Abstract
We measured in vitro activity of plazomicin, a next-generation aminoglycoside, and other aminoglycosides against 50 carbapenem-resistant Klebsiella pneumoniae strains from two centers and correlated the results with the presence of various aminoglycoside-modifying enzymes (AMEs). Ninety-four percent of strains were sequence type 258 (ST258) clones, which exhibited 5 ompK36 genotypes; 80% and 10% of strains produced Klebsiella pneumoniae carbapenemase 2 (KPC-2) and KPC-3, respectively. Ninety-eight percent of strains possessed AMEs, including AAC(6′)-Ib (98%), APH(3′)-Ia (56%), AAC(3)-IV (38%), and ANT(2″)-Ia (2%). Gentamicin, tobramycin, and amikacin nonsusceptibility rates were 40, 98, and 16%, respectively. Plazomicin MICs ranged from 0.25 to 1 μg/ml. Tobramycin and plazomicin MICs correlated with gentamicin MICs (r = 0.75 and 0.57, respectively). Plazomicin exerted bactericidal activity against 17% (1× MIC) and 94% (4× MIC) of strains. All strains with AAC(6′)-Ib were tobramycin-resistant; 16% were nonsusceptible to amikacin. AAC(6′)-Ib combined with another AME was associated with higher gentamicin, tobramycin, and plazomicin MICs than AAC(6′)-Ib alone (P = 0.01, 0.0008, and 0.046, respectively). The presence of AAC(3)-IV in a strain was also associated with higher gentamicin, tobramycin, and plazomicin MICs (P = 0.0006, P < 0.0001, and P = 0.01, respectively). The combination of AAC(6′)-Ib and another AME, the presence of AAC(3)-IV, and the presence of APH(3′)-Ia were each associated with gentamicin resistance (P = 0.0002, 0.003, and 0.01, respectively). In conclusion, carbapenem-resistant K. pneumoniae strains (including ST258 clones) exhibit highly diverse antimicrobial resistance genotypes and phenotypes. Plazomicin may offer a treatment option against strains resistant to other aminoglycosides. The development of molecular assays that predict antimicrobial responses among carbapenem-resistant K. pneumoniae strains should be a research priority.
INTRODUCTION
Carbapenem-resistant Klebsiella pneumoniae strains have emerged worldwide as important nosocomial pathogens, capable of causing infections with high rates of morbidity and mortality. Carbapenem resistance arises through production of metallo-β-lactamases (MBLs) or non-metallo-carbapenemases (such as Klebsiella pneumoniae carbapenemases [KPCs] and OXA-type carbapenemases) (1). Alternatively, strains may express extended-spectrum β-lactamases (ESBLs) or AmpC β-lactamases in conjunction with loss or decreased expression of outer membrane porins (OMPs) (2, 3). In addition to determinants of carbapenem resistance, strains possess plasmids that carry genes that attenuate susceptibility to multiple classes of antimicrobials (4). As a result, therapeutic options against carbapenem-resistant K. pneumoniae infections are extremely limited, and optimal treatment regimens remain undefined.
Aminoglycosides retain potent bactericidal activity against some but not all carbapenem-resistant K. pneumoniae strains (5). Aminoglycoside resistance among Enterobacteriaceae is mediated by multiple mechanisms, including impaired membrane permeability, efflux mechanisms, ribosomal alterations, or expression of aminoglycoside-modifying enzymes (AMEs) (6). AMEs are the most important determinant of aminoglycoside resistance among K. pneumoniae strains (7). Plazomicin, a derivative of sisomicin, is a next-generation aminoglycoside that is in clinical development for the treatment of serious infections due to carbapenem-resistant Enterobacteriaceae. It has broad-spectrum in vitro activity against Klebsiella pneumoniae and other Gram-negative bacteria, including carbapenem-resistant strains (8–12). The agent has side-chain substituents that shield it from the action of most AMEs. An exception is AAC(2)-I, an AME that renders Providencia stuartii intrinsically resistant to plazomicin (13). To date, the only mechanism of plazomicin resistance demonstrated among carbapenem-resistant K. pneumoniae strains is expression of acquired 16S rRNA methyltransferase (14). This ribosome-modifying enzyme has been primarily reported among strains in which production of New Delhi metallo-β-lactamase-1 (NDM-1) is the mechanism of carbapenem resistance (15). The vast majority of carbapenem-resistant K. pneumoniae strains in the United States are classified as sequence type 258 (ST258) clones by multilocus sequence typing (MLST), and produce KPC-2 or KPC-3 rather than NDM-1 (16–18).
In this study, we evaluated the in vitro activity of plazomicin and clinically relevant aminoglycosides against 50 carbapenem-resistant K. pneumoniae strains from unique patients at two centers, including 47 ST258 strains. We screened the strains for the presence of four common AME genes in order to assess heterogeneity in resistance mechanisms and to identify genetic markers for diminished susceptibility to one or more agents.
MATERIALS AND METHODS
Clinical strains.
Fifty K. pneumoniae clinical strains obtained from unique patients at the University of Pittsburgh Medical Center (UPMC) Presbyterian and Montefiore Hospitals, Pittsburgh, PA (n = 41), and the University of Florida (UF) Health-Shands Hospital, Gainesville, FL (n = 9), were included in this study. The strains were stored at −80°C and subcultured at least twice on Mueller-Hinton agar before use. Gentamicin, tobramycin, and amikacin were purchased from the UPMC pharmacy. Plazomicin was supplied by Achaogen, Inc. (South San Francisco, CA). Kanamycin, neomycin, and netilmicin were purchased from Sigma-Aldrich (St. Louis, MO). Stock solutions of antimicrobial agents were prepared and stored at −80°C in the XDR Pathogen Laboratory at UPMC, where experiments were conducted. MICs of the aminoglycosides were determined by the standard broth microdilution method. MICs of chloramphenicol, fluoroquinolones (ciprofloxacin, levofloxacin, and moxifloxacin), minocycline, tigecycline, and trimethoprim-sulfamethoxazole were measured using Etest strips (bioMérieux). Susceptible, intermediate, and resistant MICs were defined according to Clinical and Laboratory Standards Institute (CLSI) criteria (19); the Food and Drug Administration (FDA) breakpoint for susceptibility was used for tigecycline (≤2 μg/ml). Strains were typed by multilocus sequence typing (MLST), and mechanisms of carbapenem resistance determined using standard methods, as previously reported by our groups (20, 21). Heterogeneity among strains was demonstrated by a combination of geographic origin, MLST typing, ompK36 genotypes, AME patterns, and MICs of various antimicrobials.
Detection of AME genes.
All strains were examined by PCR for genes encoding the following common AMEs: AAC(6′)-Ib, AAC(3)-IV, ANT(2″)-Ia, and APH(3′)-Ia. Primers are listed in Table 1 (22–24). We decided a priori that PCR would be performed to evaluate for the presence of 16S rRNA methyltransferase if a strain exhibited high-level resistance to gentamicin, tobramycin, and amikacin or exhibited a plazomicin MIC of ≥4 μg/ml.
TABLE 1.
Primers used in this study
| AME | Primer direction | DNA sequence (5′→3′) | Reference |
|---|---|---|---|
| AAC(6′)-Ib | Forward | TTGCGATGCTCTATGAGTGGCTA | 22 |
| Reverse | CTCGAATGCCTGGCGTGTTT | ||
| AAC(3)-IV | Forward | TCGATGGGCAGGTACTTCTC | This study |
| Reverse | ACCGACTGGACCTTCCTTCT | ||
| ANT(2″)-Ia | Forward | ATGGACACAACGCAGGTCGC | 23 |
| Reverse | TTAGGCCGCATATCGCGACC | ||
| APH(3′)-Ia | Forward | CGAGCATCAAATGAAACTGC | 24 |
| Reverse | GCGTTGCCAATGATGTTACAG |
Time-kill assays.
Time-kill assays were performed using a final volume of 20 ml of Mueller-Hinton broth and an initial inoculum of ∼1 × 106 CFU/ml (20, 25). The plazomicin concentrations tested were 1× and 4× MIC. Bactericidal activity was defined as a ≥3-log10-CFU/ml decrease from the starting concentration. Strains that experienced bactericidal activity at any point during time-kill assays but then regrew by at least 2 log10 were defined as having regrowth. Strains exhibiting regrowth at 4× MIC were also tested at 16× and 64× MIC.
Statistical analysis.
CFU and MICs were logarithmically transformed prior to statistical analysis. Graphics and statistical analysis were performed using Microsoft Excel (Redmond, WA) and GraphPad Instat and Prism software (La Jolla, CA). Comparisons between groups of antimicrobial agents were made by Fisher's exact test for categorical variables and the Mann-Whitney test for continuous variables. Correlations between pairs of variables were assessed by calculating Spearman's rank correlation coefficient. Significance was defined as P ≤ 0.05 (two tailed).
RESULTS
Fifty carbapenem-resistant K. pneumoniae strains were tested, including 47 strains belonging to the ST258 clonal group. Among ST258 strains, there were 40 KPC-2 producers, 5 KPC-3 producers, and 2 non-KPC producers. The 3 non-ST258 strains were non-KPC producers. All KPC producers also harbored TEM-1 and SHV-12 genes. None of the strains carried NDM, IMP, VIM, or OXA-48 β-lactamase genes. Fifty-nine percent and 14% of strains were susceptible and intermediate to tigecycline, respectively. The corresponding rates for trimethoprim-sulfamethoxazole, minocycline, and chloramphenicol were 38 and 3%, 10 and 14%, and 3 and 24%, respectively. All strains were resistant to fluoroquinolones.
Aminoglycoside MICs.
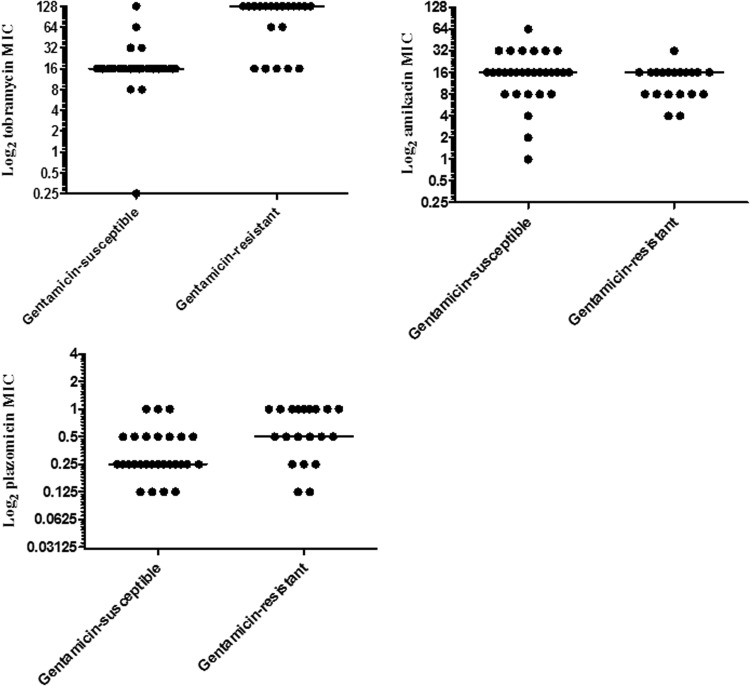
The activities of the aminoglycosides against the 50 strains are summarized in Table 2 and Fig. 1. There were no associations between ST status, presence or type of KPC, and aminoglycoside MICs. Based on CLSI susceptibility breakpoints, 40% (20/50) and 60% (30/50) of the strains were gentamicin resistant and susceptible, respectively. Strains were overwhelmingly tobramycin resistant (98% [49/50]). Amikacin MICs were within the intermediate and resistant range against 14% (7/50) and 2% (1/50) of strains, respectively. Plazomicin MICs were ≤1 μg/ml (range, 0.25 to 1 μg/ml); the MIC50 and MIC90 were 0.25 and 0.5 μg/ml, respectively. Tobramycin and plazomicin MICs correlated with gentamicin MICs by Spearman's rank test (r = 0.75, P < 0.0001, and r = 0.57, P < 0.0001, respectively). Median tobramycin and plazomicin MICs were significantly higher against gentamicin-resistant strains than gentamicin-susceptible strains (Fig. 1 and Table 2; P < 0.0001 and P = 0.0008, respectively). Ten percent (3/30) and 40% (12/30) of gentamicin-susceptible strains exhibited plazomicin MICs of 1 and ≥0.5 μg/ml, respectively, compared to 45% (9/20) and 75% (15/20) of gentamicin-resistant strains (P = 0.007 and 0.02, respectively).
TABLE 2.
In vitro susceptibility to various aminoglycosides, stratified by gentamicin susceptibility
| Aminoglycoside | Gentamicin susceptible (n = 30)a |
Gentamicin resistant (n = 20)a |
P valuec | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MIC (μg/ml) |
Resistance rate, %b | MIC (μg/ml) |
Resistance rate, %b | ||||||
| Median (range) | 50% | 90% | Median (range) | 50% | 90% | ||||
| Amikacind | 16 (1 to 64) | 16 | 32 | 23 (7/30)d | 16 (4 to 32) | 16 | 16 | 5 (1/20) | NS (0.16) |
| Kanamycin | 256 (2 to >256) | 256 | >256 | 97 (29/30) | 512 (64 to >256) | >256 | >256 | 100 (20/20) | 0.009 |
| Neomycine | 2 (0.5 to 128) | 2 | 64 | NA | 32 (1 to >256) | 32 | 512 | NA | <0.0001 |
| Netilmicin | 64 (0.25 to >64) | 64 | >64 | 97 (29/30) | >64 (32 to >64) | >64 | >64 | 100 (20/20) | 0.032 |
| Plazomicine | 0.25 (0.125 to 1) | 0.25 | 0.5 | NA | 0.5 (0.125 to 1) | 0.5 | 1 | NA | <0.0001 |
| Tobramycin | 16 (0.25 to 32) | 16 | 32 | 97 (29/30) | 32 (16 to >64) | >64 | >64 | 100 (20/20) | 0.008 |
Thirty and 20 isolates were susceptible and resistant to gentamicin, respectively. No strain was intermediate to gentamicin. 50% and 90%, MIC50 and MIC90, respectively; NA, not applicable.
Numbers in parentheses represent the number with resistance/number tested.
P values denote the difference in median MIC of respective aminoglycosides between the gentamicin-susceptible and -resistant isolates. P values were calculated using the Mann-Whitney test. NS, not significant.
Amikacin resistance rates include both intermediate and resistant strains.
Neomycin and plazomicin interpretive breakpoint MICs have not been established.
FIG 1.
In vitro susceptibility to various aminoglycosides, stratified by gentamicin susceptibility.
Amikacin MICs did not correlate with gentamicin MICs (r = −0.02, P = 0.88). Median amikacin MICs did not differ significantly against gentamicin-resistant or -susceptible strains (Fig. 1 and Table 2).
Plazomicin time-kill curves.
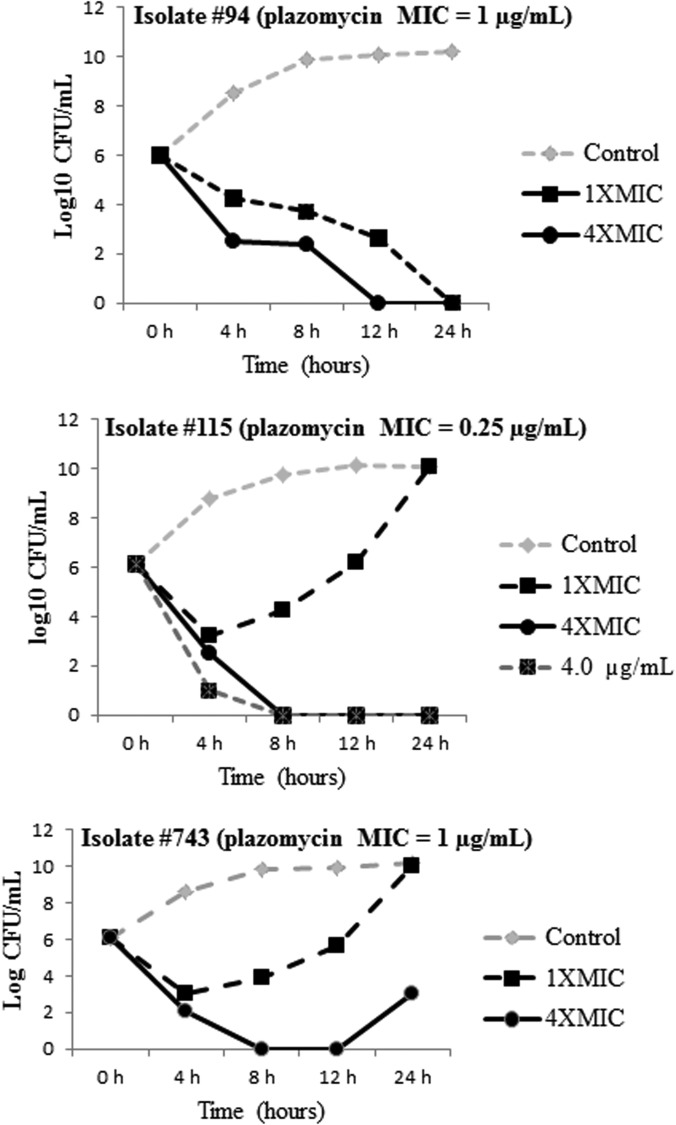
Eighteen strains were randomly selected to undergo time-kill assays. Representative kill curves for 3 strains are presented in Fig. 2. Plazomicin exhibited dose-dependent killing against all strains. At 1× MIC, bactericidal activity was evident against 17% (3/18) of the strains by 12 h. One of the three (33%) strains for which plazomicin was bactericidal exhibited subsequent regrowth by 24 h (Fig. 2). At 4× MIC, bactericidal activity was observed against 94% (17/18) of strains by 12 h; regrowth by 24 h occurred in 29% (5/17). Plazomicin showed sustained bactericidal activity at 16× MIC or 64× MIC against the 5 strains exhibiting regrowth at 4× MIC, and no regrowth was apparent.
FIG 2.
Time-kill curves of plazomicin against representative strains. Note the regrowth of the last two strains. The regrowth of strain 115 was eliminated when tested with plazomicin concentrations of 4× and 16× MIC. The regrowth of strain 743 was eliminated when tested with plazomicin at 16× MIC.
AMEs.
At least one AME was detected in 98% (49/50) of strains. AAC(6′)-Ib was the most prevalent AME, detected in each of the 49 strains that harbored an AME. The rank order of prevalence for the other AMEs was APH(3′)-Ia (56% [28/50]), AAC(3)-IV (38% [19/50]), and ANT(2″)-Ia (2% [1/50]). None of the strains met the criteria for 16S rRNA methyltransferase testing. The distribution of AMEs, the AME combinations, and their associations with aminoglycoside MICs are shown in Table 3. The one strain that did not have any of the AMEs was susceptible to all aminoglycosides (gentamicin, tobramycin, and amikacin MICs of 0.25, 0.25, and 1 μg/ml, respectively) and exhibited a plazomicin MIC of 0.25 μg/ml.
TABLE 3.
Aminoglycoside MICs stratified by presence or absence of AMEsa
| AME pattern | No. of strains | Gentamicin |
Tobramycin |
Amikacin |
Plazomicin |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Median MIC (μg/ml) |
P valueb | Median MIC (μg/ml) |
P valueb | Median MIC (μg/ml) |
P valueb | Median MIC (μg/ml) |
P valueb | ||||||
| Pattern present | Pattern absent | Pattern present | Pattern absent | Pattern present | Pattern absent | Pattern present | Pattern absent | ||||||
| No AMEs | 1 | 0.25 | NAc | NA | 0.25 | NA | NA | 1 | NA | NA | 0.25 | NA | NA |
| AAC(6′)-Ib only | 17 | 0.5 | 16 | 0.01 | 16 | 45.2 | 0.0008 | 16 | 16 | NSd (0.31) | 0.25 | 0.5 | 0.046 |
| AAC(3)-IV, AAC(6′)-Ib | 4 | 22.6 | 1 | NS (0.09) | 128 | 16 | 0.02 | 11.3 | 16 | NS (0.37) | 0.70 | 0.5 | NS (0.45) |
| APH(3′)-Ia, AAC(6′)-Ib | 12 | 0.71 | 1 | NS (0.24) | 16 | 16 | NS (0.20) | 16 | 16 | NS (0.74) | 0.35 | 0.5 | NS (0.50) |
| AAC(3)-IV, APH(3′)-Ia, AAC(6′)-Ib | 15 | 32 | 1 | 0.01 | 128 | 16 | 0.001 | 16 | 16 | NS (0.83) | 0.5 | 0.25 | 0.03 |
| ANT(2″)-Ia, APH(3′)-Ia, AAC(6′)-Ib | 1 | 32 | NA | NA | 64 | NA | NA | 16 | NA | NA | 0.5 | NA | NA |
| Any AAC(3)-IV | 19 | 32 | 0.5 | 0.0006 | 128 | 16 | <0.0001 | 16 | 16 | NS (0.58) | 0.5 | 0.25 | 0.01 |
| Any APH(3′)-Ia | 28 | 16 | 1 | 0.03 | 16 | 16 | 0.048 | 16 | 16 | NS (0.82) | 0.5 | 0.25 | NS (0.13) |
The four AMEs tested were AAC(6′)-Ia, APH(3′)-IV, AAC(3)-Ia, and ANT(2″)-Ia. AAC(6′)-Ia was present in all strains except one. On the AAC(6′)-Ib backbone, the addition of other AMEs resulted in significantly higher MICs of gentamicin, tobramycin, and plazomicin. The key AME was AAC(3)-IV, which if present, was associated with significantly higher MICs of gentamicin, tobramycin, and plazomicin. The presence of APH(3′)-Ia was not significantly associated with elevated MICs of any aminoglycosides tested. The single strain with ANT(2″)-Ia, AAC(6′)-Ib, and APH(3′)-IV exhibited higher gentamicin, tobramycin, and plazomicin MICs than strains with only AAC(6′)-Ib and APH(3′)-IV.
P values denote the statistically significant difference in log2 MICs of the aminoglycosides among the groups with and without specific AMEs.
NA, not applicable.
NS, not significant.
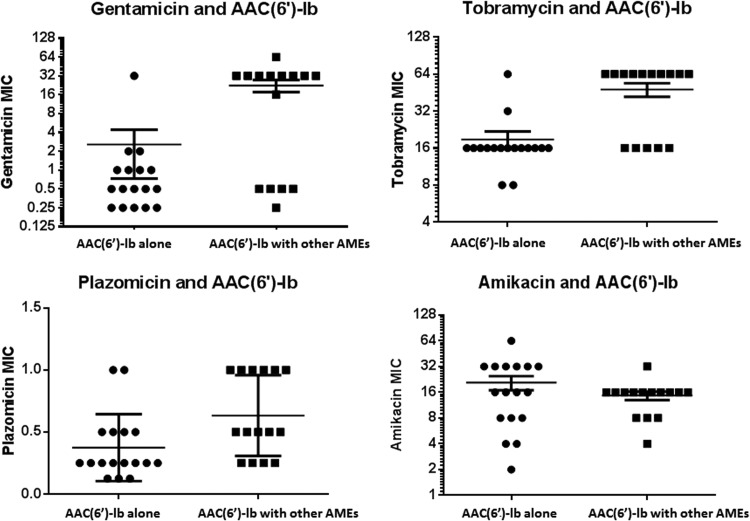
All strains with AAC(6′)-Ib were resistant to tobramycin. Ninety-four percent (16/17) and 6% (1/17) of the strains that carried AAC(6′)-Ib alone were gentamicin susceptible and resistant, respectively. The combination of AAC(6′)-Ib and at least one other AME was associated with significantly higher gentamicin, tobramycin, and plazomicin MICs than AAC(6′)-Ib alone (P = 0.01, 0.0008 and 0.046, respectively) (Table 3; Fig. 3). Among other AMEs, AAC(3)-IV made the strongest contribution to aminoglycoside MICs in combination with AAC(6′)-Ib. Gentamicin, tobramycin and plazomicin MICs were significantly higher against strains with AAC(3)-IV than against AME-carrying strains that lacked AAC(3)-IV (P = 0.0006, P < 0.0001, and P = 0.01, respectively) (Table 3; Fig. 4). Moreover, the triple combination of AAC(3)-IV, APH(3′)-Ia, and AAC(6′)-Ib was associated with significantly higher gentamicin, tobramycin, and plazomicin MICs than other AME-carrying strains (P = 0.01, 0.001, and 0.03, respectively), whereas the double combination of APH(3′)-Ia and AAC(6′)-Ib was not (P = 0.24, 0.20, and 0.50, respectively).
FIG 3.
Distribution of aminoglycoside MICs according to the presence or absence of additional AMEs [ANT(2″)-Ib, AAC(3)-IV, or APH(3′)-Ia] on the AAC(6′)-Ib backbone. The horizontal lines represent the mean MIC ± standard error. Note that the combination of AAC(6′)-Ib and ≥1 other AME was associated with significantly higher MICs of gentamicin (P = 0.01), tobramycin (P = 0.0008), and plazomicin (P = 0.046) but not amikacin (P = 0.31).
FIG 4.
Distribution of aminoglycoside MICs according to the presence or absence of AAC(3)-IV in combination with AAC(6′)-Ib. The horizontal line represents the mean MIC ± standard error. Note that the presence of AAC(3)-IV was associated with significantly higher MICs of gentamicin (P = 0.0006), tobramycin (P < 0.0001), and plazomicin (P = 0.01) but not amikacin (P = 0.58).
The presence of APH(3′)-Ia in any combination was associated with significantly higher gentamicin and tobramycin MICs than those observed in the absence of APH(3′)-Ia (P = 0.03 and 0.048, respectively); the presence of APH(3′)-Ia showed a trend toward higher plazomicin MICs (P = 0.13). The one strain with ANT(2″)-Ia also carried APH(3′)-Ia and AAC(6′)-Ib. Gentamicin and tobramycin MICs against this strain (32 and 64 μg/ml, respectively) were higher than mean MICs against strains with only APH(3′)-Ia and AAC(6′)-Ib (0.5 and 16 μg/ml, respectively).
The combination of AAC(6′)-Ib with another AME, the presence of AAC(3)-IV, and the presence of APH(3′)-Ia were each significantly associated with gentamicin resistance (P = 0.0002, 0.003, and 0.01, respectively) (Fig. 5). There were no correlations between AMEs and amikacin MICs or resistance (Table 3; Fig. 3 and 4).
FIG 5.
Associations between specific AMEs and gentamicin resistance. Gentamicin-susceptible and -resistant strains are shown as black and gray bars, respectively. The percentages of strains that were gentamicin resistant are shown above the respective bars.
Heterogeneity of carbapenem-resistant K. pneumoniae strains.
Strains were shown to be heterogeneous by a combination of geographic origin, MLST typing, ompK36 genotype, AME patterns, and MICs of various antimicrobials. Twenty-seven gentamicin-susceptible, ST258, KPC-2 producing strains exhibited five different ompK36 porin genotypes: 8 wild-type strains, 13 strains with guanine and alanine insertions at amino acids (aa) 134 and 135 (ins aa 134 to 135 GD), 3 strains with IS5 and 1 strain with IS1 insertion sequences within the promoter, 1 strain with an asparagine-asparagine-threonine-glutamic acid (NNTE) deletion at aa 84 to 87, and 1 with a guanine insertion at nucleotide (nt) position 382 (ins nt 382 G). These strains possessed various combinations of AMEs (AAC(6′)-Ib alone, AAC(6′)-Ib in combination with either AAC(3)-IV or APH(3′)-Ia, or AAC(6′)-Ib in combination with both AAC(3)-IV and APH(3′)-Ia). The strains also exhibited wide ranges of MICS for the 6 aminoglycosides tested (Table 2).
Thirteen gentamicin-resistant, ST258, KPC-2-producing strains exhibited 3 different ompK36 porin genotypes: 5 wild-type strains and 8 strains with IS5 promoter insertions with or without partial promoter deletions (n = 4 each). The strains possessed various combinations of AMEs [AAC(6′)-Ib in combination with APH(3′)-Ia or AAC(6′)-Ib in combination with both APH(3′)-Ia and AAC(3)-IV]. The strains also exhibited wide ranges of MICs for the 6 aminoglycosides tested (Table 2). The KPC-3-producing and non-KPC-producing strains (n = 5 each) differed in multilocus sequence type, ompk36 genotype, AME pattern, and/or aminoglycoside MICs.
DISCUSSION
The most notable finding of this study was that strains exhibited remarkable AME diversity. Overall, we identified six AME patterns, which correlated with different levels of aminoglycoside resistance. This heterogeneity was apparent despite the fact that 94% of strains were ST258 clones. Indeed, ST258 strains were distinct by genotypes (presence of particular molecular mechanisms of antimicrobial resistance) and/or phenotypes (antimicrobial susceptibility profiles). Our data add to a growing body of evidence that ST258 K. pneumoniae strains are highly heterogenous, despite being considered clonal by conventional molecular epidemiologic criteria (5, 26–30). We previously showed that differences in ompK36 genotypes and gene expression among ST258 strains at our center predict susceptibility or resistance to carbapenem-colistin combinations during time-kill assays (27, 29). Recent studies from our group and others have demonstrated striking diversity in the core genome of ST258 K. pneumoniae strains, including strains recovered from patients at single centers or longitudinally from the bloodstreams of individual patients (26, 28). Variations in plasmid content and number are associated with differences in multidrug resistance patterns (26). ST258 strains also exhibit variability in capsular polysaccharide, pathogenesis-related phenotypes, such as resistance to serum killing and virulence in a Galleria mellonella model of infection (30). Taken together, the data indicate that “one size fits all” approaches to identifying effective antimicrobial regimens against carbapenem-resistant K. pneumoniae strains are not likely to be useful. Rather, increased effort should be given to establishing correlations between strain genetics and responsiveness to particular antimicrobial agents and combinations (27, 29) and then using these data to define how best to utilize new drugs.
Along these lines, plazomicin demonstrated excellent activity against all carbapenem-resistant K. pneumoniae strains, regardless of AME pattern or level of resistance to other agents. Plazomicin MICs ranged from 0.25 to 1 μg/ml, concentrations at which pharmacokinetic-pharmacodynamic (PK-PD) models predicted that the overall probability of patients achieving plasma area under the concentration-time curve (AUC)/MIC ratio targets was >98% (31). Moreover, plazomicin was consistently bactericidal and inhibited regrowth during time-kill studies at ≤4 μg/ml and ≤16 μg/ml, respectively, concentrations that should be achievable within serum. Indeed, the target AUC from 0 to 24 h (AUC0–24) associated with plazomicin doses recommended in a phase 3 study of serious infections due to carbapenem-resistant Enterobacteriaceae was 262 μg · h/ml, and mean maximum concentration of drug in serum (Cmax) values ranged from 62 to 107 μg/ml (Achaogen data on file). Our findings are consistent with previous studies that showed plazomicin to be highly active against Enterobacteriaceae that do not carry 16S rRNA methyltransferase (8, 10). It is important to acknowledge that we did not test NDM-1-producing K. pneumoniae strains, which have been reported to harbor 16S rRNA methyltransferase. Nevertheless, our findings suggest that plazomicin, if used appropriately, will be an important addition to the antimicrobial armamentarium at a time of rapidly emerging antibiotic resistance among Enterobacteriaceae.
Of note, plazomicin MICs directly correlated with those of gentamicin (r = 0.57, P < 0.0001). As a result, plazomicin MICs were higher against gentamicin-resistant than gentamicin-susceptible strains (P < 0.0001). This is the first description of a correlation between MICs of plazomicin and another aminoglycoside. In fact, correlations of this sort have not been systematically investigated, in large part because plazomicin MICs run low against almost all Enterobacteriaceae regardless of their susceptibility to other aminoglycosides. At present, the association would seem to be of little clinical significance, as the difference in median plazomicin MICs was only a single 2-fold dilution and occurred at concentrations that should still achieve PK-PD targets (31). Nevertheless, the data suggest that some cross-resistance exists between plazomicin and other aminoglycosides. This situation may provide a foundation for the accumulation of additional mechanisms that further diminish susceptibility. Along related lines, 29% (5/17) of strains tested in time-kill assays exhibited regrowth in the presence of plazomicin at 4× MIC. The findings may not be clinically relevant at the current plazomicin MIC distribution, and regrowth was completely suppressed at 16× MIC. However, as MICs increase in the future, a propensity for regrowth during the course of infections in humans may create pressure for plazomicin resistance. Once plazomicin is introduced to clinical practice, providers must stay vigilant in prescribing the drug responsibly and using optimal dosing regimens to ensure that our in vitro data are not a harbinger of rapid emergence of resistance.
AAC(6′)-Ib was the backbone AME in this study, detected in 98% (49/50) of strains and associated with resistance to tobramycin but not gentamicin. The high prevalence of AAC(6′)-Ib is likely explained by the fact that the vast majority of our carbapenem-resistant K. pneumoniae isolates possessed ESBLs, as genes for AAC(6′)-Ib and ESBLs are known to localize to the same plasmid (32, 33). In general keeping with our findings, previous studies have reported that AAC(6′)-Ib affects tobramycin and amikacin susceptibility, while sparing gentamicin (34). The 6′-hydroxyethyl of plazomicin likely blocks access of AAC(6′)-Ib, thereby limiting its capacity to attenuate antimicrobial activity (13). The addition of other AMEs to the AAC(6′)-Ib backbone, however, clearly resulted in significantly higher gentamicin, tobramycin, and plazomicin MICs, as well as gentamicin resistance. In fact, the presence of AAC(6′)-Ib and another AME was the strongest predictor of gentamicin resistance (Fig. 3).
AAC(3)-IV was the key AME in conjunction with AAC(6′)-Ib. The double combination of AAC(3)-IV and AAC(6′)-Ib was present in only 8% (4/50) of the strains, but it was significantly linked to elevated tobramycin MICs and showed a trend toward elevated gentamicin MICs. In contrast, the double combination of APH(3′)-Ia and AAC(6′)-Ib was present in 24% (12/50) of strains, but it was not associated with higher MICs of any agent. The addition of AAC(3)-IV to the APH(3′)-Ia and AAC(6′)-Ib double combination, as occurred in 30% (15/50) of strains, resulted in strong associations with elevated gentamicin, tobramycin, and plazomicin MICs and gentamicin resistance. Overall, AAC(3)-IV and APH(3′)-Ia when part of any combination were both significant predictors of gentamicin resistance. AAC(3)-IV is known to confer gentamicin and tobramycin resistance among clinical strains of Escherichia coli (35), but to our knowledge, it has not been reported previously in K. pneumoniae or significantly associated with higher plazomicin MICs. Our findings suggest that the 1-N-hydroxyaminobutyric acid within plazomicin may not confer full protection from inactivation by AAC(3)-IV (12). APH(3′)-Ia cannot modify plazomicin because the drug lacks the target 3′-OH group, and it has not been linked to resistance to any of the agents in this study. Therefore, the association that we observed between the presence of APH(3′)-Ia and gentamicin resistance likely reflects the activity of other AMEs within strains. The data for ANT(2″)-Ia were too limited to comment upon its contribution to aminoglycoside MICs. ANT(2″)-Ia is always associated with other AMEs and confers resistance to gentamicin and tobramycin (36).
Amikacin MICs did not correlate with gentamicin, tobramycin, or plazomicin MICs, nor did the presence of particular AMEs correspond with higher amikacin MICs. These observations are consistent with findings that AAC(3)-IV, APH(3′)-Ia, and ANT(2″)-Ia do not impact the activity of amikacin (37, 38). We were unable to assess the previously reported impact of AAC(6′)-Ib on amikacin susceptibility since 98% (49/50) of our strains carried this enzyme. The issue was further complicated by that fact that amikacin nonsusceptibility was uncommon (14% [7/50] intermediate, 2% [1/50] resistant), but median and modal MICs (16 μg/ml) were at the upper limit of the susceptible range. Indeed, the European Committee on Antimicrobial Susceptibility Testing (EUCAST) recommends that if a member of the Enterobacteriaceae group tests as tobramycin intermediate or resistant and gentamicin and amikacin susceptible, its amikacin susceptibility status should be revised to “intermediate” (39). If we applied this rule, 3% (1/30), 93% (28/30), and 3% (1/30) of gentamicin-susceptible K. pneumoniae strains would be considered amikacin susceptible, intermediate, and resistant, respectively (rather than 77% [23/30], 20% [6/30], and 3% [1/30], respectively). The clinical validity of the EUCAST recommendation remains uncertain, and amikacin MICs below the intermediate breakpoints have been reported in numerous strains of Gram-negative bacteria that harbor AAC(6′)-Ib (40–42). The issue merits further investigation.
In conclusion, a full understanding of AMEs and other molecular mechanisms of diminished susceptibility to plazomicin and currently available aminoglycosides will allow clinicians to incorporate these agents most rationally into treatment regimens against carbapenem-resistant K. pneumoniae infections. The development of molecular assays that accurately and rapidly predict antimicrobial responses among carbapenem-resistant K. pneumoniae strains should be a top research priority.
ACKNOWLEDGMENTS
This project was supported by funding from the University of Pittsburgh Medical Center to the XDR Pathogen Laboratory and in part by grants (to Y.D. and B.N.K.) from the National Institutes of Health (R21AI107302 and 1R01AI090155, respectively). R.K.S. is supported by the National Institutes of Health through grant no. KL2TR000146. Achaogen provided plazomicin and pharmacokinetic-pharmacodynamic data from a phase 3 clinical trial.
Footnotes
Published ahead of print 27 May 2014
REFERENCES
- 1.Schultsz C, Geerlings S. 2012. Plasmid-mediated resistance in Enterobacteriaceae: changing landscape and implications for therapy. Drugs 72:1–16. 10.2165/11597960-000000000-00000 [DOI] [PubMed] [Google Scholar]
- 2.Tsai YK, Fung CP, Lin JC, Chen JH, Chang FY, Chen TL, Siu LK. 2011. Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 55:1485–1493. 10.1128/AAC.01275-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsai YK, Liou CH, Fung CP, Lin JC, Siu LK. 2013. Single or in combination antimicrobial resistance mechanisms of Klebsiella pneumoniae contribute to varied susceptibility to different carbapenems. PLoS One 8:e79640. 10.1371/journal.pone.0079640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rice LB, Carias LL, Hutton RA, Rudin SD, Endimiani A, Bonomo RA. 2008. The KQ element, a complex genetic region conferring transferable resistance to carbapenems, aminoglycosides, and fluoroquinolones in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 52:3427–3429. 10.1128/AAC.00493-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clancy CJ, Hao B, Shields RK, Chen L, Perlin DS, Kreiswirth BN, Nguyen MH. 2014. Doripenem, gentamicin, and colistin, alone and in combinations, against gentamicin-susceptible, KPC-producing Klebsiella pneumoniae strains with various ompK36 genotypes. Antimicrob. Agents Chemother. 58:3521–3525. 10.1128/AAC.01949-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aminoglycoside Resistance Study Groups. 1995. The most frequently occurring aminoglycoside resistance mechanisms—combined results of surveys in eight regions of the world. J. Chemother. 7(Suppl 2):17–30 [PubMed] [Google Scholar]
- 7.Ramirez MS, Tolmasky ME. 2010. Aminoglycoside modifying enzymes. Drug Resist. Updat. 13:151–171. 10.1016/j.drup.2010.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Endimiani AHK, Hujer AM, Armstrong ES, Choudhary Y, Aggen JB, Bonomo RA. 2009. ACHN-490, a neoglycoside with potent in vitro activity against multidrug-resistant Klebsiella pneumoniae isolates. Antimicrob. Agents Chemother. 53:4504–4507. 10.1128/AAC.00556-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landman DBE, Shah N, Kelly P, Bäcker M, Bratu S, Quale J. 2010. Activity of a novel aminoglycoside, ACHN-490, against clinical isolates of Escherichia coli and Klebsiella pneumoniae from New York City. J. Antimicrob. Chemother. 65:2123–2127. 10.1093/jac/dkq278 [DOI] [PubMed] [Google Scholar]
- 10.Livermore DM, Mushtaq S, Warner M, Zhang JC, Maharjan S, Doumith M, Woodford N. 2011. Activity of aminoglycosides, including ACHN-490, against carbapenem-resistant Enterobacteriaceae isolates. J. Antimicrob. Chemother. 66:48–53. 10.1093/jac/dkq408 [DOI] [PubMed] [Google Scholar]
- 11.Galani ISM, Daikos GL, Chrysouli Z, Poulakou G, Psichogiou M, Panagea T, Argyropoulou A, Stefanou I, Plakias G, Giamarellou H, Petrikkos G. 2012. Activity of plazomicin (ACHN-490) against MDR clinical isolates of Klebsiella pneumoniae, Escherichia coli, and Enterobacter spp. from Athens, Greece. J. Chemother. 24:191–194. 10.1179/1973947812Y.0000000015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aggen JB, Armstrong ES, Goldblum AA, Dozzo P, Linsell MS, Gliedt MJ, Hildebrandt DJ, Feeney LA, Kubo A, Matias RD, Lopez S, Gomez M, Wlasichuk KB, Diokno R, Miller GH, Moser HE. 2010. Synthesis and spectrum of the neoglycoside ACHN-490. Antimicrob. Agents Chemother. 54:4636–4642. 10.1128/AAC.00572-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Armstrong ES, Miller GH. 2010. Combating evolution with intelligent design: the neoglycoside ACHN-490. Curr. Opin. Microbiol. 13:565–573. 10.1016/j.mib.2010.09.004 [DOI] [PubMed] [Google Scholar]
- 14.Hidalgo LHK, Gutierrez B, Ovejero CM, Shukla S, Douthwaite S, Prasad KN, Woodford N, Gonzalez-Zorn B. 2013. Association of the novel aminoglycoside resistance determinant RmtF with NDM carbapenemase in Enterobacteriaceae isolated in India and the UK. J. Antimicrob. Chemother. 68:1543–1550. 10.1093/jac/dkt078 [DOI] [PubMed] [Google Scholar]
- 15.Berçot BPL, Nordmann P. 2011. Updated multiplex polymerase chain reaction for detection of 16S rRNA methylases: high prevalence among NDM-1 producers. Diagn. Microbiol. Infect. Dis. 71:442–445. 10.1016/j.diagmicrobio.2011.08.016 [DOI] [PubMed] [Google Scholar]
- 16.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 13:785–796. 10.1016/S1473-3099(13)70190-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17:1791–1798. 10.3201/eid1710.110655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. 2012. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25:682–707. 10.1128/CMR.05035-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 8th ed. Approved standard M07-A8. CLSI, Wayne, PA [Google Scholar]
- 20.Hong JH, Clancy CJ, Cheng S, Shields RK, Chen L, Doi Y, Zhao Y, Perlin DS, Kreiswirth BN, Nguyen MH. 2013. Characterization of porin expression in Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae identifies isolates most susceptible to the combination of colistin and carbapenems. Antimicrob. Agents Chemother. 57:2147–2153. 10.1128/AAC.02411-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clancy CJ, Chen L, Hong JH, Cheng S, Hao B, Shields RK, Farrell AN, Doi Y, Zhao Y, Perlin DS, Kreiswirth BN, Nguyen MH. 2013. Mutations of the ompK36 porin gene and promoter impact responses of sequence type 258, KPC-2-producing Klebsiella pneumoniae strains to doripenem and doripenem-colistin. Antimicrob. Agents Chemother. 57:5258–5265. 10.1128/AAC.01069-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC. 2006. Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 50:3953–3955. 10.1128/AAC.00915-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hujer KM, Hujer AM, Hulten EA, Bajaksouzian S, Adams JM, Donskey CJ, Ecker DJ, Massire C, Eshoo MW, Sampath R, Thomson JM, Rather PN, Craft DW, Fishbain JT, Ewell AJ, Jacobs MR, Paterson DL, Bonomo RA. 2006. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob. Agents Chemother. 50:4114–4123. 10.1128/AAC.00778-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noppe-Leclercq IWF, Haentjens S, Courcol R, Simonet M. 1999. PCR detection of aminoglycoside resistance genes: a rapid molecular typing method for Acinetobacter baumannii. Res. Microbiol. 150:317–322. 10.1016/S0923-2508(99)80057-6 [DOI] [PubMed] [Google Scholar]
- 25.Jernigan MG, Press EG, Nguyen MH, Clancy CJ, Shields RK. 2012. The combination of doripenem and colistin is bactericidal and synergistic against colistin-resistant, carbapenemase-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 56:3395–3398. 10.1128/AAC.06364-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deleo FR, Chen L, Porcella SF, Martens CA, Kobayashi SD, Porter AR, Chavda KD, Jacobs MR, Mathema B, Olsen RJ, Bonomo RA, Musser JM, Kreiswirth BN. 2014. Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc. Natl. Acad. Sci. U. S. A. 111:4988–4993. 10.1073/pnas.1321364111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clancy CJ, Chen L, Hong JH, Cheng S, Hao B, Shields RK, Farrell AN, Doi Y, Zhao Y, Perlin DS, Kreiswirth BN, Nguyen MH. 2013. Mutations of the ompK36 porin gene and promoter impact responses of sequence type 258, KPC-2-producing Klebsiella pneumoniae strains to doripenem and doripenem-colistin. Antimicrob. Agents Chemother. 57:5258–5265. 10.1128/AAC.01069-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clancy CJ, Chen L, Shields RK, Zhao Y, Cheng S, Chavda KD, Hao B, Hong JH, Doi Y, Kwak EJ, Silveira FP, Abdel-Massih R, Bogdanovich T, Humar A, Perlin DS, Kreiswirth BN, Hong Nguyen M. 2013. Epidemiology and molecular characterization of bacteremia due to carbapenem-resistant Klebsiella pneumoniae in transplant recipients. Am. J. Transplant. 13:2619–2633. 10.1111/ajt.12424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong JH, Clancy CJ, Cheng S, Shields RK, Chen L, Doi Y, Zhao Y, Perlin DS, Kreiswirth BN, Nguyen MH. 2013. Characterization of porin expression in Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae identifies isolates most susceptible to the combination of colistin and carbapenems. Antimicrob. Agents Chemother. 57:2147–2153. 10.1128/AAC.02411-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diago-Navarro E, Chen L, Passet V, Burack S, Ulacia-Hernando A, Kodiyanplakkal RP, Levi MH, Brisse S, Kreiswirth BN, Fries BC. 14 March 2014. Carbapenem-resistant Klebsiella pneumoniae exhibit variability in capsular polysaccharide and capsule associated virulence traits. J. Infect. Dis. 10.1093/infdis/jiu157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Wart SA, Forrest A, Bulik CC, Ambrose PG, Kostrub CF, Louie A, Drusano GL, Bhavnani SM. 2013. Pharmacokinetic-pharmacodynamic assessment predicts high efficacy for plazomicin against serious infections caused by carbapenem-resistant Klebsiella pneumoniae. Abstr. 53rd Intersci. Conf. Antimicrob. Agents Chemother. http://www.icaac.org/ [Google Scholar]
- 32.Chen YT, Shu HY, Li LH, Liao TL, Wu KM, Shiau YR, Yan JJ, Su IJ, Tsai SF, Lauderdale TL. 2006. Complete nucleotide sequence of pK245, a 98-kilobase plasmid conferring quinolone resistance and extended-spectrum-beta-lactamase activity in a clinical Klebsiella pneumoniae isolate. Antimicrob. Agents Chemother. 50:3861–3866. 10.1128/AAC.00456-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szabó D, Kocsis B, Rókusz L, Szentandrássy J, Katona K, Kristóf K, Nagy K. 2008. First detection of plasmid-mediated, quinolone resistance determinants qnrA, qnrB, qnrS and aac(6′)-Ib-cr in extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae in Budapest, Hungary. J. Antimicrob. Chemother. 62:630–632. 10.1093/jac/dkn206 [DOI] [PubMed] [Google Scholar]
- 34.Neonakis I, Gikas A, Scoulica E, Manios A, Georgiladakis A, Tselentis Y. 2003. Evolution of aminoglycoside resistance phenotypes of four Gram-negative bacteria: an 8-year survey in a University Hospital in Greece. Int. J. Antimicrob. Agents 22:526–531. 10.1016/S0924-8579(03)00152-3 [DOI] [PubMed] [Google Scholar]
- 35.Jakobsen L, Sandvang D, Jensen VF, Seyfarth AM, Frimodt-Møller N, Hammerum AM. 2007. Gentamicin susceptibility in Escherichia coli related to the genetic background: problems with breakpoints. Clin. Microbiol. Infect. 13:830–832. 10.1111/j.1469-0691.2007.01751.x [DOI] [PubMed] [Google Scholar]
- 36.Miro E, Grunbaum F, Gomez L, Rivera A, Mirelis B, Coll P, Navarro F. 2013. Characterization of aminoglycoside-modifying enzymes in Enterobacteriaceae clinical strains and characterization of the plasmids implicated in their diffusion. Microb. Drug Resist. 19:94–99. 10.1089/mdr.2012.0125 [DOI] [PubMed] [Google Scholar]
- 37.Ramirez MS, Tolmasky ME. 2010. Aminoglycoside modifying enzymes. Drug Resist. Updat. 13:151–171. 10.1016/j.drup.2010.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vakulenko SB, Mobashery S. 2003. Versatility of aminoglycosides and prospects for their future. Clin. Microbiol. Rev. 16:430–450. 10.1128/CMR.16.3.430-450.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leclercq R, Canton R, Brown DF, Giske CG, Heisig P, MacGowan AP, Mouton JW, Nordmann P, Rodloff AC, Rossolini GM, Soussy CJ, Steinbakk M, Winstanley TG, Kahlmeter G. 2013. EUCAST expert rules in antimicrobial susceptibility testing. Clin. Microbiol. Infect. 19:141–142. 10.1111/j.1469-0691.2011.03703.x [DOI] [PubMed] [Google Scholar]
- 40.Lindemann PC, Risberg K, Wiker HG, Mylvaganam H. 2012. Aminoglycoside resistance in clinical Escherichia coli and Klebsiella pneumoniae isolates from western Norway. APMIS 120:495–502. 10.1111/j.1600-0463.2011.02856.x [DOI] [PubMed] [Google Scholar]
- 41.Kim SY, Park YJ, Yu JK, Kim YS. 2011. Aminoglycoside susceptibility profiles of Enterobacter cloacae isolates harboring the aac(6′)-Ib gene. Korean J. Lab. Med. 31:279–281. 10.3343/kjlm.2011.31.4.279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen PJ, Jiang Y, Zhou Z, Zhang J, Yu Y, Li L. 2008. Complete nucleotide sequence of pKP96, a 67 850 bp multiresistance plasmid encoding qnrA1, aac(6′)-Ib-cr and blaCTX-M-24 from Klebsiella pneumoniae. J. Antimicrob. Chemother. 62:1252–1256. 10.1093/jac/dkn397 [DOI] [PubMed] [Google Scholar]