Abstract
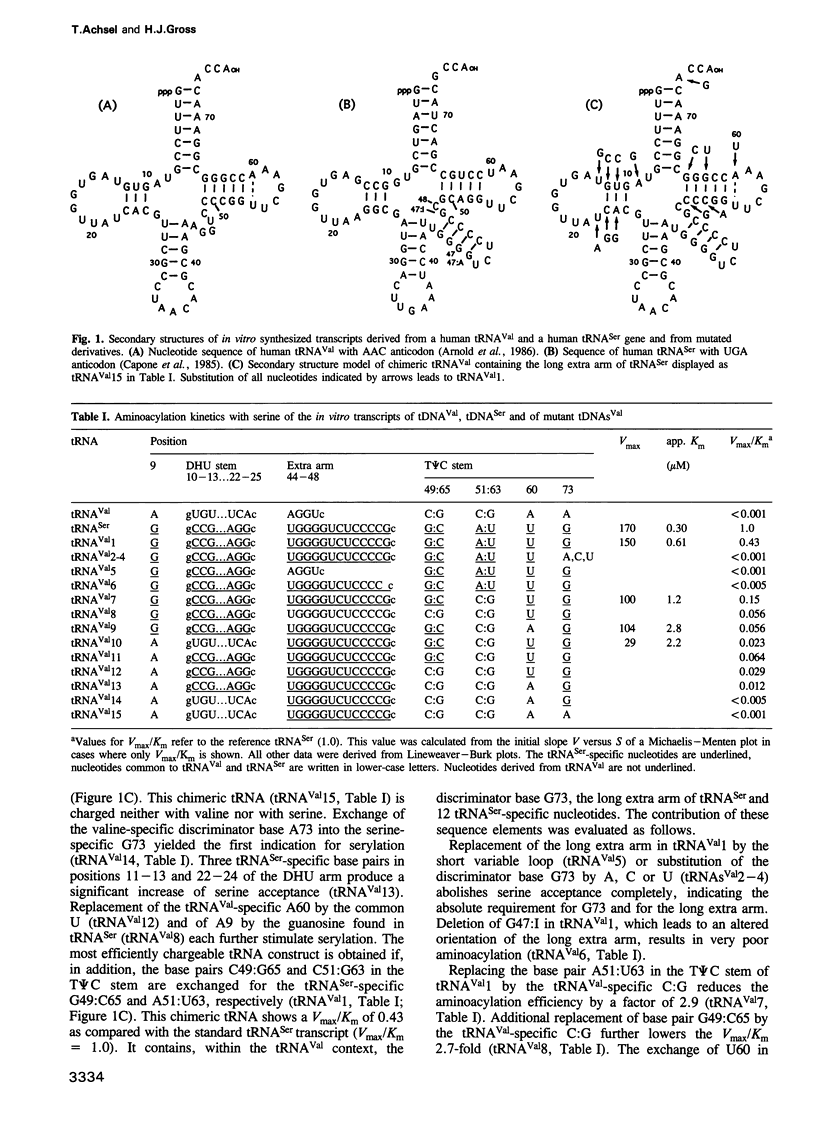
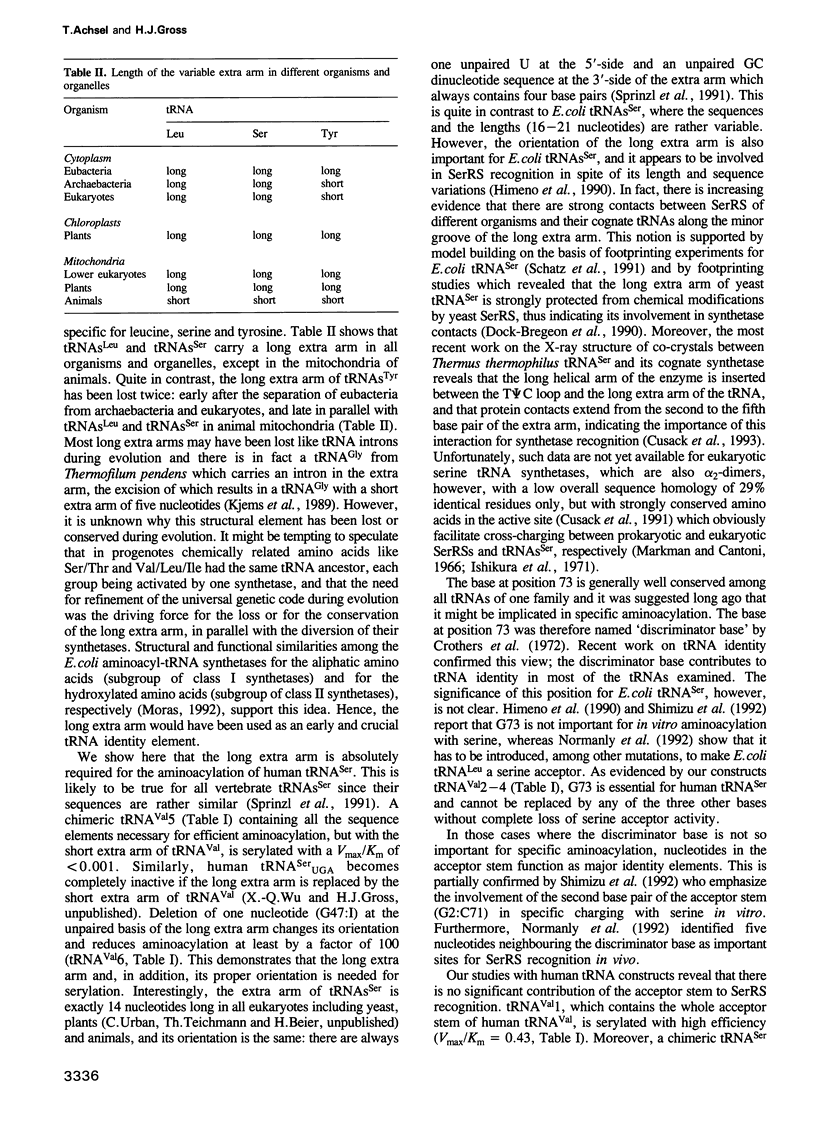
Recently, there has been much progress in understanding tRNA identity, i.e. in elucidating the sets of nucleotides that are responsible for the specific aminoacylation of a tRNA with its cognate amino acid. Interest focused, however, on tRNAs from Escherichia coli and yeast. Here we have identified the major and minor determinants of human tRNA(Ser) which were revealed by an identity switch from human tRNA(Val) to tRNA(Ser). We used in vitro transcripts and subsequent aminoacylation by HeLa S100 extract to determine the kinetic parameter Vmax/Km. The two major identity elements which are absolutely required for aminoacylation by human seryl-tRNA synthetase are the discriminator base and the long extra arm. This is in contrast to E. coli tRNA(Ser) where the discriminator base is unimportant, whereas identity determinants in the acceptor stem are required. Other sequence elements have an influence not only on serylation, but also on tRNA maturation in vitro, i.e. on pre-tRNA processing and base modification. These nucleotides are located in the DHU and the T phi C arm and are probably necessary for the proper folding of tRNAs containing a long extra arm. A34 to inosine modification depends highly on the correct three-dimensional structure of the tRNA, whereas A58 to m1A methylation does not rely on the three-dimensional folding of the substrate. This is the first tRNA identity switch involving the exchange of a short versus a long extra arm.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnold G. J., Schmutzler C., Thomann U., van Tol H., Gross H. J. The human tRNAVal gene family: organization, nucleotide sequences and homologous transcription of three single-copy genes. Gene. 1986;44(2-3):287–297. doi: 10.1016/0378-1119(86)90193-9. [DOI] [PubMed] [Google Scholar]
- Beier D., Beier H. Expression of variant nuclear Arabidopsis tRNA(Ser) genes and pre-tRNA maturation differ in HeLa, yeast and wheat germ extracts. Mol Gen Genet. 1992 May;233(1-2):201–208. doi: 10.1007/BF00587580. [DOI] [PubMed] [Google Scholar]
- Capone J. P., Sharp P. A., RajBhandary U. L. Amber, ochre and opal suppressor tRNA genes derived from a human serine tRNA gene. EMBO J. 1985 Jan;4(1):213–221. doi: 10.1002/j.1460-2075.1985.tb02338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crothers D. M., Seno T., Söll G. Is there a discriminator site in transfer RNA? Proc Natl Acad Sci U S A. 1972 Oct;69(10):3063–3067. doi: 10.1073/pnas.69.10.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack S., Härtlein M., Leberman R. Sequence, structural and evolutionary relationships between class 2 aminoacyl-tRNA synthetases. Nucleic Acids Res. 1991 Jul 11;19(13):3489–3498. doi: 10.1093/nar/19.13.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dock-Bregeon A. C., Garcia A., Giegé R., Moras D. The contacts of yeast tRNA(Ser) with seryl-tRNA synthetase studied by footprinting experiments. Eur J Biochem. 1990 Mar 10;188(2):283–290. doi: 10.1111/j.1432-1033.1990.tb15401.x. [DOI] [PubMed] [Google Scholar]
- Haumont E., Fournier M., de Henau S., Grosjean H. Enzymatic conversion of adenosine to inosine in the wobble position of yeast tRNAAsp: the dependence on the anticodon sequence. Nucleic Acids Res. 1984 Mar 26;12(6):2705–2715. doi: 10.1093/nar/12.6.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayase Y., Jahn M., Rogers M. J., Sylvers L. A., Koizumi M., Inoue H., Ohtsuka E., Söll D. Recognition of bases in Escherichia coli tRNA(Gln) by glutaminyl-tRNA synthetase: a complete identity set. EMBO J. 1992 Nov;11(11):4159–4165. doi: 10.1002/j.1460-2075.1992.tb05509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeno H., Hasegawa T., Ueda T., Watanabe K., Shimizu M. Conversion of aminoacylation specificity from tRNA(Tyr) to tRNA(Ser) in vitro. Nucleic Acids Res. 1990 Dec 11;18(23):6815–6819. doi: 10.1093/nar/18.23.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y. M., Francklyn C., Schimmel P. Molecular dissection of a transfer RNA and the basis for its identity. Trends Biochem Sci. 1989 Jun;14(6):233–237. doi: 10.1016/0968-0004(89)90033-9. [DOI] [PubMed] [Google Scholar]
- Hou Y. M., Schimmel P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988 May 12;333(6169):140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- Ishikura H., Yamada Y., Nishimura S. Structure of serine tRNA from Escherichia coli. I. Purification of serine tRNA's with different codon responses. Biochim Biophys Acta. 1971 Jan 28;228(2):471–481. doi: 10.1016/0005-2787(71)90052-9. [DOI] [PubMed] [Google Scholar]
- Jahn M., Rogers M. J., Söll D. Anticodon and acceptor stem nucleotides in tRNA(Gln) are major recognition elements for E. coli glutaminyl-tRNA synthetase. Nature. 1991 Jul 18;352(6332):258–260. doi: 10.1038/352258a0. [DOI] [PubMed] [Google Scholar]
- Jank P., Riesner D., Gross H. J. Rabbit liver tRNA1Val:II. unusual secondary structure of T psi C stem and loop due to a U54:A60 base pair. Nucleic Acids Res. 1977 Jun;4(6):2009–2200. doi: 10.1093/nar/4.6.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahnt B., Frank R., Blöcker H., Gross H. J. An efficiently transcribed human tRNA(Val) gene variant produces a stable pre-tRNA: repair of the processing defect by in vitro mutations. DNA. 1989 Jan-Feb;8(1):51–58. doi: 10.1089/dna.1.1989.8.51. [DOI] [PubMed] [Google Scholar]
- Kaçar Y., Thomann H. U., Gross H. J. The first human genes for tRNA(ArgICG), tRNA(GlyUCC), and tRNA(ThrIGU) and more tRNA(Val) pseudogenes: expression and pre-tRNA maturation in HeLa cell-free extracts. DNA Cell Biol. 1992 Dec;11(10):781–790. doi: 10.1089/dna.1992.11.781. [DOI] [PubMed] [Google Scholar]
- Kjems J., Leffers H., Olesen T., Garrett R. A. A unique tRNA intron in the variable loop of the extreme thermophile Thermofilum pendens and its possible evolutionary implications. J Biol Chem. 1989 Oct 25;264(30):17834–17837. [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. P., Dyson M. R., Mandal N., Varshney U., Bahramian B., RajBhandary U. L. Striking effects of coupling mutations in the acceptor stem on recognition of tRNAs by Escherichia coli Met-tRNA synthetase and Met-tRNA transformylase. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9262–9266. doi: 10.1073/pnas.89.19.9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makman M. H., Cantoni G. L. Studies concerning the interaction of serine soluble ribonucleic acid with seryl soluble ribonucleic acid synthetase from baker's yeast. Biochemistry. 1966 Jul;5(7):2246–2254. doi: 10.1021/bi00871a013. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Foss K., Jenkins R. A., Schneider J. Four sites in the acceptor helix and one site in the variable pocket of tRNA(Ala) determine the molecule's acceptor identity. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):9272–9276. doi: 10.1073/pnas.88.20.9272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain W. H. Transfer RNA identity. FASEB J. 1993 Jan;7(1):72–78. doi: 10.1096/fasebj.7.1.8422977. [DOI] [PubMed] [Google Scholar]
- Moras D. Structural and functional relationships between aminoacyl-tRNA synthetases. Trends Biochem Sci. 1992 Apr;17(4):159–164. doi: 10.1016/0968-0004(92)90326-5. [DOI] [PubMed] [Google Scholar]
- Nishikura K., Kurjan J., Hall B. D., De Robertis E. M. Genetic analysis of the processing of a spliced tRNA. EMBO J. 1982;1(2):263–268. doi: 10.1002/j.1460-2075.1982.tb01157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J., Abelson J. tRNA identity. Annu Rev Biochem. 1989;58:1029–1049. doi: 10.1146/annurev.bi.58.070189.005121. [DOI] [PubMed] [Google Scholar]
- Normanly J., Ollick T., Abelson J. Eight base changes are sufficient to convert a leucine-inserting tRNA into a serine-inserting tRNA. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5680–5684. doi: 10.1073/pnas.89.12.5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallanck L., Schulman L. H. Anticodon-dependent aminoacylation of a noncognate tRNA with isoleucine, valine, and phenylalanine in vivo. Proc Natl Acad Sci U S A. 1991 May 1;88(9):3872–3876. doi: 10.1073/pnas.88.9.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pütz J., Puglisi J. D., Florentz C., Giegé R. Identity elements for specific aminoacylation of yeast tRNA(Asp) by cognate aspartyl-tRNA synthetase. Science. 1991 Jun 21;252(5013):1696–1699. doi: 10.1126/science.2047878. [DOI] [PubMed] [Google Scholar]
- Rogers M. J., Söll D. Discrimination between glutaminyl-tRNA synthetase and seryl-tRNA synthetase involves nucleotides in the acceptor helix of tRNA. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6627–6631. doi: 10.1073/pnas.85.18.6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson J. R., DiRenzo A. B., Behlen L. S., Uhlenbeck O. C. Nucleotides in yeast tRNAPhe required for the specific recognition by its cognate synthetase. Science. 1989 Mar 10;243(4896):1363–1366. doi: 10.1126/science.2646717. [DOI] [PubMed] [Google Scholar]
- Sampson J. R., Uhlenbeck O. C. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz D., Leberman R., Eckstein F. Interaction of Escherichia coli tRNA(Ser) with its cognate aminoacyl-tRNA synthetase as determined by footprinting with phosphorothioate-containing tRNA transcripts. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6132–6136. doi: 10.1073/pnas.88.14.6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P. Parameters for the molecular recognition of transfer RNAs. Biochemistry. 1989 Apr 4;28(7):2747–2759. doi: 10.1021/bi00433a001. [DOI] [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. Anticodon switching changes the identity of methionine and valine transfer RNAs. Science. 1988 Nov 4;242(4879):765–768. doi: 10.1126/science.3055296. [DOI] [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. The anticodon contains a major element of the identity of arginine transfer RNAs. Science. 1989 Dec 22;246(4937):1595–1597. doi: 10.1126/science.2688091. [DOI] [PubMed] [Google Scholar]
- Schulman L. H. Recognition of tRNAs by aminoacyl-tRNA synthetases. Prog Nucleic Acid Res Mol Biol. 1991;41:23–87. [PubMed] [Google Scholar]
- Shimizu M., Asahara H., Tamura K., Hasegawa T., Himeno H. The role of anticodon bases and the discriminator nucleotide in the recognition of some E. coli tRNAs by their aminoacyl-tRNA synthetases. J Mol Evol. 1992 Nov;35(5):436–443. doi: 10.1007/BF00171822. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Dank N., Nock S., Schön A. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2127–2171. doi: 10.1093/nar/19.suppl.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomann H. U., Schmutzler C., Hüdepohl U., Blow M., Gross H. J. Genes, variant genes and pseudogenes of the human tRNA(Val) gene family. Expression and pre-tRNA maturation in vitro. J Mol Biol. 1989 Oct 20;209(4):505–523. doi: 10.1016/0022-2836(89)90590-1. [DOI] [PubMed] [Google Scholar]
- Traboni C., Ciliberto G., Cortese R. Mutations in Box B of the promoter of a eucaryotic tRNAPro gene affect rate of transcription, processing, and stability of the transcripts. Cell. 1984 Jan;36(1):179–187. doi: 10.1016/0092-8674(84)90087-4. [DOI] [PubMed] [Google Scholar]
- Vlassov V. V., Pusyriov A. T., Gross J. Evidence from chemical modification for an unusual tertiary structure of the TpseudouridineC loop in rabbit liver tRNA1Val. FEBS Lett. 1978 Oct 1;94(1):157–160. doi: 10.1016/0014-5793(78)80927-2. [DOI] [PubMed] [Google Scholar]
- Zawadzki V., Gross H. J. Rapid and simple purification of T7 RNA polymerase. Nucleic Acids Res. 1991 Apr 25;19(8):1948–1948. doi: 10.1093/nar/19.8.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]