LETTER
Cholera has reinstated its deadly grip on the planet with recent epidemics in Zimbabwe and Haiti (1, 2), along with the emergence of Vibrio cholerae having reduced susceptibility to fluoroquinolones. India also experiences frequent outbreaks caused by multidrug-resistant V. cholerae (3). We report reduced sensitivity to ciprofloxacin in V. cholerae O1 El Tor Ogawa and novel multiple mutations in the DNA topoisomerase parC gene.
Twelve V. cholerae strains were obtained from the Government Medical College Hospital, Kozhikode, Kerala, India, and serotyped as V. cholerae O1 El Tor Ogawa. Detection of the ctxA, ctxB, and tcpA genes (encoding cholera toxin and toxin-coregulated pili) (4, 5) was done by PCR, and the ctxB gene was sequenced to determine the cholera toxin genotype. Antimicrobial susceptibility testing was performed by disc diffusion (6), and the MIC of ciprofloxacin was determined by Etest (AB bioMérieux, Solna, Sweden). To check for mutations in the quinolone resistance-determining region (QRDR), the genes encoding DNA gyrase (gyrA, gyrB) and DNA topoisomerase (parC, parE) were amplified and subsequently sequenced (7).
All of the isolates possessed the ctxA and ctxB genes and the El Tor allele of tcpA. The ctxB gene sequences were similar to those of the Haitian variant of V. cholerae O1 (5). This variant has DNA sequences identical to those of the classical type of ctxB with an additional mutation at position 58 resulting in the substitution of asparagine for histidine. Antimicrobial susceptibility testing showed that the isolates were sensitive to tetracycline, erythromycin, and chloramphenicol and resistant to ampicillin, furazolidone, nalidixic acid, streptomycin, trimethoprim, cotrimoxazole, and polymyxin B. All of the strains showed reduced susceptibility to ciprofloxacin (MICs, 0.25 to 0.5 mg/liter). The QRDRs of gyrA, gyrB, parC, and parE of all of the isolates were amplified. Since all of the isolates had the same antibiotic susceptibility pattern, four were selected for sequencing of the gyrase and topoisomerase genes. Sequence analysis revealed five mutations in the parC gene, i.e., TCG to AAT (Ser-60→Asn), TAC to TTT (Tyr-65→Phe), TCG to ATC (Ser-85→Ile), GCC to TCC (Ala-128→Ser), and AAA to CGG (Lys-129→Arg) (Fig. 1A), and a single mutation in gyrA (Ser-83→Ile), while no changes were detected in the gyrB and parE genes.
FIG 1.
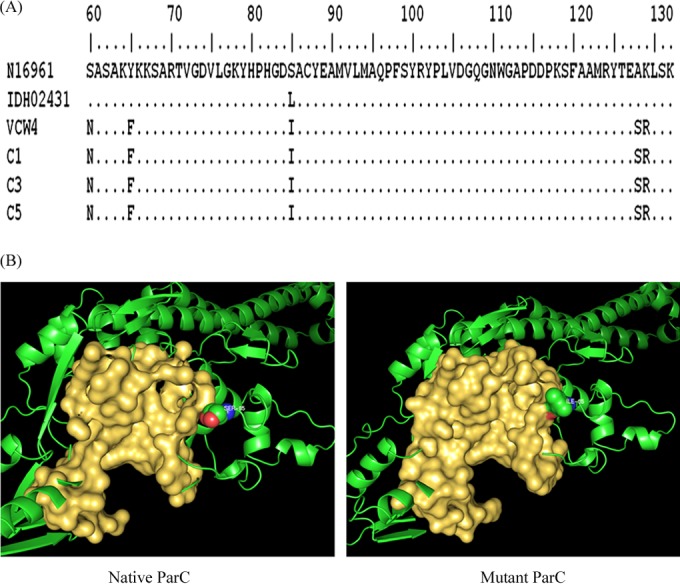
(A) Amino acid sequence alignment of the ParC subunit of V. cholerae reference El Tor strain N16961, QRDR mutant strain IDH02431, and isolates VCW4, C1, C3, and C5. Identical amino acid residues are indicated by dots. (B) Difference in the volumes of the binding pockets of the QRDRs of the native (382.9 Å3) and mutant (158.7 Å3) forms of ParC. The whole protein is represented by a cartoon, while the binding pocket is shown in a surface view and the residues are represented by spheres.
From the available literature, the most common amino acid substitution observed in the parC gene is the replacement of serine with leucine at position 85 (TCG →TTG) (8, 9), while in this study, serine was replaced with isoleucine in the same position. To understand the effect of this mutation on ciprofloxacin resistance, molecular docking analysis of ciprofloxacin on wild-type (native) and mutant ParC was performed with Autodock 4.0 as shown in Fig. 1B. In the native ParC protein, the optimal binding energy in the QRDR was −4.95 kcal/mol, whereas in the mutant protein, no ciprofloxacin binding was observed. To explain the lack of ciprofloxacin interaction affinity in the QRDR of the mutated protein, a large-scale dynamic simulation for 50 ns was performed with the Gromacs 4.5.4 package running on a Linux cluster. The root mean square fluctuation (RMSF) of the backbone residues and hydrogen bond variations between the native and mutant ParC structures were investigated with the help of g_rmsf and g_hbonds, respectively. The RMSF score revealed the presence of a higher degree of flexibility in the native protein. However, isoleucine exhibited a higher RMSF value than serine. This indicates that the mutation of S85I affects the binding conformation of ciprofloxacin. The side groups of isoleucine cause the rise in flexibility and impose constraints on the flexibility of the residues in the surrounding region, thereby preventing the binding of ciprofloxacin. A remarkable loss of hydrogen bonds was observed in the QRDR of the mutant protein. The average numbers of hydrogen bonds in the native and mutant proteins were 1.62 and 0.37, respectively. The overall results of the simulation analysis suggest that the Ser-85→Ile mutation in the parC gene has significantly contributed to the loss of stability of ciprofloxacin in the QRDR, thereby contributing to its resistance. The mutated residues, except S85I, did not exhibit a large conformational fluctuation during the simulation.
The reduced ciprofloxacin susceptibility of V. cholerae O1 Ogawa has been attributed to single amino acid changes (Ser-83→Ile in gyrA and Ser-85→Leu in parC) (10). The primary target of fluoroquinolones appears to be gyrase rather than topoisomerase (11), so the role of the multiple mutations identified in the parC gene in the present investigation in fluoroquinolone susceptibility requires careful study. The resistance pattern of our isolates is similar to that of the Haitian outbreak strain (12), but the amino acid change in position 85 and the four additional mutations detected are hitherto unknown mutations. To our knowledge, the five mutations in the parC gene described here have not been reported previously. The docking and molecular dynamic simulation implies that the Ser-85→Ile mutation in the parC gene imposes significant constraints on the flexibility of the protein, to which the observed drug resistance can be attributed. Thus, V. cholerae appears to be evolving with respect to cholera toxin and antibiotic resistance genes. Such changes call for more stringent surveillance and monitoring of the organism.
Nucleotide sequence accession numbers.
The nucleotide sequences of ctxB and parC of isolates VCW4, C1, C3, and C5 have been deposited in GenBank under accession numbers KF042305 to KF042308 and KF741367 to KF741370, respectively.
ACKNOWLEDGMENTS
We thank P. Manoj for his technical assistance in nucleotide sequencing. We are grateful to M. Radhakrishna Pillai, Director, RGCB, for the facilities provided.
We have no conflicts of interest to declare.
Footnotes
Published ahead of print 27 May 2014
REFERENCES
- 1.Mukandavire Z, Liao S, Wang J, Gaff H, Smith DL, Morris JG., Jr 2011. Estimating the reproductive numbers for the 2009 cholera outbreaks in Zimbabwe. Proc. Natl. Acad. Sci. U. S. A. 108:8767–8772. 10.1073/pnas.1019712108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2010. Update: cholera outbreak—Haiti, 2010. MMWR Morb. Mortal. Wkly. Rep. 59:1473–1479 [PubMed] [Google Scholar]
- 3.Goel AK, Jain M, Kumar P, Bhadauria S, Kmboj DV, Singh L. 2008. A new variant of Vibrio cholerae O1 El Tor causing cholera in India. J. Infect. 57:280–281. 10.1016/j.jinf.2008.06.015 [DOI] [PubMed] [Google Scholar]
- 4.Olsvik O, Wahlberg J, Petterson B, Uhlén M, Popovic T, Wachsmuth IK, Fields PI. 1993. Use of automated sequencing of polymerase chain reaction-generated amplicons to identify three types of cholera toxin subunit B in Vibrio cholerae O1 strains. J. Clin. Microbiol. 31:2–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh DV, Isac SR, Colwell RR. 2002. Development of a hexaplex PCR assay for rapid detection of virulence and regulatory genes in Vibrio cholerae and Vibrio mimicus. J. Clin. Microbiol. 40:4321–4324. 10.1128/JCM.40.11.4321-4324.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2010. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria; approved guideline—second edition, M45-A2 Clinical and Laboratory Standards Institute; Wayne, PA [Google Scholar]
- 7.Baranwal S, Dey K, Ramamurthy T, Nair GB, Kundu M. 2002. Role of active efflux in association with target gene mutations in fluoroquinolone resistance in clinical isolates of Vibrio cholerae. Antimicrob. Agents Chemother. 46:2676–2678. 10.1128/AAC.46.8.2676-2678.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kutar BM, Rajpara N, Upadhyay H, Ramamurthy T, Bhardwaj AK. 2013. Clinical isolates of Vibrio cholerae O1 El Tor Ogawa of 2009 from Kolkata, India: preponderance of SXT element and presence of Haitian ctxB variant. PLoS One 8:e56477. 10.1371/journal.pone.0056477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim HB, Wang M, Ahmed S, Park CH, LaRocque RC, Faruque AS, Salam MA, Khan WA, Qadri F, Calderwood SB, Jacoby GA, Hooper DC. 2010. Transferable quinolone resistance in Vibrio cholerae. Antimicrob. Agents Chemother. 54:799–803. 10.1128/AAC.01045-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quilici ML, Massenet D, Gake B, Bwalki B, Olson DM. 2010. Vibrio cholerae O1 variant with reduced susceptibility to ciprofloxacin, western Africa. Emerg. Infect. Dis. 16:1804–1805. 10.3201/eid1611.100568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacoby GA. 2005. Mechanisms of resistance to quinolones. Clin. Infect. Dis. 41(Suppl 2):S120–S126. 10.1086/428052 [DOI] [PubMed] [Google Scholar]
- 12.Sjölund-Karlsson M, Reimer A, Folster JP, Walker M, Dahourou GA, Batra DG, Martin I, Joyce K, Parsons MB, Boncy J, Whichard JM, Gilmour MW. 2011. Drug-resistance mechanisms in Vibrio cholerae O1 outbreak strain, Haiti, 2010. Emerg. Infect. Dis. 17:2151–2154. 10.3201/eid1711.110720 [DOI] [PMC free article] [PubMed] [Google Scholar]


