Abstract
In immunosuppressed hosts, the development of multidrug resistance complicates the treatment of cytomegalovirus (CMV) infection. Improved genotypic detection of impending drug resistance may follow from recent technical advances. A severely T-cell-depleted patient with chronic lymphocytic leukemia developed CMV pneumonia and high plasma viral loads that were poorly responsive to antiviral therapy. Serial plasma specimens were analyzed for mutant viral populations by conventional and high-throughput deep-sequencing methods. Uncharacterized mutations were phenotyped for drug resistance using recombinant viruses. Conventional genotyping detected viruses with the UL97 kinase substitution C607Y after ganciclovir treatment, a transient subpopulation of UL54 polymerase L773V mutants first detected 8 weeks after foscarnet was started, and a subpopulation of a mutant with deletion of UL54 codons 981 and 982 2 months after the addition of cidofovir. Deep sequencing of the same serial specimens revealed the same UL54 mutants sooner, along with a more complex evolution of known and newly recognized mutant subpopulations missed by conventional sequencing. The UL54 exonuclease substitutions D413N, K513R, and C539G were newly shown to confer ganciclovir-cidofovir resistance, while L773V was shown to confer foscarnet resistance and add to the ganciclovir resistance conferred by UL97 C607Y. Increased sequencing depth provided a more timely and detailed diagnosis of mutant viral subpopulations that evolved with changing anti-CMV therapy.
INTRODUCTION
Human cytomegalovirus (CMV) is a major viral opportunistic pathogen in T-cell-immunodeficient populations, including those with AIDS, transplants, and hematologic malignancies, causing diseases such as pneumonia, gastrointestinal disease, and retinitis. Prevention of CMV morbidity and mortality has been largely successful over the past 2 decades, using ganciclovir or its oral prodrug valganciclovir in a prophylaxis or pre-emptive treatment strategy designed to prevent active systemic viral infection or to suppress it promptly upon reactivation (1). In a minority of cases, the strategy fails when antiviral therapy is insufficient to shut off viral replication in the presence of adverse host factors. Over time, this may result in the accumulation of drug resistance mutations that eventually confer resistance to all licensed anti-CMV drugs, including foscarnet and cidofovir, which have the same CMV DNA polymerase target.
Because viral isolation in cell culture is uncommon in current diagnostic laboratory practice, and susceptibility testing of clinical CMV isolates lacks timely availability, genotypic testing has become the usual means for detecting drug resistance and the basis for selection of alternative therapy. This relies on the PCR amplification and sequencing of viral DNA from clinical specimens, representing portions of the viral UL97 kinase gene involved in the initial phosphorylation of ganciclovir and the UL54 DNA polymerase gene (2). Seven common UL97 kinase amino acid substitutions (M460V/I, H520Q, C592G, A594V, L595S, and C603W) are the basis for diagnosing ganciclovir resistance in a large majority of cases, but less common UL97 mutations clustered at codons 590 to 607 can sometimes be involved. UL54 mutations tend to evolve after more extensive drug exposure and commonly confer various levels of cross-resistance to other CMV antivirals (2).
Although considerable recombinant phenotyping data are available to estimate the level of resistance conferred by specific mutations and possible responsiveness to antiviral dose escalation or alternative drugs, there are technical limitations in genotypic diagnosis. The current standard Sanger dideoxy sequencing method often does not detect subpopulations of viral mutants of less than 20 to 30% (3). The increasing adoption of massively parallel high-throughput sequencing technologies is enabling the acquisition of sufficient reads of individual DNA molecules to detail the sequence subpopulations that occur throughout the CMV genome in vivo (4, 5). When focused on gene regions involved in drug resistance, it provides a cost-effective approach to deep sequencing whereby a targeted codon range is covered hundreds or thousands of times so that small variant subpopulations can be detected, as we recently showed for CMV (6). This may improve the detection of emerging drug resistance. The objective of the current study was to use this technology retrospectively to assess the resistance mutations that developed over time in a case of CMV disease with high accompanying viral loads that responded only transiently to various alternative antiviral treatments.
CASE REPORT
The patient was a 54-year-old male with a diagnosis of B-cell chronic lymphocytic leukemia (CLL) made 11 years earlier and treated with 6 cycles of cyclophosphamide, fludarabine, alemtuzumab, and rituximab (CFAR), leading to partial remission. Seven years after initial diagnosis, a matched unrelated donor stem cell transplant was unsuccessful, resulting in persistent lymphopenia followed by frank CLL relapse, which was treated again with monthly cycles of CFAR. After the second cycle, a peripheral plasma CMV load of 880 IU/ml was detected, and valganciclovir pre-emptive therapy was given for 1 month. A month after the third cycle, the viral load rose to 2 × 104 IU/ml and another month of valganciclovir was given. A fourth and delayed CFAR cycle was then given. Five weeks later, the plasma CMV load was 1.6 × 105 IU/ml while the patient was on valganciclovir. Two weeks later, the patient was admitted for fever and hypoxemia. This is the zero point of the timeline shown in Fig. 1, where serial data on viral loads, genotypes, and therapy are shown. Drug-resistant virus was suspected, and therapy was switched to intravenous foscarnet and immune globulin for possible CMV pneumonia. This resulted in worse hypoxemia and pulmonary infiltrates attributed to fluid overload. A bronchoscopy was positive only for Pneumocystis, which was treated with trimethoprim-sulfamethoxazole, and a shell vial culture for CMV was negative. Over the next several weeks, while the patient was on full-dose (90 mg/kg of body weight twice daily) foscarnet, pulmonary infiltrates gradually resolved. After 6 weeks, foscarnet dosing was reduced to once daily, and valganciclovir was resumed at treatment doses. At week 8, the patient was readmitted for diffuse pneumonia; a bronchoscopy with biopsy showed widespread presence of CMV inclusion bodies and no Pneumocystis. Resumption of twice-daily foscarnet was poorly tolerated because of pulmonary edema, hypoxemia, and electrolyte abnormalities. With rising plasma viral loads, therapy was switched to high-dose intravenous ganciclovir (10 mg/kg twice daily adjusted for renal function) in combination with intravenous cidofovir (5 mg/kg weekly). Over the next several weeks the pneumonia stabilized and improved. Cidofovir dosing was reduced to every 2 weeks, and the patient was discharged on valganciclovir. The plasma viral load then started rising sharply. The relapsed CLL remained evident, with a total T lymphocyte count of <100/μl, but ongoing CMV infection was felt to exclude further chemotherapy or marrow transplantation. Specific treatments, including antivirals, were stopped and death occurred shortly thereafter and was not directly attributed to CMV end organ disease. This case report and viral deep sequence analysis of residual diagnostic specimens were determined by the local institutional review board not to require human research approval or informed consent procedures.
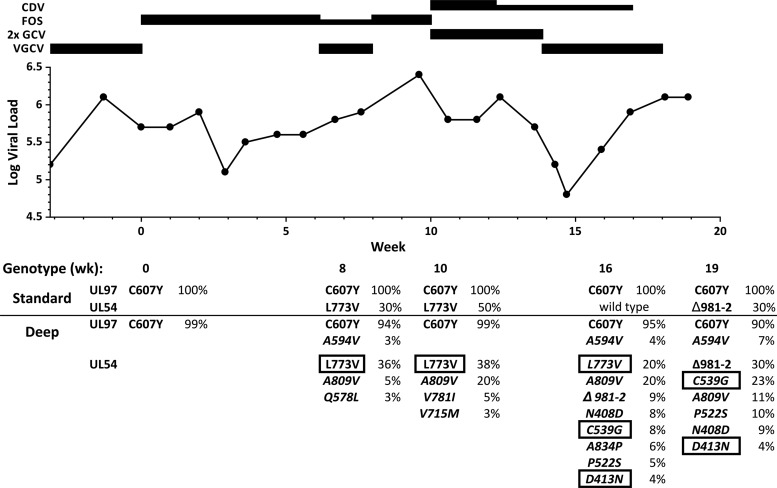
FIG 1.
Timeline of antiviral therapy, plasma viral loads, and standard and deep sequencing data. Thick and thin bars show full and reduced doses of antivirals (reduced = 90 mg foscarnet [FOS]/kg/day or 5 mg cidofovir [CDV]/kg every other week). 2× GCV = ganciclovir 10 mg/kg twice a day. All doses were adjusted for renal function. Weeks are numbered from the start of FOS treatment. Viral loads are in IU/ml, with an assay coefficient of variation 3.5% (7). Deep-sequencing data were selected from the data for AA12 in Table 1. Sequence variants shown in italics were not detected by standard sequencing. Those in boxes were newly phenotyped (Table 2).
MATERIALS AND METHODS
Viral loads and genotypic resistance testing.
Serial plasma samples were collected for routine monitoring of viral loads. Plasma viral DNA was extracted using automated equipment (Roche Magna Pure) with final aqueous elution volume equal to half the plasma input volume and used to determine CMV loads as described previously (7). Amounts of these extracts (1 to 15 μl) estimated to contain >2,000 copies of viral DNA were amplified in nested PCRs to produce sequencing templates for dideoxy sequencing (BigDye v.3; Applied Biosystems) of UL97 kinase codons 375 to 650 and UL54 DNA polymerase codons 300 to 1000. Sequences were compared with the AD169 reference sequence for variants affecting amino acid encoding. High-throughput deep sequencing was performed on the original DNA genotyping extracts stored at 4°C and later on fresh extracts of plasma specimens stored at −80°C. For the fresh extracts, DNA was extracted from 100 μl of plasma using the EZ1 virus minikit, v2.0, on the EZ1 Advanced XL platform (both from Qiagen, Germantown, MD) and eluted in 60 μl buffer AVE containing carrier RNA. Sequencing was performed using the Roche 454 GS Jr. pyrosequencing platform as recently published (6). After an initial long-range PCR to amplify ∼2-kb and ∼4-kb targets for UL97 and UL54, respectively, the PCR products were used as templates for further amplification using primers specific for overlapping ∼350-bp segments of the CMV target sequence and incorporating 454 adapter sequences and barcodes specific to individual specimens. This sequencing library preparation stage was multiplexed across specimens and primers using the Access Array system (Fluidigm). Amplicons were purified on magnetic beads and loaded in calibrated quantities for emulsion PCR and pyrosequencing data acquisition and analysis. By adjusting the extent of multiplexing, the objective was to achieve ∼1,000 reads through most of the targeted sequence of UL97 codons 399 to 655 and UL54 codons 212 to 311 and 356 to 1042.
Recombinant phenotyping.
The drug resistance phenotypes for ganciclovir, foscarnet, and cidofovir were determined after construction of cloned recombinant viral strains containing mutations detected in this case that had not been previously characterized. Mutagenesis of bacterial artificial chromosome clones of a laboratory CMV strain expressing a secreted alkaline phosphatase (SEAP) reporter gene, followed by human foreskin fibroblast cell culture assay of the drug concentration required to reduce SEAP production by 50% (EC50), was performed as previously published (8). The comparative growth fitness of the mutant strains was compared using the SEAP reporter system used previously to compare other CMV mutants (8).
RESULTS
Viral mutations detected by conventional genotyping.
No pretreatment plasma specimens were available to establish a baseline viral genotype. The first genotyping data at the time of initiation of foscarnet, after ∼4 months of valganciclovir therapy, already showed the full development of the UL97 amino acid substitution C607Y, one of the less common ganciclovir resistance mutations (9), along with the known UL97 polymorphism D605E (10). These findings persisted through all subsequent genotypic assays (Fig. 1). At week 8, a partial ∼30% subpopulation of UL54 substitution L773V was detected. This mutation was previously observed with foscarnet treatment in cell culture, but its phenotype was not confirmed by transfer into a baseline virus (11). It was redetected twice during and immediately after foscarnet was stopped but not at week 16, at which time no UL54 mutations were detected. Two months after cidofovir was started (week 19), an ∼30% subpopulation with an in-frame deletion of UL54 codons 981 and 982 was detected. This mutation is known to confer resistance to ganciclovir, foscarnet, and cidofovir (10).
Viral mutations detected by deep sequencing.
An initial round of high-throughput pyrosequencing (access array 10 [AA10]) (Table 1) was performed using residual DNA extracts from the conventional genotyping and stored at 4°C for 16 to 20 months. Results from the first available specimen (week 0) were consistent with conventional sequencing in showing a 99% population with the UL97 mutation C607Y and 100% populations with the sequence polymorphisms in UL97 of D605E and in UL54 of S655L, N685S, and N898D relative to reference strain AD169. At week 10, resistance-related subpopulations with the UL54 mutations L773V (51%), A809V (20%), and Q578H (6%) were detected. At week 16, after foscarnet was stopped, L773V had decreased in abundance, but many other known and probable resistance mutations were noted to be evolving (Table 1), none of which had been detected by conventional genotyping. Four weeks later, L773V and A809V were absent or decreasing, but the diverse UL54 mutations known or suspected to confer ganciclovir-cidofovir or triple drug resistance mostly increased in abundance.
TABLE 1.
Genotypic variants detected by deep sequencing
| Variant type | Gene | Variantb | Subpopulation % of varianta |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wk 0 |
Wk 4 | Wk 6 | Wk 8 | Wk 10 |
Wk 12 | Wk 16 |
Wk 19 |
|||||||
| AA10 | AA12 | AA12 | AA12 | AA12 | AA10 | AA12 | AA12 | AA10 | AA12 | AA10 | AA12 | |||
| Established resistance-related variants | UL97 | A594V | 3 | 4 | 4 | 6 | 7 | |||||||
| C607Y | 99 | 99 | 96 | 95 | 94 | 100 | 99 | 97 | 95 | 95 | 90 | 90 | ||
| UL54 | N408D | 4 | 8 | 8 | 9 | |||||||||
| P522S | 9 | 5 | 11 | 10 | ||||||||||
| Q578H | 6 | |||||||||||||
| Q578L | 3 | |||||||||||||
| V715 M | 3 | |||||||||||||
| V781I | 5 | |||||||||||||
| A809V | 5 | 20 | 20 | 37 | 28 | 20 | 7 | 11 | ||||||
| A834P | 3 | 6 | 7 | |||||||||||
| Δ981-2c | 13 | 9 | 31 | 30 | ||||||||||
| Newly phenotyped resistance-related variants | UL54 | D413N | 4 | 4 | 5 | 4 | ||||||||
| K513R | 4 | |||||||||||||
| C539G | 21 | 8 | 19 | 23 | ||||||||||
| L773V | 5 | 36 | 51 | 38 | 38 | 6 | 20 | |||||||
| Common baseline polymorphisms | UL97 | D605E | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| UL54 | S655L | 99 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 99 | 100 | 99 | 100 | |
| N685S | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | ||
| N898D | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | ||
| Other variantsd | A | B | C | |||||||||||
Sequencing statistics (reads per position) were as follows. For AA10, the UL54 median was 1,074 (interquartile range [IQR], 803 to 1,578; minimum, 106; maximum, 3,596), and the UL97 median, 1,052 (IQR, 767 to 1,380; minimum, 298; maximum, 2,701). For AA12, the UL54 median was 832 (IQR, 649 to 1,161; minimum, 350; maximum, 2,435) and the UL97 median was 772 (IQR, 556 to 918; minimum, 397; maximum, 1,748). Weeks are numbered as in the timeline in Fig. 1. Data from the same week were derived from independent extracts.
Variants in bold were detected by conventional genotyping.
In-frame deletion of codons 981 and 982.
A, UL97 R645H (3%) and UL54 V914A (3%); B, UL54 G971D (3%); C, UL54 P522L (3%) and F817C (3%).
To test the reproducibility of the deep-sequencing data, the same clinical specimens, and those from additional sampling dates, were independently extracted from frozen plasma, PCR amplified, and sequenced as described previously (6) (AA12) (Table 1). In comparison with AA10, there was good reproducibility of the detected resistance-related genotypes, including most of the minor subpopulations. Low-level findings in AA10 of Q578H, K513R, and several variants unrelated to drug resistance were not confirmed in AA12, whereas AA12 but not AA10 detected low levels of Q578L, V715M, and V781I. The two sequencing runs provided comparable depth of sequencing (Table 1).
Recombinant phenotyping.
Results of recombinant phenotyping are shown in Table 2. The UL54 substitution L773V conferred moderate foscarnet resistance with low-grade ganciclovir and cidofovir cross-resistance. It doubled the level of ganciclovir resistance when added to the UL97 substitution C607Y, which in the context of the polymorphism D605E, seen in this patient, conferred a lesser degree of ganciclovir resistance than the canonical substitution A594V (with or without D605E) (10). The newly phenotyped UL54 substitutions D413N, K513R, and C539G each conferred dual ganciclovir and cidofovir resistance similar to other nearby exonuclease domain mutations (2), but no foscarnet resistance.
TABLE 2.
Genotypes and phenotypes of recombinant viruses
| Virus type and strain | Substitutiona encoded in: |
Cidofovir |
Ganciclovir |
Foscarnet |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UL97 | UL54 pol | EC50 (μM)b |
nc | EC50 ratiod | EC50 (μM) |
n | EC50 ratio | EC50 (μM) |
n | EC50 ratio | ||||
| Mean | SD | Mean | SD | Mean | SD | |||||||||
| Controls | ||||||||||||||
| T3261 | 0.22 | 0.07 | 21 | 1.13 | 0.20 | 31 | 45 | 12 | 24 | |||||
| T3265 | 0.21 | 0.06 | 31 | 1.15 | 0.27 | 40 | 46 | 12 | 28 | |||||
| Known drug-resistant strains | ||||||||||||||
| T3259 | C592G | 3.17 | 0.65 | 38 | 2.8 | |||||||||
| T3982 | C607Y6 | 0.25 | 0.10 | 10 | 1.1 | 5.19 | 0.83 | 16 | 4.6 | 50 | 9 | 14 | 1.1 | |
| T3271 | A809V | 178 | 44 | 14 | 3.9 | |||||||||
| T3429 | A987G | 1.25 | 0.38 | 21 | 6.0 | |||||||||
| Newly phenotyped variants | ||||||||||||||
| T3983 | L773V | 0.52 | 0.21 | 12 | 2.5 | 3.47 | 0.53 | 17 | 3.0 | 202 | 42 | 7 | 4.4 | |
| T4118 | C607Ye | L773V | 0.59 | 0.18 | 10 | 2.8 | 10.45 | 1.89 | 9 | 9.1 | 230 | 40 | 8 | 5.0 |
| T4135 | D413N | 2.03 | 0.22 | 10 | 10 | 4.32 | 1.16 | 8 | 3.8 | 48 | 18 | 7 | 1.0 | |
| T4148 | K513R | 2.15 | 0.53 | 12 | 10 | 4.26 | 1.09 | 8 | 3.7 | 51 | 16 | 10 | 1.1 | |
| T4136 | C539G | 0.93 | 0.29 | 12 | 4.4 | 3.52 | 1.13 | 11 | 3.1 | 45 | 14 | 14 | 1.0 | |
Amino acid substitution transferred into baseline strain or clone.
Drug concentration required to reduce SEAP growth by 50% at 6 to 7 days postinfection. Values in bold indicate drug resistance (EC50 > 1.9 times the control value).
Number of assays (over at least 4 setup dates).
Ratio of EC50 to that for the matching baseline strain.
Also contains the UL97 sequence polymorphism D605E.
Growth fitness in cell culture.
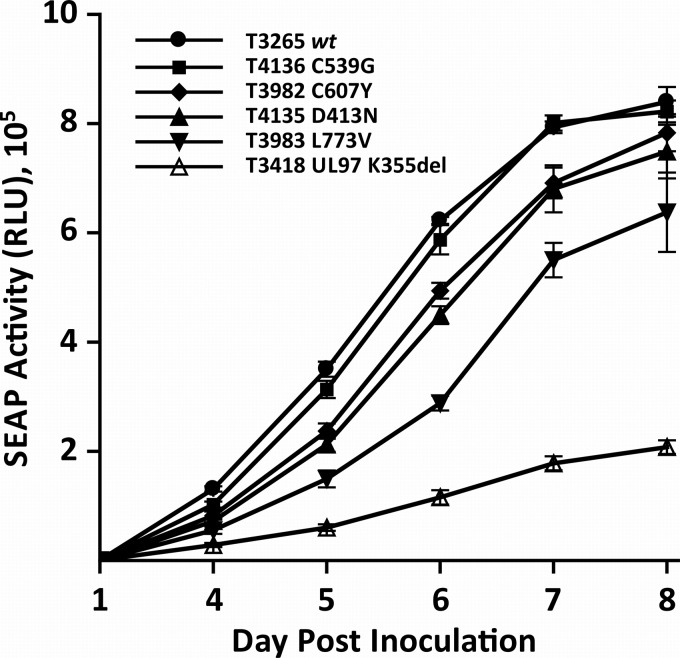
Replication curves of the mutant viral strains over a 1-week period after calibrated low-multiplicity inocula are shown in Fig. 2. The UL54 substitution L773V is significantly attenuating, whereas the UL97 substitution C607Y and the UL54 substitution D413N have a lesser growth impact.
FIG 2.
Comparative replication curves of recombinant mutant viruses. Viral stocks were inoculated in quadruplicate and sampled for supernatant SEAP activity (relative light units [RLU]) at 1 and 4 to 8 days after inoculation. Data points are means and standard deviations of 4 values. The input multiplicity of infection was calibrated to 0.01 by closely matching day 1 SEAP RLU values for each strain that ranged from 550 ± 46 to 582 ± 50 (10). The UL97 kinase K355del knockout mutant was used as a control for severe growth impairment (17).
DISCUSSION
This case illustrates the relatively rapid evolution of mutations conferring multidrug resistance in the setting of severe T-cell depletion, persistently high plasma viral loads, and transient responses to various treatments. High-throughput deep-sequencing technology gave a more timely and detailed view of the process, revealing diverse mutant subpopulations too small to be detectable by conventional Sanger sequencing. This correlated well with the treatment history, increasing plasma viral loads, and eventual treatment failure.
Following a published guideline (1), therapy was switched to full-dose intravenous foscarnet upon evidence of valganciclovir treatment failure corroborated by the genotypic finding of UL97 C607Y (9). Foscarnet was poorly tolerated, but after 4 weeks there was a fall in plasma viral load followed by a brisk rebound (Fig. 1) unresponsive to a combination of reduced-dose foscarnet and valganciclovir (9, 12). By week 8 of foscarnet treatment, conventional genotyping had shown a subpopulation of virus with the UL54 substitution L773V that conferred foscarnet resistance and increased ganciclovir resistance, thus diminishing the efficacy of valganciclovir, although this phenotype was not known at the time. Deep sequencing first detected L773V at week 6, and by weeks 8 to 10, it also showed minor components with other known foscarnet resistance substitutions, including A809V, Q578H/L, V715M, and V781I (2) (Table 1), that were not detected then or later by standard genotyping. This coincided with the onset of biopsy-proven CMV pneumonia.
Given the patient's intolerance of additional foscarnet, treatment was switched to double-dose ganciclovir and cidofovir. Ganciclovir dose escalation is described in the current clinical guideline for lower-grade resistance mutations (1), which could apply to C607Y but perhaps less so in the presence of a UL54 mutation (Table 2). After several weeks, this treatment appeared to improve the viral load (Fig. 1) and clinical condition, but the loads again increased rapidly when the double-dose ganciclovir was replaced by valganciclovir and the cidofovir dose was reduced to every other week and later stopped due to nephrotoxicity. The most notable difference between conventional and newer sequencing technology was observed at this point, where the conventional genotype showed only the UL97 substitution C607Y, whereas deep sequencing showed exonuclease substitutions N408D and P522S, which are known to confer ganciclovir-cidofovir resistance (2), D413N, K513R, and C539G, which were later confirmed to do so, and two substitutions that confer triple-drug resistance (A834P and deletion of codons 981 and 982) (2). Thus, significant genotypic cidofovir resistance was present but not detected conventionally. Further, the UL97 substitution C607Y was gradually being displaced by the canonical A594V substitution, which confers higher-level ganciclovir resistance (Table 2).
New recombinant phenotyping data generated for this study validated the UL54 L773V amino acid substitution as conferring foscarnet resistance and revealed previously undocumented low-grade ganciclovir and cidofovir cross-resistance. The mutation was sufficiently growth attenuating (Fig. 2) to keep the subpopulation at no more than ∼50%, fading more rapidly upon withdrawal of foscarnet than the well-known substitution A809V (2) (Fig. 1), and this probably accounts for its rarity in clinical practice. The new exonuclease domain substitutions D413N, K513R, and C539G confer ganciclovir-cidofovir resistance phenotypes comparable to those conferred by other established exonuclease substitutions, such as N408D and P522S, also seen in this patient (2). The level of cidofovir resistance conferred by exonuclease mutations varies but can be relatively high (10- to 20-fold) (2). This case illustrates the limitations of cidofovir in salvaging ganciclovir and foscarnet treatment failures and the urgent need for antivirals not targeting the viral DNA polymerase, such as letermovir, a viral terminase inhibitor that has undergone phase II clinical trials (13) and has a nonoverlapping drug resistance profile (14).
Deep-sequencing data were reproducible on repeated analysis of samples from the same plasma specimens (arrays 10 and 12) (Table 1), except some low-level subpopulations in the 3 to 6% range. The resistance-related mutations detected at these low levels were compatible with the antiviral treatment history. In plasma extracts stored under adverse conditions (AA10), there were some other nonreproducible subpopulations detected that did not track across specimens (Table 1), and multiple unrecognized variants coexisted in isolated samples, similar to artifacts attributable to PCR reported in a valganciclovir clinical trial (15). Although high-throughput sequencing technology has quality control concerns distinct from those of conventional genotyping (6), it is important to beware of artifacts common to both technologies that result from nonrepresentative PCR amplification from samples with insufficient copies of intact viral DNA. In this case, fresh re-extraction and amplification from properly archived plasma completely eliminated the isolated findings of unexplained sequence subpopulations while demonstrating the progression of multiple authenticated resistance mutations, including those observed at low levels. Because our data pertain to a single patient and are narrowly focused on UL97 and UL54, they cannot be directly compared to reports of multiple fluctuating baseline CMV genotypes in vivo (4, 5). Known UL97 and UL54 baseline polymorphisms (Table 1) that reflect baseline CMV genotypes remained stable as 100% sequence populations over time.
Our initial experience with high-throughput deep sequencing suggests that it provides a higher-resolution view of evolving drug resistance mutations that is preferable for optimal case management. The technology is becoming more accessible, with automation and streamlined processing of batched specimens (6). The extraordinary plasma viral loads in this case probably favored the rapid emergence of resistance within 6 to 7 weeks of starting a new drug as well as the diversity of evolved mutations. The exonuclease substitution D413N may also have accelerated the evolution of resistance mutations by impairing exonuclease proofreading, as documented with D413A (16). More experience with newer sequencing technology in cases with lower viral loads will be helpful in determining whether the simultaneous rapid evolution of so many proven resistance mutations is common in treated individuals.
ACKNOWLEDGMENTS
This work was supported by Department of Veterans Affairs grant I01-BX00925 and NIH grant R01-AI39938.
We thank Gail Marousek, Benjamin Houser, and Colleen Pollock for technical assistance. We also thank the members of the Stanford Histocompatibility, Immunogenetics, and Disease Profiling laboratory, and in particular Fumiko Yamamoto, for technical assistance.
Footnotes
Published ahead of print 2 June 2014
REFERENCES
- 1.Kotton CN, Kumar D, Caliendo AM, Asberg A, Chou S, Danziger-Isakov L, Humar A. 2013. Updated international consensus guidelines on the management of cytomegalovirus in solid-organ transplantation. Transplantation 96:333–360. 10.1097/TP.0b013e31829df29d [DOI] [PubMed] [Google Scholar]
- 2.Lurain NS, Chou S. 2010. Antiviral drug resistance of human cytomegalovirus. Clin. Microbiol. Rev. 23:689–712. 10.1128/CMR.00009-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuurman R, Demeter L, Reichelderfer P, Tijnagel J, de Groot T, Boucher C. 1999. Worldwide evaluation of DNA sequencing approaches for identification of drug resistance mutations in the human immunodeficiency virus type 1 reverse transcriptase. J. Clin. Microbiol. 37:2291–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorzer I, Guelly C, Trajanoski S, Puchhammer-Stockl E. 2010. Deep sequencing reveals highly complex dynamics of human cytomegalovirus genotypes in transplant patients over time. J. Virol. 84:7195–7203. 10.1128/JVI.00475-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Renzette N, Gibson L, Bhattacharjee B, Fisher D, Schleiss MR, Jensen JD, Kowalik TF. 2013. Rapid intrahost evolution of human cytomegalovirus is shaped by demography and positive selection. PLoS Genet. 9:e1003735. 10.1371/journal.pgen.1003735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sahoo MK, Lefterova MI, Yamamoto F, Waggoner JJ, Chou S, Holmes SP, Anderson MW, Pinsky BA. 2013. Detection of cytomegalovirus drug resistance mutations by next-generation sequencing. J. Clin. Microbiol. 51:3700–3710. 10.1128/JCM.01605-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong KM, Najjar H, Hawley M, Press RD. 2004. Quantitative real-time PCR with automated sample preparation for diagnosis and monitoring of cytomegalovirus infection in bone marrow transplant patients. Clin. Chem. 50:846–856. 10.1373/clinchem.2003.026484 [DOI] [PubMed] [Google Scholar]
- 8.Chou S. 2011. Phenotypic diversity of cytomegalovirus DNA polymerase gene variants observed after antiviral therapy. J. Clin. Virol. 50:287–291. 10.1016/j.jcv.2011.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baldanti F, Underwood MR, Talarico CL, Simoncini L, Sarasini A, Biron KK, Gerna G. 1998. The Cys607→Tyr change in the UL97 phosphotransferase confers ganciclovir resistance to two human cytomegalovirus strains recovered from two immunocompromised patients. Antimicrob. Agents Chemother. 42:444–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou S, Van Wechel LC, Lichy HM, Marousek GI. 2005. Phenotyping of cytomegalovirus drug resistance mutations by using recombinant viruses incorporating a reporter gene. Antimicrob. Agents Chemother. 49:2710–2715. 10.1128/AAC.49.7.2710-2715.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mousavi-Jazi M, Schloss L, Wahren B, Brytting M. 2003. Point mutations induced by foscarnet (PFA) in the human cytomegalovirus DNA polymerase. J. Clin. Virol. 26:301–306. 10.1016/S1386-6532(02)00046-X [DOI] [PubMed] [Google Scholar]
- 12.Mylonakis E, Kallas WM, Fishman JA. 2002. Combination antiviral therapy for ganciclovir-resistant cytomegalovirus infection in solid-organ transplant recipients. Clin. Infect. Dis. 34:1337–1341. 10.1086/340101 [DOI] [PubMed] [Google Scholar]
- 13.Chemaly RF, Ullmann AJ, Stoelben S, Richard MP, Bornhauser M, Groth C, Einsele H, Silverman M, Mullane KM, Brown J, Nowak H, Kolling K, Stobernack HP, Lischka P, Zimmermann H, Rubsamen-Schaeff H, Champlin RE, Ehninger G. 2014. Letermovir for cytomegalovirus prophylaxis in hematopoietic-cell transplantation. N. Engl. J. Med. 370:1781–1789. 10.1056/NEJMoa1309533 [DOI] [PubMed] [Google Scholar]
- 14.Goldner T, Hempel C, Ruebsamen-Schaeff H, Zimmermann H, Lischka P. 2014. Geno- and phenotypic characterization of human cytomegalovirus mutants selected in vitro after letermovir (AIC246) exposure. Antimicrob. Agents Chemother. 58:610–613. 10.1128/AAC.01794-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chou S, Boivin G, Ives J, Elston R. 2014. Phenotypic evaluation of previously uncharacterized cytomegalovirus DNA polymerase sequence variants detected in a valganciclovir treatment trial. J. Infect. Dis. 209:1219–1226. 10.1093/infdis/jit654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chou S, Marousek GI. 2008. Accelerated evolution of maribavir resistance in a cytomegalovirus exonuclease domain II mutant. J. Virol. 82:246–253. 10.1128/JVI.01787-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou S, Ercolani RJ, Marousek G, Bowlin TL. 2013. Cytomegalovirus UL97 kinase catalytic domain mutations that confer multi-drug resistance. Antimicrob. Agents Chemother. 57:3375–3379. 10.1128/AAC.00511-13 [DOI] [PMC free article] [PubMed] [Google Scholar]