Abstract
Daptomycin (DAP) is a lipopeptide antibiotic frequently used as a “last-resort” antibiotic against vancomycin-resistant Enterococcus faecium (VRE). However, an important limitation for DAP therapy against VRE is the emergence of resistance during therapy. Mutations in regulatory systems involved in cell envelope homeostasis are postulated to be important mediators of DAP resistance in E. faecium. Thus, in order to gain insights into the genetic bases of DAP resistance in E. faecium, we investigated the presence of changes in 43 predicted proteins previously associated with DAP resistance in enterococci and staphylococci using the genomes of 19 E. faecium with different DAP MICs (range, 3 to 48 μg/ml). Bodipy-DAP (BDP-DAP) binding to the cell membrane assays and time-kill curves (DAP alone and with ampicillin) were performed. Genetic changes involving two major pathways were identified: (i) LiaFSR, a regulatory system associated with the cell envelope stress response, and (ii) YycFGHIJ, a system involved in the regulation of cell wall homeostasis. Thr120→Ala and Trp73→Cys substitutions in LiaS and LiaR, respectively, were the most common changes identified. DAP bactericidal activity was abolished in the presence of liaFSR or yycFGHIJ mutations regardless of the DAP MIC and was restored in the presence of ampicillin, but only in representatives of the LiaFSR pathway. Reduced binding of BDP-DAP to the cell surface was the predominant finding correlating with resistance in isolates with DAP MICs above the susceptibility breakpoint. Our findings suggest that genotypic information may be crucial to predict response to DAP plus β-lactam combinations and continue to question the DAP breakpoint of 4 μg/ml.
INTRODUCTION
The surge of Enterococcus faecium as an important nosocomial pathogen has been associated with an expanding pandemic caused by a hospital-associated (HA) genetic clade (1, 2). Indeed, isolates belonging to this genetic lineage are frequently multidrug resistant (MDR) with high MICs of ampicillin and vancomycin (3). Daptomycin (DAP) is a cell membrane (CM)-targeting lipopeptide that has in vitro bactericidal activity against MDR E. faecium and, due to the paucity of other bactericidal options, is often used as first-line therapy despite lacking U.S. Food and Drug Administration approval for these organisms. However, one of the major problems when using DAP against enterococci is the emergence of resistance during therapy (4–6).
The mechanisms of DAP resistance in enterococci are not fully understood, but recent evidence suggests that there are several genetic pathways involved and that resistance results from a sequential and ordered mutational pathway (7–9). In Enterococcus faecalis, we have previously shown that emergence of resistance during therapy involves substitutions in three proteins: (i) LiaF, a member of the three-component regulatory system LiaFSR which, in Bacillus subtilis and other Gram-positive bacteria (10), has been shown to orchestrate the cell envelope response to stress; (ii) GdpD, a glycerophosphoryl-diester phosphodiesterase, involved in phospholipid metabolism; and (iii) Cls, a cardiolipin synthase (11). The mechanism of DAP resistance in Gram-positive bacteria was initially postulated to involve repulsion of the calcium-decorated DAP from the cell surface (12). However, we recently provided evidence that DAP resistance in an E. faecalis strain was due to diversion of DAP from the division septum associated with redistribution of CM cardiolipin (CL) microdomains, without repulsion of DAP from the cell surface (8). Of note, deletion of Ile at position 177 of LiaF was sufficient for membrane remodeling and also abolished DAP in vitro bactericidal activity (8, 13).
Genomic analyses in E. faecium, using clinical-strain pairs of DAP-susceptible and DAP-resistant isolates recovered during therapy, have identified many genetic changes associated with DAP resistance (14, 15), but the specific role of each of these mutations remains to be established. Interestingly, in a collection of clinical E. faecium isolates recovered from bacteremia, ca. 75% of DAP-“susceptible” isolates with MICs close to the established breakpoint (4 μg/ml) harbored changes in LiaFSR (16). Furthermore, recent data suggest that the combination of DAP with certain β-lactams (e.g., ampicillin and ceftaroline) restores DAP activity in vitro and in vivo by increasing the ability of the antibiotic to bind to its CM target (17–20, 41), but the mechanism or genetic basis for such synergism is unknown. Therefore, in an attempt to dissect the genetic determinants implicated in DAP resistance in E. faecium and the interaction with β-lactams, we used whole-genome sequencing of a collection of 19 unrelated clinical isolates of E. faecium with a diverse range of DAP MICs and investigated the presence of genetic changes in 43 predicted proteins previously associated with DAP resistance. Note that, although the accepted term is “DAP nonsusceptibility,” we use the term “DAP resistance” throughout for ease of presentation.
(Parts of the results of this study were presented at the 53rd Interscience Conference on Antimicrobial Agents and Chemotherapy, September 10 to 13, 2013, Denver, Colorado, USA.)
MATERIALS AND METHODS
Bacterial isolates, molecular typing, and MIC determinations.
A total of 19 clinical strains of E. faecium were included in the present study (DAP MICs, 3 to 48 μg/ml) (Table 1). We included 17 U.S. isolates from different patients and submitted to reference laboratories from diverse geographical areas. In addition, there were two E. faecium strains recovered in South America (21) before the introduction of DAP to the region (Table 1). Typing of isolates was performed using in silico multilocus sequence typing (MLST) analysis derived from the genomic sequence. Determination of DAP MICs was performed by Etest (bioMérieux, Marcy l'Etoile, France) on Mueller-Hinton agar according to the manufacturer's instructions. Etest was used for DAP since our goal was to detect small variations in DAP MICs. The MICs for each strain were determined in triplicate. Two independent observers read the results, and a third investigator was consulted whenever a disagreement was identified. MICs of other antibiotics were determined by an agar dilution method (22).
TABLE 1.
E. faecium strains included in this studya
| Strain | MIC (μg/ml)b |
van gene | MLST | BioProject accession numbers | Source or reference | |||
|---|---|---|---|---|---|---|---|---|
| VAN | TEI | DAP | AMP | |||||
| 503 | >256 | 32 | 3 | >64 | vanA | 280 | PRJNA181868 | This study |
| 504 | 1 | 0.12 | 4 | >64 | 649 | PRJNA181867 | This study | |
| 505 | 1 | 0.12 | 6 | >64 | SLVc ST39 | PRJNA181866 | This study | |
| 506 | >256 | 64 | 6 | >64 | vanA | 18 | PRJNA181865 | This study |
| 509 | 256 | 0.25 | 4 | >64 | vanB | 17 | PRJNA181864 | This study |
| 510 | 1 | 0.12 | 4 | >64 | 18 | PRJNA181863 | This study | |
| 511 | 1 | 0.25 | 8 | >64 | 17 | PRJNA181862 | This study | |
| 513 | 1 | 0.25 | 4 | >64 | 736 | PRJNA181861 | This study | |
| 514 | 1 | 0.25 | 8 | >64 | 17 | PRJNA181860 | This study | |
| 515 | 0.5 | 0.5 | 3 | >64 | 549 | PRJNA181859 | This study | |
| S447 | 256 | 16 | 3 | >64 | vanA | 203 | PRJNA181832 | 11, 15 |
| R446 | 16 | 4 | 16 | >64 | vanA | 203 | PRJNA181838 | 11, 15 |
| R501 | 256 | 8 | 32 | >64 | vanA | 17 | PRJNA181833 | 11 |
| R494 | 1 | 0.12 | 48 | >64 | 664 | PRJNA181837 | 11 | |
| R496 | 1 | 0.25 | 32 | >64 | 412 | PRJNA181836 | 11 | |
| R497 | 1 | 0.25 | 16 | >64 | 752 | PRJNA181835 | 11 | |
| R499 | 256 | 8 | 48 | >64 | vanA | 412 | PRJNA181834 | 11 |
| V689 | 256 | 64 | 4 | >64 | vanA | 736 | PRJNA181831 | 21 |
| P1190 | 256 | 128 | 3 | 64 | vanA | 125 | PRJNA181840 | 21 |
All strains (except S447 and R446, which were isolated from the same patient) had no epidemiological relationship between them and were recovered at different time points and geographical areas in the United States. Isolates V689 and P1190 were recovered in Venezuela and Peru, respectively, before DAP was introduced in these countries. The pulsed-field gel electrophoresis patterns of all strains were different and categorized as unrelated (data not shown).
VAN, vancomycin; TEI, teicoplanin; DAP, daptomycin; AMP, ampicillin.
SLV, single locus variant.
Genome sequencing and mutational analysis.
Whole-genome analysis, genomic assemblies, and annotation were performed as described previously (15). A total of 43 predicted proteins (Fig. 1; see also Table S1 in the supplemental material) were included in the analysis representing genes previously associated with DAP resistance in enterococci (11, 14, 15, 23) and E. faecium homologues of six genes associated with DAP resistance in S. aureus (mprF, pgsA, and the dlt cluster; see Tables S2 and S3 in the supplemental material) (24–27). A relevant change was defined as a nucleotide change that resulted in an amino acid substitution that was not present at the same position on the predicted protein sequences of other E. faecium genomes publicly available (independent of the DAP MIC). Sequence comparisons were performed using the multiple sequence alignment program ClustalW2 and E. faecium DO (TX16), a DAP-susceptible (MIC = 2 μg/ml) clinical strain (whose genome is sequenced and closed) (28, 29), as the template to refine comparisons. All accession numbers are shown in Table 1.
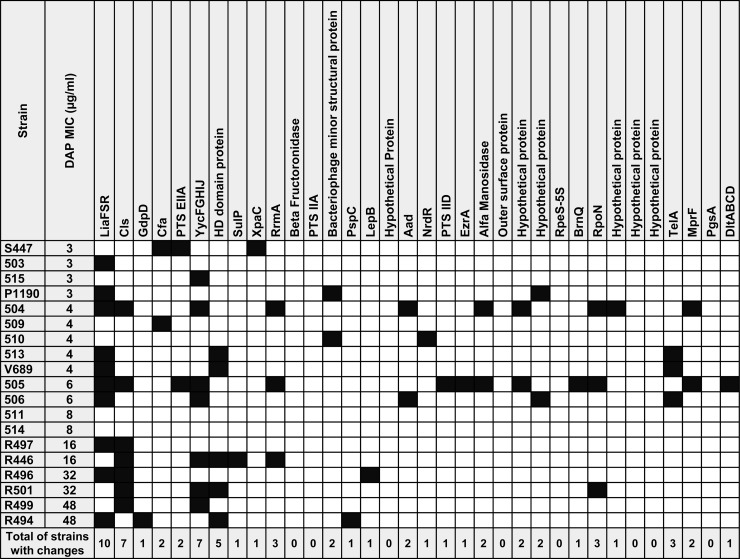
FIG 1.
Amino acid changes in 43 predicted proteins associated with DAP resistance in E. faecium isolates. Black squares indicate the presence of amino acid changes. Cls, cardiolipin synthase; GdpD, glycerophosphodiester phosphodiesterase; Cfa, cyclopropane-fatty-acyl-phospholipid synthase; PTS-EIIA, phosphotransferase system, phosphoenolpyruvate-dependent sugar EIIA 2; SulP, sulfate transporter; XpaC, 5-bromo-4-chloroindolyl phosphate hydrolysis protein; RrmA, rRNA (guanine-N1-)-methyltransferase A; PTS-IIA, PTS system fructose IIA component; PspC, phage shock protein C; LepB, signal peptidase I; Aad, aldehyde-alcohol dehydrogenase; NrdR, nicotinamide mononucleotide transporter; PTS IID, mannose/fructose/sorbose transporter subunit IID; EzrA, septation ring formation regulator; RpeS-5S, ribosomal protein S5; BrnQ, branched-chain amino acid transport protein; RpoN, RNA polymerase sigma factor 54; TelA, telurite resistance protein; MprF, lysylphosphatidylglycerol synthetase; PgsA, CDP-diacylglycerol-glycerol-3-phosphate 3-phosphatidyltransferase.
Time-kill assays and evaluation of synergism between ampicillin (AMP) and DAP.
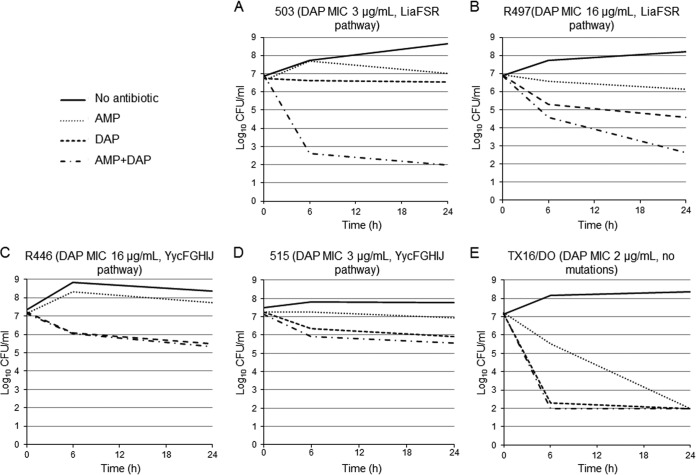
To assess the bactericidal activity of DAP against E. faecium, we used time-kill assays with two representative isolates of each of the two most common genetic pathways identified displaying/or with DAP MICs below and above the susceptibility breakpoint (4 μg/ml), respectively, as follows: (i) 503 and R497 (LiaFSR pathway representatives) with MICs of 3 and 16 μg/ml, respectively (11), and (ii) 515 and R446 (15) (YycFGHIJ pathway) with MICs of 3 and 16 μg/ml, respectively. E. faecium DO (MIC 2 μg/ml) was used as a control for these experiments. Time-kill assays were performed with an initial bacterial inoculum of 107 CFU/ml in Mueller-Hinton broth (MHB) supplemented with calcium (50 mg/liter). DAP was added at concentrations of 5× the MIC of each strain, and bacterial counts were performed at 0, 6, and 24 h. Antibiotic carryover was controlled by centrifugation (bacterial cell suspensions [1-ml samples] were centrifuged, and the pelleted bacteria were suspended in the same volume of 0.9% saline solution before plating) as described earlier (13, 30, 31). Bactericidal activity was defined as a ≥3 log10 reduction in CFU/ml at 24 h in comparison to the initial inoculum. The limit of detection was 200 CFU/ml, assuming maximum plating efficiency. We also tested the ability of AMP (64 μg/ml) to achieve synergistic activity when combined with DAP against the same strains. Synergism was defined as a decrease of ≥2 log10 CFU/ml in bacterial counts at 24 h compared to the most active single agent alone. All assays were performed in triplicate.
BODIPY-labeled daptomycin (BDP-DAP) assays.
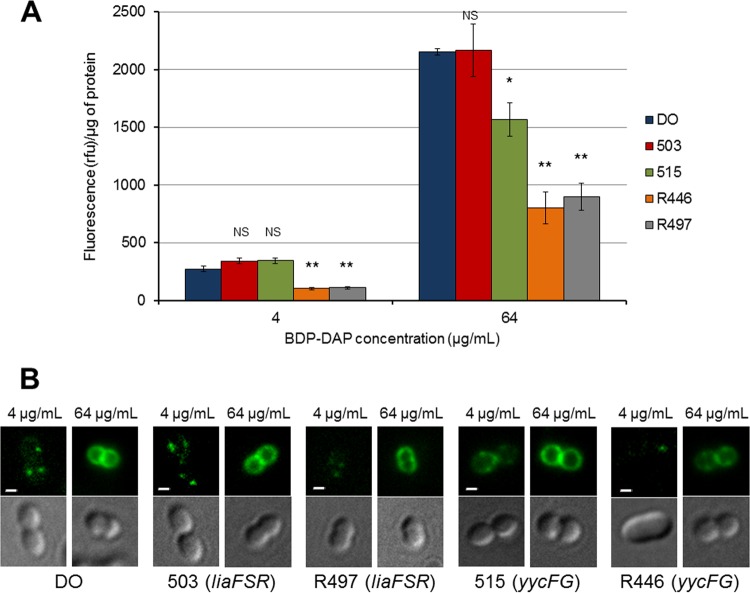
We used BDP-DAP to assess the interaction of DAP with the bacterial CM, as described before (8). The assays were performed using isolates 503, R497, R446, and 515 (representatives of the most common mutational pathways [see above]). The protocol for BDP-DAP staining followed techniques previously described (7, 8, 32, 33). Briefly, isolates were grown in Luria-Bertani (LB) broth at 37°C and incubated with BDP-DAP at two concentrations (4 and 64 μg/ml in LB broth supplemented with Ca2+ at 50 mg/liter) for 10 min in darkness. Fluorescence was assessed using a standard fluorescein isothiocyanate filter set (excitation at 490 nm and emission at 528 nm). A minimum of two independent experiments was performed for each strain on different days. In order to estimate the amount of BDP-DAP bound to E. faecium strains, the fluorescence intensity was quantitated and normalized to protein concentration of the samples, as described previously (8).
RESULTS
Changes in genes involved in cell envelope homeostasis.
We sought to identify substitutions in 43 predicted proteins previously associated with DAP resistance in enterococci and staphylococci within unrelated E. faecium isolates exhibiting a diverse range of MICs (3 to 48 μg/ml). The “control” strain for our genomic comparisons was E. faecium DO (28, 29), a DAP-susceptible clinical isolate (DAP MIC 2 μg/ml) whose genome has been sequenced and closed. The changes are shown in Fig. 1 and are summarized according to their frequency in Table S3 in the supplemental material. The most common genes affected were those encoding regulatory systems involved in cell envelope homeostasis, liaFSR and/or yycFG (including the accessory genes yycHIJ). Indeed, 10 isolates with DAP MICs ≥ 3 μg/ml harbored changes in LiaFSR, with the most common substitutions found in LiaS (the putative histidine kinase of the system [n = 9]), followed by the response regulator LiaR (n = 7) (Fig. 1; see also Table S3 in the supplemental material). A T120A substitution was often identified in LiaS (n = 7), and W73C was found in all isolates (n = 7) with changes in LiaR. Moreover, LiaST120A was always present in isolates harboring LiaRW73C, suggesting that these substitutions might have coevolved. Of note, mutations in liaF, which was previously associated with DAP resistance in E. faecalis, were found in four isolates, and each predicted change was unique (Fig. 1; see Table S3 in the supplemental material). Three of these isolates (DAP MICs of 4, 6, and 48 μg/ml) also harbored changes in LiaSR, and the remaining isolate (MIC of 32 μg/ml) harbored an I142T substitution in LiaF without concomitant changes in LiaRS. Overall, alterations in LiaFSR were identified in 50% (5 of 10 isolates) with MICs between 3 and 4 μg/ml and in 50% (5 of 10) with MICs > 4 μg/ml (Fig. 1; see Table S3 in the supplemental material). Among seven isolates harboring mutations in the YycFGHIJ system, the most commonly involved proteins were YycH, a putative signal transduction protein (n = 3), followed by YycG (sensor histidine kinase of the system [n = 2]) and YycI, a putative regulatory protein (n = 2) (15, 34, 35). Three isolates harbored mutations in both LiaFSR and YycFGHIJ systems concomitantly, suggesting that these two systems may interact to develop DAP resistance in some isolates.
Substitutions in phospholipid metabolism enzymes.
We had previously postulated that changes in enzymes involved in phospholipid metabolism were likely to appear at latest stages of the DAP resistance pathway in E. faecalis (8, 9). Interestingly, after genes predicted to function in cell envelope homeostasis, the most common gene affected was cls (n = 7), encoding a CL synthase involved in the last committed step of synthesis of CL from the precursor phosphatidylglycerol. The predicted amino acid changes were located in the phospholipase D domains (PLD1 and PLD2) and in the linker region joining the two putative transmembrane helices, as previously described (36). Of note, Cls substitutions were found mostly in isolates with high DAP MIC (>4 μg/ml [n = 6]) and only in one isolate with an MIC of 4 μg/ml (Fig. 1; see Table S3 in the supplemental material). Moreover, the changes in Cls were always observed in isolates that also had changes in one of the above-mentioned regulatory systems (LiaFSR or YycFG), supporting our previous hypothesis that Cls substitutions follow initial changes in cell envelope homeostasis and “enhance” the resistance phenotype (9). Other less frequent changes found in enzymes involved in phospholipid metabolism were (i) in the homolog of MprF (a lysylphosphatidylglycerol synthetase [n = 2]) (25, 37); (ii) Cfa, a putative cyclopropane fatty acid synthase (n = 2) (38); and (iii) GdpD a glycerophosphoryl-diester phosphodiesterase (n = 2) (11) (Fig. 1; see Table S3 in the supplemental material).
Other, less frequent mutations.
A total of five isolates with MICs between 4 and 48 μg/ml exhibited amino acid changes in a protein of unknown function that harbors an HD domain, which designates a superfamily of enzymes that possess phosphohydrolase activity and may be involved in nucleic acid metabolism and signal transduction (39). Four strains (MICs = 4 and 32 μg/ml) showed changes in the 23S rRNA methyltransferase, RrmA (40). In addition, three isolates exhibited substitutions in TelA, a putative tellurite resistance protein (Fig. 1; see also Table S3 in the supplemental material), previously associated with DAP resistance in E. faecalis (23). Changes in TelA, RrmA, and in the HD domain protein were always identified in conjunction with substitutions in LiaFSR or YycFGHIJ. Other genes altered less commonly are shown in Fig. 1 and in Table S3 in the supplemental material. Of note, we were unable to identify any changes in predicted proteins associated with DAP resistance in two DAP-resistant isolates (both with MICs of 8 μg/ml) (Fig. 1), indicating that additional genetic pathways leading to DAP resistance in E. faecium remain to be identified.
Mutational pathways influence in vitro bactericidal activity of DAP.
Our previous work (8, 9, 11, 13, 15, 16) and the current genomic analysis indicate that LiaFSR and/or YycFGHIJ are the two most common genetic pathways resulting in DAP resistance. Our previous studies in E. faecalis indicated that a single liaF mutation abolished the in vitro bactericidal activity of DAP (DAP-tolerant phenotype) (13) but did not increase the MIC above the breakpoint. Thus, we examined the bactericidal activity of DAP against E. faecium 503 (Table 1), a DAP-susceptible isolate (MIC 3 μg/ml) that harbors LiaRW73C and LiaST120A. Figure 2A shows that DAP (5× the MIC) lacked bactericidal activity against E. faecium 503 with reductions of <1 log10 CFU/ml at 24 h. A similar effect was observed against E. faecium R497 (Fig. 2B), a DAP-resistant (16 μg/ml) isolate also carrying LiaRW73C and LiaST120A but no changes in YycFGHIJ. Interestingly, for both 503 and R497, DAP bactericidal activity was restored by adding AMP (64 μg/ml), an observation consistent with previous reports (18, 41). Next, we sought to test the in vitro bactericidal activity of DAP in two representative isolates altered in the YycFG pathway (515 and R446) with MICs of 3 and 16 μg/ml, respectively. As expected, DAP did not have any killing effect against R446 (Fig. 2C) but also lacked bactericidal activity against 515 which has an MIC within the susceptible range (Fig. 2D). However, in contrast to representatives with LiaFSR changes, addition of AMP (64 μg/ml) to DAP had no synergistic effect, suggesting that an altered YycFGHIJ system is not affected by the combination of DAP and β-lactams. For the DAP-susceptible strain TX16/DO, both DAP and AMP exhibited bactericidal activity (Fig. 2E).
FIG 2.
Time-kill assays with DAP (5× the MIC) with or without AMP (64 μg/ml). Bacteria were grown in Mueller-Hinton broth (MHB) supplemented with calcium (50 mg/liter). The E. faecium strains are indicated at the top of each panel. AMP, ampicillin; DAP, daptomycin. The limit of detection was 200 CFU/ml.
Lack of DAP binding to the cell surface is the predominant mechanism of DAP resistance in E. faecium.
Two main mechanisms of DAP resistance have been postulated in enterococci: (i) electrostatic “repulsion” of calcium-decorated DAP (positively charged) from the cell surface due to a more positively charged cell envelope (23) and (ii) “diversion” of DAP from the division septum (the main DAP cell target) (only described in E. faecalis) (8). In order to gain insights into the mechanism of DAP resistance and the genetic background, we used fluorescent BDP-DAP to study the interactions of the antibiotic with the CM in representative strains of the LiaFSR or YycFGHIJ pathways, as described earlier (8). We used DAP-susceptible E. faecium DO (MIC = 2 μg/ml) as the control and performed the assays with two BDP-DAP concentrations (4 and 64 μg/ml) since DAP binding to the cell membrane target is concentration dependent (7, 8). Figure 3 shows that binding of BDP-DAP was significantly decreased with the DAP-resistant strains R497 (LiaFSR pathway) and R446 (YycFGHIJ pathway) at low and high concentrations compared to the control (E. faecium DO), suggesting that antibiotic “repulsion” may play a prominent role in resistance. In contrast, the pattern of BDP-DAP binding at low concentrations was similar to that of DO for DAP-susceptible (MIC = 3 μg/ml) 503 (with LiaFSR changes) and 515 (altered YycFGHIJ) (Fig. 3). However, at high BDP-DAP concentrations (64 μg/ml), 515 had significantly less binding to the cell membrane than DO, whereas no statistically significant difference in BDP-DAP binding was observed between 503 (LiaFSR pathway) versus DO. Our results suggest that diversion (rather than repulsion) may be the mechanism for decreased DAP killing with this isolate; however, further analyses are required to corroborate this hypothesis.
FIG 3.
BODIPY-labeled DAP (BDP-DAP) staining of E. faecium strains. (A) Fluorescence intensities of representative E. faecium strains. Cells were treated with BDP-DAP and fluorescence was normalized to cell protein content. Intensities were compared to E. faecium DO. Rfu, relative fluorescence units; *, P < 0.05; **, P < 0.001; NS, not significant. (B) BDP-DAP staining of representative E. faecium cells at concentrations of 4 μg/ml and 64 μg/ml. The top images capture bacterial cells under fluorescence microscopy (bars, 1 μm). The bottom images are the same bacterial cell in phase contrast. The mutated pathway is indicated in parentheses.
DISCUSSION
Using genomic analyses of clinical isolates with a wide variety of DAP MICs, we investigated the genetic basis of DAP resistance in E. faecium. Our findings suggest that two regulatory systems are likely to be involved in development of DAP resistance in E. faecium: (i) LiaFSR, which has been associated with the cell envelope response to cell wall acting antibiotics and antimicrobial peptides (10, 42), and (ii) YycFGHIJ, an essential two-component regulatory system characterized in several Gram-positive organisms (including E. faecalis) and shown to be involved in cell wall homeostasis and cell division (34, 43, 44, 47).
Our previous work indicated that development of DAP resistance in E. faecalis is a stepwise and ordered process (9, 11, 13). One distinct pathway involves initial mutations occurring in genes encoding the three-component regulatory system LiaFSR followed by changes in genes encoding enzymes involved in phospholipid metabolism (such as CL synthase) (9). Furthermore, we previously found that a single mutation in liaF of E. faecalis was sufficient to produce DAP tolerance and that substitutions in LiaRS were commonly found in E. faecium bloodstream isolates with MICs of ≥3 μg/ml but absent in isolates with MICs of ≤2 μg/ml (16). In the present study, we expand these observations and provide several additional lines of evidence to support the pivotal role of the LiaFSR three-component regulatory system in the pathway leading to DAP resistance in E. faecium. First, two substitutions (LiaRW73C and LiaST120A) were commonly found in unrelated clinical isolates of E. faecium with DAP MICs of ≥3 μg/ml. Moreover, identical changes in these two predicted proteins were found in two isolates (V689 and P1190) with DAP MICs of 3 μg/ml which were recovered in countries where DAP had not been introduced in clinical practice, suggesting that these mutations can be selected even without DAP exposure, as previously reported (45). Second, the presence of the same LiaRS substitutions was associated with the DAP-tolerant phenotype, as previously shown for E. faecalis. Indeed, E. faecium 503, which only harbors LiaRW73C and LiaST120A and no other changes in predicted proteins associated with DAP resistance, behaved as a DAP-resistant isolate in time-kill assays, despite of the fact that its DAP MIC is within the “susceptible” range. Our findings continue to question the stated CLSI DAP breakpoint of 4 μg/ml and suggest that 2 μg/ml would likely be a better cutoff value if considering DAP for E. faecium causing deep-seated infections.
Our genomic studies also suggest that a second pathway for DAP resistance involves changes in the YycFGHIJ system, as previously observed (15). After LiaFSR, substitutions in this system were the second most common changes observed in E. faecium with an MIC of ≥3 μg/ml. YycFGHIJ has been implicated in development of DAP nonsusceptibility in E. faecium and also in vancomycin and DAP nonsusceptibility in Staphylococcus aureus (15, 26, 46). Therefore, it is tempting to speculate that YycFGHIJ is another important regulatory network that contributes to the response to the cell membrane attack by DAP (and possibly other cell membrane-acting antimicrobials) and may be more relevant in isolates whose DAP MICs are above the breakpoint. Although our findings support a role of the YycFGHIJ in DAP nonsusceptibility, direct evidence of such contribution is still lacking and is the object of our future studies. Further evidence that the LiaFSR and YycFGHIJ systems may lead to distinct pathways in the development of DAP resistance comes from our time-kill curve assays with selected isolates representing each pathway. Recent data suggest that the addition of AMP to DAP may restore the bactericidal activity of DAP by mechanisms that are unclear (18, 41). Here, we show that the synergistic effect of the AMP plus DAP combination was seen only in isolates representing the LiaFSR pathway (503 and 497) regardless of the MIC, but it was absent with R446 or 515 (DAP-resistant and -susceptible, representing the YycFGHIJ pathway). These findings suggest that the synergistic effect is dependent on the genetic pathway and that it is not a universal phenomenon seen in all DAP-nonsusceptible E. faecium, a finding with important therapeutic implications.
Our data using the fluorescent derivative BDP-DAP also provide evidence that in DAP-nonsusceptible isolates (MICs above the E. faecalis breakpoint), reduced binding of the antibiotic molecule from the surface appears to be the main mediator of resistance regardless of the mutational pathway. However, specific genetic changes also seem to affect the interaction of DAP with the cell membrane in isolates with MICs below the breakpoint that exhibit DAP tolerance. Indeed, in E. faecium 503 (DAP MIC = 3 μg/ml) binding of BDP-DAP did not differ from that of DO (control) even at high antibiotic concentrations, similar to what it has been shown previously in E. faecalis (8). Thus, it is tempting to speculate that in E. faecium 503, LiaFSR-mediated “diversion” of DAP from lethal target sites (i.e., septum) is pivotal for the DAP tolerance phenotype. It is important to note that changes in these systems may not be mutually exclusive since two strains with mutations in both YycFGHIJ and LiaFSR were identified, adding complexity to the genetic changes associated with DAP resistance.
Finally, our findings also support our previous hypothesis that changes in phospholipid enzymes are common among DAP-resistant E. faecium and are likely to be associated with later stages of development of DAP resistance. In E. faecium, changes in CL synthase seem to be the prevailing “route” in isolates with high DAP MIC although changes in this enzyme were not found in all isolates. We had previously shown that mutations in the predicted Cls active site appear to increase the catalytic activity of the enzyme (36), which may be sufficient to optimize the DAP resistance phenotype. Although changes in many other predicted proteins were found, we speculate that these changes may play a less important role in the mutational sequence and may be minor contributors since they were found in small number of isolates. The role of these proteins in DAP resistance remains to be elucidated.
In summary, we provide genomic evidence that two major pathways (LiaFSR and YycFGHIJ) appear to be the most important changes associated with development of DAP resistance in E. faecium clinical isolates. Although the sequence of mutations appear to be complex, initial changes in regulatory systems that control the cell envelope response to stress may be of paramount importance to fully develop DAP resistance. Our findings also open the possibility of using genotypic information to identify isolates that are likely to fail DAP therapy in vivo and those in which the combination of DAP plus β-lactams may be effective.
Supplementary Material
ACKNOWLEDGMENTS
We thank Silvia Munoz-Price, James H. Jorgensen, Helio Sader, Ronald Jones, Chris Pillar, Daniel Sahm, Manuel Guzman, and Carlos Carrillo for providing the enterococcal isolates. We also thank Isabel Reyes for technical support and Jared A. Silverman and Aileen Rubio for providing BODIPY FL-labeled daptomycin.
Support for this study was provided by the Instituto Colombiano para el Desarrollo de la Ciencia y Tecnología, Francisco José de Caldas, COLCIENCIAS (graduate scholarship to L.D.), the American Society of Microbiology (Latin American Fellowship for Epidemiology to L.D.), and the Universidad El Bosque (graduate fellowships to L.D. and J.R.). C.A.A. was supported by National Institutes of Health (NIH-NIAID) grant R01 AI093749. Y.S. was supported by NIH-NIAID grant R01 AI080714, and B.E.M. was supported by NIH-NIAID grant R01 AI047923. The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 27 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02686-14.
REFERENCES
- 1.Galloway-Peña J, Roh JH, Latorre M, Qin X, Murray BE. 2012. Genomic and SNP analyses demonstrate a distant separation of the hospital and community-associated clades of Enterococcus faecium. PLoS One 7:e30187. 10.1371/journal.pone.0030187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lebreton F, van Schaik W, McGuire AM, Godfrey P, Griggs A, Mazumdar V, Corander J, Cheng L, Saif S, Young S, Zeng Q, Wortman J, Birren B, Willems RJ, Earl AM, Gilmore MS. 2013. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio 4:e00534–13. 10.1128/mBio.00534-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arias CA, Murray BE. 2012. The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 10:266–278. 10.1038/nrmicro2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arias CA, Torres HA, Singh KV, Panesso D, Moore J, Wanger A, Murray BE. 2007. Failure of daptomycin monotherapy for endocarditis caused by an Enterococcus faecium strain with vancomycin-resistant and vancomycin-susceptible subpopulations and evidence of in vivo loss of the vanA gene cluster. Clin. Infect. Dis. 45:1343–1346. 10.1086/522656 [DOI] [PubMed] [Google Scholar]
- 5.Munoz-Price LS, Lolans K, Quinn JP. 2005. Emergence of resistance to daptomycin during treatment of vancomycin-resistant Enterococcus faecalis infection. Clin. Infect. Dis. 41:565–566. 10.1086/432121 [DOI] [PubMed] [Google Scholar]
- 6.Lewis JS, Owens A, Cadena J, Sabol K, Patterson JE, Jorgensen JH. 2005. Emergence of daptomycin resistance in Enterococcus faecium during daptomycin therapy. Antimicrob. Agents Chemother. 94:1664–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pogliano J, Pogliano N, Silverman JA. 2012. Daptomycin-mediated reorganization of membrane architecture causes mislocalization of essential cell division proteins. J. Bacteriol. 194:4494–4504. 10.1128/JB.00011-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tran TT, Panesso D, Mishra NN, Mileykovskaya E, Guan Z, Munita JM, Reyes J, Diaz L, Weinstock GM, Murray BE, Shamoo Y, Dowhan W, Bayer AS, Arias CA. 2013. Daptomycin-resistant Enterococcus faecalis diverts the antibiotic molecule from the division septum and remodels cell membrane phospholipids. mBio 4:e00281–13. 10.1128/mBio.00281-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller C, Kong J, Tran TT, Arias CA, Saxer G, Shamoo Y. 2013. Adaptation of Enterococcus faecalis to daptomycin reveals an ordered progression to resistance. Antimicrob. Agents Chemother. 57:5373–5383. 10.1128/AAC.01473-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jordan S, Junker A, Helmann JD, Mascher T. 2006. Regulation of LiaRS-dependent gene expression in Bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J. Bacteriol. 188:5153–5166. 10.1128/JB.00310-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arias CA, Panesso D, McGrath DM, Qin X, Mojica MF, Miller C, Diaz L, Tran TT, Rincon S, Barbu EM, Reyes J, Roh JH, Lobos E, Sodergren E, Pasqualini R, Arap W, Quinn JP, Shamoo Y, Murray BE, Weinstock GM. 2011. Genetic basis for in vivo daptomycin resistance in enterococci. N. Engl. J. Med. 365:892–900. 10.1056/NEJMoa1011138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mishra NN, Yang SJ, Sawa A, Rubio A, Nast CC, Yeaman MR, Bayer AS. 2009. Analysis of cell membrane characteristics of in vitro-selected daptomycin-resistant strains of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53:2312–2318. 10.1128/AAC.01682-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munita JM, Tran TT, Diaz L, Panesso D, Reyes J, Murray BE, Arias CA. 2013. A liaF codon deletion abolishes daptomycin bactericidal activity against vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 57:2831–2833. 10.1128/AAC.00021-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humphries RM, Kelesidis T, Tewhey R, Rose WE, Schork N, Nizet V, Sakoulas G. 2012. Genotypic and phenotypic evaluation of the evolution of high-level daptomycin nonsusceptibility in vancomycin-resistant Enterococcus faecium. Antimicrob. Agents Chemother. 56:6051–6053. 10.1128/AAC.01318-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tran TT, Panesso D, Gao H, Roh JH, Munita JM, Reyes J, Diaz L, Lobos EA, Shamoo Y, Mishra NN, Bayer AS, Murray BE, Weinstock GM, Arias CA. 2013. Whole-genome analysis of a daptomycin-susceptible Enterococcus faecium strain and its daptomycin-resistant variant arising during therapy. Antimicrob. Agents Chemother. 57:261–268. 10.1128/AAC.01454-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munita JM, Panesso D, Diaz L, Tran TT, Reyes J, Wanger A, Murray BE, Arias CA. 2012. Correlation between mutations in liaFSR of Enterococcus faecium and MIC of daptomycin: revisiting daptomycin breakpoints. Antimicrob. Agents Chemother. 56:4354–4359. 10.1128/AAC.00509-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakoulas G, Nonejuie P, Nizet V, Pogliano J, Crum-Cianflone N, Haddad F. 2013. Treatment of high-level gentamicin-resistant Enterococcus faecalis endocarditis with daptomycin plus ceftaroline. Antimicrob. Agents Chemother. 57:4042–4045. 10.1128/AAC.02481-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sierra-Hoffman M, Iznaola O, Goodwin M, Mohr J. 2012. Combination therapy with ampicillin and daptomycin for treatment of Enterococcus faecalis endocarditis. Antimicrob. Agents Chemother. 56:6064. 10.1128/AAC.01760-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Werth BJ, Sakoulas G, Rose WE, Pogliano J, Tewhey R, Rybak MJ. 2013. Ceftaroline increases membrane binding and enhances the activity of daptomycin against daptomycin-nonsusceptible vancomycin-intermediate Staphylococcus aureus in a pharmacokinetic/pharmacodynamic model. Antimicrob. Agents Chemother. 57:66–73. 10.1128/AAC.01586-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakoulas G, Rose W, Nonejuie P, Olson J, Pogliano J, Humphries R, Nizet V. 2014. Ceftaroline restores daptomycin activity against daptomycin-nonsusceptible vancomycin-resistant Enterococcus faecium. Antimicrob. Agents Chemother. 58:1494–1500. 10.1128/AAC.02274-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panesso D, Reyes J, Rincón S, Díaz L, Galloway-Peña J, Zurita J, Carrillo C, Merentes A, Guzmán M, Adachi JA, Murray BE, Arias CA. 2010. Molecular epidemiology of vancomycin-resistant Enterococcus faecium: a prospective, multicenter study in South American hospitals. J. Clin. Microbiol. 48:1562–1569. 10.1128/JCM.02526-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clinical and Laboratory Standards Institute. 2012. MS100-S22. Performance standards for antimicrobial susceptibility testing: 21st informational supplement, vol 32 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 23.Palmer KL, Daniel A, Hardy C, Silverman J, Gilmore MS. 2011. Genetic basis for daptomycin resistance in enterococci. Antimicrob. Agents Chemother. 55:2245–2256. 10.1128/AAC.01350-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peleg AY, Miyakis S, Ward DV, Earl AM, Rubio A, Cameron DR, Pillai S, Moellering RCJ, Eliopoulos GM. 2012. Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus. PLoS One 7:e28316. 10.1371/journal.pone.0028316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang SJ, Mishra NN, Rubio A, Bayer AS. 2013. Causal role of single nucleotide polymorphisms within the mprF gene of Staphylococcus aureus in daptomycin resistance. Antimicrob. Agents Chemother. 57:5658–5664. 10.1128/AAC.01184-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bayer AS, Schneider T, Sahl HG. 2013. Mechanisms of daptomycin resistance in Staphylococcus aureus: role of the cell membrane and cell wall. Ann. N. Y. Acad. Sci. 1277:139–158. 10.1111/j.1749-6632.2012.06819.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang SJ, Kreiswirth BN, Sakoulas G, Yeaman MR, Xiong YQ, Sawa A, Bayer AS. 2009. Enhanced expression of dltABCD is associated with the development of daptomycin nonsusceptibility in a clinical endocarditis isolate of Staphylococcus aureus. J. Infect. Dis. 200:1916–1920. 10.1086/648473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nallapareddy SR, Weinstock GM, Murray BE. 2003. Clinical isolates of Enterococcus faecium exhibit strain-specific collagen binding mediated by Acm, a new member of the MSCRAMM family. Mol. Microbiol. 47:1733–1747. 10.1046/j.1365-2958.2003.03417.x [DOI] [PubMed] [Google Scholar]
- 29.Qin X, Galloway-Peña JR, Sillanpaa J, Roh JH, Nallapareddy SR, Chowdhury S, Bourgogne A, Choudhury T, Muzny DM, Buhay CJ, Ding Y, Dugan-Rocha S, Liu W, Kovar C, Sodergren E, Highlander S, Petrosino JF, Worley KC, Gibbs RA, Weinstock GM, Murray BE. 2012. Complete genome sequence of Enterococcus faecium strain TX16 and comparative genomic analysis of Enterococcus faecium genomes. BMC Microbiol. 12:135. 10.1186/1471-2180-12-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berti AD, Sakoulas G, Nizet V, Tewhey R, Rose WE. 2013. β-Lactam antibiotics targeting PBP1 selectively enhance daptomycin activity against methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 57:5005–5012. 10.1128/AAC.00594-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vignaroli C, Rinaldi C, Varaldo PE. 2011. Striking “seesaw effect” between daptomycin nonsusceptibility and beta-lactam susceptibility in Staphylococcus haemolyticus. Antimicrob. Agents Chemother. 55:2495–2496. 10.1128/AAC.00224-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hachmann AB, Angert ER, Helmann JD. 2009. Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin. Antimicrob. Agents Chemother. 53:1598–1609. 10.1128/AAC.01329-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hachmann AB, Sevim E, Gaballa A, Popham DL, Antelmann H, Helmann JD. 2011. Reduction in membrane phosphatidylglycerol content leads to daptomycin resistance in Bacillus subtilis. Antimicrob. Agents Chemother. 55:4326–4337. 10.1128/AAC.01819-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Türck M, Bierbaum G. 2012. Purification and activity testing of the full-length YycFGHI proteins of Staphylococcus aureus. PLoS One 7:e30403. 10.1371/journal.pone.0030403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szurmant H, Mohan MA, Imus PM, Hoch JA. 2007. YycH and YycI interact to regulate the essential YycFG two-component system in Bacillus subtilis. J. Bacteriol. 189:3280–3289. 10.1128/JB.01936-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davlieva M, Zhang W, Arias CA, Shamoo Y. 2013. Biochemical characterization of cardiolipin synthase mutations associated with daptomycin resistance in enterococci. Antimicrob. Agents Chemother. 57:289–296. 10.1128/AAC.01743-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friedman L, Alder JD, Silverman JA. 2006. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:2137–2145. 10.1128/AAC.00039-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grogan DW, Cronan JEJ. 1997. Cyclopropane ring formation in membrane lipids of bacteria. Microbiol. Mol. Biol. Rev. 61:429–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aravind L, Koonin EV. 1998. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci. 23:469–472. 10.1016/S0968-0004(98)01293-6 [DOI] [PubMed] [Google Scholar]
- 40.Gustafsson C, Persson BC. 1998. Identification of the rrmA gene encoding the 23S rRNA m1G745 methyltransferase in Escherichia coli and characterization of an m1G745-deficient mutant. J. Bacteriol. 180:359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakoulas G, Bayer AS, Pogliano J, Tsuji BT, Yang SJ, Mishra NN, Nizet V, Yeaman MR, Moise PA. 2012. Ampicillin enhances daptomycin- and cationic host defense peptide-mediated killing of ampicillin- and vancomycin-resistant Enterococcus faecium. Antimicrob. Agents Chemother. 56:838–844. 10.1128/AAC.05551-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jordan S, Hutchings MI, Mascher T. 2008. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol. Rev. 32:107–146. 10.1111/j.1574-6976.2007.00091.x [DOI] [PubMed] [Google Scholar]
- 43.Dubrac S, Bisicchia P, Devine KM, Msadek T. 2008. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol. Microbiol. 70:1307–1322. 10.1111/j.1365-2958.2008.06483.x [DOI] [PubMed] [Google Scholar]
- 44.Dubrac S, Boneca IG, Poupel O, Msadek T. 2007. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J. Bacteriol. 189:8257–8269. 10.1128/JB.00645-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mishra NN, Bayer AS, Moise PA, Yeaman MR, Sakoulas G. 2012. Reduced susceptibility to host-defense cationic peptides and daptomycin coemerge in methicillin-resistant Staphylococcus aureus from daptomycin-naive bacteremic patients. J. Infect. Dis. 206:1160–1167. 10.1093/infdis/jis482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mishra NN, A Rubio, CC Nast and AS Bayer 2012. Differential adaptations of methicillin-resistant Staphylococcus aureus to serial in vitro passage in daptomycin: evolution of daptomycin resistance and role of membrane carotenoid content and fluidity. Int. J. Microbiol. 2012:683450. 10.1155/2012/683450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hancock LE, Perego M. 2004. Systematic inactivation and phenotypic characterization of two-component signal transduction systems of Enterococcus faecalis V583. J. Bacteriol. 186:7951–7958. 10.1128/JB.186.23.7951-7958.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.