. Raman Spectrosc. 36:307–319. 10.1002/jrs.1323 [DOI] [Google Scholar]
- 18.Hartman KA, McDonald-Ordzie PE, Kaper JM, Prescott B, Thomas GJ., Jr 1978. Studies of virus structure by laser-Raman spectroscopy. Turnip yellow mosaic virus and capsids. Biochemistry 17:2118–2123 [DOI] [PubMed] [Google Scholar]
- 19.Li TS, Johnson JE, Thomas GJ., Jr 1993. Raman dynamic probe of hydrogen exchange in bean pod mottle virus: base-specific retardation of exchange in packaged ssRNA. Biophys. J. 65:1963–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li TS, Chen ZG, Johnson JE, Thomas GJ., Jr 1990. Structural studies of bean pod mottle virus, capsid, and RNA in crystal and solution states by laser Raman spectroscopy. Biochemistry 29:5018–5026. 10.1021/bi00473a004 [DOI] [PubMed] [Google Scholar]
- 21.Prescott B, Sitaraman K, Argos P, Thomas GJ., Jr 1985. Protein-RNA interactions in belladonna mottle virus investigated by laser Raman spectroscopy. Biochemistry 24:1226–1231. 10.1021/bi00326a026 [DOI] [PubMed] [Google Scholar]
- 22.Overman SA, Aubrey KL, Vispo NS, Cesareni G, Thomas GJ., Jr 1994. Novel tyrosine markers in Raman spectra of wild-type and mutant (Y21M and Y24M) Ff virions indicate unusual environments for coat protein phenoxyls. Biochemistry 33:1037–1042. 10.1021/bi00171a001 [DOI] [PubMed] [Google Scholar]
- 23.Overman SA, Thomas GJ., Jr 1995. Raman spectroscopy of the filamentous virus Ff (fd, fl, M13): structural interpretation for coat protein aromatics. Biochemistry 34:5440–5451. 10.1021/bi00016a015 [DOI] [PubMed] [Google Scholar]
- 24.Tuma R, Bamford J, Bamford D, Russell M, Thomas GJ., Jr 1996. Structure, interactions and dynamics of PRD1 virus II. Organization of the viral membrane and DNA. J. Mol. Biol. 257:102–115. 10.1006/jmbi.1996.0150 [DOI] [PubMed] [Google Scholar]
- 25.Fish SR, Hartman KA, Fuller MT, King J, Thomas GJ. 1980. Investigation of secondary structures and macromolecular interactions in bacteriophage p22 by laser Raman spectroscopy. Biophys. J. 32:234–327. 10.1016/S0006-3495(80)84945-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aubrey KL, Casjens SR, Thomas GJ., Jr 1992. Secondary structure and interactions of the packaged dsDNA genome of bacteriophage P22 investigated by Raman difference spectroscopy. Biochemistry 31:11835–11842. 10.1021/bi00162a023 [DOI] [PubMed] [Google Scholar]
- 27.Reilly KE, Thomas GJ., Jr 1994. Hydrogen exchange dynamics of the P22 virion determined by time-resolved Raman spectroscopy. Effects of chromosome packaging on the kinetics of nucleotide exchanges. J. Mol. Biol. 241:68–82 [DOI] [PubMed] [Google Scholar]
- 28.Hogle JM, Chow M, Filman DJ. 1985. Three-dimensional structure of poliovirus at 2.9 Å resolution. Science 229:1358–1365. 10.1126/science.2994218 [DOI] [PubMed] [Google Scholar]
- 29.Walter TS, Ren J, Tuthill TJ, Rowlands DJ, Stuart DI, Fry EE. 2012. A plate-based high-throughput assay for virus stability and vaccine formulation. J. Virol. Methods 185:166–170. 10.1016/j.jviromet.2012.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abraham G, Colonno RJ. 1984. Many rhinovirus stereotypes share the same cellular receptor. J. Virol. 51:340–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493–497 [Google Scholar]
- 32.Porterfield JZ, Zlotnick A. 2010. A simple and general method for determining the protein and nucleic acid content of viruses by UV absorbance. Virology 407:281–288. 10.1016/j.virol.2010.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 13:1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 34.McGregor S, Mayer HD. 1968. Biophysical studies on rhinovirus and poliovirus. I. Morphology of viral ribonucleoprotein. J. Virol. 2:149–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren J, Wang X, Hu Z, Gao Q, Sun Y, Li X, Porta C, Walter TS, Gilbert RJ, Zhao Y, Axford D, Williams M, McAuley K, Rowlands DJ, Yin W, Wang J, Stuart DI, Rao Z, Fry EE. 2013. Picornavirus uncoating intermediate captured in atomic detail. Nat. Commun. 4:1929. 10.1038/ncomms2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Peng W, Ren J, Hu Z, Xu J, Lou Z, Li X, Yin W, Shen X, Porta C, Walter TS, Evans G, Axford D, Owen R, Rowlands DJ, Wang J, Stuart DI, Fry EE, Rao Z. 2012. A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nat. Struct. Mol. Biol. 19:424–429. 10.1038/nsmb.2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Gelder J, De Gussem K, Vandenabeele P, Moens L. 2007. Reference database of Raman spectra of biological molecules. J. Raman Spectrosc. 38:1133–1147. 10.1002/jrs.1734 [DOI] [Google Scholar]
- 38.Matthäus C, Bird B, Miljkovic M, Chernenko T, Romeo M, Diem M. 2008. Infrared and Raman microscopy in cell biology. Methods Cell Biol. 89:275–308. 10.1016/S0091-679X(08)00610-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hobro AJ, Rouhi M, Blanch EW, Conn GL. 2007. Raman and Raman optical activity (ROA) analysis of RNA structural motifs in domain I of the EMCV IRES. Nucleic Acids Res. 35:1169–1177. 10.1093/nar/gkm012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hobro AJ, Standley DM, Ahmad S, Smith NI. 2013. Deconstructing RNA: optical measurement of composition and structure. Phys. Chem. Chem. Phys. 15:13199–13208. 10.1039/c3cp52406j [DOI] [PubMed] [Google Scholar]
- 41.Sane SA, Cramer SM, Przybycien TM. 1999. A holistic approach to protein secondary structure characterization using amide I band Raman spectroscopy. Anal. Biochem. 269:255–272. 10.1006/abio.1999.4034 [DOI] [PubMed] [Google Scholar]
- 42.Barth A, Zscherp C. 2002. What vibrations tell about proteins. Q. Rev. Biophys. 35:369–430. 10.1017/S0033583502003815 [DOI] [PubMed] [Google Scholar]
- 43.Siamwiza MN, Lord RC, Chen MC, Takamatsu T, Harada I, Matsuura H, Shimanouchi T. 1975. Interpretation of the doublet at 850 and 830 cm-1 in the Raman spectra of tyrosyl residues in proteins and certain model compounds. Biochemistry 14:4870–4876. 10.1021/bi00693a014 [DOI] [PubMed] [Google Scholar]
- 44.Miura T, Takeuchi H, Harada I. 1988. Characterization of individual tryptophan side chains in proteins using Raman spectroscopy and hydrogen-deuterium exchange kinetics. Biochemistry 27:88–94. 10.1021/bi00401a015 [DOI] [PubMed] [Google Scholar]
- 45.Schlamadinger DE, Gable JE, Kim JE. 2009. Hydrogen bonding and solvent polarity markers in the UV resonance Raman spectrum of tryptophan: application to membrane proteins. J. Phys. Chem. B 113:14769–14778. 10.1021/jp905473y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaminaka S, Imamura Y, Shingu M, Kitagawa T, Toyoda T. 1999. Studies of bovine enterovirus structure by ultraviolet resonance Raman spectroscopy. J. Virol. Methods 77:117–123. 10.1016/S0166-0934(98)00153-0 [DOI] [PubMed] [Google Scholar]
- 47.Miura T, Takeuchi H, Harada I. 1989. Tryptophan Raman bands sensitive to hydrogen bonding and side-chain conformation. J. Raman Spectrosc. 20:667–671. 10.1002/jrs.1250201007 [DOI] [Google Scholar]
- 48.Takeuchi H. 2003. Raman structural markers of tryptophan and histidine side chains in proteins. Biopolymers 72:305–317. 10.1002/bip.10440 [DOI] [PubMed] [Google Scholar]
- 49.Hernández B, Baumruk V, Leulliot N, Gouyette C, Huynh-Dinh T, Ghomi M. 2003. Thermodynamic and structural features of ultrastable DNA and RNA hairpins. J. Mol. Struct. 651–653:67–74. 10.1016/S002202860(02)00627-0 [DOI] [Google Scholar]
- 50.Park ES, Boxer SG. 2002. Origins of the sensitivity of molecular vibrations to electric fields: carbonyl and nitrosyl stretches in model compounds and proteins. J. Phys. Chem. B 106:5800–5806. 10.1021/jp0203043 [DOI] [Google Scholar]
- 51.Taha M, Ming-Jer L. 2010. Interactions of TRIS [tris(hydroxymethyl)aminomethane] and related buffers with peptide backbone: thermodynamic characterization. Chem. Phys. 12:12840–12850. 10.1039/c0cp00253d [DOI] [PubMed] [Google Scholar]
- 52.Allison SD, Dong A, Carpenter JF. 1996. Counteracting effects of thiocyanate and sucrose on chymotrypsinogen secondary structure and aggregation during freezing, drying, and rehydration. Biophys. J. 71:2022–2032. 10.1016/S0006-3495(96)79400-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu X, Liu Q, Benavides-Montano JA, Nicola AV, Aston DE, Rasco BA, Aguilar HC. 2013. Detection of receptor-induced glycoprotein conformational changes on enveloped virions by using confocal micro-Raman spectroscopy. J. Virol. 87:3130–3142. 10.1128/JVI.03220-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shanmukh S, Jones L, Driskell J, Zhao Y, Dluhy R, Tripp RA. 2006. Rapid and sensitive detection of respiratory virus molecular signatures using a silver nanorod array SERS substrate. Nano Lett. 6:2630–2636. 10.1021/nl061666f [DOI] [PubMed] [Google Scholar]
- 55.Joisson C, Kuster F, Plaué S, Van Regenmortel MH. 1993. Antigenic analysis of bean pod mottle virus using linear and cyclized synthetic peptides. Arch. Virol. 128:299–317. 10.1007/BF01309441 [DOI] [PubMed] [Google Scholar]
- 56.Li TS, Chen ZG, Johnson JE, Thomas GJ., Jr 1992. Conformations, interactions, and thermostabilities of RNA and proteins in bean pod mottle virus: investigation of solution and crystal structures by laser Raman spectroscopy. Biochemistry 31:6673–6682. 10.1021/bi00144a006 [DOI] [PubMed] [Google Scholar]
- 57.Prestrelski SJ, Tedeschi N, Arakawa T, Carpenter JF. 1993. Dehydration-induced conformational transitions in proteins and their inhibition by stabilizers. Biophys. J. 65:661–671. 10.1016/S0006-3495(93)81120-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Filman DJ, Wien MW, Cunningham JA, Bergelson JM, Hogle JM. 1998. Structure determination of echovirus 1. Acta Crystallogr. 54:1261–1272 [DOI] [PubMed] [Google Scholar]
- 59.He Y, Liu S, Li J, Lu H, Qi Z, Liu Z, Debnath AK, Jiang S. 2008. Conserved salt bridge between the N- and C-terminal heptad repeat regions of the human immunodeficiency virus type 1 gp41 core structure is critical for virus entry and inhibition. J. Virol. 82:11129–11139. 10.1128/JVI.01060-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rachakonda PS, Veit M, Korte T, Ludwig K, Böttcher C, Huang Q, Schmidt MF, Herrmann A. 2007. The relevance of salt bridges for the stability of the influenza virus hemagglutinin. FASEB J. 21:995–1002. 10.1096/fj.06-7052hyp [DOI] [PubMed] [Google Scholar]
- 61.Dolja VV, Boyko VP, Agranovsky AA, Koonin EV. 1991. Phylogeny of capsid proteins of rod-shaped and filamentous RNA plant viruses: two families with distinct patterns of sequence and probably structure conservation. Virology 184:79–86. 10.1016/0042-6822(91)90823-T [DOI] [PubMed] [Google Scholar]
- 62.Gertsman I, Fu CY, Huang R, Komives EA, Johnson JE. 2010. Critical salt bridges guide capsid assembly, stability, and maturation behavior in bacteriophage HK97. Mol. Cell. Proteomics 9:1752–1763. 10.1074/mcp.M000039-MCP201 [DOI] [PMC free article] [PubMed] [Google Scholar]


