Abstract
A carbapenem-resistant Acinetobacter pittii strain carrying an OXA-24-like enzyme was isolated in northern Spain in 2008. Sequence analysis confirmed the presence of the novel blaOXA-207 gene flanked by the site-specific XerC/XerD-like recombination binding sites and showing a unique Gly222Val substitution compared to OXA-24. Cloning and kinetic analysis showed that OXA-207 presents a reduction in the catalytic efficiency against carbapenems and a noticeable increase for oxacillin.
TEXT
Acinetobacter pittii is a member of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex. This species is ecologically diverse and can produce serious infections and even hospital outbreaks (1–3). Although this microorganism remains susceptible to many antimicrobial agents used in clinical practice (4), carbapenem-resistant A. pittii isolates carrying oxacillinase genes have been reported (5, 6). Oxacillinases are chromosomal or plasmid-encoded class D β-lactamases with heterogeneous structural and biochemical properties (7), and of these β-lactamases, the carbapenem-hydrolyzing class D β-lactamases (CHDLs) are mainly found in A. baumannii (8). Eventually, acquired CHDLs can also be found in other non-A. baumannii species, compromising the therapeutic options for patients infected by these pathogens (9, 10). The aim of this study was to characterize a novel OXA-24 variant in a carbapenem-resistant A. pittii isolate from Spain.
In April 2008, a carbapenem-resistant Gram-negative coccobacillus was recovered from a bronchial aspirate of a patient admitted to the Medical Intensive Care Unit in the Hospital Universitario Marqués de Valdecilla (HUMV), Santander, Spain. The isolate (HUMV-1588) was initially identified by the automated system MicroScan Walk-Away (Siemens Healthcare Diagnostic Inc., West Sacramento, CA) as Acinetobacter baumannii/Acinetobacter haemolyticus. Definitive identification of HUMV-1588 as A. pittii was confirmed by amplified ribosomal DNA restriction analysis (ARDRA) and sequencing of partial regions of RNA polymerase β subunit (rpoB) gene (GenBank accession number JQ838184), as previously published (11–13). The MICs of different antimicrobial agents were determined by using Etest strips (bioMérieux, Marcy l'Étoile, France) according to the manufacturer's recommendations, and results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) (14). The A. pittii HUMV-1588 strain was resistant to imipenem and meropenem (MIC of 32 μg/ml for both) and intermediate to cefepime (MIC of 16 μg/ml) but maintained susceptibility to ceftazidime and cefotaxime (MICs ranging from 2 to 4 μg/ml), as shown in Table 1. High MICs (MICs of >256 μg/ml) were observed for the aminopenicillins, isoxazolilpenicillins, ureidopenicillins, and aztreonam. The most active antimicrobial agents tested were tobramycin and trimethoprim-sulfamethoxazole (MIC of 0.06 μg/ml for both). Excellent results were also obtained for tigecycline and colistin (MICs of 0.125 and 0.25 μg/ml, respectively), the few options to treat infections caused by multidrug-resistant Acinetobacter species isolates. Of the tetracyclines, minocycline (MIC, 0.125 μg/ml) was more active than doxycycline (MIC, 0.25 μg/ml) and tetracycline (MIC, 2 μg/ml). Interesting, the HUMV-1588 strain was resistant to ciprofloxacin (MIC, 16 μg/ml) but maintained susceptibility to levofloxacin (MIC, 2 μg/ml).
TABLE 1.
Antimicrobial susceptibility profiles of A. pittii and A. baylyi determined by broth microdilution
| Antimicrobial agent(s) | MIC (μg/ml) of antimicrobial agent(s) ina: |
|||
|---|---|---|---|---|
| A. pittii (blaOXA-207) | A. baylyi ADP1(pET-RA+blaOXA-24) | A. baylyi ADP1(pET-RA+blaOXA-207) | A. baylyi ADP1(pET-RA) | |
| Ampicillin | 4,096 | 1,024 | 512 | 4 |
| Ampicillin-sulbactamb | 16/8 | 16/8 | 16/8 | 2/1 |
| Piperacillin | >512 | 256 | 256 | 8 |
| Piperacillin-tazobactamc | >256/8 | 256/8 | 256/8 | 16/8 |
| Cefoxitin | >256 | 24 | 24 | 24 |
| Ceftazidime | 2 | 2 | 2 | 2 |
| Cefotaxime | 4 | 4 | 4 | 4 |
| Cefepime | 16 | 4 | 4 | 1 |
| Aztreonam | >256 | 4 | 8 | 2 |
| Imipenem | 32 | 32 | 16 | 0.125 |
| Meropenem | 32 | 32 | 16 | 0.125 |
Antimicrobial susceptibility profiles of the OXA-207-producing A. pittii strain, the host strain A. baylyi ADP1 with the plasmid pET-RA alone, and A. baylyi ADP1 transformants harboring the plasmid pET-RA with the blaOXA-24 or blaOXA-207 gene. The MICs of the antimicrobial agents used in combinations are shown in the same order as the drugs (e.g., the MIC of ampicillin or piperacillin is shown before the slash).
Ampicillin-sulbactam was used at a 2:1 ratio.
Tazobactam was used at a fixed concentration of 8 μg/ml.
Genomic DNA from A. pittii HUMV-1588 was extracted, and multiplex PCR assays were performed to investigate the presence of CHDLs and metallo-β-lactamase (MβLs) encoding genes, as previously reported (15–17). The PCR results showed that the HUMV-1588 strain carried a blaOXA-24-like gene. Based on that, primers Pre-OXA-24.A (5′AYTTCGBATAAYSSCCATTATGTTAAATTAAAAGA3′) and Pre-OXA-24.B (5′ATTTCGYATAASGYGTATTATGTTAATTTTAGAAA3′), amplifying the entire blaOXA-24-like genes and surrounding regions, including its flanking region composed of XerC/XerD-like recombination sites, were designed according to previously reported sequences (18, 19). PCR conditions were as follows: (i) 95°C for 5 min; (ii) 35 cycles, with 1 cycle consisting of 95°C for 30 s, 45°C for 30 s, and 72°C for 2 min; (iii) 72°C for 7 min. PCR products were analyzed on a 1% (wt/vol) agarose gel stained with ethidium bromide. PCR products were purified by using a High Pure PCR product purification kit (Roche Diagnostics, Mannheim, Germany), and bidirectional DNA sequencing was performed by Macrogen Inc. (Seoul, South Korea). Sequencing of a 967-bp amplicon showed the presence of an open reading frame of 828 bp which encoded a product predicted to have 99.0% amino acid identity with OXA-24. This blaOXA-24-like gene presented a G→T mutation at nucleotide 665, causing a unique Gly222Val change, and the β-lactamase was designated OXA-207 (GenBank accession number JQ838185). In addition, the blaOXA-207 gene was flanked by the site-specific recombination XerC-XerD-like binding sites. Although the XerC/XerD-mediated recombination mechanism is not well understood, it seems that XerC/XerD recombination sites are key structures associated with the spreading of blaOXA-24-like genes (19, 20). Overall, it is clear that the OXA-24 variants are able to cause resistance to carbapenems by themselves, since their expression is not influenced by the presence of additional promoters brought by insertion sequences, as happens with the majority of CHDLs (7, 8, 20).
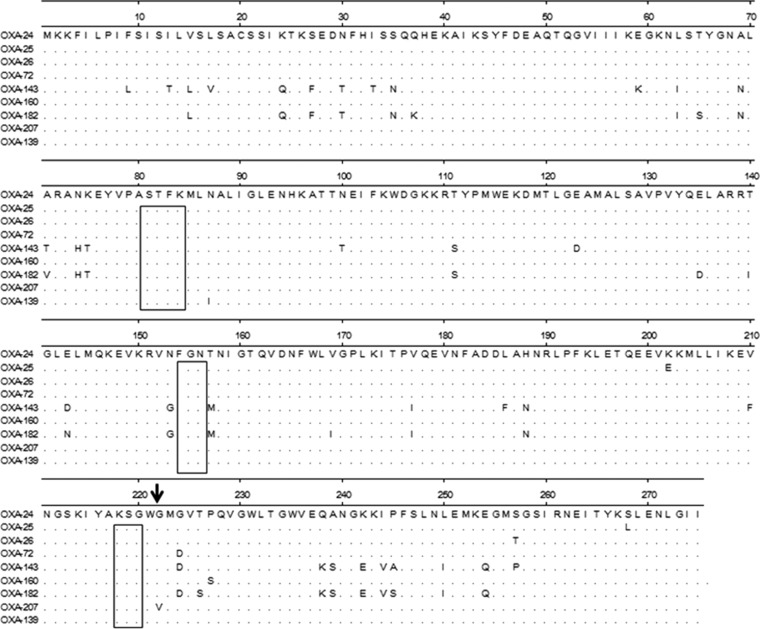
A comparison of amino acid sequences of the seven OXA-24 variants known at this time, and the closely related OXA-143 and OXA-182 enzymes included in the OXA-143 group, is shown in Fig. 1. The three conserved motifs (STKF, FGN, and KSG) responsible for the formation of the active site are identical among the three CHDL groups. The unique Gly222Val substitution present in OXA-207 is near the third conserved motif KSG and adjacent to Met-223, which has been defined as a very important residue from a structural point of view for the biochemical properties of the active site of the OXA-24 enzyme, also called OXA-40 (21, 22). A previous analysis of the crystal structure of OXA-24 showed that the specificity for carbapenems is determined by a hydrophobic barrier that is associated with the amino acid residues Tyr-112 and Met-223 (21, 22) that define a tunnel across the active site of OXA-24 and orientate the access of carbapenem to the catalytic binding site (21). The small size of the carbapenems and the hydrophobic feature of the tunnel-like channel are appropriate to accommodate these drugs rather than the oxacillin (22).
FIG 1.
Comparison of the amino acid sequences of the seven OXA-24 variants (OXA-24, OXA-25, OXA-26, OXA-72, OXA-139, OXA-160, and OXA-207) with the related CHDL groups (OXA-143 and OXA-182), using the ClustalW multiple-sequence alignment program. All sequences were obtained with the accession numbers provided by the β-Lactamase Classification and Amino Acid Sequences website (http://www.lahey.org/Studies/). The amino acid sequences of OXA-24 and OXA-40 are identical (25, 26). Conserved motifs are boxed. Conserved amino acids are indicated by periods. The Gly222Val substitution in the predicted amino acid sequence of OXA-207 is indicated by a black arrow.
In order to compare the kinetic features of OXA-24 and OXA-207, the blaOXA-24 and blaOXA-207 genes were amplified in parallel by PCR with the primers OXA24/207-Fow-XbaI (Fow stands for forward) (5′-CCCTCTAGAATGAAAAAATTTATACTTCC-3′) and OXA24/207-Rev-NcoI (Rev stands for reverse) (5′-CCCCCATGGTTAAATGATTCCAAGATTTTC-3′). The purified amplicons were cloned between XbaI and NcoI restriction sites in pET-RA plasmid harboring a rifampin resistance gene (GenBank accession number HM219006). The constructions were used to transform Acinetobacter baylyi ADP1 cells by electroporation (25 μF, 200 Ω, 2.5 kV), with a Gene Pulser II (Bio-Rad, Richmond, CA). Transformants were selected on LB plates supplemented with 50 μg/ml of ampicillin and 50 μg/ml of rifampin, and the presence of CHDL genes was confirmed by sequencing. The MICs of ampicillin (Sigma, St. Louis, MO, USA), imipenem (Merck, Whitehouse Station, NJ), and meropenem (AstraZeneca, London, United Kingdom) were determined by the broth microdilution method (23) for the A. pittii HUMV-1588 and A. baylyi ADP1 strains, as well as for the A. baylyi ADP1 transformants. Other antibiotics were determined by Etest strips. Data are presented in Table 1. The MICs of ampicillin, imipenem, and meropenem for A. baylyi ADP1 expressing the blaOXA-207 gene increased to values similar to those obtained by A. baylyi ADP1 expressing the blaOXA-24 gene and A. pittii HUMV-1588. To purify OXA-207, the corresponding gene was cloned into the pGEX-6P-1 vector with BamHI and EcoRI restriction sites to produce translational fusions with the glutathione S-transferase (GST) gene. The primers used for amplification of the gene lacking the signal peptide are OXA24/207-pepBamHI (5′-AAGGATCCTCTATTAAAACTAAATCTGAAG-3′) and OXA24/207-RevEcoRI (5′-AAAGAATTCTTAAATGATTCCAAGATTTTC-3′). The OXA-207 enzyme was purified to homogeneity with the GST gene fusion system (Amersham Pharmacia Biotech, Europe GmbH) in accordance with the manufacturer's instructions and with other publications (21). The mature purified protein, lacking the GST fusion protein, appeared on SDS-polyacrylamide gels as a band of 29.1 kDa (≥95% purity). The concentrations of the purified proteins were determined by a protein assay (Bio-Rad, Richmond, CA). The additional nonnatural residues added to the mature protein were Gly-Pro-Leu-Gly-Ser. As the procedure to purify β-lactamase enzymes is always the same, the OXA-24 protein used as a comparator in our work, harbored the same identical five amino acids. The protein OXA-207 was subjected to further biochemical studies with ampicillin, oxacillin, imipenem, and meropenem. The antibiotic molar extinction coefficients in the spectrophotometric assays and the wavelengths used for measurements for ampicillin, oxacillin, imipenem, and meropenem were the same as previously published (21).
Biochemical studies (kcat and Km) were carried out in triplicate in phosphate-buffered saline (PBS) buffer with bovine serum albumin (BSA) (20 μg/ml) at 25°C in a Nicolet Evolution 300 spectrophotometer (Thermo Electron Corporation, Waltham, MA, USA) and quartz cuvettes with an optical path length of 0.2 cm. The data were analyzed with Vision Pro software (Thermo Electron Corporation). The kinetic parameters for nitrocefin were determined from the initial rates by Hanes-Woolf linearization of the Henri-Michaelis-Menten equation. The Km values for ampicillin, oxacillin, imipenem, and meropenem were calculated as Ki values in competitive assays with nitrocefin (Oxoid, Basingstoke, Hants, United Kingdom) as the substrate. The Vmax was calculated by considering an antibiotic concentration 10 times the Km, and the kcat values were obtained under zero-order conditions (substrate concentration ≫ Km). The catalytic efficiencies for ampicillin observed in the OXA-24 and OXA-207 enzymes are similar, but there was an increase in the Km for OXA-207. Although both enzymes showed high affinities for imipenem and meropenem, OXA-24 efficiently hydrolyzed meropenem and imipenem, hydrolyzing meropenem and imipenem 7 and 2.5 times better than OXA-207 (Table 2), respectively. However, the OXA-207 enzyme (kcat/Km = 8,934) shows high rates of hydrolysis for oxacillin compared to the OXA-24 enzyme (kcat/Km = 202).
TABLE 2.
Kinetic parameters of purified β-lactamases OXA-24 and OXA-207a
| Antimicrobial agent | OXA-24b |
OXA-207 |
||||
|---|---|---|---|---|---|---|
| kcat (s−1)c | Km (μM) | kcat/Km (mM−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (mM−1 s−1) | |
| Ampicillin | 223 (92.9) | 80 (15) | 2,75 | 337 (6.7) | 151 (15.4) | 2,232 |
| Oxacillin | 257 (0.086) | 1,272 (222) | 202 | 383 (28.5) | 43 (8.3) | 8,907 |
| Imipenem | 1.62 (0.47) | 0.58 (0.084) | 2,786 | 0.41 (0.01) | 0.37 (0.07) | 1,108 |
| Meropenem | 0.37 (0.1) | 0.0099 (0.0014) | 37,869 | 0.043 (0.002) | 0.008 (0.001) | 5,375 |
Data are the means (standard deviations shown in parentheses) of three independent experiments.
Data for OXA-24 were obtained from reference 21.
Estimated kcat value.
Santillana et al. (21) determined the crystal structure of OXA-24 at a resolution of 2.5 Å and then resolved the biochemical mechanism toward carbapenems of this enzyme. The structure shows that carbapenem's substrate specificity is determined by a hydrophobic barrier that is established through the specific arrangement of the Tyr-112 and Met-223 side chains, which define a tunnel-like entrance to the active site. The importance of these residues was further confirmed by mutagenesis studies. In previous biochemical studies performed with OXA-24 (21) when Met-223 was replaced with Ala in OXA-24 either alone or in combination with the replacement Tyr112Ala (double mutant), a reduction in the catalytic efficiency against carbapenems was observed (21). However, analysis of each independent residue showed that Tyr-112 is more important than Met-223 ing conferring high MICs for imipenem and meropenem in the bacterial strain expressing OXA-24. It seems that the Tyr-112 residue is essential for catalytic efficiency against carbapenems, because it hooks the 6-α-hydroxyethyl group of these β-lactam antibiotics before catalysis and introduces the antibiotic molecule onto the active site.
With respect to oxacillin, the activity of OXA-24 toward this antibiotic (kcat/Km = 200) was very low with other oxacillinases (21). This may be due to the bulky methylphenylisoxazolyl moiety of the oxacillin molecule located at the left position of the β-lactam ring. Therefore, access of this substrate to the active site in the wild-type (WT) OXA-24 enzyme may be difficult, because of steric restrictions and the architecture of the catalytic binding site.
It is interesting that the Gly222Val substitution observed in OXA-207 occurs at the amino acid position preceding Met-223, whose role in substrate binding and catalysis was previously studied by mutagenesis as discussed above (21). Although the Gly222Val substitution observed in our study may alter the tunnel-like entrance used for the carbapenems in the active center of OXA-207, this change seems to be insufficient to abrogate the entrance of these antibiotics into the active site of the enzyme, as Tyr-112 is present in OXA-207 and may still help orientate carbapenem molecules to the active site of OXA-207 but with less efficiency than the wild-type OXA-24 enzyme. However, this change seems sufficient to abrogate any steric restriction for oxacillin, thus facilitating the entrance of this antibiotic to the OXA-207 active center, with kcat/Km ratios (mM−1 s−1) for oxacillin and OXA-24 and OXA-207 of 202 and 8,907, respectively.
OXA-24 is the most disseminated CHDL in A. baumannii clinical isolates in Spain (18, 24), and this is the first report of an OXA-24 variant in a non-A. baumannii species in Spain. Although OXA-207 more weakly hydrolyzed meropenem than imipenem compared to OXA-24, this CHDL significantly contributes to the resistance to both carbapenems in the A. pittii HUMV-1588 strain. In addition, the presence of the same XerC/XerD structure flanking the novel OXA-207 found in A. pittii HUMV-1588, as well as in the OXA-24-producing A. baumannii clones identified in our hospital during the last 5 years (unpublished data), indicate a possible gene transfer from A. baumannii to A. pittii.
In conclusion, we have identified a carbapenem-resistant A. pittii clinical isolate harboring the novel OXA-207 enzyme in Spain. Our findings suggest that the unique mutation present in the OXA-207 enzyme changed its catalytic activity against carbapenems and oxacillin. The emergence of this CHDL in A. pittii is a concern, as it would drastically compromise the use of β-lactams to treat infections caused by this pathogen.
ACKNOWLEDGMENTS
This work was partially supported by the Ministerio de Economía y Competitividad, Instituto de Salud Carlos III - cofinanced by the European Development Regional Fund “A way to achieve Europe” ERDF, the Spanish Network for the Research in Infectious Diseases (REIPI RD12/0015), and by the Fondo de Investigación Sanitaria (PI080209, PS09/00687, and PI12/00552). We are grateful to the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), which gave a PDEE grant to R.C. (protocol 4149/08-4).
We gratefully acknowledge the assistance of Sophia Mooney in the preparation of the manuscript.
We have no conflicts of interest to report.
Footnotes
Published ahead of print 2 June 2014
REFERENCES
- 1.Nemec A, Krizova L, Maixnerova M, van der Reijden TJ, Deschaght P, Passet V, Vaneechoutte M, Brisse S, Dijkshoorn L. 2011. Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex with the proposal of Acinetobacter pittii sp. nov. (formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov. (formerly Acinetobacter genomic species 13TU). Res. Microbiol. 162:393–404. 10.1016/j.resmic.2011.02.006 [DOI] [PubMed] [Google Scholar]
- 2.Chu YW, Leung CM, Houang ET, Ng KC, Leung CB, Leung HY, Cheng AF. 1999. Skin carriage of acinetobacters in Hong Kong. J. Clin. Microbiol. 37:2962–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Idzenga D, Schouten MA, van Zanten AR. 2006. Outbreak of Acinetobacter genomic species 3 in a Dutch intensive care unit. J. Hosp. Infect. 63:485–487. 10.1016/j.jhin.2006.03.014 [DOI] [PubMed] [Google Scholar]
- 4.Turton JF, Shah J, Ozongwu C, Pike R. 2010. Incidence of Acinetobacter species other than A. baumannii among clinical isolates of Acinetobacter: evidence for emerging species. J. Clin. Microbiol. 48:1445–1449. 10.1128/JCM.02467-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans BA, Hamouda A, Towner KJ, Amyes SG. 2010. Novel genetic context of multiple blaOXA-58 genes in Acinetobacter genospecies 3. J. Antimicrob. Chemother. 65:1586–1588. 10.1093/jac/dkq180 [DOI] [PubMed] [Google Scholar]
- 6.Wang H, Guo P, Sun H, Wang H, Yang Q, Chen M, Xu Y, Zhu Y. 2007. Molecular epidemiology of clinical isolates of carbapenem-resistant Acinetobacter spp. from Chinese hospitals. Antimicrob. Agents Chemother. 51:4022–4028. 10.1128/AAC.01259-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poirel L, Naas T, Nordmann P. 2010. Diversity, epidemiology, and genetics of class D beta-lactamases. Antimicrob. Agents Chemother. 54:24–38. 10.1128/AAC.01512-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poirel L, Nordmann P. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12:826–836. 10.1111/j.1469-0691.2006.01456.x [DOI] [PubMed] [Google Scholar]
- 9.Fernández-Cuenca F, Rodríguez-Martínez JM, Gómez-Sánchez MA, de Alba PD, Infante-Martínez V, Pascual A. 2012. Production of a plasmid-encoded OXA-72 β-lactamase associated with resistance to carbapenems in a clinical isolate Acinetobacter junii. Int. J. Antimicrob. Agents 39:93–94. 10.1016/j.ijantimicag.2011.07.017 [DOI] [PubMed] [Google Scholar]
- 10.Mendes RE, Bell JM, Turnidge JD, Castanheira M, Jones RN. 2009. Emergence and widespread dissemination of OXA-23, -24/40 and -58 carbapenemases among Acinetobacter spp. in Asia-Pacific nations: report from the SENTRY Surveillance Program. J. Antimicrob. Chemother. 63:55–59 [DOI] [PubMed] [Google Scholar]
- 11.Vaneechoutte M, Dijkshoorn L, Tjernberg I, Elaichouni A, de Vos P, Claeys G, Verschraegen G. 1995. Identification of Acinetobacter genomic species by amplified ribosomal DNA restriction analysis. J. Clin. Microbiol. 33:11–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.La Scola B, Gundi VA, Khamis A, Raoult D. 2006. Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J. Clin. Microbiol. 44:827–832. 10.1128/JCM.44.3.827-832.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gundi VA, Dijkshoorn L, Burignat S, Raoult D, La Scola B. 2009. Validation of partial rpoB gene sequence analysis for the identification of clinically important and emerging Acinetobacter species. Microbiology 155:2333–2341. 10.1099/mic.0.026054-0 [DOI] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing - 24th ed. Informational supplement M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 15.Cayô R, Rodríguez MC, Espinal P, Fernández-Cuenca F, Ocampo-Sosa AA, Pascual A, Ayala JA, Vila J, Martínez-Martínez L. 2011. Analysis of genes encoding penicillin-binding proteins in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 55:5907–5913. 10.1128/AAC.00459-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendes RE, Kiyota KA, Monteiro J, Castanheira M, Andrade SS, Gales AC, Pignatari AC, Tufik S. 2007. Rapid detection and identification of metallo-beta-lactamase-encoding genes by multiplex real-time PCR assay and melt curve analysis. J. Clin. Microbiol. 45:544–547. 10.1128/JCM.01728-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, Amyes SG, Livermore DM. 2006. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27:351–353. 10.1016/j.ijantimicag.2006.01.004 [DOI] [PubMed] [Google Scholar]
- 18.Lopez-Otsoa F, Gallego L, Towner KJ, Tysall L, Woodford N, Livermore DM. 2002. Endemic carbapenem resistance associated with OXA-40 carbapenemase among Acinetobacter baumannii isolates from a hospital in northern Spain. J. Clin. Microbiol. 40:4741–4743. 10.1128/JCM.40.12.4741-4743.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Andrea MM, Giani T, D'Arezzo S, Capone A, Petrosillo N, Visca P, Luzzaro F, Rossolini GM. 2009. Characterization of pABVA01, a plasmid encoding the OXA-24 carbapenemase from Italian isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 53:3528–3533. 10.1128/AAC.00178-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merino M, Acosta J, Poza M, Sanz F, Beceiro A, Chaves F, Bou G. 2010. OXA-24 carbapenemase gene flanked by XerC/XerD-like recombination sites in different plasmids from different Acinetobacter species isolated during a nosocomial outbreak. Antimicrob. Agents Chemother. 54:2724–2727. 10.1128/AAC.01674-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santillana E, Beceiro A, Bou G, Romero A. 2007. Crystal structure of the carbapenemase OXA-24 reveals insights into the mechanism of carbapenem hydrolysis. Proc. Natl. Acad. Sci. U. S. A. 104:5354–5359. 10.1073/pnas.0607557104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider KD, Ortega CJ, Renck NA, Bonomo RA, Powers RA, Leonard DA. 2011. Structures of the class D carbapenemase OXA-24 from Acinetobacter baumannii in complex with doripenem. J. Mol. Biol. 406:583–594. 10.1016/j.jmb.2010.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically - 9th ed. Approved standard M7-A9. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 24.Acosta J, Merino M, Viedma E, Poza M, Sanz F, Otero JR, Chaves F, Bou G. 2011. Multidrug-resistant Acinetobacter baumannii harboring OXA-24 carbapenemase, Spain. Emerg. Infect. Dis. 17:1064–1067. 10.3201/eid/1706.091866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bou G, Oliver A, Martínez-Beltrán J. 2000. OXA-24, a novel class D beta-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob. Agents Chemother. 44:1556–1561. 10.1128/AAC.44.6.1556-1561.2000 (Erratum, 50: 2280, 2006, doi:10.1128/AAC.00514-06). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Héritier C, Poirel L, Aubert D, Nordmann P. 2003. Genetic and functional analysis of the chromosome-encoded carbapenem-hydrolyzing oxacillinase OXA-40 of Acinetobacter baumannii. Antimicrob. Agents Chemother. 47:268–273. 10.1128/AAC.47.1.268-273.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]