Abstract
Antimicrobial peptides (AMPs) are garnering attention as possible alternatives to antibiotics. Here, we describe the antimicrobial properties of epinecidin-1 against a multidrug-resistant clinical isolate of P. aeruginosa (P. aeruginosa R) and a P. aeruginosa strain from ATCC (P. aeruginosa ATCC 19660) in vivo. The MICs of epinecidin-1 against P. aeruginosa R and P. aeruginosa ATCC 19660 were determined and compared with those of imipenem. Epinecidin-1 was found to be highly effective at combating peritonitis infection caused by P. aeruginosa R or P. aeruginosa ATCC 19660 in mouse models, without inducing adverse behavioral effects or liver or kidney toxicity. Taken together, our results indicate that epinecidin-1 enhances the rate of survival of mice infected with the bacterial pathogen P. aeruginosa through both antimicrobial and immunomodulatory effects.
INTRODUCTION
Sepsis and septic shock develop as complications of hospitalization in intensive care units (ICUs) and result in high health care costs. In most cases, the offending pathogens are multidrug resistant (MDR), and their resistance to available antibiotics makes treatment difficult (1). This emerging problem may be combated through a greater understanding of the pathogenesis underlying MDR bacterium-induced sepsis and the identification of novel antimicrobial agents. The opportunistic Gram-negative bacterium Pseudomonas aeruginosa is a frighteningly dangerous pathogen that can give rise to both localized and systemic infections, including burn wound infections, pneumonia, abdominal infections, chronic ulcers, cystic fibrosis, and sepsis (2–5). There have been several outbreaks of MDR or even panresistant P. aeruginosa worldwide, and these have been associated with high morbidity and mortality (6). Strains of P. aeruginosa have developed resistance to many antibiotics, including imipenem, cephalosporins, piperacillin-tazobactam, aztreonam, carbapenems, ciprofloxacin, and aminoglycosides (7, 8). Recent reports of intestinal carriage of imipenem-resistant, Gram-negative bacilli in ICU patients have necessitated the search for new antimicrobial agents, such as eukaryotic antimicrobial peptides (AMPs) (9–11).
AMPs are key components of the innate immune response in all multicellular organisms (12, 13). AMPs have been predicted to facilitate the development of new, powerful antimicrobial drugs on the basis of their mechanism of action, which involves permeation of bacterial membranes through the following models: the barrel-stave, Shai-Matsuzaki-Huang, aggregate, and carpet and toroidal pore models (14–17). However, the action of AMPs is not limited to the disruption of bacterial membranes, as they have also been reported to translocate across the bacterial cytoplasmic membrane to inhibit various cellular processes, including synthesis of cell walls and DNA/RNA, translation, and protein folding (18–20).
The central role of AMPs in host defense makes them potential candidates for the treatment of infections (21–23), and the effects of several AMPs on P. aeruginosa infection have been analyzed in vitro (21). Recently, we identified a cDNA sequence from Epinephelus coioides that encodes a peptide named epinecidin-1; we found that epinecidin-1 is active against Gram-negative and -positive bacteria, viruses, Candida albicans, and Trichomonas vaginalis in vitro and in vivo (23–26). Morphological changes in bacteria treated with epinecidin-1 suggest that the peptide disrupts membranes (24) and may thus prevent or delay the development of microbial resistance. In addition to its direct antibacterial effects, epinecidin-1 appears to regulate the host innate immune system in response to Vibrio vulnificus infection in zebrafish and also mediates cytokine secretion in response to bacterial infection in mice (27, 28). Consequently, epinecidin-1, like other AMPs, possesses several important activities, including broad antibacterial, antiendotoxin, antitumor, antiviral, and immunomodulatory properties.
We speculated that the host-friendly epinecidin-1 may be suitable for use as an antibacterial agent against P. aeruginosa infection. In this study, we examined the antibacterial and immunomodulatory effects of epinecidin-1 using a mouse model of infection with susceptible P. aeruginosa isolates or imipenem-resistant clinical isolates of P. aeruginosa as a first step toward the in vivo use of epinecidin-1 as an alternative to antibiotics. We report that epinecidin-1 effectively rescued mice from imipenem-resistant P. aeruginosa-mediated death and the peptide possesses anti-inflammatory and antibacterial activities.
MATERIALS AND METHODS
Bacteria and cells.
The Pseudomonas aeruginosa R strain is a clinical isolate from stool obtained from Taipei City Hospital (Heping Fuyou Branch); it is resistant to meropenem, imipenem, ciprofloxacin, cefotaxime, levofloxacin, and ampicillin-sulbactam. A second P. aeruginosa strain, ATCC 19660, was purchased from the American Type Culture Collection (ATCC; Manassas, VA). All bacterial strains were identified by routine laboratory methods and stored in 20% (vol/vol) glycerol at −80°C. Mueller-Hinton broth was used as the culture medium.
Peptides, reagents, and antibodies.
Reagents and chemicals were purchased from Sigma (St. Louis, MO). Standard laboratory powders of clarithromycin (catalog no. C9742; Sigma, St. Louis, MO) and imipenem (catalog no. 1337809; USP, Rockville, MD) were used and prepared according to the guidelines of the CLSI. Epinecidin-1 (H-GFIFHIIKGLFHAGKMIHGLV-OH) was synthesized by solid-phase peptide synthesis and purified by reverse-phase high-performance liquid chromatography to a grade of >98.19% by GL Biochemistry (catalog no. 080571; Shanghai, China). For pharmacokinetic studies, the epinecidin-1 peptide was purchased from Genesis Biotech Inc. (purity, 100%; catalog no. GCP0822A95; Taipei, Taiwan). Synthetic epinecidin-1 was dissolved in 0.8% phosphate-buffered saline (PBS; pH 7.4) for all experiments. Interleukin-6 (IL-6; murine IL-6 mini-enzyme-linked immunosorbent assay [mini-ELISA] development kit; catalog no. 900-M50) and tumor necrosis factor alpha (TNF-α; murine TNF-α ELISA development kit; lot no. 0510054; catalog no. 900-K54) were purchased from Peprotech (Rocky Hill, NJ). Pseudomonas aeruginosa serotype 5c antibody (PAS) SD6930 (catalog no. ab69232) and IL-1β antibody (catalog no. ab8320) were purchased from Abcam.
Bacterial infection model in vivo.
Male C57BL/6 mice (age, 8 to 10 weeks; weight, 20.21 ± 0.10 g) were housed under standard conditions of light, temperature, and water and food availability. All experiments were approved by the laboratory animal ethics committee of Southern Taiwan University. Mice were injected intraperitoneally with 107 CFU P. aeruginosa per mouse. Ten minutes after P. aeruginosa injection, mice were injected intraperitoneally with clarithromycin (0.01 mg/g mouse body weight), imipenem (0.01 mg/g mouse body weight), or epinecidin-1 (0.005 mg/g mouse body weight). In a second set of experiments, mice were given intraperitoneal injections of epinecidin-1 (0.005 mg/g mouse body weight) at 10, 60, 120, 180, or 360 min after P. aeruginosa injection. The survival rate and status were recorded every 24 h for up to 168 h. The strains used were P. aeruginosa ATCC 19660 and P. aeruginosa R. The R indicates MDR. To examine bacterial dissemination, mice were sacrificed at 48 h after injection with antibiotics or epinecidin-1, and the bacterial numbers in blood, peritoneum, spleen, liver, and mesenteric lymph nodes were recorded. Experiments were performed as previously described (29). Colony counts from the diluted bacterial solutions were expressed relative to those at the start of treatment. These experiments consisted of four groups, and each group contained 27 mice (with 10 mice each being used for two independent time points and 7 being used for the third time point).
In vivo toxicity and pharmacokinetics.
For acute toxicity studies, epinecidin-1 was dissolved in PBS and administered as intramuscular bolus injections in the left thigh (5, 25, 50, 75, or 100 mg/kg). Mice were observed for signs of systemic toxicity (see the footnotes to Table 2). For pharmacokinetic analyses, epinecidin-1 was dissolved in sterile PBS and injected intravenously, subcutaneously, or intraperitoneally into healthy Wistar rats (body weight, 276.46 ± 1.35 g; BioLASCO, Taipei, Taiwan). Two Wistar rats were used for each time point. Two soft tubes (BioLASCO) were inserted into the carotid artery and jugular vein of each rat. Each Wistar rat received single doses of 25 μg peptide per rat for pharmacokinetic experiments. About 1 to 3 ml of blood was taken from the carotid artery at 10, 20, 30, 60, 120, and 180 min. Control blood was obtained from untreated rats. The blood samples were centrifuged, and sera were collected. The concentration of epinecidin-1 in sera was determined by liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS), following a previously published protocol and procedure (23).
TABLE 2.
Gross toxicity in mice following intramuscular injection of epinecidin-1a
| Dose (mg/kg) | No. of mice, effect after receiving epinecidin-1a |
|---|---|
| 5 | 3, no effect |
| 25 | 3, no effect |
| 50 | 3, no effect |
| 75 | 2, no effect; 1, toxicity level 1 |
| 100 | 2, no effect; 1, toxicity level 1 |
Toxicity level 1, narrowing of eyes. Most mice recovered by 5 h after treatment.
Determination of endotoxin and cytokine levels in vivo.
To determine the concentrations of endotoxin, P. aeruginosa, IL-1β, IL-6, and TNF-α in plasma, 0.2 ml of blood was collected from the tail vein at 0, 2, 6, 12, 24, and 48 h after injection (antibiotics or epinecidin-1) and transferred to tubes containing EDTA tripotassium salt. Endotoxin concentrations were measured by the Limulus amebocyte lysate test (Cape Cod, Inc.), according to the manufacturer's protocol. The endotoxin concentrations for the standard curve ranged from 0.005 endotoxin units (EU)/ml to 50 EU/ml. The sensitivity (λ) of the assay was defined as the lowest concentration used in the standard curve. Endotoxin standards (0, 0.015, 0.03, 0.06, 0.125, 0.25, and 0.5 endotoxin units/ml) were analyzed in each run, and the concentrations of endotoxin in the experimental groups were calculated by comparison with the standard curve. The P. aeruginosa concentrations in mouse sera were measured using an ELISA with a P. aeruginosa antibody. The plasma concentrations of IL-1β, IL-6, and TNF-α were determined using immunosorbent assays, in accordance with the manufacturer's instructions. All experimental results were compared with the standard curve to determine the amounts of cytokines present. All samples were analyzed in triplicate.
Microarray analysis and real-time PCR.
The TRIzol reagent was used to extract RNA from the livers of mice treated with epinecidin-1 or PBS alone. Two independent RNA replicates were obtained and stored separately. RNA from mice treated with epinecidin-1 was individually compared to RNA from PBS-treated control mice. Total RNA was isolated and quantified using a NanoDrop ND-1000 spectrophotometer (Wilmington, DE). Total RNA (1.5 μg) from each sample was amplified and labeled with cyanine-3 (Cy3), according to the 1-color labeling protocol of a low-RNA-input linear amplification kit (Agilent Technologies, Santa Clara, CA). The resulting cDNA samples were hybridized to 4 × 44K mouse oligonucleotide microarrays using Agilent reagents and protocols and following our previously published methods (26). All data were entered into Agilent GeneSpring GX7 software (Agilent Technologies) and Pathway Studio software (Ariadne Genomics, Rockville, MD) for analysis and data mining. Genes with an expression level change of ≥2-fold (average ratio of biological duplicates, ≥2 or ≤0.5) were considered to be significantly affected by treatment. The gene list was converted to a log2 ratio and analyzed using Pathway Studio (version 6.2) software. Genes which showed a direct relationship with the entities based on protein expression or regulation were mapped using the Adriane ontology. After microarray analysis, total RNA was isolated from mouse liver and spleen and purified using a Qiagen RNeasy kit. Reverse transcription was performed with an iScript cDNA synthesis kit (Epicentre), according to the manufacturer's recommendations. Real-time PCR was used to analyze gene expression, in accordance with standard protocols, with SYBR green (Toyobo, Japan), 2× SYBR green PCR buffer, 0.5 μl cDNA, and 500 nM specific forward and reverse primers (primer sequences are provided in Table S1 in the supplemental material). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as the reference gene. Quantitative PCR (qPCR) was performed under the following conditions: 40 cycles of 10 min at 95°C, 15 s at 95°C, and 1 min at 60°C. The threshold cycle (CT) number was calculated with ABI software. Relative transcript quantities were calculated using the ΔCT method with Ef1a as the internal reference gene. ΔCT is the difference between the threshold cycle number of the gene of interest and the threshold cycle number of GAPDH. Real-time PCR was performed in triplicate for each experimental group.
Statistical analyses.
Three biologically independent replicates were performed for each experiment, and each replicate was performed in triplicate. Univariate analysis of variance (ANOVA), performed with SPSS software (Chicago, IL), was used to identify significant differences between treatments. Error bars represent the standard deviation or standard error of the mean (SEM). Differences were defined to be significant at a P value of <0.05 or <0.01. Different letters indicate significant differences between groups, while the same letter indicates no difference between groups.
Microarray data accession number.
Microarray data are available under GEO accession number GSE57976.
RESULTS
Epinecidin-1 enhances the survival of mice infected with P. aeruginosa and exhibits in vivo bacteriostatic properties against P. aeruginosa.
We first analyzed the in vitro antimicrobial activity of epinecidin-1 using the broth microdilution method. Both the P. aeruginosa ATCC 19660 strain and the MDR P. aeruginosa R strain showed susceptibility to epinecidin-1, with MIC90s (the MIC at which 90% of growth was inhibited) of 50 and 3.12 μg/ml, respectively (see Table S2 in the supplemental material). The MIC90 of epinecidin-1 was higher than that of imipenem for ATCC 19660 (50 versus 3.12 μg/ml, respectively), whereas the MIC of epinecidin-1 was lower than that of imipenem for P. aeruginosa R (3.12 versus 200 μg/ml, respectively). Transmission electron micrographs revealed membrane disruption and a lighter electron density in the cytoplasm of epinecidin-1-treated P. aeruginosa cells than untreated cells (see Fig. S1 in the supplemental material). This finding suggests that the bactericidal activity of this peptide is mediated through membrane lysis.
We proceeded to investigate the bactericidal effects of epinecidin-1 in vivo, by monitoring the survival of mice infected with P. aeruginosa prior to treatment with epinecidin-1. All untreated mice infected with either P. aeruginosa ATCC 19660 or P. aeruginosa R died within 72 h of infection, whereas cotreatment with epinecidin-1 decreased the mortality rate. At 7 days after ATCC 19660 infection, the survival rates were 100%, 88.4%, and 0% for mice treated with imipenem (0.01 mg/g), epinecidin-1 (0.005 mg/g), and clarithromycin (0.01 mg/g), respectively. On the other hand, at 7 days after P. aeruginosa R infection, the survival rates were 88.4%, 16.1%, and 0% for mice treated with epinecidin-1, imipenem, and clarithromycin, respectively (Fig. 1A). The rates of lethality by 48 h in the untreated groups were 93.3% and 88.6% in mice infected with ATCC 19660 and R, respectively, and treatment with imipenem or epinecidin-1 significantly decreased the rate of mortality (Table 1). Bacteriologic evaluation revealed that untreated mice infected with either strain exhibited 100% positive blood cultures and a high level of bacterial colonization (with the numbers of CFU/g being no lower than 106) for all organs tested (Table 1). Epinecidin-1 treatment significantly reduced the bacterial burden in all examined organs compared to that for the untreated controls (P < 0.05); clarithromycin and imipenem were less effective in mice infected with ATCC 19660 (Table 1). In the R infection model, the bacterial burdens in all examined organs were also reduced to a significantly greater extent by epinecidin-1 than by the tested antibiotics. These data indicate that epinecidin-1 can efficiently control the multiplication of ATCC 19660 and R in the organs of infected mice. To determine the curative potential, mice were first injected with P. aeruginosa ATCC 19660 or P. aeruginosa R and then injected with epinecidin-1 (0.005 mg/g) 10, 60, 120, 180, or 360 min later. At these injection times, the P. aeruginosa ATCC 19660 experimental groups exhibited survival rates of 93.3%, 73.3%, 60.0%, 46.6%, and 33.3%, respectively, while the P. aeruginosa R experimental groups exhibited survival rates of 93.3%, 60.0%, 60.0%, 46.6%, and 20.0%, respectively (Fig. 1B). The survival rates of mice treated with epinecidin-1 were consistently greater than those of untreated mice (PBS-treated control mice). These data indicate that immediate application of epinecidin-1 (0.005 mg/g) is important to prevent severe infection. Application within 10 to 120 min of P. aeruginosa infection enabled epinecidin-1 to act as an effective curative agent.
FIG 1.
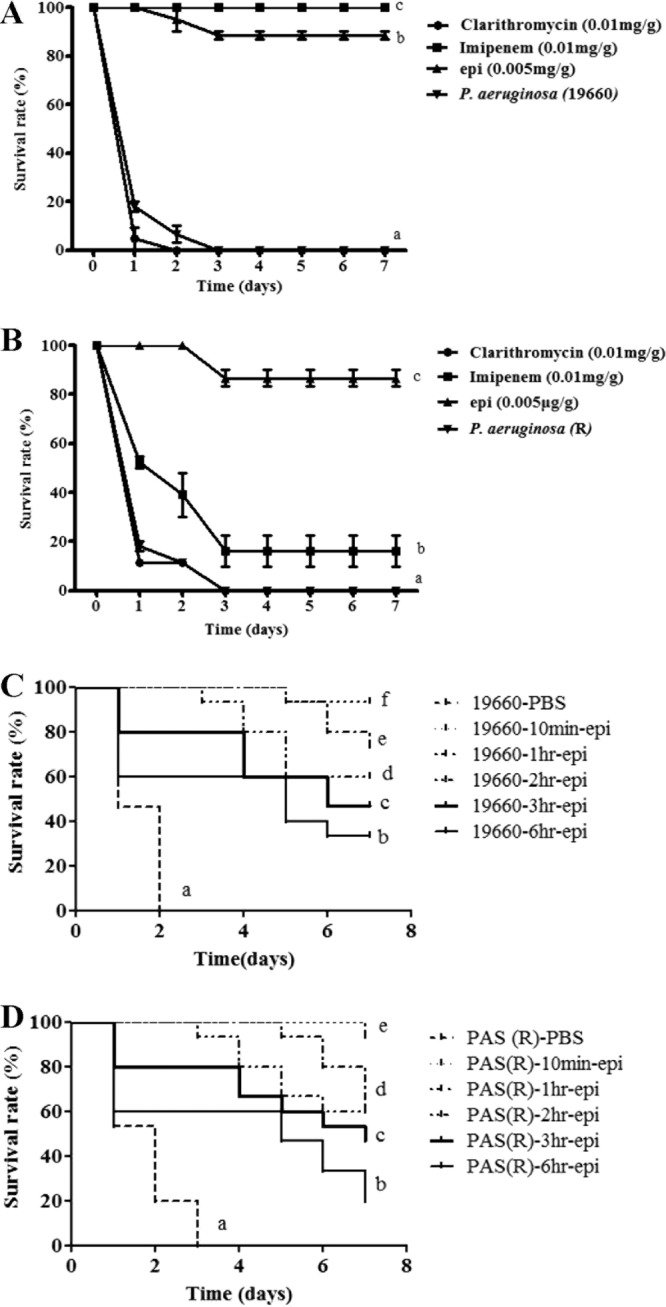
Effects of epinecidin-1 (epi) treatment on mice infected with P. aeruginosa R or P. aeruginosa ATCC 19660. (A and B) Mice were injected with P. aeruginosa ATCC 19660 (A) or P. aeruginosa R (B), and independent groups (n = 27) were subsequently injected with epinecidin-1, clarithromycin, or imipenem. The survival rate was monitored on a daily basis for up to 7 days. (C and D) To determine the curative potential, mice were first injected with P. aeruginosa ATCC 19660 (C) or P. aeruginosa R (D) and then with epinecidin-1 (0.005 mg/g) 10, 60, 120, 180, or 360 min later. At these injection times, the P. aeruginosa ATCC 19660 experimental groups exhibited survival rates of 93.3%, 73.3%, 60.0%, 46.6%, and 33.3%, respectively, while the P. aeruginosa R experimental groups exhibited survival rates of 93.3%, 60.0%, 60.0%, 46.6%, and 20.0%, respectively.
TABLE 1.
Effect of epinecidin-1, clarithromycin, and imipenem on mouse survival following intraperitoneal injection of 107 CFU of P. aeruginosa ATCC 19660 or P. aeruginosa Ra
| Strain and treatment | % lethality | Mean ± SD bacterial count (CFU/ml) in: |
||||
|---|---|---|---|---|---|---|
| Blood | Peritoneum | Spleen | Liver | Mesenteric lymph nodes | ||
| P. aeruginosa ATCC 19660 | ||||||
| No treatment | 93.34C | 8.9 × 106 ± 2.4 × 106B | 2.6 × 109 ± 1.0 × 109B | 2.8 × 109 ± 1.6 × 109B | 3.3 × 109 ± 1.0 × 109B | 4.3 × 109 ± 2.1 × 109B |
| Clarithromycin (0.01 mg/g) | 95.34D | 8.9 × 106 ± 1.0 × 106B | 3.4 × 109 ± 1.4 × 109B | 2.6 × 109 ± 1.2 × 109B | 3.6 × 109 ± 2.0 × 109B | 4.3 × 109 ± 1.0 × 109B |
| Imipenem (0.01 mg/g) | 0A | 0A | 1.7 × 106 ± 1.0 × 106A | 2.2 × 106 ± 0.6 × 106A | 2.8 × 106 ± 1.0 × 106A | 1.8 × 106 ± 1.0 × 106A |
| Epinecidin-1 (5 μg/g) | 5B | 0A | 3.1 × 106 ± 1.2 × 106A | 1.3 × 106 ± 1.0 × 106A | 3.8 × 106 ± 1.4 × 106A | 1.3 × 106 ± 1.6 × 106A |
| P. aeruginosa R | ||||||
| No treatment | 88.6C | 7.9 × 106 ± 1.4 × 106B | 4.6 × 109 ± 1.6 × 109C | 2.8 × 109 ± 1.6 × 109C | 3.3 × 109 ± 1.0 × 109B | 4.3 × 109 ± 2.1 × 109B |
| Clarithromycin (0.01 mg/g) | 88.6C | 8.4 × 106 ± 1.0 × 106B | 2.4 × 109 ± 1.4 × 109C | 2.6 × 109 ± 1.2 × 109C | 3.6 × 109 ± 2.0 × 109B | 4.3 × 109 ± 1.0 × 109B |
| Imipenem (0.01 mg/g) | 60.97B | 1.7 × 106 ± 1.0 × 106B | 4.7 × 107 ± 1.0 × 107B | 1.2 × 107 ± 0.2 × 107B | 3.6 × 109 ± 2.0 × 109B | 1.7 × 106 ± 1.0 × 106A |
| Epinecidin-1 (5 μg/g) | 0A | 0A | 4.1 × 105 ± 1.0 × 105A | 5.3 × 106 ± 1.8 × 106A | 3.5 × 107 ± 1.4 × 107A | 2.4 ×06 ± 1.0 × 106A |
Lethality was monitored for 48 h following the injection of epinecidin-1 or antibiotics. Different letters indicate a significant difference between two groups.
Toxicity and pharmacokinetics.
We did not observe systemic toxic effects of intramuscularly injected epinecidin-1 at the highest tested concentration of 100 mg/kg or lower concentrations. One of the three epinecidin-1-treated mice exhibited eye narrowing (level 1) after injection of 75 mg/kg or 100 mg/kg of epinecidin-1 (Table 2). Intraperitoneal injections of epinecidin-1 at dosages consisting of the 50% effective dose (ED50; 50 μg/mice) or ED100 (150 μg/mice) did not induce any side effects, as determined by measurement of biochemical factors; no significant changes in the levels of glutamic oxaloacetic transaminase (GOT), glutamic pyruvic transaminase (GPT), blood urea nitrogen (BUN), creatinine (CRE), total bilirubin (TBIL), or uric acid (UA) were observed (see Table S3 in the supplemental material). We next investigated the pharmacokinetic properties of epinecidin-1 administered through different routes. We delivered epinecidin-1 at a dose of 25 μg/rat by intravenous, subcutaneous, or intraperitoneal injection into healthy Wistar rats. The serum concentration of epinecidin-1 delivered by intravenous injection decreased over time, from 3,833 ng/ml at 10 min after injection to 950 ng/ml at 180 min (Fig. 2). However, the concentration of free epinecidin-1 in the blood did not exceed 1,100 ng/ml between 10 and 180 min after subcutaneous or intraperitoneal injection. These results indicate that intravenous injection results in an epinecidin-1 bioavailability of 15.3%, while subcutaneous and intraperitoneal injection results in an epinecidin-1 (or single degradation product) bioavailability of 4.4% at 10 min after injection.
FIG 2.
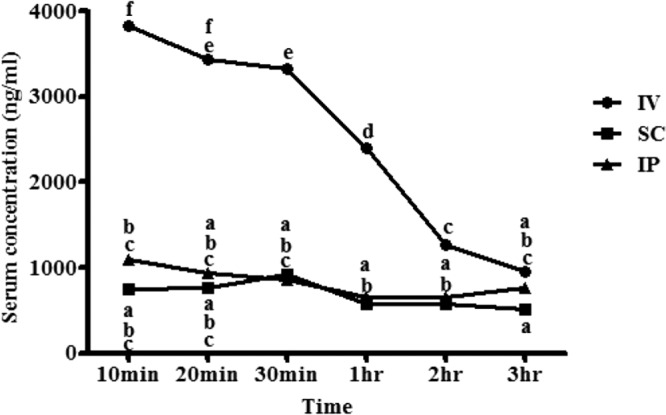
Serum pharmacokinetics of epinecidin-1, following administration of single doses by intravenous (IV), subcutaneous (SC), or intraperitoneal (IP) injection into healthy Wistar rats. Each symbol represents the mean concentration from two rats. Data with different letters differ significantly (P < 0.05) between time points.
Anti-inflammatory effect of epinecidin-1 on endotoxin and cytokine levels.
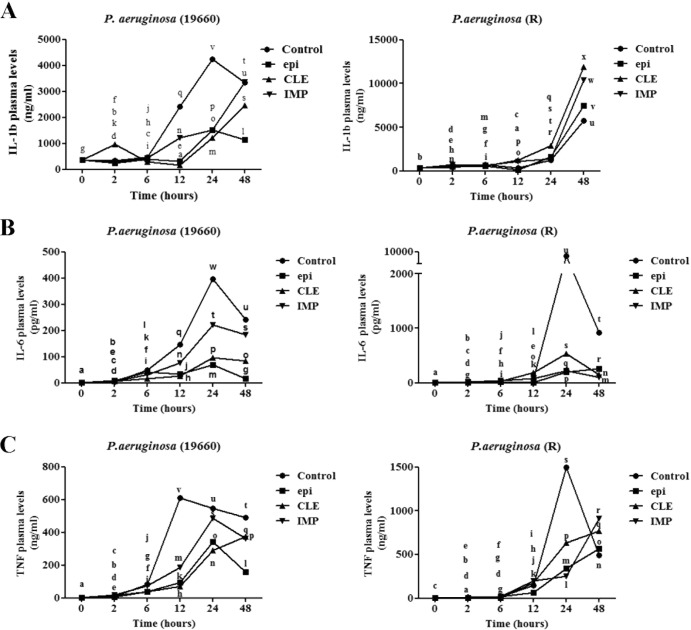
We subsequently investigated the effect of epinecidin-1 and antibiotics on bacterial endotoxins. To this end, mice infected with P. aeruginosa were treated with 5 μg/g epinecidin-1, 10 μg/g clarithromycin, or 10 μg/g imipenem, and blood samples were taken at 2, 6, 12, 24, and 48 h during the treatment period. Control mice were infected with P. aeruginosa but left untreated. Plasma concentrations of endotoxin and P. aeruginosa serotype 5c antibody (PAS) were consistently higher in control mice than in mice treated with epinecidin-1 or antibiotics over a 48-h period (Fig. 3). In addition, plasma levels of endotoxin and PAS were generally higher for mice infected with the R strain than mice infected with the ATCC 19660 strain. Furthermore, mice treated with antibiotics had significantly higher plasma concentrations of endotoxin and PAS than epinecidin-1-treated mice at 48 h postinjection. Of the inflammatory mediators, IL-6, IL-1β, and TNF-α are of particular importance, because they play a major role in coordinating proinflammatory mechanisms. As shown in Fig. 4 (left), epinecidin-1 significantly decreased the concentrations of IL-6, IL-1β, and TNF-α in the blood of mice infected with ATCC 19660. Conversely, treatment with epinecidin-1 or antibiotics significantly increased the plasma concentrations of IL-1β and TNF-α in mice infected with the R strain, with mean peak levels being achieved at 48 h postinjection. Overall, epinecidin-1 exhibited stronger antimicrobial activity than the tested antibiotics and induced a greater reduction in plasma endotoxin and cytokine levels.
FIG 3.
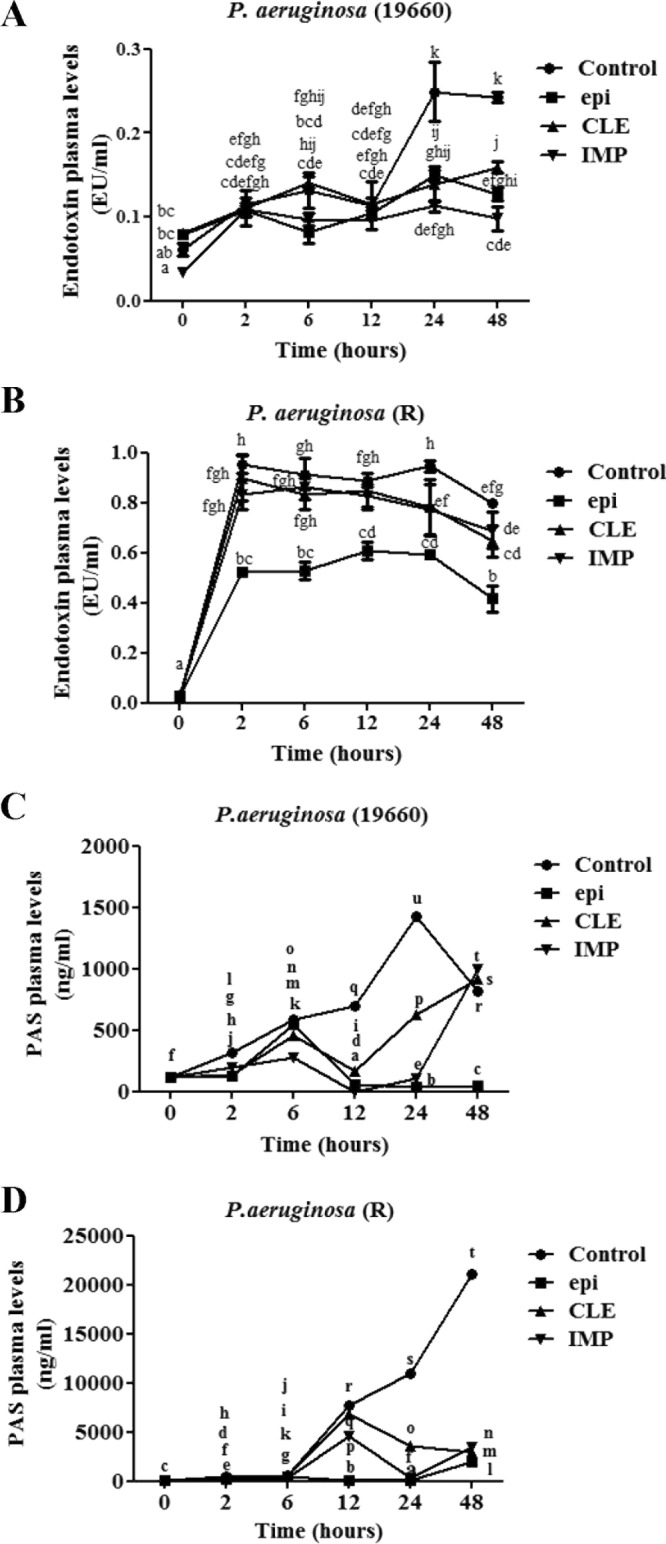
Plasma levels of endotoxins and PAS (P. aeruginosa) in mice infected with P. aeruginosa R or ATCC 19660 after treatment with antibiotics or epinecidin-1. epi, epinecidin-1; CLE, clarithromycin; IMP, imipenem. Data with different letters differ significantly (P < 0.05) between treatments.
FIG 4.
Plasma levels of IL-1β, IL-6, and TNF-α. Sera for ELISA were removed from the tail vein at 0, 2, 6, 12, 24, and 48 h during the treatment period. Data with different letters differ significantly (P < 0.05) between treatments.
Gene expression profiles are altered by injection of epinecidin-1.
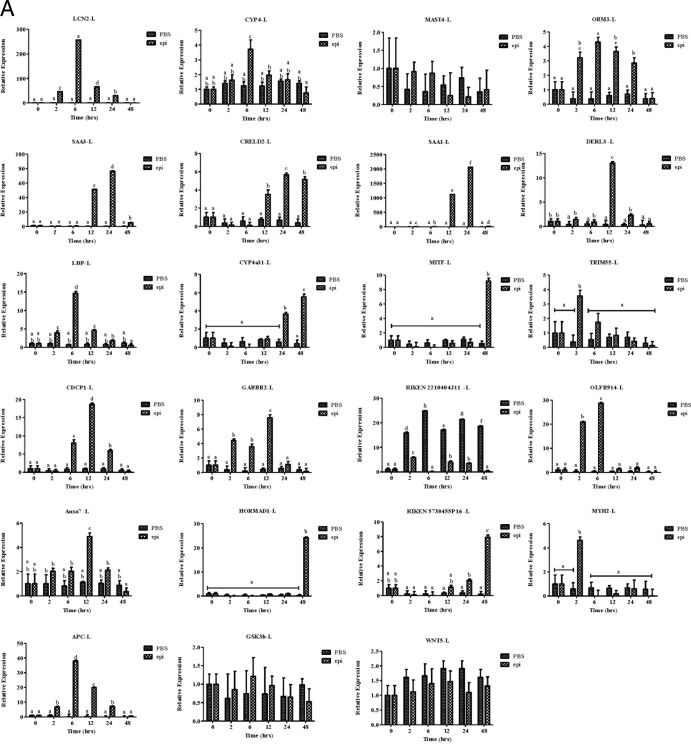
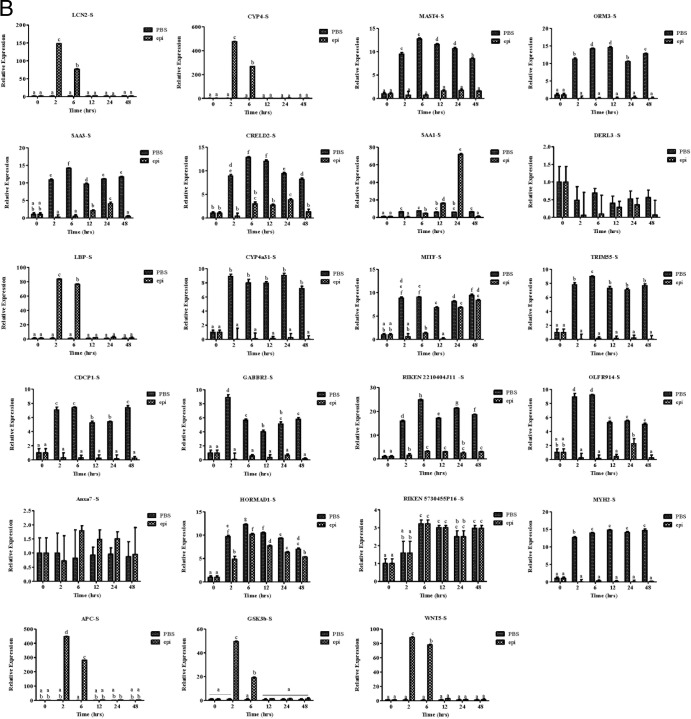
Finally, we used microarray analysis to examine the effect of epinecidin-1 on gene expression profiles in mouse. Experimental mice were treated with epinecidin-1 for 24 h, while control mice were untreated. RNA was subsequently extracted from the liver, and three biological replicates were analyzed. Of the probes examined, 1,796 exhibited a multiple of change of >2.0 and a P value of <0.05. Of these 1,796 probes, 759 probes (∼42%) were downregulated and 1,037 probes (57.7%) were upregulated between epinecidin-1-treated and control mice (a detailed list is provided in Data Set S1 in the supplemental material). Several of the downregulated genes are well-characterized proinflammatory cytokines, chemokines, and associated genes, suggesting that epinecidin-1 negatively regulates several immune-related genes. On the other hand, genes induced by epinecidin-1 are related to several signal transduction pathways, metabolic processes, protein complex assembly, protein complex biogenesis, and protein folding, among other processes (Table 3). We used qPCR to confirm the up- or downregulation of selected genes in the liver (Fig. 5A) and spleen (Fig. 5B) (see Table S1 in the supplemental material). In the liver, epinecidin-1 treatment increased the expression of MITF (after 6 h of treatment) and RIKEN cDNA 2210404J11 (6 to 48 h) but decreased the expression of SAA1, SAA3, LCN2, OLFR914, and APC. Conversely, in the spleen, the expression levels of ORM3, SAA3, CYP4a31, TRIM55, RIKEN cDNA 2210404J11, OLFR914, and MYH2 were increased by epinecidin-1 treatment, while the expression levels of CYP4, LBP, APC, GSK3b, and WNT5 were decreased by epinecidin-1 treatment (2 to 48 h). These results indicate that sepsis- and inflammation-oriented genes are suppressed by epinecidin-1.
TABLE 3.
Genes up- and downregulated by epinecidin-1 in mice, as determined by microarray analysis
| GO no. | Function | Count (no. of genes) | P value |
|---|---|---|---|
| GO:0007166 | Cell surface receptor-linked signal transduction | 200 | 0.03687171 |
| GO:0007186 | G-protein-coupled receptor protein signaling pathway | 159 | 0.01237965 |
| GO:0016071 | mRNA metabolic process | 30 | 0.06115789 |
| GO:0034621 | Cellular macromolecular complex subunit organization | 28 | 0.01568345 |
| GO:0034622 | Cellular macromolecular complex assembly | 27 | 0.00627889 |
| GO:0006461 | Protein complex assembly | 26 | 0.01967148 |
| GO:0070271 | Protein complex biogenesis | 26 | 0.01967148 |
| GO:0032940 | Secretion by cell | 24 | 0.00674446 |
| GO:0046903 | Secretion | 24 | 0.04300132 |
| GO:0008380 | RNA splicing | 21 | 0.08089444 |
| GO:0006457 | Protein folding | 16 | 0.03637995 |
| GO:0010817 | Regulation of hormone levels | 16 | 0.03861235 |
| GO:0016042 | Lipid catabolic process | 15 | 0.0956397 |
| GO:0007018 | Microtubule-based movement | 14 | 0.02684929 |
| GO:0003001 | Generation of a signal involved in cell-cell signaling | 13 | 0.01552742 |
| GO:0006887 | Exocytosis | 13 | 0.0916722 |
| GO:0033365 | Protein localization in organelle | 12 | 0.08372683 |
| GO:0046942 | Carboxylic acid transport | 12 | 0.09359128 |
| GO:0015931 | Nucleobase, nucleoside, nucleotide, and nucleic acid transport | 11 | 0.03636812 |
| GO:0034504 | Protein localization in nucleus | 10 | 0.0278588 |
| GO:0032269 | Negative regulation of cellular protein metabolic process | 10 | 0.06888556 |
| GO:0006022 | Aminoglycan metabolic process | 8 | 0.0742951 |
| GO:0006953 | Acute-phase response | 7 | 0.01744936 |
| GO:0046879 | Hormone secretion | 7 | 0.06848175 |
| GO:0009914 | Hormone transport | 7 | 0.07531571 |
| GO:0030072 | Peptide hormone secretion | 6 | 0.09087306 |
| GO:0006970 | Response to osmotic stress | 5 | 0.02885352 |
| GO:0010741 | Negative regulation of protein kinase cascade | 5 | 0.09817185 |
| GO:0030433 | Endoplasmic reticulum-associated protein catabolic process | 4 | 0.06021506 |
FIG 5.
Epinecidin-1 modulates gene expression profiles in mice. Adult mice were injected with epinecidin-1, while controls were untreated. After 24 h, total RNA was isolated from the liver (L) (A) and spleen (S) (B) and reverse transcribed for use in real-time qPCR analysis.
DISCUSSION
Severe sepsis and septic shock remain the leading causes of multiple organ failure and mortality in surgical intensive care units. Early antibiotic therapy has become problematic with the increased number of infections caused by MDR bacteria, with Gram-negative bacteria such as Klebsiella pneumoniae and Pseudomonas aeruginosa being the most frequently cultured pathogens (30). There is thus an urgent need to identify new strategies to treat these infections. One possibility is the use of antimicrobial peptides (AMPs), such as the newly identified epinecidin-1. Epinecidin-1 has been reported to have antisepsis, antitumor, antivirus, and immunomodulatory activities and may be useful in combating MDR infections (26, 27, 31–33).
This study aimed to evaluate the potential clinical use of epinecidin-1, whose effects were compared with those of conventional antibiotics, which are generally the last line of defense against infections caused by MDR P. aeruginosa. Infected mice immediately treated with 0.005 mg/g of epinecidin-1 were most effectively protected against sepsis. Another AMP, A3-APO, also demonstrated a protective effect when it was administered to mice immediately after infection (34). No toxic or other side effects were observed following treatment of mice with epinecidin-1, even at relatively high concentrations (5 to 100 mg/kg). Moreover, epinecidin-1 exhibits antibacterial activity greater than that of other AMPs identified from grouper or tilapia to date (30, 35). In addition, 50 μg/ml epinecidin-1 caused hemolysis in human erythrocytes, but not in tilapia erythrocytes (Fig. S2 in the supplemental material). Tilapia erythrocytes may somehow be resistant to its hemolytic activity. Although high hemolytic activity is a potential barrier to the application of epinecidin-1, our systemic toxic effect analysis revealed that injection of 50 mg/kg epinecidin-1 does not cause serious side effects in mice. Intraperitoneal injection of mice with 50 mg/kg of another AMP, the A3-APO dimer, is lethal, as it causes hemolysis (36). The A3-APO dimer is thought to be degraded into shorter fragments within the mouse body, generating the active metabolite (37). Most peptides, including defensins, protegrins, and clavanins, are inactivated in the presence of blood or have reduced antimicrobial activities at pHs of <7 (38, 39). Proteolytic degradation and sequestration by serum are two major hurdles for the in vivo application of AMPs (40). Currently, we do not know whether epinecidin-1 forms a peptide dimer or cleavage products in the blood. However, our pharmacokinetic analysis revealed a steady decrease in serum epinecidin-1 levels when epinecidin-1 was administered by the subcutaneous and intraperitoneal routes. Consequently, it seems that to maintain bioactivity epinecidin-1 may require stringent conditions similar to those encountered at the onset of clinical treatment.
Epinecidin-1 demonstrated potent bactericidal activity when it was administered to mice after challenge with P. aeruginosa. Epinecidin-1 treatment resulted in 100% clearance of P. aeruginosa ATCC 19660 and P. aeruginosa R bacteria from blood after 48 h. Moreover, the numbers of CFU of the ATCC 19660 and R strains decreased in the peritoneum, spleen, liver, and mesenteric lymph nodes after 48 h of epinecidin-1 treatment. Epinecidin-1 treatment was also previously reported to reduce the bacterial (Riemerella anatipestifer) load in duck (41). Taken together, these findings highlight the utility of epinecidin-1 in treating P. aeruginosa-infected mice.
The data reported here illustrate the potential antiendotoxin properties of epinecidin-1. We used our in vivo system to demonstrate that intraperitoneal administration of 0.005 mg/g of epinecidin-1 was effective at treating an MDR infection, increasing the survival rate, and reducing endotoxin and PAS plasma levels compared to the results obtained with the antibiotic-treated groups. The inhibition of lipopolysaccharide-induced cytokine/chemokine release by epinecidin-1 suggests that the peptide may bind this agonist, as has been demonstrated for other AMPs (42). TNF-α is an indicator of endotoxin shock and tissue injury during bacterial infection (43). We report here that epinecidin-1 rapidly blocked P. aeruginosa-stimulated TNF release in mouse blood, and temporal analyses of mouse experiments have suggested that peptide-macrophage interactions are the critical determinant of the TNF blockade. Alternatively, circulation of the peptide to the entire mouse body may be the critical determinant of the TNF blockade, as the peptide may kill bacteria in every organ, thereby decreasing the bacterial load and decreasing PAS plasma levels.
Epinecidin-1 treatment also affected the expression of several immune-responsive genes. This may result in modulation of the mouse immune response, possibly enhancing the survival of mice with bacterial infections. Notably, epinecidin-1 upregulated orosomucoid 3 expression in mice; orosomucoid interacts with bacterial lipopolysaccharides and protects against sepsis, suggesting that it may help mediate the effect of epinecidin-1 on bacterial infection (44). Monitoring of urinary excretion of orosomucoid (as protein/creatinine ratios) may provide a window for the clinically relevant, real-time observation of changes in acute inflammatory processes.
Epinecidin-1 also affected the expression of other genes, including those encoding serum amyloid A (SAA) protein and lipocalin 2. SAA proteins, which represent a family of low-molecular-weight acute-phase proteins, are secreted by hepatocytes in response to infection and inflammatory stimuli (45). Neutrophil gelatinase-associated lipocalin is secreted by injured kidney cells and also by activated neutrophils in response to bacterial infections (38). Lipocalin 2 is either a marker of deactivated macrophages or deactivates macrophages upon attenuating the early inflammatory response and bacterial clearance, which impairs the survival of mice with pneumococcal pneumonia (38). Epinecidin-1 also modulated several genes associated with transcription factors, interleukins, and interferons, which may facilitate control of the immune response at the level of transcription.
In this paper, we have demonstrated the potent antimicrobial activities of epinecidin-1 against P. aeruginosa strains (an MDR clinical isolate and the ATCC 19660 strain) in vitro and in vivo. Epinecidin-1 killed the P. aeruginosa strains rapidly, indicative of a high therapeutic index in the mouse peritonitis sepsis model. Epinecidin-1 treatment significantly increased the survival rate of these mice compared to that of untreated controls or mice treated with antibiotics. Importantly, the dose of epinecidin-1 used in these experiments did not induce liver, kidney, or behavioral defects. Furthermore, the rapid induction of bacterial death by epinecidin-1 decreased TNF-α production in mice, by reducing endotoxin and plasma PAS levels. In addition, epinecidin-1 modulated the expression of immune-responsive genes, such as ORM3, SAA3, TRIM55, CYP4a31, MYH2, OLFR914, and CXCL13. Therefore, the epinecidin-1 peptide is a promising candidate for treating MDR infections caused by various families of microbes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Duncan Wright at the Institute of Cellular and Organismic Biology editorial office for manuscript editing.
This work was partially supported by research funding from the National Science Council (99-2313-B-001-002-MY3; Taiwan) and partially by research funding from the Marine Research Station, Institute of Cellular and Organismic Biology, Academia Sinica (Jiaushi, Taiwan), to Jyh-Yih Chen. We thank the core facility (Ching-Chun Lin) of the Institute of Cellular and Organismic Biology, Academia Sinica (Taipei, Taiwan), for assistance with microarray data analysis.
Footnotes
Published ahead of print 12 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02958-14.
REFERENCES
- 1.Toufekoula C, Papadakis V, Tsaganos T, Routsi C, Orfanos SE, Kotanidou A, Carrer DP, Raftogiannis M, Baziaka F, Giamarellos-Bourboulis EJ. 2013. Compartmentalization of lipid peroxidation in sepsis by multidrug-resistant gram-negative bacteria: experimental and clinical evidence. Crit. Care 17:R6. 10.1186/cc11930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Delden C. 2007. Pseudomonas aeruginosa bloodstream infections: how should we treat them? Int. J. Antimicrob. Agents 30:S71–S75. 10.1016/j.ijantimicag.2007.06.015 [DOI] [PubMed] [Google Scholar]
- 3.Page MG, Heim J. 2009. Prospects for the next anti-Pseudomonas drug. Curr. Opin. Pharmacol. 9:558–565. 10.1016/j.coph.2009.08.006 [DOI] [PubMed] [Google Scholar]
- 4.Senati M, Polacco M, Grassi VM, Carbone A, De-Giorgio F. 2013. Child abuse followed by fatal systemic Pseudomonas aeruginosa infection. Leg. Med. (Tokyo) 15:28–31. 10.1016/j.legalmed.2012.08.001 [DOI] [PubMed] [Google Scholar]
- 5.Haja Mydin H, Corris PA, Nicholson A, Perry JD, Meachery G, Marrs EC, Peart S, Fagan C, Lordan JL, Fisher AJ, Gould FK. 2012. Targeted antibiotic prophylaxis for lung transplantation in cystic fibrosis patients colonised with Pseudomonas aeruginosa using multiple combination bactericidal testing. J. Transplant. 2012:135738. 10.1155/2012/135738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsueh PR, Tseng SP, Teng LJ, Ho SW. 2005. Pan-drug-resistant Pseudomonas aeruginosa causing nosocomial infection at a university hospital in Taiwan. Clin. Microbiol. Infect. 11:670–681. 10.1111/j.1469-0691.2005.01196.x [DOI] [PubMed] [Google Scholar]
- 7.Barnea Y, Carmeli Y, Kuzmenko B, Gur E, Hammer-Munz O, Navon-Venezia S. 2006. The establishment of a Pseudomonas aeruginosa-infected burn-wound sepsis model and the effect of imipenem treatment. Ann. Plast. Surg. 56:674–679. 10.1097/01.sap.0000203984.62284.7a [DOI] [PubMed] [Google Scholar]
- 8.Ahn JY, Song JY, Yun YS, Jeong G, Choi IS. 2006. Protection of Staphylococcus aureus-infected septic mice by suppression of early acute inflammation and enhanced antimicrobial activity by ginsan. FEMS Immunol. Med. Microbiol. 46:187–197. 10.1111/j.1574-695X.2005.00021.x [DOI] [PubMed] [Google Scholar]
- 9.Armand-Lefèvre L, Angebault C, Barbier F, Hamelet E, Defrance G, Ruppé E, Bronchard R, Lepeule R, Lucet JC, El Mniai A, Wolff M, Montravers P, Plésiat P, Andremont A. 2013. Emergence of imipenem-resistant gram-negative bacilli in intestinal flora of intensive care patients. Antimicrob. Agents Chemother. 57:1488–1495. 10.1128/AAC.01823-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saugar JM, Alarcón T, López-Hernández S, López-Brea M, Andreu D, Rivas L. 2002. Activities of polymyxin B and cecropin A-melittin peptide CA(1-8)M(1-18) against a multiresistant strain of Acinetobacter baumannii. Antimicrob. Agents Chemother. 46:875–878. 10.1128/AAC.46.3.875-878.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saugar JM, Rodríguez-Hernández MJ, de la Torre BG, Pachón-Ibañez ME, Fernández-Reyes M, Andreu D, Pachón J, Rivas L. 2006. Activity of cecropin A-melittin hybrid peptides against colistin-resistant clinical strains of Acinetobacter baumannii: molecular basis for the differential mechanisms of action. Antimicrob. Agents Chemother. 50:1251–1256. 10.1128/AAC.50.4.1251-1256.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowdish DM, Davidson DJ, Hancock RE. 2005. A re-evaluation of the role of host defence peptides in mammalian immunity. Curr. Protein Pept. Sci. 6:35–51. 10.2174/1389203053027494 [DOI] [PubMed] [Google Scholar]
- 13.Reddy KV, Yedery RD, Aranha C. 2004. Antimicrobial peptides: premises and promises. Int. J. Antimicrob. Agents 24:536–547. 10.1016/j.ijantimicag.2004.09.005 [DOI] [PubMed] [Google Scholar]
- 14.Bobone S, Roversi D, Giordano L, De Zotti M, Formaggio F, Toniolo C, Park Y, Stella L. 2012. The lipid dependence of antimicrobial peptide activity is an unreliable experimental test for different pore models. Biochemistry 51:10124–10126. 10.1021/bi3015086 [DOI] [PubMed] [Google Scholar]
- 15.Wu M, Maier E, Benz R, Hancock RE. 1999. Mechanism of interaction of different classes of cationic antimicrobial peptides with planar bilayers and with the cytoplasmic membrane of Escherichia coli. Biochemistry 38:7235–7242. 10.1021/bi9826299 [DOI] [PubMed] [Google Scholar]
- 16.Yang L, Weiss TM, Lehrer RI, Huang HW. 2000. Crystallization of antimicrobial pores in membranes: magainin and protegrin. Biophys. J. 79:2002–2009. 10.1016/S0006-3495(00)76448-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hallock KJ, Lee DK, Ramamoorthy A. 2003. MSI-78, an analogue of the magainin antimicrobial peptides, disrupts lipid bilayer structure via positive curvature strain. Biophys. J. 84:3052–3060. 10.1016/S0006-3495(03)70031-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeung AT, Gellatly SL, Hancock RE. 2011. Multifunctional cationic host defence peptides and their clinical applications. Cell. Mol. Life Sci. 68:2161–2176. 10.1007/s00018-011-0710-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hancock RE, Lehrer R. 1998. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 16:82–88. 10.1016/S0167-7799(97)01156-6 [DOI] [PubMed] [Google Scholar]
- 20.David SA, Awasthi SK, Balaram P. 2000. The role of polar and facial amphipathic character in determining lipopolysaccharide-binding properties in synthetic cationic peptides. J. Endotoxin Res. 6:249–256. 10.1179/096805100101532117 [DOI] [PubMed] [Google Scholar]
- 21.Sainath Rao S, Mohan KV, Atreya CD. 2013. A peptide derived from phage display library exhibits antibacterial activity against E. coli and Pseudomonas aeruginosa. PLoS One 8:e56081. 10.1371/journal.pone.0056081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hancock RE, Sahl HG. 2006. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24:1551–1557. 10.1038/nbt1267 [DOI] [PubMed] [Google Scholar]
- 23.Pan CY, Chen JY, Cheng YS, Chen CY, Ni IH, Sheen JF, Pan YL, Kuo CM. 2007. Gene expression and localization of the epinecidin-1 antimicrobial peptide in the grouper (Epinephelus coioides), and its role in protecting fish against pathogenic infection. DNA Cell Biol. 26:403–413. 10.1089/dna.2006.0564 [DOI] [PubMed] [Google Scholar]
- 24.Nijnik A, Madera L, Ma S, Waldbrook M, Elliott MR, Easton DM, Mayer ML, Mullaly SC, Kindrachuk J, Jenssen H, Hancock RE. 2010. Synthetic cationic peptide IDR-1002 provides protection against bacterial infections through chemokine induction and enhanced leukocyte recruitment. J. Immunol. 184:2539–2550. 10.4049/jimmunol.0901813 [DOI] [PubMed] [Google Scholar]
- 25.Wang YD, Kung CW, Chi SC, Chen JY. 2010. Inactivation of nervous necrosis virus infecting grouper (Epinephelus coioides) by epinecidin-1 and hepcidin 1-5 antimicrobial peptides, and downregulation of Mx2 and Mx3 gene expressions. Fish Shellfish Immunol. 28:113–120. 10.1016/j.fsi.2009.10.001 [DOI] [PubMed] [Google Scholar]
- 26.Huang HN, Pan CY, Rajanbabu V, Chan YL, Wu CJ, Chen JY. 2011. Modulation of immune responses by the antimicrobial peptide, epinecidin (Epi)-1, and establishment of an Epi-1-based inactivated vaccine. Biomaterials 32:3627–3636. 10.1016/j.biomaterials.2011.01.061 [DOI] [PubMed] [Google Scholar]
- 27.Lee SC, Pan CY, Chen JY. 2012. The antimicrobial peptide, epinecidin-1, mediates secretion of cytokines in the immune response to bacterial infection in mice. Peptides 36:100–108. 10.1016/j.peptides.2012.04.002 [DOI] [PubMed] [Google Scholar]
- 28.Pan CY, Wu JL, Hui CF, Lin CH, Chen JY. 2011. Insights into the antibacterial and immunomodulatory functions of the antimicrobial peptide, epinecidin-1, against Vibrio vulnificus infection in zebrafish. Fish Shellfish Immunol. 31:1019–1025. 10.1016/j.fsi.2011.09.001 [DOI] [PubMed] [Google Scholar]
- 29.Cirioni O, Silvestri C, Ghiselli R, Orlando F, Riva A, Gabrielli E, Mocchegiani F, Cianforlini N, Trombettoni MM, Saba V, Scalise G, Giacometti A. 2009. Therapeutic efficacy of buforin II and rifampin in a rat model of Acinetobacter baumannii sepsis. Crit. Care Med. 37:1403–1407. 10.1097/CCM.0b013e31819c3e22 [DOI] [PubMed] [Google Scholar]
- 30.Pan CY, Lee SC, Rajanbabu V, Lin CH, Chen JY. 2012. Insights into the antibacterial and immunomodulatory functions of tilapia hepcidin (TH)2-3 against Vibrio vulnificus infection in mice. Dev. Comp. Immunol. 36:166–173. 10.1016/j.dci.2011.06.013 [DOI] [PubMed] [Google Scholar]
- 31.Huang TC, Chen JY. 2013. Proteomic and functional analysis of zebrafish after administration of antimicrobial peptide epinecidin-1. Fish Shellfish Immunol. 34:593–598. 10.1016/j.fsi.2012.11.032 [DOI] [PubMed] [Google Scholar]
- 32.Lin WJ, Chien YL, Pan CY, Lin TL, Chen JY, Chiu SJ, Hui CF. 2009. Epinecidin-1, an antimicrobial peptide from fish (Epinephelus coioides) which has an antitumor effect like lytic peptides in human fibrosarcoma cells. Peptides 30:283–290. 10.1016/j.peptides.2008.10.007 [DOI] [PubMed] [Google Scholar]
- 33.Cirioni O, Wu G, Li L, Orlando F, Silvestri C, Ghiselli R, Shen Z, Gabrielli E, Brescini L, Lezoche G, Provinciali M, Guerrieri M, Giacometti A. 2011. S-thanatin in vitro prevents colistin resistance and improves its efficacy in an animal model of Pseudomonas aeruginosa sepsis. Peptides 32:697–701. 10.1016/j.peptides.2011.01.016 [DOI] [PubMed] [Google Scholar]
- 34.Ostorhazi E, Rozgonyi F, Sztodola A, Harmos F, Kovalszky I, Szabo D, Knappe D, Hoffmann R, Cassone M, Wade JD, Bonomo RA, Otvos L., Jr 2010. Preclinical advantages of intramuscularly administered peptide A3-APO over existing therapies in Acinetobacter baumannii wound infections. J. Antimicrob. Chemother. 65:2416–2422. 10.1093/jac/dkq337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qu H, Chen B, Peng H, Wang K. 2013. Molecular cloning, recombinant expression, and antimicrobial activity of EC-hepcidin3, a new four-cysteine hepcidin isoform from Epinephelus coioides. Biosci. Biotechnol. Biochem. 77:103–110. 10.1271/bbb.120600 [DOI] [PubMed] [Google Scholar]
- 36.Szabo D, Ostorhazi E, Binas A, Rozgonyi F, Kocsis B, Cassone M, Wade JD, Nolte O, Otvos L., Jr 2010. The designer proline-rich antibacterial peptide A3-APO is effective against systemic Escherichia coli infections in different mouse models. Int. J. Antimicrob. Agents 35:357–361. 10.1016/j.ijantimicag.2009.10.015 [DOI] [PubMed] [Google Scholar]
- 37.Noto PB, Abbadessa G, Cassone M, Mateo GD, Agelan A, Wade JD, Szabo D, Kocsis B, Nagy K, Rozgonyi F, Otvos L., Jr 2008. Alternative stabilities of a proline-rich antibacterial peptide in vitro and in vivo. Protein Sci. 17:1249–1255. 10.1110/ps.034330.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warszawska JM, Gawish R, Sharif O, Sigel S, Doninger B, Lakovits K, Mesteri I, Nairz M, Boon L, Spiel A, Fuhrmann V, Strobl B, Müller M, Schenk P, Weiss G, Knapp S. 2013. Lipocalin 2 deactivates macrophages and worsens pneumococcal pneumonia outcomes. J. Clin. Invest. 123:3363–3372. 10.1172/JCI67911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng KC, Lee SH, Hour AL, Pan CY, Lee LH, Chen JY. 2012. Five different piscidins from Nile tilapia, Oreochromis niloticus: analysis of their expressions and biological functions. PLoS One 7:e50263. 10.1371/journal.pone.0050263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.López-Rojas R, Docobo-Pérez F, Pachón-Ibáñez ME, de la Torre BG, Fernández-Reyes M, March C, Bengoechea JA, Andreu D, Rivas L, Pachón J. 2011. Efficacy of cecropin A-melittin peptides on a sepsis model of infection by pan-resistant Acinetobacter baumannii. Eur. J. Clin. Microbiol. Infect. Dis. 30:1391–1398. 10.1007/s10096-011-1233-y [DOI] [PubMed] [Google Scholar]
- 41.Pan CY, Chow TY, Yu CY, Yu CY, Chen JC, Chen JY. 2010. Antimicrobial peptides of an anti-lipopolysaccharide factor, epinecidin-1, and hepcidin reduce the lethality of Riemerella anatipestifer sepsis in ducks. Peptides 31:806–815. 10.1016/j.peptides.2010.01.013 [DOI] [PubMed] [Google Scholar]
- 42.Rosenfeld Y, Shai Y. 2006. Lipopolysaccharide (endotoxin)-host defense antibacterial peptides interactions: role in bacterial resistance and prevention of sepsis. Biochim. Biophys. Acta 1758:1513–1522. 10.1016/j.bbamem.2006.05.017 [DOI] [PubMed] [Google Scholar]
- 43.Mandard N, Sodano P, Labbe H, Bonmatin JM, Bulet P, Hetru C, Ptak M, Vovelle F. 1998. Solution structure of thanatin, a potent bactericidal and fungicidal insect peptide, determined from proton two-dimensional nuclear magnetic resonance data. Eur. J. Biochem. 256:404–410. 10.1046/j.1432-1327.1998.2560404.x [DOI] [PubMed] [Google Scholar]
- 44.Moore DF, Rosenfeld MR, Gribbon PM, Winlove CP, Tsai CM. 1997. Alpha-1-acid (AAG, orosomucoid) glycoprotein: interaction with bacterial lipopolysaccharide and protection from sepsis. Inflammation 21:69–82. 10.1023/A:1027342909423 [DOI] [PubMed] [Google Scholar]
- 45.Tkalčević VI, Hrvačić B, Pašalić I, Eraković-Haber V, Glojnarić I. 2011. Immunomodulatory effects of azithromycin on serum amyloid A production in lipopolysaccharide-induced endotoxemia in mice. J. Antibiot. (Tokyo) 64:515–517. 10.1038/ja.2011.14 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.